Abstract
Clostridium botulinum is a genetically diverse Gram-positive bacterium producing extremely potent neurotoxins (botulinum neurotoxins A through G [BoNT/A-G]). The complete genome sequences of three strains harboring only the BoNT/A1 nucleotide sequence are publicly available. Although these strains contain a toxin cluster (HA+ OrfX−) associated with hemagglutinin genes, little is known about the genomes of subtype A1 strains (termed HA− OrfX+) that lack hemagglutinin genes in the toxin gene cluster. We sequenced the genomes of three BoNT/A1-producing C. botulinum strains: two strains with the HA+ OrfX− cluster (69A and 32A) and one strain with the HA− OrfX+ cluster (CDC297). Whole-genome phylogenic single-nucleotide-polymorphism (SNP) analysis of these strains along with other publicly available C. botulinum group I strains revealed five distinct lineages. Strains 69A and 32A clustered with the C. botulinum type A1 Hall group, and strain CDC297 clustered with the C. botulinum type Ba4 strain 657. This study reports the use of whole-genome SNP sequence analysis for discrimination of C. botulinum group I strains and demonstrates the utility of this analysis in quickly differentiating C. botulinum strains harboring identical toxin gene subtypes. This analysis further supports previous work showing that strains CDC297 and 657 likely evolved from a common ancestor and independently acquired separate BoNT/A1 toxin gene clusters at distinct genomic locations.
INTRODUCTION
Clostridium botulinum bacteria are genetically diverse, Gram-positive anaerobic organisms that produce botulinum neurotoxins (BoNTs), which can cause a severe neuroparalytic illness (botulism) in humans and animals. C. botulinum can be divided into 4 different groups (I to IV) based on 16S rRNA genes and metabolic profiles (1, 2). Members of group I are proteolytic and produce BoNT types A, B, and F. Members of group II are nonproteolytic and produce BoNT types B, E, and F. Most human cases of botulism are associated with groups I and II (2–5). In contrast, cases of botulism among animals are usually caused by group III, which produces BoNT types C and D (6). Group IV produces BoNT G, which is the least-studied toxin and has not been clearly implicated in either human or animal botulism cases (7). Other rare Clostridium species (i.e., C. butyricum and C. baratii) are also capable of producing BoNT types E and F, respectively (8–10).
Botulinum toxin is among the most poisonous substances known; the estimated lethal dose of crystalline type A toxin for a 70-kg human is between 0.09 to 0.15 μg intravenously or intramuscularly, 0.70 to 0.90 μg by inhalation, and 70 μg orally (11). Because of its high toxicity, BoNT poses a significant risk to humans and it represents a possible biological warfare agent (11, 12). Therefore, BoNTs are listed as the highest-risk (category A) threat agents by the U.S. Centers for Disease Control and Prevention (CDC) (http://emergency.cdc.gov/bioterrorism). Despite being dangerous biohazard agents, BoNTs also have therapeutic applications, such as in the treatment of various muscle spasm disorders and for cosmetic purposes (13–15).
Genes responsible for BoNT production are located within a neurotoxin cluster arranged into two different conformations (3, 16). One conformation contains hemagglutinin (HA) genes (ha17, ha33, and ha70), and the other contains orfX genes, which have unknown function (orfX1, orfX2, and orfX3) (2–4, 17). The toxin clusters in subtype A1 strains are categorized as HA+ OrfX− (containing HA but not OrfX genes) or HA− OrfX+ BoNT (containing OrfX but not HA genes) (4).
Recently, several strains (CDC297, CDC51348, CDC1882, CDC1903, and CDC5328) were reported to have a HA− OrfX+ toxin gene cluster producing BoNT/A1 (4). These strains were shown to be highly clonal (4). In separate studies, strain CDC297 was shown to be more similar to the subtype Ba4 strain 657 than to other subtype A1-producing strains (HA+ OrfX−) by amplified fragment length polymorphism (AFLP) (2) and multilocus sequence typing (MLST) (18). Carter et al. (2009) also reported that two additional HA− OrfX+ A1 strains (F9604A and MUL0109ASA) (19) also clustered with Bf and Ba4 strains when a hybridization microarray using ATCC 3502 as the template was performed for 61 C. botulinum and C. sporogenes strains. The organization of the HA− OrfX+ A1 toxin cluster of those strains reported by Raphael et al. (2008) was shown by PCR to be the same as that of strain CDC51303 (4). In one strain (CDC5328), the toxin gene cluster was localized to the chromosome (20), although its precise location remained unknown. In contrast, the HA− OrfX+ BoNT/A4 cluster location in strain 657 (a bivalent strain producing toxins subtypes A4 and B5) is located on a plasmid (17, 20).
Several studies have been conducted to elucidate relationships among various HA− OrfX+ A1 strains in the context of the broader C. botulinum phylogeny (3, 4, 20, 21). However, no genome sequence has been produced for any of these unique HA− OrfX+ A1 strains. Also, the genomic location and surrounding areas of this BoNT cluster remain unknown. In the present study, we sequenced the genome of strain CDC297, a representative strain of the HA− OrfX+ A1 group, along with two other BoNT/A-producing C. botulinum strains. We then compared these newly sequenced genomes to other previously sequenced group I C. botulinum genomes by reference-free genome-wide single-nucleotide-polymorphism (SNP) analysis.
MATERIALS AND METHODS
Bacterial strains.
The bacterial strains used in this study (32A, 69A, and CDC297) are listed in Table 1. These C. botulinum strains were grown on Trypticase-peptone-glucose-yeast extract (TPGY) and cooked meat medium (CMM) and incubated anaerobically at 37°C for 72 h. CMM cultures from each sample were inoculated by evenly spreading 100 μl on solid botulinum selective medium (BSM) and incubated anaerobically at 37°C for 72 h. Single colonies were inoculated in TPGY medium for DNA extraction.
TABLE 1.
Characteristics of the C. botulinum strains used in this study
| Strain | Subtype (no. of SNP differences)d | Cluster type | Toxin cluster locationc | Origin |
|---|---|---|---|---|
| CDC297 | A1 (5)a,b | HA− OrfX+a,b | Chr-arsC | Human |
| 32A | A1 (0)a | HA+ OrfX−a | Chr-oppA | Human |
| 69A | A1 (0)a | HA+ OrfX−a | Chr-oppA | Spinach |
Determined in this study.
From Raphael et al., 2008 (4).
Chr-arsC, chromosome in arsC operon; Chr-oppA, chromosome in the oppA-brnQ operon.
Compared to the A1 toxin gene from C. botulinum ATCC 3502.
DNA extraction and quantification.
Genomic DNA from each strain was isolated from overnight cultures using the DNeasy Blood & Tissue kit (Qiagen, Valencia, CA). The quality of the DNA was checked using a NanoDrop 1000 (Thermo Scientific, Rockford, IL), and the concentration was determined using a Qubit double-stranded DNA BR assay kit and a Qubit fluorometer (Life Technologies, Grand Island, NY) according to each manufacturer's instructions.
WGS, contig assembly, and annotation.
For whole-genome sequencing (WGS), the genomes were sequenced using Ion Torrent (PGM) sequencing with the 200-bp template (Ion OneTouch 200 Template kit v2 DL) and sequencing (Ion PGM Sequencing 200 kit v2) kits (Life Technologies) according to the manufacturer's instructions at approximately 20× coverage. The libraries were constructed using 2 μg of genomic DNA previously sheared using a Covaris E220 Focused-ultrasonicator (Covaris, Inc., Woburn, MA) according to a protocol to generate an average of 200 bp and the SPRIworks Library Preparation System III (Beckman Coulter, Indianapolis, IN). These libraries were bar coded using the Ion Xpress Barcode Adapters 17-32 kit (Life Technologies). The resultant libraries were diluted to the recommended concentration determined by quantitative PCR (qPCR; Life Technologies) and used as the templates for emulsion PCR (emPCR) according to each manufacturer's instructions. The enriched emPCR results were loaded onto a 318 chip and sequenced using the Ion PGM 200 sequencing kit according to the manufacturer's instructions. The initial analysis and identification of each strain were performed using a reference mapping approach (CLC Genomics Workbench software version 6; CLC bio, Germantown, MD, USA) against all C. botulinum genomes available in the NCBI database.
In addition, genomic reads for all the strains were de novo assembled using Newbler (454 Life Sciences, Roche, Branford, CT). Toxin cluster identification was performed using a reference mapping approach (CLC Genomics Workbench software version 6). The contigs were reorganized by Mauve analysis (http://gel.ahabs.wisc.edu/mauve), using the C. botulinum genome identified as the closest relative: for example, 32A and 69A were most closely matched to C. botulinum A strain ATCC 3502, and thus contigs were reorganized using this genome as the reference. In the case of CDC297, the closest match was C. botulinum Ba strain 657, and contigs were reorganized using this genome as the reference.
In silico MLST phylogenetic analysis.
MLST analysis was conducted using seven loci (aroE, mdh, aceK, oppB, rpoB, recA, and hsp) described previously for type A C. botulinum strains (18). The sequence for each allele was obtained using a reference mapping approach (CLC Genomics Workbench software version 6). Numbers for alleles and sequence types (STs) were assigned according to the MLST database created for C. botulinum (http://pubmlst.org/cbotulinum/). The maximum likelihood tree was constructed based on the Kimura 2-parameter model (22) using concatenated sequences of the seven genes with MEGA5 software (23) with 1,000 bootstrap replicates.
Phylogenomic analysis.
Two analyses were performed. First, a broad set of Clostridium isolates were analyzed to provide an evolutionary context for the group I strains. This taxon set included one strain each for C. novyi, C. perfringens, and C. tetani and 20 diverse strains for C. botulinum. A matrix including all SNPs was determined using the program kSNP v2 (24) (k-mer length, 25), which uses a k-mer approach to SNP recovery. Because these genomes were so divergent, they shared very few core SNPs, so all of the SNPs were used for the broader analysis. The all-SNP matrix was used to reconstruct the maximum likelihood phylogeny using RAxML (25) with the following parameters: -N autoMRE -f a -m GTRCAT. The second analysis focused on group I C. botulinum strains. Our taxon sampling included 3 newly sequenced strains in addition to 14 publicly available genomes (see Table S1 in the supplemental material) comprising a data set of 17 draft and complete genomes. A core SNP matrix was determined with kSNP, using the same k-mer length of 25. The ML phylogeny was reconstructed using RAxML and rooted with C. botulinum subtype A3 strain Loch Maree (see Fig. 2). There are several advantages to using kSNP, which is a reference-free method of identifying genome-wide SNPs. First, the kSNP approach to gathering reference-free SNPs for downstream phylogenetic analysis has been shown to be effective for microbial-scale phylogenomic analysis (26, 27). Second, it is not sensitive to assembly error because the putative SNPs are extracted from k-mers (one SNP per 25-mer in our analysis), which effectively eliminates any influence of assembly. Third, the algorithm behind SNP identification is especially conservative to common pyrosequencing errors, such as homopolymeric runs commonly observed for 454 and Ion Torrent. If an insertion or deletion is present in a region, the resulting 25-mer will no longer match the other homologous k-mer regions, and thus the SNP will appear to be missing in that taxon. In addition, kSNP omits k-mers where all SNPs are in the center of a homopolymer repeat (24). The approach we have used is conservative, and repeat regions do not factor into our analysis, as the k-mers comprising such regions will not be unique and thus are not used in the analysis.
FIG 2.
Maximum likelihood phylogeny of C. botulinum group I from a 25,555-bp core SNP matrix. Strains are listed in Table 1 and in Table S1 in the supplemental material. Bootstrap support is 100% for all branches (see Fig. S2 in the supplemental material). The numbers above each branch and after each taxon ID (in parentheses) are the numbers of SNPs unique to that lineage/strain. In green font are the strains sequenced in this study.
In silico toxin cluster analysis and location.
In order to assess the organization of the toxin gene cluster and its insertion site in strain CDC297, the contig containing the HA− OrfX+ BoNT/A1 cluster was annotated using the RAST server (28). The contig containing the HA− OrfX+ BoNT/A1 cluster was also annotated in strain NCTC 2916 (accession number NZ_ABDO02000001). Both regions were compared to the chromosomally located arsR-containing region in strain 657 (accession number CP001083.1).
Nucleotide sequence accession numbers.
The draft genome sequences of the C. botulinum strain are available in GenBank under accession numbers AQPT00000000 (strain 32A, CFSAN002367), AQPU00000000 (strain CDC297, CFSAN002368), and AQPV00000000 (strain 69A CFSAN002369).
RESULTS AND DISCUSSION
Draft genome assemblies.
Draft genomes were generated for the three C. botulinum type A1 strains analyzed in this study: 32A, 69A, and CDC297. These draft genomes provided coverage of 26×, 18×, and 21×, respectively. Contigs for each strain were de novo assembled, and the number of contigs, sequence lengths, and other assembly statistics for each strain are summarized in Table 2. The genome sizes for 32A, 69A, and CDC297 varied between 3.6 Mb and 3.9 Mb, which is similar to what has been described for other group I C. botulinum strains (see Table S1 in the supplemental material). None of these strains appear to carry any known plasmids, as BLAST analysis of all contigs showed no match to plasmids in GenBank.
TABLE 2.
Summary report of the de novo assembly of the three strains analyzed
| Characteristic | Value for strain: |
||
|---|---|---|---|
| 32A | 69A | CDC297 | |
| Size (bp) | 3,856,420 | 3,629,296 | 3,848,285 |
| GC content (%) | 28.1 | 28.1 | 28.3 |
| No. of contigs | 164 | 130 | 238 |
| N50a | 28,829 | 27,597 | 31,642 |
| Minimum contig size (bp) | 483 | 482 | 446 |
| Maximum contig size (bp) | 176,269 | 161,786 | 121,302 |
| Avg contig size (bp) | 15,550 | 15,509 | 16,445 |
N50 is calculated by summarizing the lengths of the biggest contigs until 50% of the total genome length has been reached. The minimum contig length in this set is the number that is usually used to report the N50 value of a de novo assembly. It is a measure of assembly quality.
A fast comparison by genome reference mapping to other C. botulinum sequences in GenBank (www.ncbi.nlm.nih.gov), using CLC Genomics Workbench (CLC Bio), identified strains 32A and 69A as being similar to type A1 strains of C. botulinum, such as strain ATCC 3502 (data not shown). In contrast, CDC297 was identified as being most similar to the C. botulinum Ba strain 657. This finding confirms prior reports suggesting that strains CDC297 and Ba 657 are highly related (4, 18).
Phylogenetic analysis of C. botulinum genomes.
Analysis by in silico multilocus sequence typing (MLST) determined that strains 32A, 69A, and CDC297 belonged to two previously described STs (see Table S2 in the supplemental material). A comparison with other STs described for C. botulinum group I strains in the MLST database for C. botulinum (http://pubmlst.org/cbotulinum/) showed that strains 69A and 32A were indistinguishable from most subtype A1 strains (ST1). However, MLST analysis showed that CDC297 belongs to ST18. The C. botulinum MLST database currently holds only 2 isolates in ST18: one type A1 (CDC5328) and one type B (89E00061-2). A maximum likelihood tree including selected strains with similar and/or related STs showed that ST18 (CDC297) and ST7 (Ba 657) were highly related (Fig. 1). Moreover, ST18 and ST7 differed by only 1 SNP in the hsp allele.
FIG 1.
In silico MLST analysis of the strains sequenced in this study. Maximum likelihood tree using concatenated sequences for seven loci (3,619 bp) for the 3 strains sequenced in this study and the closest relatives available in the C. botulinum MLST database and rooted on one A3 strain (CDC/A3). The bootstrap values are shown on the branches. In red font are the strains sequenced in this study.
We were able to further elucidate what was observed by MLST using both a reference-free genome-wide SNP analysis for the broader-context C. botulinum phylogeny and the more narrowly focused analysis for C. botulinum group I. A 271,571-bp all-SNP matrix was used to reconstruct the broader Clostridium phylogeny (see Fig. S1 in the supplemental material). This tree reveals C. botulinum to be highly diverse and polyphyletic (see Fig. S1 in the supplemental material). C. novyi is nested within paraphyletic C. botulinum group III. C. perfringens is sister to C. botulinum group II, and C. botulinum group I appears to be more closely related to C. tetani than to other C. botulinum groups. Based on this broader-context phylogeny, we were able to choose an appropriate outgroup for our narrowly focused C. botulinum group I analysis: C. botulinum A3 Loch Maree. A 25,555-bp core-SNP matrix was used to reconstruct the group I core SNP phylogeny (Fig. 2), which revealed five highly supported lineages. Our newly sequenced C. botulinum strains 32A and 69A cluster with other A1 strains in lineage 5 that contain the toxin gene cluster (HA+ OrfX− BoNT). However, C. botulinum strain CDC297 clustered in lineage 2 along with strains that have a variety of BoNT toxin gene types (A1, A4, F, and B) and toxin gene clusters (Fig. 2).
The analysis of the strain CDC297 genome supports previous analyses by AFLP (2) placing this strain in close relationship with strain Ba 657 and suggests that both strains, as well as C. botulinum Bf, arose from a common ancestor. Furthermore, the presence of different toxin gene clusters among these strains with related genomic backgrounds supports previous reports that different toxin gene clusters may have been acquired through horizontal transfer (16, 29).
Most lineages were represented by more than one strain, except lineage 1, which contained a single strain, Loch Maree. The existence of 5 lineages within C. botulinum group I indicates a high diversity among members of this group (Fig. 2). The sources of this diversity become clearer through WGS, which documents the phages and plasmids that augment their genome. Similar patterns were observed in comparative analyses of strains of C. botulinum group I producing different toxin types using a DNA microarray containing 94% of predictive coding sequences (CDS) of C. botulinum ATCC 3502 (21). Most of these lineages contained strains exhibiting a variety of different toxin types or cluster conformations, further increasing the diversity already observed in their core genomes (unique SNPs for each lineage) and pangenome. The only exceptions were lineages 1 and 5; as previously discussed, in our analysis, lineage 1 contained only a single strain, while lineage 5 was composed of 5 strains, each of which exhibited the same toxin subtype (A1) and toxin cluster (HA+ OrfX−). This suggests that, in contrast to other lineages (such as 2, 3, and 4, which probably diverged before their characteristic toxin clusters were acquired), lineage 5 strains diverged recently from their common ancestor after acquiring the toxin cluster. More strains belonging to lineage 5 need to be sequenced in order to understand the confidence level of these findings (21).
Many of our findings help explain why microarray comparisons of CDC297 (4), F9604A and MUL0109ASA (19), and similar strains to ATCC 3502 (4, 19) have detected so many insertion/deletions, although the strains contained a BoNT/A1 gene; both the toxin cluster organization and the genomic backgrounds of these strains were not the same. We show that strain CDC297 and probably others in the same clonal group (based on their genetic similarity) (4) are more closely related to C. botulinum Ba strain 657, which is quite divergent from HA+ OrfX− BoNT/A1-producing C. botulinum strains, such as Hall and ATCC 19397. The genetic relationship between HA− OrfX+ BoNT/A1 strain CDC297 (together with the 4 other genetic related strains) (4) and the two other reported HA− OrfX+ BoNT/A1 strains, F9604A and MUL0109ASA (19), is unknown; however, they all appear to be highly related since they all clustered with C. botulinum Ba strain 657 and Bf strains (4, 19). Our analyses indicate that strains CDC297, Ba 657, and Bf are more related and probably evolved from a common ancestor. The future availability of sequence genomes for all the other HA− OrfX+ BoNT/A1 strains will help clarify how related they are to each other.
Comparative genomics: C. botulinum strain CDC297.
In addition to analyzing the phylogenetic placement of the three strains sequenced in this study, we identified differences in their genomic compositions. Moreover, we compared the genomic composition of strain CDC297 with the previously reported genome sequence of Ba strain 657.
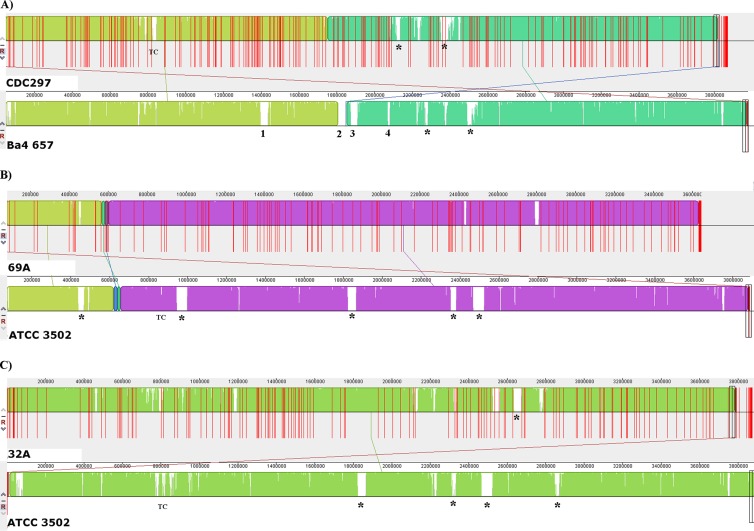
Strains 32A and 69A share a high level of similarity with other fully sequenced C. botulinum representatives, for example, ATCC 3502 (21) (Fig. 3B and C). In our analyses, CDC297 showed a high resemblance to the Ba 657 strain in chromosome content, although major differences were observed in numerous sites, including within the toxin cluster location. The difference between the sizes of strains CDC297 and Ba 657 may be attributed to the presence of several additional phages in Ba 657, resulting in a relatively smaller chromosome for CDC297, by ∼129,509 bp (∼3,848,285 versus 3,977,794 bp, respectively). Comparison of the genome sequences between these two strains revealed phage-related gene content differences (Fig. 3A). For example, we identified (i) a phage in Ba after the nimB (5-nitroimidazole antibiotic resistance protein) position between bp 1,396,660 and 1,436,600 of the Ba genome, (ii) another phage positioned between bp 1,810,000 and 1,853,700, (iii) another phage positioned between bp 1,874,000 and 1,910,000, and (iv) a genome region between bp 2,071,080 and 2,080,748. All of these phage-related genes were absent in strain CDC297. Also, Ba 657 carried two plasmids, while CDC297 lacked plasmids. Other differences in sequence regions are illustrated in Fig. 3A.
FIG 3.
Comparison of the draft genome of each strain from this study with its closest available genome in GenBank by Mauve analysis. Each genome is laid out in a horizontal track, and homologous segments are indicated in the same color and connected across genomes. Respective scales show the sequence coordinates in base pairs. A colored similarity plot is shown for each genome, the height of which is proportional to the level of sequence identity in that region. Red lines indicate the ends of each contig. (A) CDC297 versus Ba strain 657; (B) strain 69A versus strain ATCC 3502; (C) strain 32A versus strain ATCC 3502. TC, toxin cluster. The main differences observed between strains CDC297 and Ba are numbered 1 to 4. *, insertion/deletion regions differing between compared genomes.
Identification of the neurotoxin cluster genes and their arrangements in CDC297, 32A, and 69A.
Previous studies have identified five different A toxin types (3, 4, 30). The neurotoxin cluster organization and location were determined for each strain (Table 1). According to Hill et al. (16), type A toxin clusters in the chromosome are typically located at one of two sites: the arsC operon and the oppA-brnQ operon. In the three strains examined in this study, the toxin clusters were found in single large contigs (66,869 bp for CDC297, 56,882 bp for 32A, and 90,368 bp for 69A) containing mostly chromosome-borne genes. Furthermore, in 32A and 69A the toxin cluster was located in the oppA-brnQ operon and in the case of CDC297 in the arsC operon; both operons are located in the C. botulinum chromosome. These results, together with the lack of observed plasmids and previous analyses performed on CDC297 (20), allow us to place the genomic location of these clusters in the chromosome of all strains analyzed.
The nucleotide sequences of the BoNT/A1 genes determined in this study were compared to previously reported BoNT/A1 sequences. The BoNT/A1 gene from CDC297 differed from that found in C. botulinum strain ATCC 3502 at five different sites, confirming previous results from Sanger sequencing (4). In contrast, the genes from strains 32A and 69A were identical to that from C. botulinum strain ATCC 3502. The HA− OrfX+ BoNT/A1 toxin gene cluster (14,360 bp) of CDC297 was different at only 6 sites compared to the same cluster described for strain CDC5328 (3) (see Table S3 in the supplemental material). The HA+ OrfX− BoNT/A1 toxin gene clusters of strains 32A and 69A were identical to the one in C. botulinum strain ATCC 3502.
Analysis of the region containing the toxin cluster in CDC297.
Sequencing of strain CDC297 gave us the unique opportunity to examine the integration site of its unusual subtype A1 toxin cluster. This region was not identical in CDC297 and in Ba 657 (its presumed closest relative) because the toxin genes were found in the chromosome and in a plasmid, respectively. The toxin cluster for CDC297 was found in the arsC operon, which is consistent with reports for other toxin clusters: HA− OrfX+ BoNT/F in C. botulinum strain Langeland, HA− OrfX+ A1 in C. botulinum strain NCTC 2916, and HA− OrfX+ A2 in C. botulinum strain Kyoto (16). However, we found that the gene following the toxin gene cluster was lycA, which is similar to that found in strain NCTC 2916 (which contains a BoNT/A1 gene with different nucleotide variations from those in strain CDC297). Closer analyses of the chromosomal location of the toxin region showed that CDC297 and NCTC 2916 appeared to be identical in gene organization, and both differed greatly from strain Ba 657 (Fig. 4). The toxin cluster of CDC297 was located in the arsC operon (chromosome), while Ba 657, which is bivalent, had both its toxin clusters located in the plasmid pCLJ (accession number NC_012654).
FIG 4.
Schematic representation of the region containing the toxin cluster (HA− OrfX+ BoNT/A1) genes and surrounding genes found in type A1 strain CDC297 and type A(B) strain NCTC 2916 compared to the same region in type Ba4 strain 657. The toxin gene cluster in these strains is inserted into the ars operon. C. botulinum Ba4 strain 657 lacks the presence of a toxin gene cluster in this region, and the organization of the ars operon is divergent compared to that of strains NCTC 2916 and CDC297. The dashed line between arsR and thyA in strain 657 represents omitted genes present at positions 841,046 to 845,955, which do not share similarity to intervening genes present in strain NCTC 2916 or CDC297. Vertical dotted lines indicate similar genes between the strains shown.
Despite the phylogenetic differences between CDC297 and NCTC 2916, Fig. 4 shows a high similarity of their toxin clusters and surrounding genes, suggesting that this entire region was acquired by C. botulinum strain CDC297 or NCTC 2916 via horizontal transfer from a similar source. It is noteworthy that most strains containing a toxin cluster in the arsC site also contain the entire arsenate resistance operon (arsC-1, arsR, arsD, arsA, and arsB) (16). This is similar to what has been observed for all HA+ OrfX− A1 strains; other strains lacking a toxin cluster in this position also lack most of the genes in the ars operon (16). Strains without these arsenic resistance genes are more sensitive to arsenic (31). It is possible that the gain of the HA− OrfX+ BoNT toxin cluster might have occurred using the ars operon as a homologous insertion region. This would, in turn, explain the observed lack of toxin clusters in that location among those strains without the entire ars operon and suggest the possibility that C. botulinum subtype A1 strains with the HA+ OrfX− toxin gene cluster (located at the oppA site) are capable of acquiring the HA− OrfX+ BoNT toxin cluster, since their ars operons are intact. As an example, NCTC 2916 strain contains the HA− OrfX+ BoNT/A1 toxin gene cluster at the ars operon and the HA+ OrfX− BoNT/B toxin gene cluster at the oppA site (16).
Conclusions.
This study reports the use of whole-genome SNP sequence analysis for discrimination of C. botulinum group I strains and demonstrates the utility of this analysis in quickly differentiating C. botulinum strains harboring identical toxin gene subtypes. It also shows that C. botulinum group I can be divided further into 5 different lineages. These findings reinforce previous theories about genomic plasticity more specifically in C. botulinum and the likely acquisition of BoNT clusters by horizontal transfer and shows that phages are implicated in this plasticity as has been observed for other C. botulinum group I strains (21). Additional WGS analyses may help to further differentiate C. botulinum strains previously reported to be indistinguishable by pulsed-field gel electrophoresis (PFGE) and MLST (4, 18).
This work highlights the utility of genome-wide SNPs instead of traditional MLST for C. botulinum subtyping. In this study, we identified 3,817 unique SNPs differentiating lineage 2 from the other C. botulinum group I lineages. Furthermore, we identified 2 orders of magnitude more SNPs that differentiated strain CDC297 from strain Ba657, whose sequence types (STs) differed by only a single SNP. WGS data analyses provided valuable insights about the nature of other food-borne bacterial pathogens, including the Salmonella enterica serotype Montevideo outbreak in 2010 (32, 33), Vibrio cholerae in Haiti in 2010 (34), Escherichia coli O104:H4 in Germany in 2011 (35), and Salmonella Enteritidis in the United States in 2010 (36). Because C. botulinum is polyphyletic, care should be taken to focus on specific monophyletic lineages for epidemiological tracking of this bacterium. Reference-free SNP analysis is well suited to this organism since (i) complete reference genomes are lacking for several serotypes/subtypes and (ii) there are genome organizational differences even among very similar strains encoding the same toxin subtype (37). Continued sequencing of C. botulinum genomes isolated from many different sources and over several years will help establish a more comprehensive database allowing improved source tracking as well as a clearer understanding of the evolution of this important food-borne pathogen.
Supplementary Material
ACKNOWLEDGMENTS
This project was supported by FDA Foods Program intramural funds.
We thank Lili Fox Vélez for her editorial assistance on the manuscript and James Pettengill for his useful comments.
The findings and conclusions in this report are those of the authors and do not necessarily represent the official position of the Centers for Disease Control and Prevention or the Food and Drug Administration.
Footnotes
Published ahead of print 24 January 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.03934-13.
REFERENCES
- 1.Collins MD, East AK. 1998. Phylogeny and taxonomy of the food-borne pathogen Clostridium botulinum and its neurotoxins. J. Appl. Microbiol. 84:5–17. 10.1046/j.1365-2672.1997.00313.x [DOI] [PubMed] [Google Scholar]
- 2.Hill KK, Smith TJ, Helma CH, Ticknor LO, Foley BT, Svensson RT, Brown JL, Johnson EA, Smith LA, Okinaka RT, Jackson PJ, Marks JD. 2007. Genetic diversity among botulinum neurotoxin-producing clostridial strains. J. Bacteriol. 189:818–832. 10.1128/JB.01180-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jacobson MJ, Lin G, Raphael B, Andreadis J, Johnson EA. 2008. Analysis of neurotoxin cluster genes in Clostridium botulinum strains producing botulinum neurotoxin serotype A subtypes. Appl. Environ. Microbiol. 74:2778–2786. 10.1128/AEM.02828-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Raphael BH, Luquez C, McCroskey LM, Joseph LA, Jacobson MJ, Johnson EA, Maslanka SE, Andreadis JD. 2008. Genetic homogeneity of Clostridium botulinum type A1 strains with unique toxin gene clusters. Appl. Environ. Microbiol. 74:4390–4397. 10.1128/AEM.00260-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sachdeva A, Defibaugh-Chavez SL, Day JB, Zink D, Sharma SK. 2010. Detection and confirmation of Clostridium botulinum in water used for cooling at a plant producing low-acid canned foods. Appl. Environ. Microbiol. 76:7653–7657. 10.1128/AEM.00820-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Skarin H, Hafstrom T, Westerberg J, Segerman B. 2011. Clostridium botulinum group III: a group with dual identity shaped by plasmids, phages and mobile elements. BMC Genomics 12:185. 10.1186/1471-2164-12-185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Terilli RR, Moura H, Woolfitt AR, Rees J, Schieltz DM, Barr JR. 2011. A historical and proteomic analysis of botulinum neurotoxin type/G. BMC Microbiol. 11:232. 10.1186/1471-2180-11-232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hall JD, McCroskey LM, Pincomb BJ, Hatheway CL. 1985. Isolation of an organism resembling Clostridium barati which produces type F botulinal toxin from an infant with botulism. J. Clin. Microbiol. 21:654–655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McCroskey LM, Hatheway CL, Fenicia L, Pasolini B, Aureli P. 1986. Characterization of an organism that produces type E botulinal toxin but which resembles Clostridium butyricum from the feces of an infant with type E botulism. J. Clin. Microbiol. 23:201–202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McCroskey LM, Hatheway CL, Woodruff BA, Greenberg JA, Jurgenson P. 1991. Type F botulism due to neurotoxigenic Clostridium baratii from an unknown source in an adult. J. Clin. Microbiol. 29:2618–2620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Arnon SS, Schechter R, Inglesby TV, Henderson DA, Bartlett JG, Ascher MS, Eitzen E, Fine AD, Hauer J, Layton M, Lillibridge S, Osterholm MT, O'Toole T, Parker G, Perl TM, Russell PK, Swerdlow DL, Tonat K. 2001. Botulinum toxin as a biological weapon: medical and public health management. JAMA 285:1059–1070. 10.1001/jama.285.8.1059 [DOI] [PubMed] [Google Scholar]
- 12.Atlas RM. 2002. Bioterrorism: from threat to reality. Annu. Rev. Microbiol. 56:167–185. 10.1146/annurev.micro.56.012302.160616 [DOI] [PubMed] [Google Scholar]
- 13.Dressler D. 2008. Botulinum toxin drugs: future developments. J. Neural Transm. 115:575–577. 10.1007/s00702-007-0863-9 [DOI] [PubMed] [Google Scholar]
- 14.Pickett A, Perrow K. 2011. Towards new uses of botulinum toxin as a novel therapeutic tool. Toxins (Basel) 3:63–81. 10.3390/toxins3010063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Segura-Aguilar J, Kostrzewa RM. 2006. Neurotoxins and neurotoxicity mechanisms. An overview. Neurotox. Res. 10:263–287. 10.1007/BF03033362 [DOI] [PubMed] [Google Scholar]
- 16.Hill KK, Xie G, Foley BT, Smith TJ, Munk AC, Bruce D, Smith LA, Brettin TS, Detter JC. 2009. Recombination and insertion events involving the botulinum neurotoxin complex genes in Clostridium botulinum types A, B, E and F and Clostridium butyricum type E strains. BMC Biol. 7:66. 10.1186/1741-7007-7-66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Smith TJ, Hill KK, Foley BT, Detter JC, Munk AC, Bruce DC, Doggett NA, Smith LA, Marks JD, Xie G, Brettin TS. 2007. Analysis of the neurotoxin complex genes in Clostridium botulinum A1-A4 and B1 strains: BoNT/A3, /Ba4 and /B1 clusters are located within plasmids. PLoS One 2:e1271. 10.1371/journal.pone.0001271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jacobson MJ, Lin G, Whittam TS, Johnson EA. 2008. Phylogenetic analysis of Clostridium botulinum type A by multi-locus sequence typing. Microbiology 154:2408–2415. 10.1099/mic.0.2008/016915-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Carter AT, Paul CJ, Mason DR, Twine SM, Alston MJ, Logan SM, Austin JW, Peck MW. 2009. Independent evolution of neurotoxin and flagellar genetic loci in proteolytic Clostridium botulinum. BMC Genomics 10:115. 10.1186/1471-2164-10-115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Marshall KM, Bradshaw M, Pellett S, Johnson EA. 2007. Plasmid encoded neurotoxin genes in Clostridium botulinum serotype A subtypes. Biochem. Biophys. Res. Commun. 361:49–54. 10.1016/j.bbrc.2007.06.166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sebaihia M, Peck MW, Minton NP, Thomson NR, Holden MT, Mitchell WJ, Carter AT, Bentley SD, Mason DR, Crossman L, Paul CJ, Ivens A, Wells-Bennik MH, Davis IJ, Cerdeno-Tarraga AM, Churcher C, Quail MA, Chillingworth T, Feltwell T, Fraser A, Goodhead I, Hance Z, Jagels K, Larke N, Maddison M, Moule S, Mungall K, Norbertczak H, Rabbinowitsch E, Sanders M, Simmonds M, White B, Whithead S, Parkhill J. 2007. Genome sequence of a proteolytic (Group I) Clostridium botulinum strain Hall A and comparative analysis of the clostridial genomes. Genome Res. 17:1082–1092. 10.1101/gr.6282807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kimura M. 1980. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 16:111–120. 10.1007/BF01731581 [DOI] [PubMed] [Google Scholar]
- 23.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28:2731–2739. 10.1093/molbev/msr121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gardner S, Slezak T. 2010. Scalable SNP analyses of 100+ bacterial or viral genomes. J. Forensic Res. 1:107. 10.4172/2157-7145.1000107 [DOI] [Google Scholar]
- 25.Stamatakis A, Hoover P, Rougemont J. 2008. A rapid bootstrap algorithm for the RAxML Web servers. Syst. Biol. 57:758–771. 10.1080/10635150802429642 [DOI] [PubMed] [Google Scholar]
- 26.Sahl JW, Gillece JD, Schupp JM, Waddell VG, Driebe EM, Engelthaler DM, Keim P. 2013. Evolution of a pathogen: a comparative genomics analysis identifies a genetic pathway to pathogenesis in Acinetobacter. PLoS One 8:e54287. 10.1371/journal.pone.0054287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Timme RE, Pettengill JB, Allard MW, Strain E, Barrangou R, Wehnes C, Van Kessel JS, Karns JS, Musser SM, Brown EW. 2013. Phylogenetic diversity of the enteric pathogen Salmonella enterica subsp. enterica inferred from genome-wide reference-free SNP characters. Genome Biol. Evol. 5:2109–2123. 10.1093/gbe/evt159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, Edwards RA, Formsma K, Gerdes S, Glass EM, Kubal M, Meyer F, Olsen GJ, Olson R, Osterman AL, Overbeek RA, McNeil LK, Paarmann D, Paczian T, Parrello B, Pusch GD, Reich C, Stevens R, Vassieva O, Vonstein V, Wilke A, Zagnitko O. 2008. The RAST server: rapid annotations using subsystems technology. BMC Genomics 9:75. 10.1186/1471-2164-9-75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Skarin H, Segerman B. 2011. Horizontal gene transfer of toxin genes in Clostridium botulinum: involvement of mobile elements and plasmids. Mob. Genet. Elements 1:213–215. 10.4161/mge.1.3.17617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Carter AT, Pearson BM, Crossman LC, Drou N, Heavens D, Baker D, Febrer M, Caccamo M, Grant KA, Peck MW. 2011. Complete genome sequence of the proteolytic Clostridium botulinum type A5 (B3′) strain H04402 065. J. Bacteriol. 193:2351–2352. 10.1128/JB.00072-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lindstrom M, Hinderink K, Somervuo P, Kiviniemi K, Nevas M, Chen Y, Auvinen P, Carter AT, Mason DR, Peck MW, Korkeala H. 2009. Comparative genomic hybridization analysis of two predominant Nordic group I (proteolytic) Clostridium botulinum type B clusters. Appl. Environ. Microbiol. 75:2643–2651. 10.1128/AEM.02557-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Allard MW, Luo Y, Strain E, Li C, Keys CE, Son I, Stones R, Musser SM, Brown EW. 2012. High resolution clustering of Salmonella enterica serovar Montevideo strains using a next-generation sequencing approach. BMC Genomics 13:32. 10.1186/1471-2164-13-32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bakker HC, Switt AI, Cummings CA, Hoelzer K, Degoricija L, Rodriguez-Rivera LD, Wright EM, Fang R, Davis M, Root T, Schoonmaker-Bopp D, Musser KA, Villamil E, Waechter H, Kornstein L, Furtado MR, Wiedmann M. 2011. A whole-genome single nucleotide polymorphism-based approach to trace and identify outbreaks linked to a common Salmonella enterica subsp. enterica serovar Montevideo pulsed-field gel electrophoresis type. Appl. Environ. Microbiol. 77:8648–8655. 10.1128/AEM.06538-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chin C-S, Sorenson J, Harris JB, Robins WP, Charles RC, Jean-Charles RR, Bullard J, Webster DR, Kasarskis A, Peluso P, Paxinos EE, Yamaichi Y, Calderwood SB, Mekalanos JJ, Schadt EE, Waldor MK. 2011. The origin of the Haitian cholera outbreak strain. N. Engl. J. Med. 364:33–42. 10.1056/NEJMoa1012928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rasko DA, Webster DR, Sahl JW, Bashir A, Boisen N, Scheutz F, Paxinos EE, Sebra R, Chin CS, Iliopoulos D, Klammer A, Peluso P, Lee L, Kislyuk AO, Bullard J, Kasarskis A, Wang S, Eid J, Rank D, Redman JC, Steyert SR, Frimodt-Moller J, Struve C, Petersen AM, Krogfelt KA, Nataro JP, Schadt EE, Waldor MK. 2011. Origins of the E. coli strain causing an outbreak of hemolytic-uremic syndrome in Germany. N. Engl. J. Med. 365:709–717. 10.1056/NEJMoa1106920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Allard MW, Luo Y, Strain E, Pettengill J, Timme R, Wang C, Li C, Keys CE, Zheng J, Stones R, Wilson MR, Musser SM, Brown EW. 2013. On the evolutionary history, population genetics and diversity among isolates of Salmonella Enteritidis PFGE pattern JEGX01.0004. PLoS One 8:e55254. 10.1371/journal.pone.0055254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fang PK, Raphael BH, Maslanka SE, Cai S, Singh BR. 2010. Analysis of genomic differences among Clostridium botulinum type A1 strains. BMC Genomics 11:725. 10.1186/1471-2164-11-725 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.