Abstract
Corrinoids are essential cofactors of reductive dehalogenases in Dehalococcoides mccartyi, an important bacterium in bioremediation, yet sequenced D. mccartyi strains do not possess the complete pathway for de novo corrinoid biosynthesis. Pelosinus sp. and Desulfovibrio sp. have been detected in dechlorinating communities enriched from contaminated groundwater without exogenous cobalamin corrinoid. To investigate the corrinoid-related interactions among key members of these communities, we constructed consortia by growing D. mccartyi strain 195 (Dhc195) in cobalamin-free, trichloroethene (TCE)- and lactate-amended medium in cocultures with Desulfovibrio vulgaris Hildenborough (DvH) or Pelosinus fermentans R7 (PfR7) and with both in tricultures. Only the triculture exhibited sustainable dechlorination and cell growth when a physiological level of 5,6-dimethylbenzimidazole (DMB), the lower ligand of cobalamin, was provided. In the triculture, DvH provided hydrogen while PfR7 provided corrinoids to Dhc195, and the initiation of dechlorination and Dhc195 cell growth was highly dependent on the growth of PfR7. Corrinoid analysis indicated that Dhc195 imported and remodeled the phenolic corrinoids produced by PfR7 into cobalamin in the presence of DMB. Transcriptomic analyses of Dhc195 showed the induction of the CbiZ-dependent corrinoid-remodeling pathway and BtuFCD corrinoid ABC transporter genes during corrinoid salvaging and remodeling. In contrast, another operon annotated to encode a putative iron/cobalamin ABC transporter (DET1174-DET1176) was induced when cobalamin was exogenously provided. Interestingly, a global upregulation of phage-related genes was observed when PfR7 was present. These findings provide insights into both the gene regulation of corrinoid salvaging and remodeling in Dhc195 when it is grown without exogenous cobalamin and microbe-to-microbe interactions in dechlorinating microbial communities.
INTRODUCTION
Chlorinated solvents such as tetra- and trichloroethene (PCE/TCE) have been among the most common subsurface contaminants in the United States for decades (1, 2). In situ bioremediation is an effective, economical, and environmentally friendly technology for treating chlorinated solvents (2, 3). Dehalococcoides mccartyi is the only known bacterium capable of carrying out respiratory reductive dechlorination of chloroethenes to nontoxic ethene and plays a key role in the bioremediation of contaminated groundwater (4, 5).
As cofactors of reductive dehalogenases (RDases), the enzymes responsible for reductive dechlorination, corrinoids (e.g., cobalamin) are essential nutrients for supporting D. mccartyi growth and dechlorination. Genomic analyses of sequenced D. mccartyi strains reveal a lack of complete corrinoid biosynthesis pathways, rendering D. mccartyi incapable of synthesizing corrinoids de novo (6, 7). Exogenous cobalamin (a specific corrinoid also known as vitamin B12) is generally added to grow D. mccartyi in isolation (8). Cobalamin, 5-methylbenzimidazolylcobamide ([5-MeBza]Cba), and 5-methoxybenzimidazolyl cobamide ([5-OMeBza]Cba) are the functional corrinoids that can be used by D. mccartyi strain 195 (Dhc195) directly (9). D. mccartyi strains also possess corrinoid salvaging and remodeling pathways and have exhibited versatile abilities to import nonfunctional corrinoids and replace the original lower ligand with 5,6-dimethylbenzimidazole (DMB) to form cobalamin (9).
It has long been recognized that D. mccartyi exhibits faster dechlorination and more robust growth when it is grown in mixed communities than when grown in isolation, possibly due to the supply of nutrients and growth factors to D. mccartyi by other community members (5, 6, 10). In turn, D. mccartyi promotes fermentation reactions by consuming hydrogen and potentially mitigates toxicity by transforming chlorinated solvents. For in situ bioremediation applications, D. mccartyi growth is stimulated by injecting organics such as lactate, methanol, glucose, or whey into the subsurface to promote microbial fermentation that generates the hydrogen and acetate required by D. mccartyi. In addition, a number of microorganisms commonly detected in D. mccartyi-containing dechlorinating communities are reported to produce a variety of corrinoids, for example, acetogenic bacteria from the genera Clostridium, Acetobacterium, Desulfovibrio, Sporomusa, Eubacterium (11–15), and Geobacter (16) and methanogenic archaea (17–19).
In order to study specific corrinoid-related interactions within dechlorinating communities, studies have been conducted with nonsyntrophic cocultures containing D. mccartyi strains together with Geobacter sulfurreducens, Methanosarcina barkeri strain Fusaro, and Sporomusa sp. strain KB-1 providing hydrogen and acetate in cobalamin-free medium. Dechlorination in these cocultures proceeded only when DMB was provided as a source of cobalamin lower ligand, indicating that corrinoid-salvaging and remodeling were occurring (16, 20). However, whether sustained long-term growth would be supported by the corrinoid salvaging and remodeling activities in syntrophic dechlorinating cultures is unknown. In addition, the underlying transcriptional responses of D. mccartyi performing specific corrinoid salvaging and remodeling functions have not yet been reported.
In this study, we aim to investigate the corrinoid-related interactions between D. mccartyi and the supportive microorganisms that commonly occur in dechlorinating communities by constructing syntrophic consortia growing with lactate as the primary electron donor and without exogenous corrinoids. Previous studies demonstrated that Dhc195 can be grown in defined syntrophic consortia with lactate-fermenting Desulfovibrio vulgaris Hildenborough (DvH) amended with cobalamin (21). In addition, bacteria related to Pelosinus spp., which belong to the same family as the phenolic corrinoid producer Sporomusa ovata (12, 22), were found to be the most abundant species in dechlorinating enrichments grown without exogenous cobalamin (23). Therefore, in this study, syntrophic consortia were constructed by growing Dhc195 with Pelosinus fermentans R7 (PfR7) and DvH as a simplified community. The abilities of DvH and PfR7 to provide corrinoids to Dhc195 in cobalamin-free medium were tested. The dechlorination activity, cell growth, hydrogen, and organic acids were quantified, and specific corrinoids were identified in the consortia with sustained growth. Transcriptomic analysis was performed to elucidate transcriptional responses of Dhc195 within the various consortia.
MATERIALS AND METHODS
Bacterial cultures and growth conditions.
Dhc195 was generously donated by Steven Zinder, Cornell University. Desulfovibrio vulgaris Hildenborough (DvH) (ATCC 29579) and Pelosinus fermentans R7 (PfR7) (ATCC BAA-1133) were obtained from the American Type Culture Collection (ATCC). DvH pure culture was maintained on lactate and sulfate, while PfR7 pure culture was maintained on lactate using basal medium with the pH adjusted to 7.5. The composition of the basal medium has been described elsewhere (24). Cocultures Dhc195/DvH and Dhc195/PfR7 were constructed using Dhc195, DvH, and PfR7 isolates in 160-ml sealed serum bottles containing a 60-ml N2-CO2 (90:10, vol/vol) headspace and 100 ml of the same basal medium amended with ∼0.7 mmol of lactate as the electron donor and carbon source, ∼77 μmol of TCE as the electron acceptor, and 74 nM (ca. 100 μg liter−1) cobalamin, as previously described (21). Tricultures were constructed by inoculating 1% pregrown Dhc195/DvH and 2% pregrown PfR7 into the same medium, one with 74 nM B12 amendment [Dhc195/DvH/PfR7(+B12)] and one without B12 but with 36 nM DMB [Dhc195/DvH/PfR7(+DMB)]. All cultures were incubated at 34°C in the dark without shaking. Dechlorination activities, hydrogen production, and organic acid concentrations were quantified after six subculturing events (3%, vol/vol). All experiments were performed with biological triplicates.
DNA extraction and cell growth quantification.
Cells from 1.5 ml of culture sampled from each of the three biological replicates were collected by centrifugation (15,000 × g for 10 min at 4°C), and genomic DNA (gDNA) was extracted using a DNeasy Blood and Tissue Kit (Qiagen, Valencia, CA) according to the manufacturer's instructions. Quantitative PCR (qPCR) was applied to quantify cell densities using primer sets targeting the 16S rRNA gene of each microorganism. Briefly, each 20-μl reaction mixture contained 2.5 μl of gDNA sample or 10-fold serially diluted standard, 1× fast SYBR green master mix (Applied Biosystems, Foster City, CA), and 0.625 μM forward and reverse primers (see Table S1 in the supplemental material). gDNA of Dhc195, DvH, and PfR7 isolates quantified by Nanophotometer P-300 (Implen, Inc., Westlake Village, CA) was used as the standard for qPCR, the cell density of which was determined using the following equation (25): cell density (in cells/ml) = DNA concentration (in g/ml) × {[(6.023 × 1023 cells/mol)/(660 g of DNA/bp/mol)]/genome size [in bp]}. The genome sizes for Dhc195, DvH, and PfR7 are 1.5 Mbp (6), 3.6 Mbp (26), and 4.9 Mbp (27), respectively.
RNA extraction.
RNA was extracted from cell pellets collected from ∼100 ml of culture using the acid phenol (pH 4.3)-chloroform method (28). Cell pellets were resuspended in 250 μl of lysis buffer (50 mM sodium acetate, 10 mM EDTA, pH 5.1), 100 μl of 10% sodium dodecyl sulfate, and 1.0 ml of buffer-equilibrated phenol (pH 4.3) (Sigma-Aldrich, St. Louis, MO). Cells were lysed by bead beating with a Mini Bead Beater (Biospec Products) for 2 min, and the aqueous lysate was extracted twice with 1 volume of acid (pH 4.3) phenol-chloroform-isoamyl alcohol (25:24:1) and once with 1 volume of chloroform-isoamyl alcohol (24:1) (Sigma-Aldrich). RNA was then precipitated by adding 0.5 volume of 7.5 M ammonium acetate and 2 volumes of 100% ethanol. The precipitate was collected by centrifugation (21,000 × g for 30 min at 4°C), washed once with 80% ethanol, vacuum dried, and resuspended in 100 μl of diethyl pyrocarbonate (DEPC)-treated water (Bio-Express, Kaysville, UT). DNA contamination was removed by DNase I treatment using a Turbo DNA-free kit (Ambion, Life Technologies, Grand Island, NY) according to the manufacturer's instructions. Purified RNA was stored at −80°C prior to further use.
Transcriptomic microarray and data analysis.
Microarrays targeting the genomes of four D. mccartyi strains (i.e., strains CBDB1, BAV1, 195, and VS) were designed and produced by Affymetrix (Santa Clara, CA) as previously described (29). Information regarding the microarray platform (GPL10838) was previously deposited in the NCBI Gene Expression Omnibus (GEO) database (29). cDNA was synthesized from 9 μg of RNA and then fragmented, labeled, and hybridized to each chip. The hybridized chips were stained, washed, and then scanned with an Affymetrix Scan 3000 scanner (Affymetrix). All procedures were performed according to the protocol outlined in chapter 4-5 of the Affymetrix GeneChip Expression Analysis Technical Manual (30). Three replicate arrays were analyzed for each condition.
Microarray data analyses were performed as described previously (21, 31) using Affymetrix GeneChip software (Affymetrix) and the MAS5 algorithm. Each microarray was normalized by scaling the signal intensities of the positive-control spike mix, as described in section 3 of the Affymetrix GeneChip Expression Analysis Technical Manual (30), to a target signal intensity of 2,500 to allow comparison between microarrays. The two-condition comparison was performed using the R statistical program (www.r-project.org) with packages available from www.bioconductor.org (32) as previously described (33, 34). The Benjamini-Hochberg procedure (35) was applied to control the false-discovery rate (FDR) below 0.05. Genes with signal intensities greater than 250 were considered to be actively transcribed. In addition, only genes with absolute hybridization signal intensities greater than 250 for at least one condition and with more than 2-fold changes between two conditions could be considered to be significantly regulated and used for further analyses. The term “gene expression” used in this study specifically refers to the transcription of genes into RNA.
Analytical methods.
Chloroethenes and ethene were analyzed using an Agilent 7890A gas chromatograph equipped with a flame ionization detector (FID) (Agilent Technologies, Santa Clara, CA) as described elsewhere (23). Hydrogen was analyzed by gas chromatography with a reductive gas detector (Trace Analytical, Menlo Park, CA) as described previously (15). The total amounts of chloroethenes and ethene and the aqueous hydrogen concentration were calculated using the total mass balance equation and Henry's law constant as previously described (36). Organic acids were analyzed by high-performance liquid chromatography (HPLC) using a UVD 170S UV detector as described previously (24).
Corrinoids and benzimidazoles were extracted and concentrated according to a method described previously (9). Twelve corrinoids and three benzimidazoles were targeted using an Agilent 6410 liquid chromatography-tandem mass spectrometry (LC-MS/MS) system with an Agilent Eclipse Plus C18 column (1.8-μm particle size, 3.0 by 50 mm; Agilent Technologies, Santa Clara, CA) as described elsewhere (9).
Microarray data accession number.
The microarray data in this study were deposited in the NCBI Gene Expression Omnibus database (http://www.ncbi.nlm.nih.gov/geo/) under accession number GSE45533.
RESULTS
Growth of Dhc195 in defined consortia with and without exogenous cobalamin.
Cocultures containing Dhc195 and DvH (Dhc195/DvH) or Dhc195 and PfR7 (Dhc195/PfR7), as well as the triculture containing all three organisms (Dhc195/DvH/PfR7), were constructed. TCE dechlorination capabilities of these consortia grown with lactate or hydrogen as the electron donor, with or without the addition of cobalamin or DMB, were then investigated (see Table S2 in the supplemental material). Dhc195/DvH cocultures were able to dechlorinate TCE in medium amended with lactate only when cobalamin was added, indicating that DvH is able to supply Dhc195 with hydrogen and acetate but not with corrinoids. In contrast, Dhc195/PfR7 cocultures were able to dechlorinate TCE in cobalamin-free, lactate-amended medium as long as hydrogen and DMB were supplied, suggesting that PfR7 can produce sufficient amounts of corrinoids and acetate for Dhc195 but lacks the ability to produce hydrogen. In the triculture, as expected, Dhc195 was able to grow and dechlorinate TCE with only DMB and lactate added to the medium. A model of the ecological interactions among the three microorganisms is shown in Fig. 1.
FIG 1.
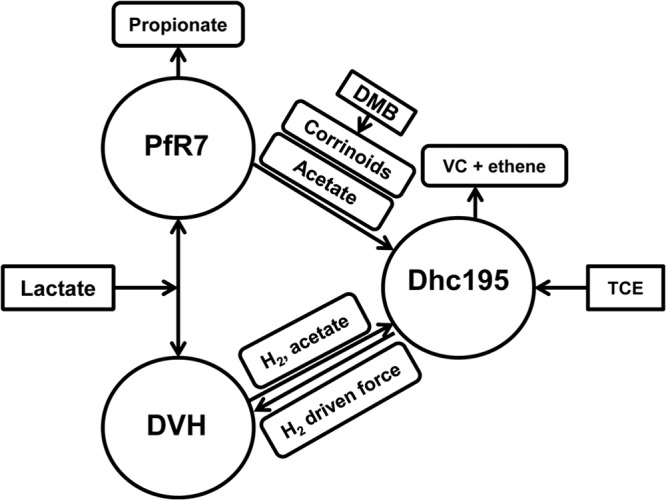
Ecological interactions among Dhc195, DvH, and PfR7 in a defined consortium.
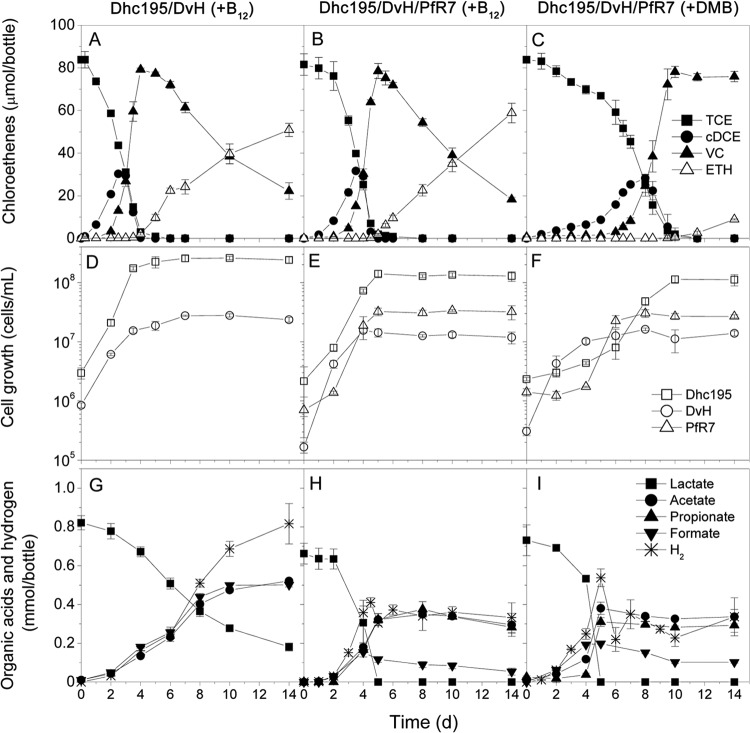
Lactate was used as the electron donor for the consortia for two reasons: (i) it promotes syntrophic growth among the chosen strains by stimulating fermentation and (ii) it is more commonly used for biostimulation than hydrogen gas since it is a more convenient and less expensive electron donor for subsurface injection. Further physiological analysis was done with the three lactate-amended consortia [i.e., Dhc195/DvH(+B12), Dhc195/DvH/PfR7(+B12), and Dhc195/DvH/PfR7(+DMB)], which exhibited stable and reproducible growth for greater than six subculturing events. Dhc195 in the Dhc195/DvH/PfR7(+B12) culture exhibited almost the same dechlorination performance as Dhc195/DvH(+B12) with a very short lag (Fig. 2A and B), whereas Dhc195 in the Dhc195/DvH/PfR7(+DMB) culture exhibited a lag of approximately 5 days (Fig. 2C). However, after the 5-day lag, TCE was dechlorinated rapidly within 4 days (Fig. 2C). A lower ethene-to-vinyl chloride (VC) ratio was observed after 14 days in the Dhc195/DvH/PfR7(+DMB) triculture than in the other two consortia (Fig. 2C). This was likely due to the dechlorination lag time since an extended incubation of Dhc195/DvH/PfR7(+DMB) resulted in an ethene-to-VC ratio similar to that of the other two cultures (data not shown).
FIG 2.
TCE dechlorination, cell growth, and organic acids in different defined consortia: Dhc195/DvH(+B12) (A, D, and G) Dhc195/DvH/PfR7(+B12) (B, E, and H), and Dhc195/DvH/PfR7(+DMB) (C, F, and I).
Similar to the TCE dechlorination, the cell growth of Dhc195 in the Dhc195/DvH/PfR7(+DMB) culture also exhibited a 4-day lag, tracking with the growth lag of PfR7 (Fig. 2F). This growth lag did not occur in the B12-amended co- and tricultures (Fig. 2D and E), suggesting that PfR7 had no significant effect on Dhc195 growth in the presence of exogenous cobalamin. The strong dependence of Dhc195 on the growth of PfR7 in the Dhc195/DvH/PfR7(+DMB) triculture is probably due to corrinoid exchange. The growth patterns of DvH were similar in all three consortia, suggesting that growth is unaffected by the presence or absence of exogenous cobalamin or PfR7. Cell numbers at the end of the feeding cycle were similar for the two tricultures, regardless of B12 amendment.
Dhc195's requirement for hydrogen as an electron donor for reductive dechlorination is fulfilled by the fermentation of lactate by DvH in all of the defined consortia (Fig. 1). Hydrogen, acetate, and formate were the observed products of lactate fermentation by DvH in the Dhc195/DvH(+B12) coculture (Fig. 2G), according to stoichiometric reaction 1 shown in Table 1. Compared to reactions 3 to 5 in Table 1, the energy generated under standard conditions at pH 7 from reaction 1 is rather low (−5.3 kJ/mol), with the aqueous hydrogen threshold for generating adequate energy to synthesize one ATP (−32 kJ/mol) at 2 mM (calculated using the equation for reaction 1). Therefore, a syntrophic interaction forms between DvH and Dhc195 (Fig. 1), with the latter consuming hydrogen for reductive dechlorination (reactions 4 and 5 in Table 1), thereby lowering the hydrogen partial pressure and maintaining continuous energy-positive lactate fermentation. Incomplete lactate fermentation and hydrogen accumulation were observed in the Dhc195/DvH(+B12) coculture (Fig. 2G), reflecting the electron acceptor (TCE and cis-dichloroethene [cDCE]) limitation in this coculture. The lactate and hydrogen would continue to be consumed if the electron acceptor TCE was added again. PfR7 fermented lactate to acetate and propionate in a 1:2 ratio (reaction 3 in Table 1; see also Fig. S1 in the supplemental material) as previously reported (22, 37). Hydrogen was too low to be quantitatively detected in PfR7 grown on lactate as the sole electron donor in this study, which is consistent with the failure of the Dhc195/PfR7 coculture to grow with lactate as the sole electron donor. In the tricultures containing PfR7, lactate was depleted within 5 days, and acetate, propionate, and hydrogen accumulated during that time. The slight decrease of hydrogen after 5 days was tied to dechlorination (Fig. 2H and I), with a final rebound tied to formate fermentation described by reaction 2 in Table 1. Although DvH and PfR7 compete for lactate in the tricultures, they maintained a balanced lactate consumption (according to propionate, acetate, and formate production in reactions 1 to 3; ∼0.3 and 0.4 mmol lactate for DvH and PfR7, respectively) during every feeding cycle in this study (see Table S3 in the supplemental material).
TABLE 1.
Reactions and roles of each member in the sustainable dechlorinating consortium Dhc195/DvH/PfR7(+DMB)
| Strain | Reaction no. | Stoichiometric equationb | Interacting strains | Role |
|---|---|---|---|---|
| DvH | 1a | CH3CHOHCOO− (lactate) + H2O → CH3COO− (acetate) + HCOO− (formate) + H+ + H2 ΔG = −5.3 kJ/rxn | Dhc195, PfR7 | Supply Dhc195 with H2 and acetate |
| 2a | HCOO− (formate) + H2O → H2 + HCO3− ΔG°′ = 1.4 kJ/rxn | Compete with PfR7 for lactate | ||
| PfR7 | 3a | 3CH3CHOHCOO− (lactate) → CH3COO− (acetate) + 2CH3CH2COO− (propionate) + H+ + HCO3− ΔG°′ = −164.8 kJ/rxn | Dhc195, DvH | Supply Dhc195 with corrinoids and acetate; compete with DvH for lactate |
| Dhc195 | 4c | C2HCl3 (TCE) + 2H2 → C2H3Cl (VC) + 2H+ + 2Cl− ΔG°′ = −149.7 kJ/rxn | DvH, PfR7 | Lower H2 partial pressure for DvH |
| 5c | C2HCl3 (TCE) + 3H2 → C2H4 (ethene) + 3H+ + 3Cl− ΔG°′ = −219.9 kJ/rxn | Lower chloroethene toxicity for DvH and PfR7 |
Corrinoid production and modification.
Corrinoids generated in the DvH and PfR7 pure cultures, as well as in the defined consortia, were identified and quantified at the end of incubation. Corrinoids were not detected when DvH was grown alone in lactate/sulfate-amended B12-free medium (data not shown), whereas p-cresolylcobamide ([p-Cre]Cba), phenolylcobamide ([Phe]Cba) and cobinamide (Cbi) were generated by PfR7 grown in lactate-amended, B12-free medium (Table 2). When PfR7 was grown with 36 nM DMB [PfR7(+DMB)], it was also able to produce cobalamin by incorporating the added DMB (Table 2), which was ∼13% of the total detected corrinoids. Cobalamin, Cbi, and [p-Cre]Cba were detected in the three dechlorinating consortia, while [Phe]Cba was not detected (Fig. 3). In the Dhc195/DvH/PfR7(+DMB) triculture, cobalamin production was double that observed in PfR7(+DMB) (Table 2). When normalized to cell number, corrinoid produced per cell of PfR7 grown in the Dhc195/DvH/PfR7(+DMB) culture was slightly higher than when PfR7 was grown alone (Table 2). Notably, supernatant-associated [p-Cre]Cba and Cbi were detected in PfR7 and Dhc195/DvH/PfR7(+B12) cultures but not in the Dhc195/DvH/PfR7(+DMB) culture (Fig. 3).
TABLE 2.
Corrinoid-producing capacity of PfR7 grown in isolation with and without DMB and in the Dhc195/DvH/PfR7(+DMB) triculture
| Culture | Corrinoid production (nmol/100 ml of culture) |
PfR7 cell density (108 cells/100 ml of culture) | Corrinoid-producing capacity (nmol/108 cells) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Cobalamin |
[p-Cre]Cba |
[Phe]Cba |
Cbi |
Total |
||||||||
| Cella | Supb | Cell | Sup | Cell | Sup | Cell | Sup | Cell | Sup | |||
| PfR7 | 0 | 0 | 0.21 | 0.025 | 0.10 | 0 | 3.0 × 10−3 | 0.010 | 0.31 | 0.035 | 0.40 | 0.86 |
| PfR7(+DMB) | 0.040 | 0.014 | 0.20 | 0.024 | 0.023 | 0 | 0.010 | 0.016 | 0.27 | 0.054 | 0.62 | 0.52 |
| Dhc195/DvH/PfR7(+DMB) | 0.088 | 3.5 × 10−3 | 0.25 | 0 | 0 | 0 | 0.012 | 0 | 0.35 | 3.5 × 10−3 | 0.27 | 1.3 |
Biomass-associated amount.
Supernatant-associated amount.
FIG 3.
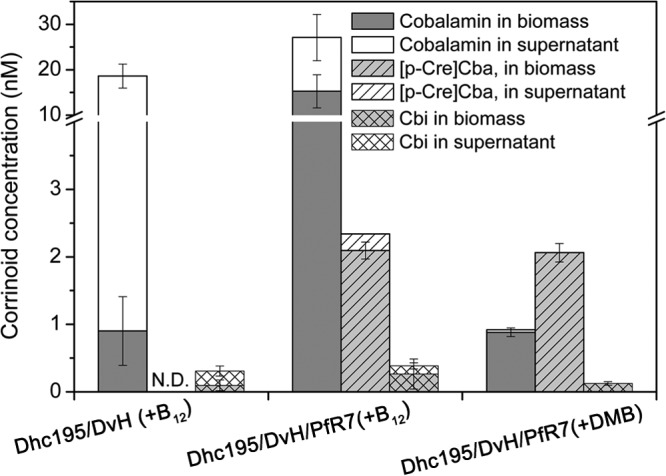
Corrinoid production in different defined consortia. Cobalamin (74 nM) was added to Dhc195/DvH(+B12) and Dhc195/DvH/PfR7(+B12) cultures, and DMB (36 nM) was added to the Dhc195/DvH/PfR7(+DMB) culture. ND, not detected.
Transcriptomic comparison of Dhc195 grown in co- and tricultures.
In order to understand the gene regulation of Dhc195 caused by the presence of PfR7 only, we first compared the transcriptome of Dhc195 in the Dhc195/DvH/PfR7(+B12) culture to that in Dhc195/DvH(+B12) (Fig. 4A). No differential gene expression was observed in the corrinoid-uptake and modification genes in Dhc195 since cobalamin was added in both cultures while a large number of phage-related genes were significantly upregulated, and genes involved in both amino acid synthesis and transport and nitrogen metabolism were downregulated in the triculture (Fig. 4A; see also Table S4 in the supplemental material). The upregulated phage-related genes covered almost all viral regions in the genome, including the three replicated integrated element (IE) regions (i.e., IE III, IV, and VI) and IE VII (see Table S4).
FIG 4.
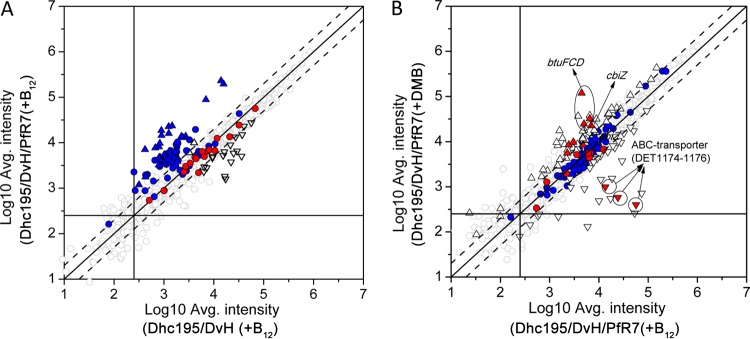
Global transcriptomic analysis of Dhc195 grown in Dhc195/DvH/PfR7(+B12) triculture compared to Dhc195/DvH(+B12) coculture (A) and Dhc195/DvH/PfR7(+DMB) triculture compared to Dhc195/DvH/PfR7(+B12) triculture (B). Dashed lines indicate 2-fold differences. Solid lines indicate an average intensity of 250 on the x or y axis (genes with signal intensities greater than 250 were considered to be actively transcribed). Upward- and downward-pointing triangles indicate up- and downregulation, respectively; red indicates corrinoid-related genes, and blue indicates phage-related genes. Differential expression is determined by the following criteria: FDR of <0.05, >2-fold change, and >250 signal intensity under at least one of the two conditions.
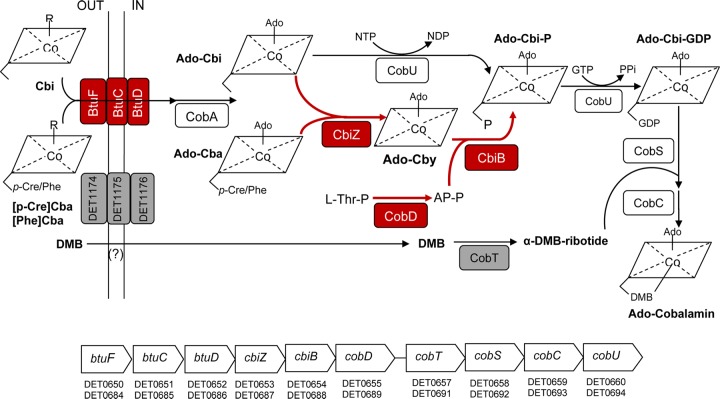
The transcriptional responses of Dhc195 to cobalamin-limited and -unlimited conditions were then investigated by comparing Dhc195/DvH/PfR7(+DMB) to Dhc195/DvH/PfR7(+B12) (Fig. 4B). A total of 74 genes were differentially expressed in the Dhc195/DvH/PfR7(+DMB) triculture compared to the Dhc195/DvH/PfR7(+B12) triculture, with 30 upregulated and 44 downregulated. Corrinoid-related genes were among the most highly regulated. Genes involved in corrinoid remodeling such as duplicated genes cbiZ (DET0653/DET0687), cbiB (DET0654/DET0688), and cobD (DET0655/DET0689) exhibited significant upregulation in Dhc195/DvH/PfR7(+DMB) compared to B12-amended coculture and triculture (Fig. 5). Two genes, cbiA (DET0128) and cbiP (DET0936) involved in the last three steps of corrin ring biosynthesis, were also significantly upregulated (Fig. 5; see also Fig. S2A in the supplemental material). Unexpectedly, the lower-ligand activation and attachment genes cobT (DET0657), cobS (DET0658), and cobC (DET0659) were downregulated in the Dhc195/DvH/PfR7(+DMB) triculture (Fig. 5; see also Fig. S2B). Notably, one cobalamin ABC transporter operon (btuFCD, DET0650-DET0652/DET0684-DET0686) was upregulated 4- to 27-fold, while another operon predicted to encode an Fe3+/cobalamin ABC transporter (DET1174-DET1176) was downregulated 20- to 167-fold in the Dhc195/DvH/PfR7(+DMB) triculture compared to Dhc195/DvH/PfR7(+B12) (Fig. 5). Using a published program, Infernal 1.1rc4 (38), nine genes (DET0125, DET0314, DET0650-DET0651/DET0684-DET0685, DET0657/DET0691, and DET1167) were identified as having putative upstream cobalamin riboswitch sequences, among which only those encoding proteins involved in corrinoid transport (DET0650-DET0651/DET0684-DET0685) and lower-ligand activation (DET0657/DET0691) exhibited differential expression.
FIG 5.
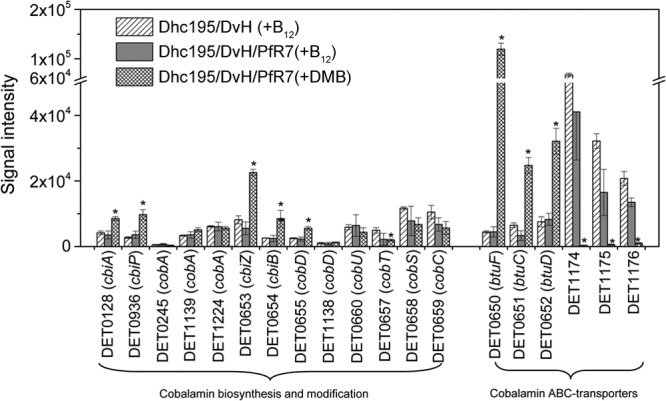
Transcriptional levels of corrinoid-related genes of Dhc195 grown in Dhc195/DvH(+B12), Dhc195/DvH/PfR7(+B12), and Dhc195/DvH/PfR7(+DMB) cultures. The asterisks indicate differential gene expression with the following criteria: FDR of <0.05, >2-fold change, and signal intensity of >250 in at least one of the three cultures.
Among other differentially expressed genes were DET1483 and DET1485 in the tryptophan biosynthesis operon, which were upregulated in Dhc195/DvH/PfR7(+DMB) triculture compared to those in Dhc195/DvH/PfR7(+B12), while almost the entire operon (DET0461-DET0468) involved in biosynthesis of chorismate, the precursor of aromatic amino acids, was downregulated (see Table S4 in the supplemental material). Other downregulated genes in the Dhc195/DvH/PfR7(+DMB) triculture with predicted functions include a peptide ABC transporter (DET1490-DET1494), hydrogenase HymC (DET0730), and oxidoreductase (DET0736) (see Table S4). In addition, no significant regulation was observed for the key functional RDase genes, which is consistent with the observed similar dechlorination and cell growth of Dhc195 among the three consortia.
DISCUSSION
In this study, we investigated the corrinoid-related interactions between Dhc195, DvH, and PfR7 in defined consortia without exogenous cobalamin. The syntrophic relationships among the three bacteria in the Dhc195/DvH/PfR7(+DMB) culture provide a simplified representation of interactions associated with a lactate-stimulated dechlorinating microbial community (23). Major syntrophic interactions within this community include the following (Fig. 1): (i) the generation of hydrogen and acetate by lactate-fermenting DvH, providing Dhc195 with electron donor and carbon source; (ii) the production of acetate by lactate-fermenting PfR7, providing Dhc195 with a carbon source; (iii) hydrogen consumption by Dhc195, maintaining positive energetics for lactate fermentation of DvH; and (iv) the generation by PfR7 of corrinoids that could be salvaged and remodeled by Dhc195 for use as essential cofactors.
Pelosinus is a recently described genus that is commonly found associated with subsurface microbial communities stimulated with lactate for in situ bioremediation of metals (22, 27). This study is the first to identify PfR7 as a corrinoid producer with the capability of synthesizing phenolic cobamides (i.e., [p-Cre]Cba and [Phe]Cba) de novo. The ability of PfR7 to produce phenolic cobamides is shared by the closely related bacterium Sporomusa ovata (12, 39), and both belong to the family Veillonellaceae. Bioinformatic analysis on the draft genome of PfR7 (http://www.ncbi.nlm.nih.gov/genome/12963) indicates that PfR7 possesses all of the genes required for de novo corrinoid biosynthesis (see Table S5 in the supplemental material). PfR7 also possesses corrinoid-dependent methionine synthase and methylmalonyl-coenzyme A (CoA) mutase enzymes. The latter is necessary for the fermentation of lactate to acetate and propionate by Pelosinus spp. through the methylmalonyl-CoA pathway (37, 40). Therefore, the phenolic cobamides produced by PfR7 likely serve as the corrinoid cofactors of these corrinoid-dependent enzymes. The ability of Pelosinus spp. to carry out de novo corrinoid biosynthesis and their observed predominance in lactate-amended TCE-dechlorinating enrichments (23) suggest their important roles as corrinoid suppliers in bioremediation sites.
Here, no corrinoids were detected when DvH was grown on lactate/sulfate without exogenous cobalamin although the presence of an almost complete corrinoid biosynthesis pathway in the DvH genome indicates its corrinoid-producing potential (see Table S5 in the supplemental material). Two corrinoids, guanylcobamide and hypoxanthylcobamide, were previously identified in DvH culture grown on lactate/sulfate (13), and corrinoid production has also been detected by unspecific but more sensitive cobalamin bioassay (41). It is possible that corrinoids synthesized by DvH, if any, were either insufficient for LC-MS/MS detection or present in other nontargeted forms. The insufficient corrinoid production by DvH also resulted in the observation that the Dhc195/DvH coculture was unable to grow without exogenous cobalamin even though DMB was added. However, it is possible that other growth conditions (e.g., different electron donor/acceptor types and concentrations) not tested in this study might result in more active corrinoid production by DvH.
Given the growth suppression caused by high levels of DMB lower ligand to corrinoid-producing microorganisms, such as S. ovata (the half-maximal inhibitory concentration [IC50] of DMB is 0.67 μM) (39) and M. barkeri (42), a physiological level (nM) of DMB was used in this study to investigate corrinoid salvaging and remodeling during sustained growth. Corrinoid-producing microorganisms are often capable of incorporating exogenous lower-ligand bases to generate the corresponding corrinoids, a process known as guided biosynthesis (13, 39). We find that cobalamin could be formed by PfR7 through guided biosynthesis when DMB was provided, and 35% (0.14 nM) of the formed cobalamin was released to the supernatant. However, by assuming that the same amount of cobalamin was produced through guided biosynthesis by PfR7 in the Dhc195/DvH/PfR7(+DMB) culture as in the PfR7(+DMB) culture, the double amount of cobalamin in Dhc195/DvH/PfR7(+DMB) relative to that in the PfR7(+DMB) culture indicates that Dhc195 also meets its need of cobalamin by salvaging and remodeling nonfunctional corrinoids (i.e., [p-Cre]Cba and Cbi) produced by PfR7. This salvaging and remodeling process is also supported by the absence of [p-Cre]Cba and Cbi in the supernatant of the Dhc195/DvH/PfR7(+DMB) culture.
It has been shown that DMB is a key requirement for D. mccartyi strains to effectively salvage and remodel other corrinoids into cobalamin (9, 16, 20). DMB is commonly produced by non-D. mccartyi community members to support corrinoid remodeling by D. mccartyi in TCE-dechlorinating enrichments without exogenous cobalamin (23). Although DMB biosynthesis under aerobic conditions has been well studied (43), little is known about DMB-producing organisms or the synthesis pathways employed under anaerobic conditions. What has been reported from studies growing the anaerobic, cobalamin-producing bacterium Eubacterium limosum with labeling compounds is that the C-4, C-2, C-3a, C-7a, N-1, and N-3 positions of the DMB molecule originate from threose, formate, glycine, and glutamine in that bacterium (11, 44, 45). Some of these precursors are also building blocks in purine nucleotide biosynthesis (11).
The Dhc195 genome possesses both of the known corrinoid-salvaging pathways (46): (i) the CbiZ-dependent pathway that uses Cbi or cobamides as a substrate and (ii) the CobU-dependent pathway that uses Cbi as the sole substrate (Fig. 6). In the Dhc195/DvH/PfR7(+DMB) culture, the observed induction of Dhc195's CbiZ-dependent corrinoid-salvaging and -remodeling pathway genes cbiZ, cbiB, and cobD (Fig. 5 and 6) is consistent with the decrease of supernatant-associated corrinoids and the increase of cobalamin observed in the corrinoid profile. The lack of observed regulation of the cobU gene may be explained by its dual function in Cbi salvaging and the conversion of adenosylcobinamide phosphate to adenosylcobinamide-GDP for lower-ligand attachment (Fig. 6). A previous study with Dhc195 grown with limited and excess cobalamin also showed upregulation of cbiZ and cbiB in response to cobalamin limitation (34), suggesting that these genes could be used as diagnostic biomarkers of this condition. In addition, the upregulation of genes for the last three steps of corrin ring biosynthesis, cbiA and cbiP in Dhc195/DvH/PfR7(+DMB), suggests that the salvage of corrinoid precursors from the de novo biosynthesis of PfR7 might also be employed by Dhc195.
FIG 6.
Transcriptional regulation of genes involved in corrinoid-salvaging and -remodeling pathways of Dhc195. Red and gray boxes indicate up- and downregulation, respectively; clear boxes indicate active gene expression without regulation. R, methyl (-CH3) or adenosyl (-Ado); Ado-Cba, adenosylcobamide; Ado-Cby, adenosylcobyric acid; Ado-Cbi-P, adenosylcobinamide-phosphate; L-Thr-P, l-threonine phosphate; AP-P, (R)-1-aminopropan-2-yl phosphate.
Another notable finding in this study is the differential expression of the putative corrinoid ABC transport systems of Dhc195 grown in consortia under different corrinoid conditions. BtuFCD is a confirmed corrinoid ABC transporter (47), and a duplicate set of genes homologous to btuFCD is present in the Dhc195 genome (DET0650-DET0652/DET0684-DET0686) located upstream of the duplicated corrinoid-salvaging and lower-ligand attachment operon (Fig. 6). In this study, the substantial upregulation of DET0650-DET0652/DET0684-DET0686 without exogenous cobalamin suggests that it is responsive to cobalamin limitation, similar to previous results with Dhc195 in isolation (34) and with the D. mccartyi-containing ANAS enrichment (an enrichment culture derived from a site at the Alameda Navel Air Station) (48). This gene expression pattern is presumably due to the transcriptional regulation by a putative cobalamin riboswitch that is located upstream of this operon (49). Another operon annotated to encode a putative Fe3+/cobalamin transporter (DET1174-DET1176) was significantly downregulated (20- to 167-fold) in the absence of cobalamin amendment. The genes in this operon do not share significant homology to btuFCD, yet they are also located downstream of a putative cobalamin riboswitch sequence (DET1167), albeit with six genes intervening. The upregulation of these genes in the presence of cobalamin and their proximity to a cobalamin riboswitch suggest that this operon might have a cobalamin-related function. The btuFCD genes and DET1174-DET1176 could thus serve as biomarkers, with upregulation of btuFCD and downregulation of DET1174-DET1176 indicating the corrinoid-salvaging condition. Further investigation of the functions of these transporters may lead to better understanding of corrinoid-related interactions between microbes.
An unexpected finding in this study is that the presence of PfR7 in the tricultures prompts a significant upregulation of the phage-related Dhc195 genes. However, no regulation of phage-related genes was observed in the Dhc195/DvH(+B12) coculture compared to Dhc195 grown in isolation (21). A previous study revealed that a number of phage-related genes in Dhc195 are significantly upregulated when the growth phase transits from late exponential phase to early stationary phase (33). Similarly, a D. mccartyi phage was induced in the dechlorinating KB-1 culture when TCE was omitted (50). This suggests that the phage-related genes in D. mccartyi might be induced under environmental stress. However, the nature of the stress imposed on Dhc195 by PfR7, if any, is unclear.
In summary, we have shown that in a syntrophic defined consortium containing Dhc195, DvH, and PfR7 amended with a physiological level (nM) of DMB but without exogenous cobalamin, PfR7 successfully sustains the growth of Dhc195 by generating corrinoids for Dhc195 to salvage and remodel. The concentration of added DMB in the consortium does not exhibit growth suppression on PfR7. Transcriptomic analysis of Dhc195 showed an induction of the CbiZ-dependent corrinoid remodeling pathway and a putative corrinoid transporter during growth in the Dhc195/DvH/PfR7(+DMB) triculture, suggesting that corrinoid salvaging and remodeling are important mechanisms employed by Dhc195 under cobalamin limitation in the presence of alternate corrinoid-producing organisms. Given that all sequenced D. mccartyi strains possess the same putative genes involved in the corrinoid-salvaging and -remodeling pathways (9), similar gene regulation patterns would be expected for the other D. mccartyi strains. These findings highlight the potential roles played by Pelosinus spp. in supporting D. mccartyi growth and in chlorinated solvent bioremediation and enrich our knowledge of the ecological interactions within dechlorinating communities, particularly between corrinoid auxotrophs and corrinoid producers. Results of this study also suggest potential indicators, such as corrinoids and DMB molecules, as well as genes for corrinoid transport, salvaging, and remodeling (i.e., btuFCD, cbiZ, cobD, cbiB), for use in monitor cobalamin availability within dechlorinating communities involved in bioremediation processes.
Supplementary Material
ACKNOWLEDGMENTS
We thank Andrew Han for the identification of putative cobalamin riboswitches in Dhc195.
This research was supported by NIEHS Superfund project P42ES004705.
Footnotes
Published ahead of print 24 January 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.03477-13.
REFERENCES
- 1.Doherty RE. 2000. A history of the production and use of carbon tetrachloride, tetrachloroethylene, trichloroethylene and 1,1,1-trichloroethane in the United States. Part 2: trichloroethylene and 1,1,1-trichloroethane. J. Environ. Forensics 1:83–93. 10.1006/enfo.2000.0011 [DOI] [Google Scholar]
- 2.Stroo HF, Leeson A, Marqusee JA, Johnson PC, Ward CH, Kavanaugh MC, Sale TC, Newell CJ, Pennell KD, Lebron CA, Unger M. 2012. Chlorinated ethene source remediation: lessons learned. Environ. Sci. Technol. 46:6438–6447. 10.1021/es204714w [DOI] [PubMed] [Google Scholar]
- 3.Lemming G, Hauschild MZ, Chambon J, Binning PJ, Bulle C, Margni M, Bjerg PL. 2010. Environmental impacts of remediation of a trichloroethene-contaminated site: life cycle assessment of remediation alternatives. Environ. Sci. Technol. 44:9163–9169. 10.1021/es102007s [DOI] [PubMed] [Google Scholar]
- 4.Maymó-Gatell X, Chien YT, Gossett JM, Zinder SH. 1997. Isolation of a bacterium that reductively dechlorinates tetrachloroethene to ethene. Science 276:1568–1571. 10.1126/science.276.5318.1568 [DOI] [PubMed] [Google Scholar]
- 5.He JZ, Ritalahti KM, Yang KL, Koenigsberg SS, Löffler FE. 2003. Detoxification of vinyl chloride to ethene coupled to growth of an anaerobic bacterium. Nature 424:62–65. 10.1038/nature01717 [DOI] [PubMed] [Google Scholar]
- 6.Seshadri R, Adrian L, Fouts DE, Eisen JA, Phillippy AM, Methe BA, Ward NL, Nelson WC, Deboy RT, Khouri HM, Kolonay JF, Dodson RJ, Daugherty SC, Brinkac LM, Sullivan SA, Madupu R, Nelson KT, Kang KH, Impraim M, Tran K, Robinson JM, Forberger HA, Fraser CM, Zinder SH, Heidelberg JF. 2005. Genome sequence of the PCE-dechlorinating bacterium Dehalococcoides ethenogenes. Science 307:105–108. 10.1126/science.1102226 [DOI] [PubMed] [Google Scholar]
- 7.Schipp CJ, Marco-Urrea E, Kublik A, Seifert J, Adrian L. 2013. Organic cofactors in the metabolism of Dehalococcoides mccartyi strains. Philos. Trans. R. Soc. Lond. B Biol. Sci. 368:20120321. 10.1098/rstb.2012.0321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Löffler FE, Yan J, Ritalahti KM, Adrian L, Edwards EA, Konstantinidis KT, Müller JA, Fullerton H, Zinder SH, Spormann AM. 2013. Dehalococcoides mccartyi gen. nov., sp. nov., obligately organohalide-respiring anaerobic bacteria relevant to halogen cycling and bioremediation, belong to a novel bacterial class, Dehalococcoidia classis nov., order Dehalococcoidales ord. nov. and family Dehalococcoidaceae fam. nov., within the phylum Chloroflexi. Int. J. Syst. Evol. Microbiol. 63:625–635. 10.1099/ijs.0.034926-0 [DOI] [PubMed] [Google Scholar]
- 9.Yi S, Seth EC, Men YJ, Allen RH, Alvarez-Cohen L, Taga ME. 2012. Versatility in corrinoid salvaging and remodeling pathways supports the corrinoid-dependent metabolism of Dehalococcoides mccartyi. Appl. Environ. Microbiol. 78:7745–7752. 10.1128/AEM.02150-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Distefano TD, Gossett JM, Zinder SH. 1992. Hydrogen as an electron donor for dechlorination of tetrachloroethene by an anaerobic mixed culture. Appl. Environ. Microbiol. 58:3622–3629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Munder M, Vogt JRA, Vogler B, Renz P. 1992. Biosynthesis of vitamin B12 in anaerobic bacteria Experiments with Eubacterium limosum on the incorporation of d-[1-13C]erythrose and [13C]formate into the 5,6-dimethylbenzimidazole moiety. Eur. J. Biochem. 204:679–683. 10.1111/j.1432-1033.1992.tb16681.x [DOI] [PubMed] [Google Scholar]
- 12.Stupperich E, Eisinger HJ, Krautler B. 1988. Diversity of corrinoids in acetogenic bacteria p-cresolylcobamide from Sporomusa ovata, 5-methoxy-6-methylbenzimidazolylcobamide from Clostridium formicaceticum and vitamin B12 from Acetobacterium woodii. Eur. J. Biochem. 172:459–464. 10.1111/j.1432-1033.1988.tb13910.x [DOI] [PubMed] [Google Scholar]
- 13.Guimaraes DH, Weber A, Klaiber I, Vogler B, Renz P. 1994. Guanylcobamide and hypoxanthylcobamide: corrinoids formed by Desulfovibrio vulgaris. Arch. Microbiol. 162:272–276. 10.1007/BF00301850 [DOI] [Google Scholar]
- 14.Duhamel M, Edwards EA. 2007. Growth and yields of dechlorinators, acetogens, and methanogens during reductive dechlorination of chlorinated ethenes and dihaloelimination of 1,2-dichloroethane. Environ. Sci. Technol. 41:2303–2310. 10.1021/es062010r [DOI] [PubMed] [Google Scholar]
- 15.Freeborn RA, West KA, Bhupathiraju VK, Chauhan S, Rahm BG, Richardson RE, Alvarez-Cohen L. 2005. Phylogenetic analysis of TCE-dechlorinating consortia enriched on a variety of electron donors. Environ. Sci. Technol. 39:8358–8368. 10.1021/es048003p [DOI] [PubMed] [Google Scholar]
- 16.Yan J, Ritalahti KM, Wagner DD, Loffler FE. 2012. Unexpected specificity of interspecies cobamide transfer from Geobacter spp. to organohalide-respiring Dehalococcoides mccartyi strains. Appl. Environ. Microbiol. 78:6630–6636. 10.1128/AEM.01535-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Renz P. 1999. Biosynthesis of the 5,6-dimethylbenzimidazole moiety of cobalamin and of the other bases found in natural corrinoids, p 557–575 In Banerjee R. (ed), Chemistry and biochemistry of B12. John Wiley & Sons, Inc., New York, NY [Google Scholar]
- 18.Guerrero-Barajas C, Field JA. 2006. Enhanced anaerobic biotransformation of carbon tetrachloride with precursors of vitamin B12 biosynthesis. Biodegradation 17:317–329. 10.1007/s10532-005-9001-2 [DOI] [PubMed] [Google Scholar]
- 19.Stupperich E, Krautler B. 1988. Pseudo vitamin B12 or 5-hydroxybenzimidazolyl cobamide are the corrinoids found in methanogenic bacteria. Arch. Microbiol. 149:268–271. 10.1007/BF00422016 [DOI] [Google Scholar]
- 20.Yan J, Im J, Yang Y, Loffler FE. 2013. Guided cobalamin biosynthesis supports Dehalococcoides mccartyi reductive dechlorination activity. Philos. Trans. R. Soc. Lond. B Biol. Sci. 368:20120320. 10.1098/rstb.2012.0320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Men Y, Feil H, VerBerkmoes NC, Shah MB, Johnson DR, Lee PKH, West KA, Zinder SH, Andersen GL, Alvarez-Cohen L. 2012. Sustainable syntrophic growth of Dehalococcoides ethenogenes strain 195 with Desulfovibrio vulgaris Hildenborough and Methanobacterium congolense: global transcriptomic and proteomic analyses. ISME J. 6:410–421. 10.1038/ismej.2011.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shelobolina ES, Nevin KP, Blakeney-Hayward JD, Johnsen CV, Plaia TW, Krader P, Woodard T, Holmes DE, VanPraagh CG, Lovley DR. 2007. Geobacter pickeringii sp. nov., Geobacter argillaceus sp. nov. and Pelosinus fermentans gen. nov., sp. nov., isolated from subsurface kaolin lenses. Int. J. Syst. Evol. Microbiol. 57:126–135 [DOI] [PubMed] [Google Scholar]
- 23.Men YJ, Lee PKH, Harding KC, Alvarez-Cohen L. 2013. Characterization of four TCE-dechlorinating microbial enrichments grown with different cobalamin stress and methanogenic conditions. Appl. Microbiol. Biotechnol. 97:6439–6450. 10.1007/s00253-013-4896-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.He JZ, Holmes VF, Lee PKH, Alvarez-Cohen L. 2007. Influence of vitamin B12 and cocultures on the growth of Dehalococcoides isolates in defined medium. Appl. Environ. Microbiol. 73:2847–2853. 10.1128/AEM.02574-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.He JZ, Ritalahti KM, Aiello MR, Löffler FE. 2003. Complete detoxification of vinyl chloride by an anaerobic enrichment culture and identification of the reductively dechlorinating population as a Dehalococcoides species. Appl. Environ. Microbiol. 69:996–1003. 10.1128/AEM.69.2.996-1003.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Heidelberg JF, Seshadri R, Haveman SA, Hemme CL, Paulsen IT, Kolonay JF, Eisen JA, Ward N, Methe B, Brinkac LM, Daugherty SC, Deboy RT, Dodson RJ, Durkin AS, Madupu R, Nelson WC, Sullivan SA, Fouts D, Haft DH, Selengut J, Peterson JD, Davidsen TM, Zafar N, Zhou LW, Radune D, Dimitrov G, Hance M, Tran K, Khouri H, Gill J, Utterback TR, Feldblyum TV, Wall JD, Voordouw G, Fraser CM. 2004. The genome sequence of the anaerobic, sulfate-reducing bacterium Desulfovibrio vulgaris Hildenborough. Nat. Biotechnol. 22:554–559. 10.1038/nbt959 [DOI] [PubMed] [Google Scholar]
- 27.Brown SD, Podar M, Klingeman DM, Johnson CM, Yang ZK, Utturkar SM, Land ML, Mosher JJ, Hurt RA, Phelps TJ, Palumbo AV, Arkin AP, Hazen TC, Elias DA. 2012. Draft genome sequences for two metal-reducing Pelosinus fermentans strains isolated from a Cr(VI)-contaminated site and for type strain R7. J. Bacteriol. 194:5147–5148. 10.1128/JB.01174-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Johnson DR, Lee PKH, Holmes VF, Alvarez-Cohen L. 2005. An internal reference technique for accurately quantifying specific mRNAs by real-time PCR with a application to the tceA reductive dehalogenase gene. Appl. Environ. Microbiol. 71:3866–3871. 10.1128/AEM.71.7.3866-3871.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee PKH, Cheng D, Hu P, West KA, Dick GJ, Brodie EL, Andersen GL, Zinder SH, He JZ, Alvarez-Cohen L. 2011. Comparative genomics of two newly isolated Dehalococcoides strains and an enrichment using a genus microarray. ISME J. 5:1014–1024. 10.1038/ismej.2010.202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Affymetrix. 2009. GeneChip expression analysis technical manual. Affymetrix, Santa Clara, CA: http://media.affymetrix.com/support/downloads/manuals/expression_analysis_manual.pdf [Google Scholar]
- 31.West KA, Johnson DR, Hu P, DeSantis TZ, Brodie EL, Lee PKH, Feil H, Andersen GL, Zinder SH, Alvarez-Cohen L. 2008. Comparative genomics of “Dehalococcoides ethenogenes” 195 and an enrichment culture containing unsequenced “Dehalococcoides” strains. Appl. Environ. Microbiol. 74:3533–3540. 10.1128/AEM.01835-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge YC, Gentry J, Hornik K, Hothorn T, Huber W, Iacus S, Irizarry R, Leisch F, Li C, Maechler M, Rossini AJ, Sawitzki G, Smith C, Smyth G, Tierney L, Yang JYH, Zhang JH. 2004. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 5:R80. 10.1186/gb-2004-5-10-r80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Johnson DR, Brodie EL, Hubbard AE, Andersen GL, Zinder SH, Alvarez-Cohen L. 2008. Temporal transcriptomic microarray analysis of “Dehalococcoides ethenogenes” strain 195 during the transition into stationary phase. Appl. Environ. Microbiol. 74:2864–2872. 10.1128/AEM.02208-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Johnson DR, Nemir A, Andersen GL, Zinder SH, Alvarez-Cohen L. 2009. Transcriptomic microarray analysis of corrinoid responsive genes in Dehalococcoides ethenogenes strain 195. FEMS Microbiol. Lett. 294:198–206. 10.1111/j.1574-6968.2009.01569.x [DOI] [PubMed] [Google Scholar]
- 35.Benjamini Y, Hochberg Y. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. B 57:289–300 [Google Scholar]
- 36.Johnson DR, Lee PKH, Holmes VF, Fortin AC, Alvarez-Cohen L. 2005. Transcriptional expression of the tceA gene in a Dehalococcoides-containing microbial enrichment. Appl. Environ. Microbiol. 71:7145–7151. 10.1128/AEM.71.11.7145-7151.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Beller HR, Han RY, Karaoz U, Lim H, Brodie EL. 2013. Genomic and physiological characterization of the chromate-reducing, aquifer-derived firmicute Pelosinus sp. strain HCF1. Appl. Environ. Microbiol. 79:63–73. 10.1128/AEM.02496-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nawrocki EP, Kolbe DL, Eddy SR. 2009. Infernal 1.0: inference of RNA alignments. Bioinformatics 25:1335–1337. 10.1093/bioinformatics/btp157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mok KC, Taga ME. 2013. Growth inhibition of Sporomusa ovata by incorporation of benzimidazole bases into cobamides. J. Bacteriol. 195:1902–1911. 10.1128/JB.01282-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Banerjee R, Ragsdale SW. 2003. The many faces of vitamin B12: catalysis by cobalamin-dependent enzymes. Annu. Rev. Biochem. 72:209–247. 10.1146/annurev.biochem.72.121801.161828 [DOI] [PubMed] [Google Scholar]
- 41.Lobo SAL, Brindley AA, Romao CV, Leech HK, Warren MJ, Saraiva LM. 2008. Two distinct roles for two functional cobaltochelatases (CbiK) in Desulfovibrio vulgaris Hildenborough. Biochemistry 47:5851–5857. 10.1021/bi800342c [DOI] [PubMed] [Google Scholar]
- 42.Ryzhkova EP, Briukhanov AL. 2009. Effect of a corrinoid on Methanosarcina barkeri DNA synthesis. Mikrobiologiia 78:5–11 [PubMed] [Google Scholar]
- 43.Taga ME, Larsen NA, Howard-Jones AR, Walsh CT, Walker GC. 2007. BluB cannibalizes flavin to form the lower ligand of vitamin B12. Nature 446:449–453. 10.1038/nature05611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lamm L, Heckmann G, Renz P. 1982. Biosynthesis of vitamin B12 in anaerobic bacteria. Mode of incorporation of glycine into the 5,6-dimethylbenzimidazole moiety in Eubacterium limosum. Eur. J. Biochem. 122:569–571 [PubMed] [Google Scholar]
- 45.Vogt JR, Renz P. 1988. Biosynthesis of vitamin B12 in anaerobic bacteria. Experiments with Eubacterium limosum on the origin of the amide groups of the corrin ring and of N-3 of the 5,6-dimethylbenzimidazole part. Eur. J. Biochem. 171:655–659 [DOI] [PubMed] [Google Scholar]
- 46.Escalante-Semerena JC. 2007. Conversion of cobinamide into adenosylcobamide in bacteria and archaea. J. Bacteriol. 189:4555–4560. 10.1128/JB.00503-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Korkhov VM, Mireku SA, Locher KP. 2012. Structure of AMP-PNP-bound vitamin B12 transporter BtuCD-F. Nature 490:367–372. 10.1038/nature11442 [DOI] [PubMed] [Google Scholar]
- 48.West KA, Lee PK, Johnson DR, Zinder SH, Alvarez-Cohen L. 2013. Global gene expression of Dehalococcoides within a robust dynamic TCE-dechlorinating community under conditions of periodic substrate supply. Biotechnol. Bioeng. 110:1333–1341. 10.1002/bit.24819 [DOI] [PubMed] [Google Scholar]
- 49.Vitreschak AG, Rodionov DA, Mironov AA, Gelfand MS. 2003. Regulation of the vitamin B12 metabolism and transport in bacteria by a conserved RNA structural element. RNA 9:1084–1097. 10.1261/rna.5710303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Waller AS, Hug LA, Mo K, Radford DR, Maxwell KL, Edwards EA. 2012. Transcriptional analysis of a Dehalococcoides-containing microbial consortium reveals prophage activation. Appl. Environ. Microbiol. 78:1178–1186. 10.1128/AEM.06416-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Madigan MT, Martinko JM, Stahl DA, Clark DP. 2012. Brock biology of microorganisms, 13th ed. Pearson Education, Inc., San Francisco, CA [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.