Abstract
Streptococcus agalactiae, also known as group B Streptococcus (GBS), is a primary colonizer of the anogenital mucosa of up to 40% of healthy women and an important cause of invasive neonatal infections worldwide. Among the 10 known capsular serotypes, GBS type III accounts for 30 to 76% of the cases of neonatal meningitis. In recent years, the ability of GBS to form biofilm attracted attention for its possible role in fitness and virulence. Here, a new in vitro biofilm formation protocol was developed to guarantee more stringent conditions, to better discriminate between strong-, low-, and non-biofilm-forming strains, and to facilitate interpretation of data. This protocol was used to screen the biofilm-forming abilities of 366 GBS clinical isolates from pregnant women and from neonatal infections of different serotypes in relation to medium composition and pH. The results identified a subset of isolates of serotypes III and V that formed strong biofilms under acidic conditions. Importantly, the best biofilm formers belonged to serotype III hypervirulent clone ST-17. Moreover, the abilities of proteinase K to strongly inhibit biofilm formation and to disaggregate mature biofilms suggested that proteins play an essential role in promoting GBS biofilm initiation and contribute to biofilm structural stability.
INTRODUCTION
Streptococcus agalactiae, also known as group B Streptococcus (GBS), is a leading cause of invasive neonatal infections worldwide. It is a common colonizer of the gastrointestinal and urogenital tracts of up to 40% of healthy individuals (1). However, under certain circumstances, GBS can become a life-threatening pathogen causing invasive infections in human neonates (2, 3). Early-onset group B streptococcal disease occurs in infants less than 7 days old, and late-onset disease (LOD) occurs in infants between 7 and 89 days old. GBS is usually transmitted from mothers to newborns during childbirth (4), but it can also penetrate the human placenta (5), and in the case of LOD, it can be nosocomially acquired.
Historically, GBS isolates have been classified into 10 different serotypes according to their capsular polysaccharide composition (6, 7). Multiple surveillance studies have indicated that all serotypes are able to colonize the vagina and perianal region of pregnant women, but five serotypes (Ia, Ib, II, III, and V) are predominant and are also the most frequent in human infections (8–12). In particular, serotype III accounts for 30 to 76% of neonatal disease cases (13, 14). The use of multilocus sequence typing (MLST) allowed the classification of GBS isolates independently from their capsular serotypes and the identification of the bacterial genogroups more often associated with invasive infections in newborns (15). Serotype III isolates of a particular genotype cluster, sequence type 17 (ST-17), disproportionately cause late-onset GBS disease (15–19) and more frequently cause meningitis than other STs do (20). The precise mechanism by which ST-17 causes LOD more frequently than other STs do is not well understood, although recent evidence indicates that ST-17 displays a conserved specific combination of the secreted and surface-exposed proteins (21, 22).
Biofilm is known to facilitate the colonization by and the persistence of a large variety of bacterial and fungal species and to support the dissemination of virulent clones (23, 24). Organisms within biofilms can withstand nutrient deprivation, pH changes, oxygen radicals, disinfectants, and antibiotics better than planktonic organisms can (25). The majority of the species of the Streptococcus genus have been shown to form biofilm, while just a limited number of studies have demonstrated GBS biofilm formation in vitro (26–29). The glucose concentration in the culture medium was shown to modulate biofilm formation by GBS, although conflicting data have been reported regarding the biofilm-forming capacities of isolates of different serotypes and the correlation between biofilm formation and pH (26, 27, 29). We hypothesized that these contradictory results could be due to the absence of in vitro protocols that allow clear discrimination between strong and weak biofilm formers and unambiguous establishment of the role of bacterial culture conditions.
In the present work, a new in vitro biofilm formation protocol was used to evaluate the abilities of a large collection of GBS isolates to produce biofilm under different growth conditions. The protocol permitted us to clearly demonstrate that GBS biofilm formation is enhanced at acid pH and to identify a subset of serotype III strains of ST-17 as strong biofilm formers. The contributions of DNA, capsule, and proteins to the induction of bacterial adherence were also investigated.
MATERIALS AND METHODS
Strains.
A total of 366 S. agalactiae isolates of eight different serotypes (Ia, n = 58; Ib, n = 18; II, n = 28; III, n = 156; IV, n = 10; V, n = 57; VIII, n = 3; IX, n = 13) and nontypeable strains (n = 23) were included in this study. Among these were 357 vaginorectal isolates obtained from pregnant women (n = 272) and isolates from neonates (n = 85, of which 64 were obtained from a sterile site) in Belgium, Bulgaria, the Czech Republic, Denmark, Germany, Great Britain, Italy, and Spain. These isolates were collected during the DEVANI (Design of a vaccine against Neonatal Infections) project supported by the European Commission Seventh Framework. Strains CJB111 (serotype V), 515 (serotype Ia), COH1 (serotype III), H36B (serotype Ib), 18RS21 (serotype II), A909 (serotype Ia), and D136C (serotype III) were kindly provided by Dennis Kasper (Harvard Medical School, Boston, MA). Strain 2603 V/R (serotype V) (30) was obtained from the Istituto Superiore di Sanità. The COH1 unencapsulated mutant carries a deletion of the cpsE gene in the capsule locus (31) and was kindly provided by M. Cieslewicz (Channing Laboratory, Brigham and Women's Hospital, Harvard Medical School, Boston, MA).
Serotype and ST-17 identification.
GBS strains were typed by the latex agglutination method (Strep-B-Latex kit; Statens Serum Institut, Copenhagen, Denmark) as described by Afshar et al. (32). ST-17 identification was performed for all of the 156 serotype III strains tested. PCR amplification and sequencing of the internal fragments of seven housekeeping genes, namely, adhP, atr, glcK, glnA, pheS, sdhA, and tkt, were performed as described previously (15). Assignment to ST-17 was performed at the GBS MLST website (http://pubmlst.org/sagalactiae/). The strains showing at least an allele sequence not corresponding to the ST-17 profile were classified as non-ST-17 strains.
Growth experiments.
Four clinical isolates (three biofilm-forming strains and one non-biofilm-forming strain) grown overnight at 37°C in Todd-Hewitt broth (THB) at pH 7.8 were diluted to an optical density at 600 nm (OD600) of 0.05 in optical tubes containing 10 ml of pH 7.8 THB, pH 7.8 THB supplemented with 1% glucose, or pH 5.0 THB. The tubes were then incubated without shaking at 37°C and the OD600 was measured for 8 to 10 h. Each experiment was performed in triplicate.
Standard biofilm formation protocol.
The standard biofilm formation protocol was performed as already described (26). In brief, S. agalactiae strains were streaked onto blood agar plates and grown for 18 h at 37°C.
GBS strains grown overnight in pH 7.8 THB were diluted 1:20 in THB and THB supplemented with 1% glucose and used to inoculate (100 μl/well) 96-well polystyrene microtiter plates (Costar; Corning Inc., Corning, NY). Plates were incubated without shaking at 37°C for 18 h aerobically in 5% CO2. The supernatant was removed, and the wells were subjected to three cycles of washing with 200 μl of double-distilled H2O (ddH2O) to remove unattached bacteria. A crystal violet (CV) assay and a 2,3-bis(2-methoxy-4-nitro-5-sulfophenyl)-5-[(phenylamino)carbonyl]-2H-tetrazolium hydroxide (XTT) assay were then performed to estimate bacterial biomass and cell viability, respectively.
New biofilm formation protocol.
A new protocol for in vitro biofilm formation was set up. S. agalactiae strains were streaked onto blood agar plates and grown at 37°C for 18 h. A bacterial suspension in THB was diluted to an OD600 of 0.05 and used to inoculate (200 μl/well) 96-well polystyrene microtiter plates (Costar; Corning Inc., Corning, NY). The preliminary protocol evaluation was performed with THB and THB supplemented with 1% glucose, as already used in the standard biofilm formation protocol. The other media used to investigate the role of pH in biofilm formation were (i) RPMI GlutaMAX (Gibco-Life Technologies, Milan, Italy), (ii) RPMI GlutaMAX supplemented with 1% glucose, (iii) RPMI GlutaMAX and THB both acidified to pH 5.0, and (iv) THB supplemented with 1% glucose and buffered at pH 7.8 with the addition of HEPES (20 to 200 mM) or Tris-HCl (20 to 200 mM). The plate was sealed to limit oxygen exchange and shaken at 60 rpm at 37°C to reduce bacterial deposition. Following 8 h of adhesion at 37°C, the plates were washed to remove loosely adherent cells and the supernatant was replaced with 200 μl of fresh medium. After 15 h at 37°C, the medium was removed and the wells were subjected to three cycles of washing with 200 μl of phosphate-buffered saline (PBS) to remove unattached bacteria. CV and XTT assays were then performed to estimate bacterial biomass and cell viability, respectively.
CV assay.
The wells were stained for 10 min with 200 μl of a 0.5% (wt/vol) solution of CV (Sigma-Aldrich, Inc., St. Louis, MO). After rinsing with ddH2O, bound dye was released from the stained cells with 30% glacial acetic acid. Biofilm formation was quantified by measuring the OD540 of the solution with a microplate reader (Infinite M200; Tecan). Samples showing an OD540 higher than 1 were diluted 5 and 20 times in water, and the absorbance reading was repeated. The measured values were subtracted from the blank and then multiplied by the dilution factors. Each assay was performed in triplicate. Wells filled with growth medium were included as negative controls.
XTT viability assay.
XTT is a tetrazolium derivative cleaved to an orange-colored formazan product by mitochondrial dehydrogenase in viable cells (33). The XTT solution was prepared by dissolving 0.5 mg XTT (Sigma-Aldrich, Inc., St. Louis, MO) in 1 ml of PBS and then supplementing it with 2.5 μl of a 10 mM menadione stock solution (dissolved in acetone). A 150-μl volume of XTT-menadione solution was added to each well. Plates were incubated in the dark for 3 h at 37°C and then centrifuged for 20 min at 4,000 rpm. One-hundred-microliter volumes of the supernatant were transferred to the wells of a new 96-well flat-bottom plate, and the absorbance at 490 nm was measured with a microplate reader (Infinite M200; Tecan).
CLSM.
GBS biofilms obtained by both the standard protocol (26) and the new protocol used in this study were visualized by confocal laser scanning microscopy (CLSM). S. agalactiae strains were inoculated (0.8 ml/well) into a Lab-Tek II eight-well 1.5 cover glass (VWR, Rochester, NY) containing THB or THB supplemented with 1% glucose and incubated as already described in the new biofilm formation protocol paragraph. Adherent bacteria were stained for 30 min with LIVE/DEAD BacLight fluorescent stain (Molecular Probes, Eugene, OR) and fixed with 2% formaldehyde for 30 min at room temperature. Samples were analyzed with a Zeiss LSM710 confocal microscope by using a Plan-Apochromat 40×/1.3 objective. SYTO 9 fluorescence, corresponding to live bacteria, was acquired in the green channel (492 to 572 nm), and propidium iodide fluorescence, which does not penetrate viable bacterial cells, was acquired in the red channel (566 to 719 nm). Images were acquired by using Zen 2008 software and modified with Volocity (Improvision, Lexington, MA). ImageJ (http://rsbweb.nih.gov/ij/) and COMSTAT2 (http://www.comstat.dk) were used to evaluate the biomass and maximum and mean thicknesses of the three-dimensional biofilm images acquired by CLSM (34).
Enzymatic inhibition/eradication of biofilms.
The MIC, minimal biofilm inhibition concentration (MBIC), and minimal biofilm eradication concentration (MBEC) were measured in 96-well polystyrene microtiter plates (Costar; Corning Inc., Corning, NY) with 0.4 to 200 μg/ml proteinase K and a maximum of 200 μg/ml DNase. For MBIC determination, the biofilm formation assays were performed as described previously, with THB supplemented with 1% glucose and proteinase K or DNase. For MBEC determination, a 24-h-old biofilm grown in the absence of proteinase K or DNase was washed twice with PBS and further incubated in THB supplemented with 1% glucose containing proteinase K or DNase for 3 h at 37°C. Biofilm was quantified by CV assay.
Quantification of capsular polysaccharides.
Capsular polysaccharides were extracted from strain COH1 and four strong/weak biofilm-forming strains (two expressing type III and two expressing type V capsular polysaccharide) and quantified by the resorcinol-hydrochloric acid assay as described earlier by Svennerholm (35).
Bacteria were inoculated into 15 ml of THB or pH 5.0 THB and grown to an OD600 of 1.8 or 1.0, respectively. The cells were pelleted, washed twice with PBS, suspended in 0.8 M NaOH, and incubated for 48 h at 37°C. Following neutralization with HCl, the insoluble material was removed by centrifugation; the supernatant was transferred to an Amicon Ultra-10 (Millipore, Bedford, MA), concentrated to 0.20 ml, and then perfused two times with 1 ml of ddH2O.
A final volume of 1.5 ml of supernatant was analyzed to determine the amount of extracted polysaccharide. Briefly, 500 μl of resorcinol-HCl reagent (2% [wt/vol] aqueous resorcinol solution added to concentrated HCl and 0.1 M CuSO4) was added to 500 μl of an extracted polysialic acid sample, which was then heated in a boiling oil bath for 20 min. The sialic acid (N-acetylneuraminic acid [NeuNAC]) released reacts with resorcinol in the presence of copper sulfate under reducing conditions to give a blue-purple color. After it cooled to room temperature, the absorbance at 564 nm was measured. NeuNAC concentrations were calculated from standard curves obtained with NeuNAC standards (range, 5 to 25 μg/ml) and converted to total GBS saccharide (conversion factor = molecular weight of NeuNAC/molecular weight of repeat unit GBS polysaccharide).
Statistical analysis.
Statistical analysis of biofilm formation by the panel of 366 GBS strains was performed with GraphPad (GraphPad Software, La Jolla, CA). The significance of differences in the relative amounts of biofilm produced by the different S. agalactiae strains under each test condition was assessed by univariate analysis of variance (ANOVA). Significance was assigned to P values of ≤0.05.
RESULTS
A novel in vitro biofilm formation protocol.
The in vitro protocols used to date to determine the biofilm formation abilities of different GBS isolates resulted in small differences between the strains tested and did not allow clear discrimination between strong and weak biofilm-forming GBS strains. Here, a novel experimental protocol was implemented to overcome this limitation. The main differences from the previously described standard protocol (see Materials and Methods) (26) were (i) replacement of culture medium with fresh medium after 8 h of incubation to remove nonadherent bacteria and (ii) incubation of the plates under shaking conditions to minimize nonspecific bacterial deposition on the bottom of the plate.
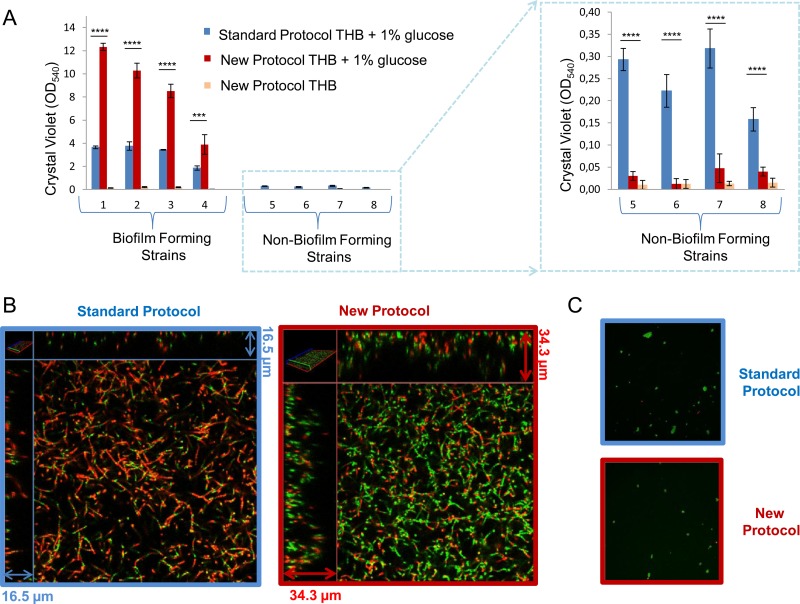
The two in vitro protocols were compared by testing four GBS strains previously shown to be strong biofilm formers and four weak biofilm formers or non-biofilm formers (26). Biofilm formation was investigated by growing the bacteria in THB in the absence or presence of glucose and measuring bacterial biomass by staining with CV as described in Materials and Methods. In the absence of glucose, no biofilm formation by any of the strains tested was observed with the new protocol (Fig. 1A), as already reported by Rinaudo et al. (26), who used the standard protocol. Moreover, biofilm formation increased when glucose was added to the culture medium in both protocols. By using the new protocol in the presence of glucose, CV assay values 2- to 4-fold higher than those achieved with the standard procedure were obtained in the case of strong biofilm formers (strains 1 to 4 in Fig. 1A), while the CV assay values of weak biofilm formers were drastically reduced (strains 5 to 8 in Fig. 1A).
FIG 1.
Comparison of biofilm formation protocols. (A) Biofilm formation abilities of four strong biofilm-forming strains and four weak biofilm formers or non-biofilm formers produced by the standard protocol and the new protocol. The GBS strains were grown in THB or in THB supplemented with 1% glucose. Surface-attached cells were CV stained and quantified by measuring the OD540. The mean values and standard deviations of three independent experiments are shown. Asterisks denote statistically significant difference determined by Student's t test (***, P < 0.001; ****, P < 0.0001). (B) CLSM of biofilms formed by a strong biofilm-forming strain with the standard protocol and the new protocol in THB in the presence of 1% glucose. (C) CLSM of the biofilm produced by a non-biofilm-forming strain with the standard protocol and the new protocol in THB in the presence of 1% glucose. Biofilms were stained with a LIVE/DEAD viability kit. Live and dead cells are green and red, respectively.
Confocal microscopy analysis of biofilms produced by a representative strong biofilm-forming strain confirmed that the new protocol permitted the formation of a thicker (average thickness, 17.1 versus 2.1 μm) and more homogeneous biofilm than that obtained by the standard protocol (Fig. 1B). The biofilms produced by the new protocol not only show a higher biovolume (8.9 versus 1.1 μm3/μm2) but also contain a higher percentage of viable cells, as suggested by the different green/red ratios in Fig. 1B and by the results obtained with the XTT cell viability assay (see Fig. S1 in the supplemental material). The same analysis with a representative non-biofilm-forming strain confirmed that CV assay values of 0.3 to 0.5 do not correspond to homogeneous biofilms (Fig. 1C). Overall, these results suggest that the newly developed protocol allows better discrimination between strong biofilm formers and non-biofilm-forming strains.
An acidic pH promotes biofilm formation by S. agalactiae.
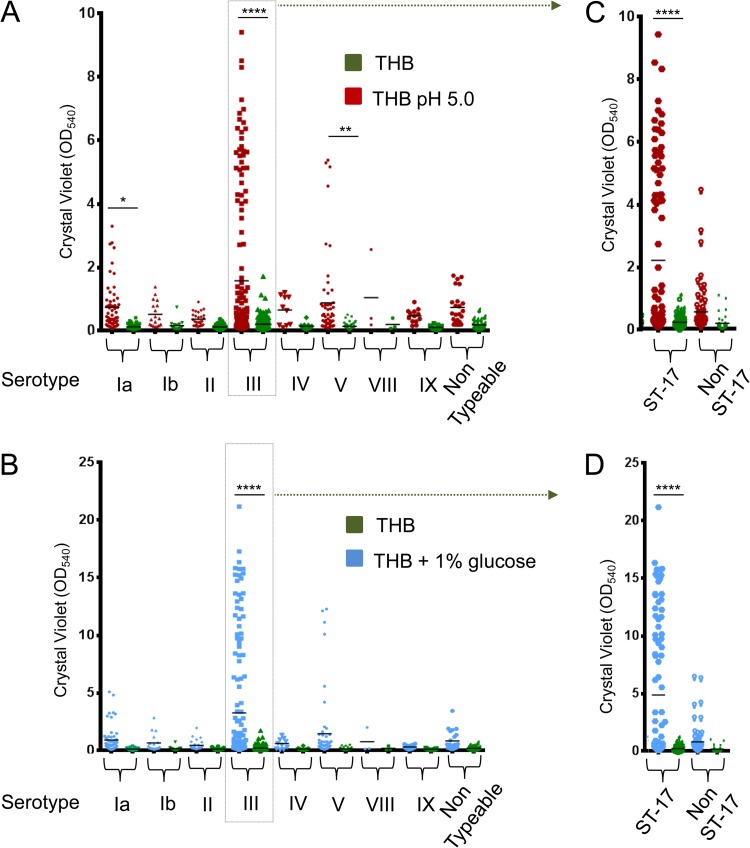
To clarify the role of pH in biofilm formation by the different GBS serotypes, biofilm assays were performed with a selection 366 S. agalactiae isolates of eight serotypes grown in THB (starting pH, 7.8) with or without 1% glucose and in THB at pH 5.0 without supplemented glucose. Similar to the results shown in Fig. 1, no significant biofilm formation by any of the isolates was detected in the presence of THB at neutral pH. On the other hand, both the use of an acidified medium (Fig. 2A) and the addition of glucose at neutral pH (Fig. 2B) resulted in a general increase in biofilm formation, with some of the isolates reaching very high CV assay values. In the absence of glucose, a 3- to 8-fold increase in the mean CV assay value was observed for isolates of all of the serotypes grown at pH 5.0 in comparision with the same strain grown at neutral pH.
FIG 2.
Effects of an acidic pH and glucose on GBS biofilm formation. Shown are the biofilm-forming abilities of 366 GBS clinical isolates of eight different serotypes and nontypeable strains grown in THB (green dots) or pH 5.0 THB (red dots) (A) or in THB (green dots) or THB supplemented with 1% glucose (red dots) (B). Panels C and D focus on serotype III strains clustered into ST-17 and non-ST-17 groups. Biofilm formation was evaluated by CV assay. Each dot represents the mean value of three independent experiments performed with each isolate. Asterisks denote statistically significant differences determined by ANOVA (*, P < 0.05; **, P < 0.01; ****, P < 0.0001).
The lower CV assay values observed under low-pH conditions in the absence of glucose than at pH 7.8 in the presence of glucose are most probably due to slower bacterial growth at a lower starting pH (see Fig. S2 in the supplemental material). However, no significant growth rate differences between biofilm-forming and non-biofilm-forming strains were observed in each of the media tested (see Fig. S2 in the supplemental material).
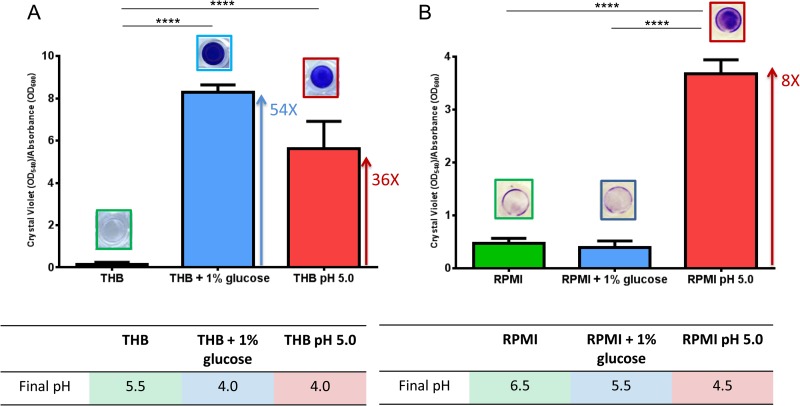
As already reported for S. pyogenes (34), the production of organic acids associated with the metabolism of glucose could induce a pH decrease and be the direct cause of the observed effect on GBS biofilm formation in glucose-rich medium. To better investigate the kinetics of biofilm formation in the presence of glucose and at low pH, we carried out time course biofilm assays by using a representative GBS that is a strong biofilm former and three different types of medium (pH 7.8 THB, pH 7.8 THB supplemented with 1% glucose, and pH 5.0 THB without glucose). A significant increment in the CV assay value (see blue and red solid lines in Fig. S3A in the supplemental material) was already observed after 4.5 to 5 h of incubation in pH 7.8 THB with 1% glucose, corresponding to a drop in pH to values lower than 5.0 (see blue and red dotted lines in Fig. S3A in the supplemental material), or in pH 5.0 THB without glucose. On the contrary, no biofilm formation was observed after the same strain was incubated in pH 7.8 THB, where the culture pH never reached values below 5.0 (see green dotted line in Fig. S3A in the supplemental material). A similar pH profile was also detected for a representative non-biofilm-forming strain in all tested media (see Fig. S3B in the supplemental material). To confirm that pH 5.0 is the signal sensed by the bacteria to start biofilm formation, a representative biofilm-forming GBS was grown in nutrition-limited RPMI GlutaMAX medium either in the presence of 1% glucose or at pH 5.0. In this case, differently from THB (Fig. 3A), the pH of the culture did not drop below 5.0 in the presence of glucose and no significant biofilm formation was observed. In contrast, significant biofilm formation (CV assay value/OD600 ratio of 3.6) was observed in the presence of RPMI GlutaMAX at pH 5.0 (Fig. 3B). Moreover, biofilm-forming GBS bacteria grown in THB supplemented with glucose and buffered with Tris or HEPES at concentrations that do not affect bacterial growth showed significantly reduced biofilm formation at final pH values higher than 4.5, in agreement with data obtained with RPMI (see Fig. S4 in the supplemental material).
FIG 3.
An acidic pH and not glucose induces GBS biofilm formation. Comparison of the biofilm formation abilities of a strong biofilm-forming strain grown in THB, THB supplemented with 1% glucose, or pH 5.0 THB (A) or in RPMI, RPMI supplemented with 1% glucose, or pH 5.0 RPMI (B). Biofilm formation was evaluated by CV assay. Bacterial growth in planktonic form was evaluated by measuring OD600. pH values were measured with pH test strips (pH increment, 0.2). To normalize the biofilm-forming ability with cell growth, the y axis shows the ratio of CV assay values to OD600 values. The mean values and standard deviations of three independent experiments are shown. Asterisks denote statistically significant difference, determined by Student's t test (****, P < 0.0001).
Most strong biofilm-forming GBS bacteria belong to the hypervirulent ST-17 lineage.
Looking for a relationship between the different serotypes and biofilm production capacity, the data on the different isolates presented in Fig. 2 were clustered on the basis of their capsular serotypes. The highest mean CV assay value increase and the highest number of strong biofilm-forming strains (CV assay values of >3) were found to belong to serotype III (Fig. 2; Tables 1 and 2), although not all type III strains appeared to be good biofilm formers. In fact, none of the two GBS type III strains tested whose genome sequences were already available, COH1 and D136C, showed a significant ability to produce biofilm, either at an acidic pH or in the presence of glucose (see Fig. S5 in the supplemental material). Among the 156 type III isolates tested, 41 (26.4%) and 35 (22.4%) formed biofilms with CV assay values of >3 in the presence of glucose and at low pH, respectively.
TABLE 1.
Effects of pH 5.0 THB versus THB on biofilm formation by different GBS serotypesa
| Serotype | Mean CV assay value |
No. (%) of strains with CV assay values of >3 | MeanpH 5.0 THB/meanTHB | Mean difference | Adjusted P value | |
|---|---|---|---|---|---|---|
| pH 5.0 THB | THB | |||||
| Ia | 0.74 | 0.13 | 1 (8.6) | 5.69 | 0.61 | 0.0481 |
| Ib | 0.52 | 0.17 | 0 (0) | 3.06 | 0.35 | 0.9851 |
| II | 0.37 | 0.13 | 0 (0) | 2.85 | 0.24 | 0.9952 |
| III | 1.58 | 0.21 | 35 (22.4) | 7.52 | 1.37 | <0.0001 |
| IV | 0.66 | 0.14 | 0 (0) | 4.71 | 0.52 | 0.9740 |
| V | 0.88 | 0.14 | 4 (7.0) | 6.29 | 0.74 | 0.0083 |
| VIII | 1.05 | 0.20 | 0 (0) | 5.25 | 0.85 | 0.9864 |
| IX | 0.47 | 0.11 | 0 (0) | 4.27 | 0.36 | 0.9938 |
| Nontypeable | 0.74 | 0.21 | 0 (0) | 3.52 | 0.53 | 0.6693 |
| III (ST-17) | 2.20 | 0.22 | 33 (36.3) | 10.00 | 1.98 | <0.0001 |
| III (non-ST-17) | 0.55 | 0.18 | 2 (3.1) | 3.06 | 0.37 | 0.3184 |
GBS biofilm formation was quantified by CV staining. Note that the percentage of biofilm formers with CV assay values of >3 is referred to the number of strains of each serotype (e.g., 156 for serotype III). Significant increments of mean CV assay values are in bold.
TABLE 2.
Effects of THB plus 1% glucose versus THB on biofilm formation by different GBS serotypesa
| Serotype | Mean CV assay value |
No. (%) of strains with CV assay values of >3 | MeanTHB + 1% glucose/meanTHB | Mean difference | Adjusted P value | |
|---|---|---|---|---|---|---|
| THB + 1% glucose | THB | |||||
| Ia | 0.91 | 0.13 | 5 (8.6) | 7.00 | 0.78 | 0.6254 |
| Ib | 0.66 | 0.17 | 0 (0) | 3.88 | 0.49 | 0.9995 |
| II | 0.45 | 0.13 | 0 (0) | 3.46 | 0.32 | >0.9999 |
| III | 3.26 | 0.21 | 41 (26.3) | 15.52 | 3.05 | <0.0001 |
| IV | 0.61 | 0.14 | 0 (0) | 4.36 | 0.47 | >0.9999 |
| V | 1.46 | 0.14 | 6 (10.5) | 10.43 | 1.32 | 0.0568 |
| VIII | 0.76 | 0.20 | 0 (0) | 3.80 | 0.56 | >0.9999 |
| IX | 0.31 | 0.11 | 0 (0) | 2.82 | 0.20 | >0.9999 |
| Nontypeable | 0.83 | 0.21 | 1 (4.3) | 3.95 | 0.62 | 0.9900 |
| III (ST-17) | 4.88 | 0.22 | 37 (40.6) | 22.27 | 4.68 | <0.0001 |
| III (non-ST-17) | 0.81 | 0.18 | 4 (6.1) | 4.37 | 0.62 | 0.9187 |
For details, see Table 1, footnote a.
We subsequently investigated whether the subset of type III strong biofilm-forming strains belonged to the hypervirulent ST-17 clone (Fig. 2C and D). More than 36% of the 91 ST-17 strains showed CV assay values of >3 in the presence of glucose (Table 2) or at low pH (Table 1), while less than 6.1% of the 65 non-ST-17 strains showed CV assay values of >3 under the same conditions. Also, the mean CV assay value and the mean increment of the ST-17 strains were higher than those of the serotype III strains, while the values of the non-ST-17 group were comparable to those of the other serotypes (Tables 1 and 2), confirming a strong correlation between the hypervirulent ST-17 clone and GBS biofilm formation.
The possible relationship between strong biofilm formation and GBS virulence was further investigated by clustering the data presented in Fig. 2 on the basis of GBS strain origins. A higher frequency of strong biofilm formers was observed in the 85 neonatal isolates than in the 272 in the colonizing group (18 versus 8%, P = 0.12). This difference was even larger when the colonizing group was compared with the subset of infective neonatal isolates obtained from a sterile site (27 versus 8%, P = 0.017). These results are clearly associated with the higher prevalence of ST-17 in the neonatal groups (40 out of 64 infective neonatal strains versus 50 out of 272 in the colonizing group). All four non-type-III strong biofilm formers belong to serotype V, and of these, two were isolated from infected neonates.
Role of capsule expression in GBS biofilm formation.
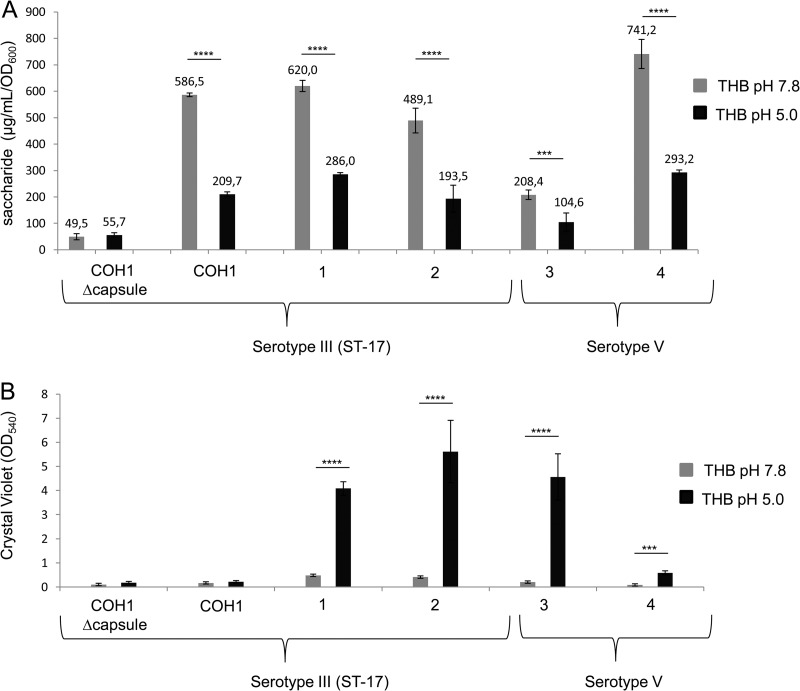
To verify the possible involvement of differential capsule expression during biofilm formation at low pH, the sialic acid contents of four biofilm-forming strains (two type III ST-17 and two type V isolates) were estimated by Svennerholm's method (35) under conditions activating a planktonic or sessile GBS lifestyle. The amount of sialic acid produced by GBS grown at pH 5.0 was 50 to 60% lower than that produced by bacteria grown at pH 7.8 (Fig. 4A), in agreement with results already reported in the literature (36). In the biofilm-forming strains, capsule reduction corresponded to increased CV assay values (Fig. 4B), suggesting an inverse relationship between capsule expression and biofilm formation. However, a comparable reduction in the amount of sialic acid was also observed at pH 5.0 for COH1, a non-biofilm-forming ST-17 strain (Fig. 4), suggesting that capsule reduction per se is not sufficient to induce biofilm formation in the entire ST-17 clone.
FIG 4.
Correlation between capsule expression and pH. Shown is an evaluation of the capsule amounts (A) and biofilm formation (B) of four serotype III and two serotype V biofilm-forming and non-biofilm-forming strains at pH 7.8 (gray) and pH 5.0 (black). Capsular polysaccharides were isolated and quantified with the resorcinol-hydrochloric acid assay by Svennerholm's method. Surface-attached cells were CV stained, and the OD540 was measured. Unencapsulated COH1 was used as a control. The mean values and standard deviations of three independent experiments are shown. Asterisks denote statistically significant differences determined by Student's t test (***, P < 0.001; ****, P < 0.0001).
Proteins play a significant role in GBS biofilm formation and maintenance.
To understand what type of low-pH-induced factors could determine GBS biofilm formation and structural stability, biofilm inhibition and eradication in six strong biofilm-forming strains (three ST-17 strains expressing type III and three expressing type V capsular polysaccharide) were evaluated in the presence of proteinase K and DNase. The maximum concentrations of proteinase K (200 μg/ml) and DNase (200 μg/ml) used in MBIC and MBEC determinations do not affect planktonic bacterial growth (MIC, ≥200 μg/ml). Almost total inhibition and eradication of 24-h-old biofilms were observed for all of the strains tested in the presence of 3 μg/ml proteinase K (Fig. 5). The same effect was also observed when the concentration of proteinase K was decreased to 0.4 μg/ml (see Fig. S6 in the supplemental material). The addition of 200 μg/ml DNase resulted in low inhibition and partial disruption of the biofilm, but the effect was weaker than that observed with proteinase K (Fig. 5).
FIG 5.
Enzymatic inhibition or eradication of GBS biofilm. Shown are the inhibition (A) and eradication (B) of biofilms formed by three serotype III and three serotype V biofilm-forming strains by proteinase K or DNase. Surface-attached cells were quantified by CV assay. The mean values and standard deviations of three independent experiments are shown.
These results suggest that an acidic pH could be important, specifically in ST-17 strains, to regulate the expression and/or promote the exposure of surface-associated proteins, inducing bacterial adhesion and contributing to biofilm structural stability.
DISCUSSION
Recent studies have demonstrated GBS biofilm formation in vitro (26–29), although the data regarding the effects of pH and medium composition are controversial. In a recent study, Ho et al. (37) found that a low pH induced biofilm formation in nutrient-limited medium (M9YE) but not in THB. Borges et al. (27), Kaur et al. (28), and Yang et al. (38) found that GBS produced a greater amount of biofilm at pH 6.5 than at pH 4.2, and Konto-Ghiorghi et al. (29) reported that a uniform biofilm was produced only on LB and RPMI 1640 supplemented with 1% glucose and not on THB. Manetti et al. (39) observed that in S. pyogenes, the presence of glucose resulted in autoacidification of the medium and consequently biofilm formation. Rinaudo et al. (26) demonstrated that the presence of 1% glucose in THB induces biofilm formation by GBS.
A major limitation of the static protocol used to screen for biofilm formation in most studies is the absence of the fluid circulation encountered in the host. Konto-Ghiorghi et al. (29) showed that a protocol under low-flow conditions was preferable to a static-condition protocol for GBS adherence to epithelial cells. To approach the conditions of the laminar-flow chamber system while maintaining the throughput of multiwell-based protocols, we developed an in-batch in vitro protocol including a medium replacement step. The biofilm produced by strong biofilm-forming strains with the new protocol was thicker and more homogeneous than that obtained with the standard protocol (26) (Fig. 1). In contrast, weak biofilm-forming or non-biofilm-forming strains produced much-reduced biofilms with this protocol (Fig. 1). Appling the new protocol to a large number of GBS clinical isolates, we obtained unequivocal evidence that an acidic pH induces GBS biofilm formation in both nutrition-rich (THB) and nutrition-limited (RPMI) environments (Fig. 3).
These data clarify previous observations regarding the role of pH during biofilm formation by GBS and revealed, for the first time, a significant divergence between different GBS serotypes.
Interestingly, the majority of the strong biofilm-forming GBS bacteria are serotype III ST-17 strains, suggesting an ST-biofilm correlation (Fig. 2). A higher frequency of strong biofilm formers was also observed among the infective neonatal isolates than in the colonizing group included in our strain collection. This result is clearly associated with the higher prevalence of ST-17 in our neonatal group (Fig. 2). In fact, the ST-17 GBS lineage is more prevalent in infected neonates than in other lineages, particularly during late-onset disease, and is considered a highly virulent clone (18, 20, 21, 40). A high heterogeneity in the ability to produce biofilm was observed not only between serotypes but also within the very homogeneous hypervirulent ST-17 clone (40, 41) (Fig. 2).
A general reduction of the capsule amount was observed in biofilm-forming serotype III and V strains in response to a pH decrease (Fig. 4). Although this capsule reduction correlated with increased biofilm formation by some, but not all, of the ST-17 isolates tested, such as COH1 (Fig. 4), capsule downregulation at pH 5.0 is not sufficient per se to ensure biofilm formation by all ST-17 strains. If the presence of the capsule favors bacterial escape from complement-mediated killing (42) in genital tracts with a low pH, where capsule formation is downregulated, the biofilm lifestyle of ST-17 strains may represent an alternative strategy to guarantee their persistence.
Proteinase K, at a concentration that does not affect cell growth, inhibited biofilm formation and induced biofilm detachment (Fig. 5), suggesting that proteins play a major role in promoting bacterial adhesion and biofilm structural stability (Fig. 5). These results suggest that an acidic pH may be important in unmasking surface-associated proteins that promote adhesion by biofilm-forming bacterial strains. Alternatively, an acidic pH may upregulate the expression of some surface-associated proteins specifically in biofilm-forming GBS bacteria. It has already been shown that an acidic pH can modulate the expression of a large number of proteins in GBS, including the proteins involved in surface adhesion (43, 44). Several proteins have already been reported to be important for ST-17 strain adhesion to solid surfaces or attachment to host cells or extracellular matrix (21, 42, 45, 46). Our data suggest that the contradictory reports about the effects of pH on GBS adherence to epithelial cells (44, 47–49), like previous reports about the effects of pH on biofilm formation, could be due to the limited number of GBS strains tested and the methods used.
Further efforts are necessary to (i) understand if the effects of pH on biofilm formation by different GBS serotypes correlate with the ability to adhere to vaginal cells and (ii) identify those proteins unmasked or differently regulated by a low pH in strong biofilm formers and non-biofilm formers (e.g., COH1) of the ST-17 lineage. The identification of those proteins that promote cell adhesion in biofilm-forming GBS strains will lead to a better understanding of the mechanism of biofilm formation by GBS and allow the design of new therapeutic approaches against this pathogen.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to R. Cozzi, C. D. Rinaudo, and R. Rosini for helpful discussions about GBS. We thank S. Bonacci for his support in ST-17 determination and C. Brettoni for her support in the experimental design.
The DEVANI project was supported by the European Commission Seventh Framework (grant agreement 200481).
We have no competing interests.
The contributing members of the DEVANI Study Group were P. Melin (Belgium), A. Decheva and B. Petrunov (Bulgaria), P. Kriz (Czech Republic), R. Berner, A. Büchele, M. Hufnagel, and M. Kunze (Germany), R. Creti, L. Badassari, A. Berardi, and G. Orefici (Italy), J. R. Granger and M. De La Rosa Fraile (Spain), and B. Afshar and A. Efstratiou (United Kingdom).
Footnotes
Published ahead of print 31 January 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.03627-13.
REFERENCES
- 1.Wilkinson HW. 1978. Group B streptococcal infection in humans. Annu. Rev. Microbiol. 32:41–57. 10.1146/annurev.mi.32.100178.000353 [DOI] [PubMed] [Google Scholar]
- 2.Fry RM. 1938. Prevention and control of puerperal sepsis: bacteriological aspects. BMJ 2:340–342. 10.1136/bmj.2.4049.340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schuchat A. 1999. Group B streptococcus. Lancet 353:51–56. 10.1016/S0140-6736(98)07128-1 [DOI] [PubMed] [Google Scholar]
- 4.Schuchat A. 1998. Epidemiology of group B streptococcal disease in the United States: shifting paradigms. Clin. Microbiol. Rev. 11:497–513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Whidbey C, Harrell MI, Burnside K, Ngo L, Becraft AK, Iyer LM, Aravind L, Hitti J, Waldorf KM, Rajagopal L. 2013. A hemolytic pigment of group B Streptococcus allows bacterial penetration of human placenta. J. Exp. Med. 210:1265–1281. 10.1084/jem.20122753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lindahl G, Stalhammar-Carlemalm M, Areschoug T. 2005. Surface proteins of Streptococcus agalactiae and related proteins in other bacterial pathogens. Clin. Microbiol. Rev. 18:102–127. 10.1128/CMR.18.1.102-127.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Slotved HC, Kong F, Lambertsen L, Sauer S, Gilbert GL. 2007. Serotype IX, a proposed new Streptococcus agalactiae serotype. J. Clin. Microbiol. 45:2929–2936. 10.1128/JCM.00117-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barcaite E, Bartusevicius A, Tameliene R, Kliucinskas M, Maleckiene L, Nadisauskiene R. 2008. Prevalence of maternal group B streptococcal colonisation in European countries. Acta Obstet. Gynecol. Scand. 87:260–271. 10.1080/00016340801908759 [DOI] [PubMed] [Google Scholar]
- 9.Phares CR, Lynfield R, Farley MM, Mohle-Boetani J, Harrison LH, Petit S, Craig AS, Schaffner W, Zansky SM, Gershman K, Stefonek KR, Albanese BA, Zell ER, Schuchat A, Schrag SJ, Active Bacterial Core Surveillance/Emerging Infections Program Network 2008. Epidemiology of invasive group B streptococcal disease in the United States, 1999-2005. JAMA 299:2056–2065. 10.1001/jama.299.17.2056 [DOI] [PubMed] [Google Scholar]
- 10.Savoia D, Gottimer C, Crocilla' C, Zucca M. 2008. Streptococcus agalactiae in pregnant women: phenotypic and genotypic characters. J. Infect. 56:120–125. 10.1016/j.jinf.2007.11.007 [DOI] [PubMed] [Google Scholar]
- 11.Diedrick MJ, Flores AE, Hillier SL, Creti R, Ferrieri P. 2010. Clonal analysis of colonizing group B Streptococcus, serotype IV, an emerging pathogen in the United States. J. Clin. Microbiol. 48:3100–3104. 10.1128/JCM.00277-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Madzivhandila M, Adrian PV, Cutland CL, Kuwanda L, Schrag SJ, Madhi SA. 2011. Serotype distribution and invasive potential of group B streptococcus isolates causing disease in infants and colonizing maternal-newborn dyads. PLoS. One. 6:e17861. 10.1371/journal.pone.0017861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ho YR, Li CM, Su HP, Wu JH, Tseng YC, Lin YJ, Wu JJ. 2007. Variation in the number of tandem repeats and profile of surface protein genes among invasive group B streptococci correlates with patient age. J. Clin. Microbiol. 45:1634–1636. 10.1128/JCM.00122-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.von Both U, John A, Fluegge K, Siedler A, Berner R. 2008. Molecular epidemiology of invasive neonatal Streptococcus agalactiae isolates in Germany. Pediatr. Infect. Dis. J. 27:903–906. 10.1097/INF.0b013e318178d1ff [DOI] [PubMed] [Google Scholar]
- 15.Jones N, Bohnsack JF, Takahashi S, Oliver KA, Chan MS, Kunst F, Glaser P, Rusniok C, Crook DW, Harding RM, Bisharat N, Spratt BG. 2003. Multilocus sequence typing system for group B Streptococcus. J. Clin. Microbiol. 41:2530–2536. 10.1128/JCM.41.6.2530-2536.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bisharat N, Jones N, Marchaim D, Block C, Harding RM, Yagupsky P, Peto T, Crook DW. 2005. Population structure of group B streptococcus from a low-incidence region for invasive neonatal disease. Microbiology 151(Pt 6):1875–1881. 10.1099/mic.0.27826-0 [DOI] [PubMed] [Google Scholar]
- 17.Jones N, Oliver KA, Barry J, Harding RM, Bisharat N, Spratt BG, Peto T, Crook DW. 2006. Enhanced invasiveness of bovine-derived neonatal sequence type 17 group B Streptococcus is independent of capsular serotype. Clin. Infect. Dis. 42:915–924. 10.1086/500324 [DOI] [PubMed] [Google Scholar]
- 18.Lin FY, Whiting A, Adderson E, Takahashi S, Dunn DM, Weiss R, Azimi PH, Philips JB, III, Weisman LE, Regan J, Clark P, Rhoads GG, Frasch CE, Troendle J, Moyer P, Bohnsack JF. 2006. Phylogenetic lineages of invasive and colonizing strains of serotype III group B streptococci from neonates: a multicenter prospective study. J. Clin. Microbiol. 44:1257–1261. 10.1128/JCM.44.4.1257-1261.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Luan SL, Granlund M, Sellin M, Lagergard T, Spratt BG, Norgren M. 2005. Multilocus sequence typing of Swedish invasive group B Streptococcus isolates indicates a neonatally associated genetic lineage and capsule switching. J. Clin. Microbiol. 43:3727–3733. 10.1128/JCM.43.8.3727-3733.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Manning SD, Springman AC, Lehotzky E, Lewis MA, Whittam TS, Davies HD. 2009. Multilocus sequence types associated with neonatal group B streptococcal sepsis and meningitis in Canada. J. Clin. Microbiol. 47:1143–1148. 10.1128/JCM.01424-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tazi A, Disson O, Bellais S, Bouaboud A, Dmytruk N, Dramsi S, Mistou MY, Khun H, Mechler C, Tardieux I, Trieu-Cuot P, Lecuit M, Poyart C. 2010. The surface protein HvgA mediates group B streptococcus hypervirulence and meningeal tropism in neonates. J. Exp. Med. 207:2313–2322. 10.1084/jem.20092594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brochet M, Couve E, Zouine M, Vallaeys T, Rusniok C, Lamy MC, Buchrieser C, Trieu-Cuot P, Kunst F, Poyart C, Glaser P. 2006. Genomic diversity and evolution within the species Streptococcus agalactiae. Microbes Infect. 8:1227–1243. 10.1016/j.micinf.2005.11.010 [DOI] [PubMed] [Google Scholar]
- 23.Wang S, Niu C, Shi Z, Xia Y, Yaqoob M, Dai J, Lu C. 2011. Effects of ibeA deletion on virulence and biofilm formation of avian pathogenic Escherichia coli. Infect. Immun. 79:279–287. 10.1128/IAI.00821-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Norice CT, Smith FJ, Jr, Solis N, Filler SG, Mitchell AP. 2007. Requirement for Candida albicans Sun41 in biofilm formation and virulence. Eukaryot. Cell 6:2046–2055. 10.1128/EC.00314-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jefferson KK. 2004. What drives bacteria to produce a biofilm? FEMS Microbiol. Lett. 236:163–173. 10.1111/j.1574-6968.2004.tb09643.x [DOI] [PubMed] [Google Scholar]
- 26.Rinaudo CD, Rosini R, Galeotti CL, Berti F, Necchi F, Reguzzi V, Ghezzo C, Telford JL, Grandi G, Maione D. 2010. Specific involvement of pilus type 2a in biofilm formation in group B Streptococcus. PLoS One 5:e9216. 10.1371/journal.pone.0009216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Borges S, Silva J, Teixeira P. 2012. Survival and biofilm formation by group B streptococci in simulated vaginal fluid at different pHs. Antonie Van Leeuwenhoek 101:677–682. 10.1007/s10482-011-9666-y [DOI] [PubMed] [Google Scholar]
- 28.Kaur H, Kumar P, Ray P, Kaur J, Chakraborti A. 2009. Biofilm formation in clinical isolates of group B streptococci from north India. Microb. Pathog. 46:321–327. 10.1016/j.micpath.2009.04.004 [DOI] [PubMed] [Google Scholar]
- 29.Konto-Ghiorghi Y, Mairey E, Mallet A, Dumenil G, Caliot E, Trieu-Cuot P, Dramsi S. 2009. Dual role for pilus in adherence to epithelial cells and biofilm formation in Streptococcus agalactiae. PLoS Pathog. 5:e1000422. 10.1371/journal.ppat.1000422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tettelin H, Masignani V, Cieslewicz MJ, Eisen JA, Peterson S, Wessels MR, Paulsen IT, Nelson KE, Margarit I, Read TD, Madoff LC, Wolf AM, Beanan MJ, Brinkac LM, Daugherty SC, DeBoy RT, Durkin AS, Kolonay JF, Madupu R, Lewis MR, Radune D, Fedorova NB, Scanlan D, Khouri H, Mulligan S, Carty HA, Cline RT, Van Aken SE, Gill J, Scarselli M, Mora M, Iacobini ET, Brettoni C, Galli G, Mariani M, Vegni F, Maione D, Rinaudo D, Rappuoli R, Telford JL, Kasper DL, Grandi G, Fraser CM. 2002. Complete genome sequence and comparative genomic analysis of an emerging human pathogen, serotype V Streptococcus agalactiae. Proc. Natl. Acad. Sci. U. S. A. 99:12391–12396. 10.1073/pnas.182380799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cieslewicz MJ, Kasper DL, Wang Y, Wessels MR. 2001. Functional analysis in type Ia group B Streptococcus of a cluster of genes involved in extracellular polysaccharide production by diverse species of streptococci. J. Biol. Chem. 276:139–146. 10.1074/jbc.M005702200 [DOI] [PubMed] [Google Scholar]
- 32.Afshar B, Broughton K, Creti R, Decheva A, Hufnagel M, Kriz P, Lambertsen L, Lovgren M, Melin P, Orefici G, Poyart C, Radtke A, Rodriguez-Granger J, Sørensen UB, Telford J, Valinsky L, Zachariadou L, Members of the DEVANI Study Group. Efstratiou A. 2011. International external quality assurance for laboratory identification and typing of Streptococcus agalactiae (group B streptococci). J. Clin. Microbiol. 49:1475–1482. 10.1128/JCM.02365-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Roehm NW, Rodgers GH, Hatfield SM, Glasebrook AL. 1991. An improved colorimetric assay for cell proliferation and viability utilizing the tetrazolium salt XTT. J. Immunol. Methods 142:257–265. 10.1016/0022-1759(91)90114-U [DOI] [PubMed] [Google Scholar]
- 34.Heydorn A, Nielsen AT, Hentzer M, Sternberg C, Givskov M, Ersbøll BK, Molin S. 2000. Quantification of biofilm structures by the novel computer program COMSTAT. Microbiology 146(Pt 10):2395–2407 [DOI] [PubMed] [Google Scholar]
- 35.Svennerholm L. 1957. Quantitative estimation of sialic acids. II. A colorimetric resorcinol-hydrochloric acid method. Biochim. Biophys. Acta 24:604–611 [DOI] [PubMed] [Google Scholar]
- 36.Källman J, Schollin J, Schalen C, Erlandsson A, Kihlstrom E. 1998. Impaired phagocytosis and opsonisation towards group B streptococci in preterm neonates. Arch. Dis. Child. Fetal Neonatal Ed. 78:F46–F50. 10.1136/fn.78.1.F46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ho YR, Li CM, Yu CH, Lin YJ, Wu CM, Harn IC, Tang MJ, Chen YT, Shen FC, Lu CY, Tsai TC, Wu JJ. 2013. The enhancement of biofilm formation in group B streptococcal isolates at vaginal pH. Med. Microbiol. Immunol. 202:105–115. 10.1007/s00430-012-0255-0 [DOI] [PubMed] [Google Scholar]
- 38.Yang Q, Porter AJ, Zhang M, Harrington DJ, Black GW, Sutcliffe IC. 2012. The impact of pH and nutrient stress on the growth and survival of Streptococcus agalactiae. Antonie Van Leeuwenhoek 102:277–287. 10.1007/s10482-012-9736-9 [DOI] [PubMed] [Google Scholar]
- 39.Manetti AG, Koller T, Becherelli M, Buccato S, Kreikemeyer B, Podbielski A, Grandi G, Margarit I. 2010. Environmental acidification drives S. pyogenes pilus expression and microcolony formation on epithelial cells in a FCT-dependent manner. PLoS One 5(11):e13864. 10.1371/journal.pone.0013864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bellais S, Six A, Fouet A, Longo M, Dmytruk N, Glaser P, Trieu-Cuot P, Poyart C. 2012. Capsular switching in group B Streptococcus CC17 hypervirulent clone: a future challenge for polysaccharide vaccine development. J. Infect. Dis. 206:1745–1752. 10.1093/infdis/jis605 [DOI] [PubMed] [Google Scholar]
- 41.Sørensen UB, Poulsen K, Ghezzo C, Margarit I, Kilian M. 2010. Emergence and global dissemination of host-specific Streptococcus agalactiae clones. mBio 1:e00178-10. 10.1128/mBio.00178-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Areschoug T, Waldemarsson J, Gordon S. 2008. Evasion of macrophage scavenger receptor A-mediated recognition by pathogenic streptococci. Eur. J. Immunol. 38:3068–3079. 10.1002/eji.200838457 [DOI] [PubMed] [Google Scholar]
- 43.Santi I, Grifantini R, Jiang SM, Brettoni C, Grandi G, Wessels MR, Soriani M. 2009. CsrRS regulates group B Streptococcus virulence gene expression in response to environmental pH: a new perspective on vaccine development. J. Bacteriol. 191:5387–5397. 10.1128/JB.00370-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Park SE, Jiang S, Wessels MR. 2012. CsrRS and environmental pH regulate group B Streptococcus adherence to human epithelial cells and extracellular matrix. Infect. Immun. 80:3975–3984. 10.1128/IAI.00699-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sharma P, Lata H, Arya DK, Kashyap AK, Kumar H, Dua M, Ali A, Johri AK. 2013. Role of pilus proteins in adherence and invasion of Streptococcus agalactiae to the lung and cervical epithelial cells. J. Biol. Chem. 288:4023–4034. 10.1074/jbc.M112.425728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Seifert KN, Adderson EE, Whiting AA, Bohnsack JF, Crowley PJ, Brady LJ. 2006. A unique serine-rich repeat protein (Srr-2) and novel surface antigen (epsilon) associated with a virulent lineage of serotype III Streptococcus agalactiae. Microbiology 152(Pt 4):1029–1040. 10.1099/mic.0.28516-0 [DOI] [PubMed] [Google Scholar]
- 47.Botta GA. 1979. Hormonal and type-dependent adhesion of group B streptococci to human vaginal cells. Infect. Immun. 25:1084–1086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zawaneh SM, Ayoub EM, Baer H, Cruz AC, Spellacy WN. 1979. Factors influencing adherence of group B streptococci to human vaginal epithelial cells. Infect. Immun. 26:441–447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tamura GS, Kuypers JM, Smith S, Raff H, Rubens CE. 1994. Adherence of group B streptococci to cultured epithelial cells: roles of environmental factors and bacterial surface components. Infect. Immun. 62:2450–2458 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.