Abstract
The biofilm-producing abilities of potentially human-pathogenic serotypes of Escherichia coli from the ovine reservoir were studied at different temperatures and on different surfaces. A possible influence of the hydrophobicity of the bacterial cells, as well as the presence of two virulence factors, the Shiga toxin-encoding (Stx) bacteriophage and the eae gene, was also studied. A total of 99 E. coli isolates of serotypes O26:H11, O103:H2, and O103:H25 isolated from sheep feces were included. The results show that isolates of all three E. coli serotypes investigated can produce biofilm on stainless steel, glass, and polystyrene at 12, 20, and 37°C. There was a good general correlation between the results obtained on the different surfaces. E. coli O103:H2 isolates produced much more biofilm than those of the other two serotypes at all three temperatures. In addition, isolates of serotype O26:H11 produced more biofilm than those of O103:H25 at 37°C. The hydrophobicity of the isolates varied between serotypes and was also influenced by temperature. The results strongly indicated that hydrophobicity influenced the attachment of the bacteria rather than their ability to form biofilm once attached. Isolates with the eae gene produced less biofilm at 37°C than isolates without this gene. The presence of a Stx bacteriophage did not influence biofilm production. In conclusion, our results show that potentially human-pathogenic E. coli from the ovine reservoir can form biofilm on various surfaces and at several temperatures relevant for food production and handling.
INTRODUCTION
Diarrheagenic strains of Escherichia coli are the most common bacterial pathogens implicated in diarrhea worldwide, causing disease ranging from mild diarrhea to hemorrhagic colitis. The life-threatening hemolytic-uremic syndrome (HUS) is caused by Shiga toxin-producing E. coli (STEC) (1). Two main virulence factors of STEC are the production of Shiga toxins (Stx) and the attaching and effacing mechanism of the locus of enterocyte effacement (LEE), which includes the eae gene encoding the outer membrane cell adhesion-mediating protein intimin (2). Sequence and antigenic polymorphisms in the C terminus of intimin are used to define a number of distinct intimin types which appear to influence the site of human intestinal mucosal colonization (3–6). STEC can harbor one or more stx genes, which are encoded by inducible lambda phages integrated into their genomes, and the entire phage and specific regions within the phage can be gained or lost through horizontal gene transfer (7).
STEC O157 is the serogroup most commonly identified in human outbreaks. However, six non-O157 STEC serogroups, namely, O26, O45, O103, O111, O121, and O145, have also attracted significant attention because they have been increasingly associated with serious outbreaks. It has been reported that these six STEC serogroups, often referred to as “the big six,” were responsible for approximately 70% of all non-O157 STEC infections from 1983 to 2002 in the United States (8). In particular, serotype O26:H11 has been regarded as one of the most severe non-O157 STEC due to its enhanced virulence and ability to cause diarrhea and HUS (9).
The reported annual incidence rate of STEC infections in humans in Norway has varied between 0 and 9.9 per 100,000 inhabitants for the last 20 years. STEC O157 has been the serogroup most frequently isolated from reported cases, followed by STEC O103, STEC O26 and STEC O145 (10). Nationwide STEC outbreaks caused by STEC serotypes O157:H7, O103:H25, and O103:H2 have been registered in Norway. The most severe outbreak was in 2006, the outbreak strain being E. coli O103:H25, with genes encoding both Stx2 and intimin (11). Sheep were implicated as the original reservoir for the outbreak strain, as fermented mutton sausages were shown to be the outbreak food source. It was also suggested that strains from the reservoir possibly could have contaminated the meat handling facilities and that a subsequent cross contamination of food products was the cause of the outbreak (12).
Food-borne microorganisms can attach to and form biofilm on various food contact surfaces (13). Bacterial biofilms are defined as microbial sessile communities that are attached to a substance, to an interface, or to each other (14). In the biofilm, the cells are embedded in a self-produced matrix and protected against environmental stress, e.g., disinfectants (14–17). A correlation between biofilm-forming abilities and persistence in food and feed-producing facilities has been reported for Salmonella enterica (18) and Listeria monocytogenes (19), and it has been suggested that biofilms in food-producing environments may serve as reservoirs for bacteria that may contaminate the food products. Furthermore, we have previously shown that dissemination of virulence genes like stx can occur in E. coli biofilm (20). Consequently, biofilms can also be environments for the evolution of new pathogenic E. coli isolates.
Most studies on the biofilm-forming potentials of E. coli have been performed on the laboratory strain K-12 and on isolates of the serogroup O157. Little is known about the biofilm-forming abilities of other E. coli serogroups. The aim of the present study was to investigate the biofilm-producing abilities of potentially human-pathogenic serotypes of E. coli from the ruminant reservoir at different temperatures and on different surfaces relevant for food production and handling. The serotypes included were O103:H2, O103:H25, and O26:H11. A possible influence on biofilm production of the hydrophobicity of the bacterial cells, as well as the presence of the two virulence genes stx and eae, was also studied.
MATERIALS AND METHODS
Bacterial isolates.
The E. coli isolates were originally collected during a national survey of E. coli in sheep in Norway during the years 2006 and 2007. First, a total of 511 isolates (142 E. coli O26 and 369 E. coli O103) was selected from this survey and investigated for eae, stx, and H type, as previously described (21, 22). Among these isolates, 99 isolates of serotypes O26:H11 (n = 25), O103:H2 (n = 42), and O103:H25 (n = 32) were semirandomly selected for the present study, making sure that isolates from as many counties as possible were included within each serotype. The E. coli O26:H11 and O103:H2 isolates came from 13 counties, and O103:25 isolates from 10 counties. In the national survey, all E. coli O26:H11 and O103:H25 isolates were eae positive, whereas both eae-positive and eae-negative E. coli O103:H2 isolates were identified. This was also reflected in our study, which included 23 eae-negative and 19 eae-positive isolates. Due to the very low number of stx-positive isolates in the national survey, we chose to include only stx-negative isolates in order to have properly sized research groups in our study. The importance of stx in biofilm production was studied by constructing stx-positive strains by incorporation of a Stx bacteriophage.
All strains were stored at −80°C in brain heart infusion broth (BHI; Difco, BD, Franklin Lakes, NJ, USA) supplemented with 15% glycerin (Merck KGaA, Darmstadt, Germany) and recovered on blood agar (sheep blood) at 37.0 ± 1.0°C overnight. The bacterial cultures were then transferred into Luria-Bertani broth (LB; Merck) and incubated statically overnight at 37.0 ± 1.0°C (mean ± standard deviation [SD]). LB without NaCl (LBwo/NaCl; Bacto-tryptone 10 g/liter, yeast extract 5 g/liter) was used as the test broth in the biofilm assays.
Genotyping.
Pulsed-field gel electrophoresis (PFGE) was carried out using XbaI (Sigma-Aldrich, St. Louis, MO, USA) restriction endonuclease as described by Sekse et al. (21). Subtyping of eae for detection of eaeβ and eaeε was previously described by Zhang and coworkers (23), and subtyping of eaeγ2/θ was performed as described by Blanco et al. (24), with a change in annealing temperature to 52°C.
Construction of stx-positive E. coli isolates.
The Stx2-encoding bacteriophage Φ731 (Δstx2::cat) (hereinafter called Φ731) was inserted into the chromosome of E. coli isolates and confirmed to be stable and functioning, as described previously (20). The original bacteriophage was carried by an E. coli O103:H25 strain from a Norwegian HUS patient (25).
Biofilm production on polystyrene (microtiter plate assay).
Biofilm production on polystyrene was measured in the microtiter plate assay and performed as described previously (18) using 96-well Nunc microtiter plates with Nunclon (Nunc A/S, Roskilde, Denmark). In short, 30-μl amounts of an overnight culture were added to 100 μl LBwo/NaCl in the wells of the microtiter plate (in three parallel assays). The microtiter plates were incubated statically for 2 days at 20.0 ± 1.0°C and 37.0 ± 1.0°C and for 7 days at 12.0 ± 1.0°C. After incubation, the wells were washed with tap water, stained with 1% crystal violet solution (Sigma-Aldrich, St. Louis, MO, USA) for 30 min at room temperature, and washed at least three times to remove excess dye. The remaining dye was dissolved in ethanol-acetone (70:30, vol/vol) for 10 min at room temperature, and the optical density at 595 nm (OD595) (Multiscan MS; Thermo Fisher Scientific, Inc., Waltham, MA, USA) was measured. For each strain, the result was calculated by subtracting the median OD595 of the three parallel assays of the control (test broth only) from the median OD595 of the three parallel assays of the sample.
Biofilm production on glass slides and stainless steel coupons.
Overnight cultures were inoculated (1 ml) into sterile centrifuge tubes (Greiner bio-one GmbH, Frickenhausen, Germany) containing 9 ml LBwo/NaCl. An autoclaved microscope slide (76 by 26 mm; Menzel GmbH + CoKG, Braunschweig, Germany) or an autoclaved stainless steel (AISI304, 2B) coupon of approximately the same size was placed in each tube. The tubes were incubated at 20.0 ± 1.0°C or 37.0 ± 1.0°C for 3 days or at 12.0 ± 1.0°C for 5 days. During incubation, biofilm was formed on both sides of the microscope slides or steel coupons at the liquid-air interface. After incubation, the slides/coupons were transferred to a test tube with 1% crystal violet solution (Sigma-Aldrich) for staining of the biofilm for 30 min. Excess crystal violet solution was rinsed from the slides/coupons in tap water. The isolates were then examined visually and given a score from 0 to 3 according to the amount of stained biofilm observed.
Hydrophobicity.
Hydrophobicity was determined using the microbial adhesion to hydrocarbons (MATH) assay as described by Mattos-Guaraldi et al. (26), with minor modifications. Briefly, 10-μl amounts of overnight cultures were mixed with 10 ml LB broth in 50-ml tubes and incubated for7 days at 12°C, 3 days at 20°C, or 24 h at 37°C. The bacteria were washed once with 5 ml phosphate-buffered saline (PBS) (pH 7.2) at the test temperature (for 10 min at 3,000 rpm) and resuspended in PBS (to an OD600 of 0.6 to 0.8, using a Hitachi U-1100 spectrophotometer). Then, 2-ml amounts of diluted bacterial suspension were transferred to glass tubes, and each was overlaid with 400 μl hexadecane (ReagentPlus, 99%; Sigma-Aldrich). The mixtures were vigorously vortexed for 1 min and incubated for 30 min to allow phase separation, and then the OD600 of the lower, aqueous phase was recorded. Controls consisted of untreated bacterial suspensions. The percentage of hydrophobicity for each strain was calculated according to the following formula: % hydrophobicity = [(OD600 untreated sample − OD600 aqueous phase)/OD600 untreated sample] × 100. Each strain was tested in three independent experiments at all temperatures.
Statistical analyses.
The computer program JMP 9.0.0 (2010; SAS Institute, Inc., Cary, NC, USA) was used for all statistical calculations. For comparing responses when the data were normally distributed, analysis of variance (ANOVA) and the Tukey-Kramer honestly significant difference (HSD) test for multiple comparisons of means were used. When data were not normally distributed, responses were compared by using the Wilcoxon each pair test for multiple nonparametric comparisons. Differences were considered statistically significant if the P value was <0.05.
RESULTS
Genotypes.
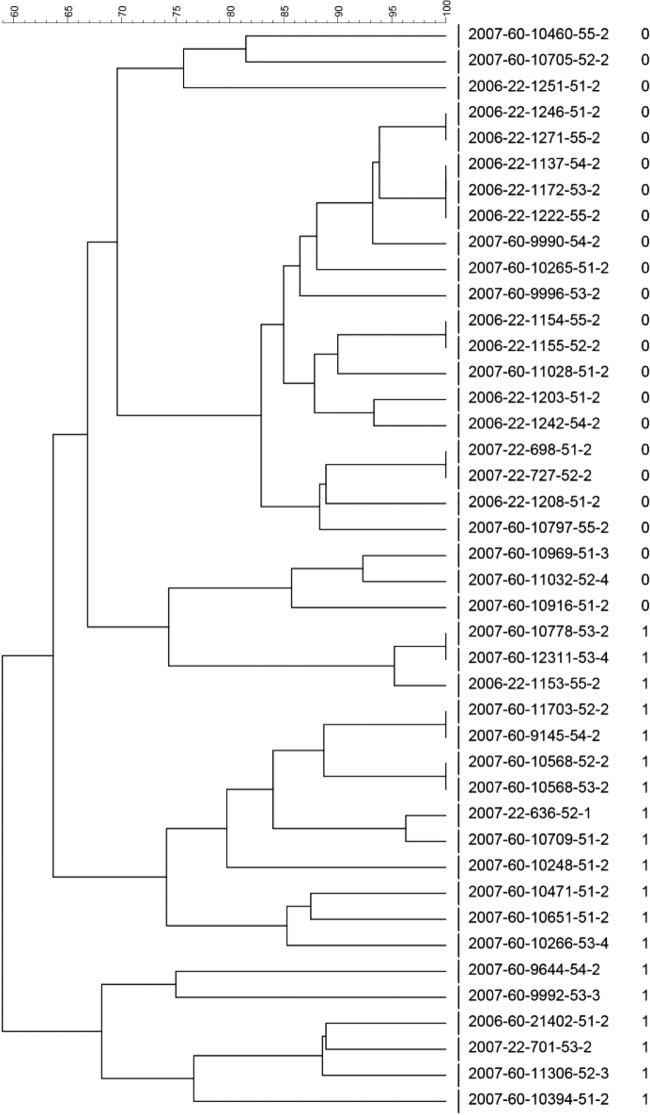
All of the eae-positive E. coli O103:H2 isolates had the eaeε gene, all of the O103:H25 isolates carried the eaeγ2/θ gene, and all of the O26:H11 isolates carried the eaeβ gene. Dendrograms based on the results from PFGE of all the isolates displayed a high variability within each serotype (Fig. 1, 2, and 3). Within the serotype O103:H2, eae-positive and -negative isolates showed a tendency toward clustering separately (Fig. 1). All eae-negative isolates except three had PFGE profiles with higher similarities to profiles of other eae-negative isolates than to those of eae-positive isolates. However, as many as 13 of the eae-positive isolates displayed higher similarities to eae-negative isolates than to the remaining 6 eae-positive isolates.
FIG 1.
Dendrogram with percent similarity of PFGE profiles of E. coli O103:H2 isolates (isolate numbers are given for all isolates). In addition, the number 0 or 1 to the right of the isolate numbers signifies the absence or presence of the eae gene, respectively.
FIG 2.
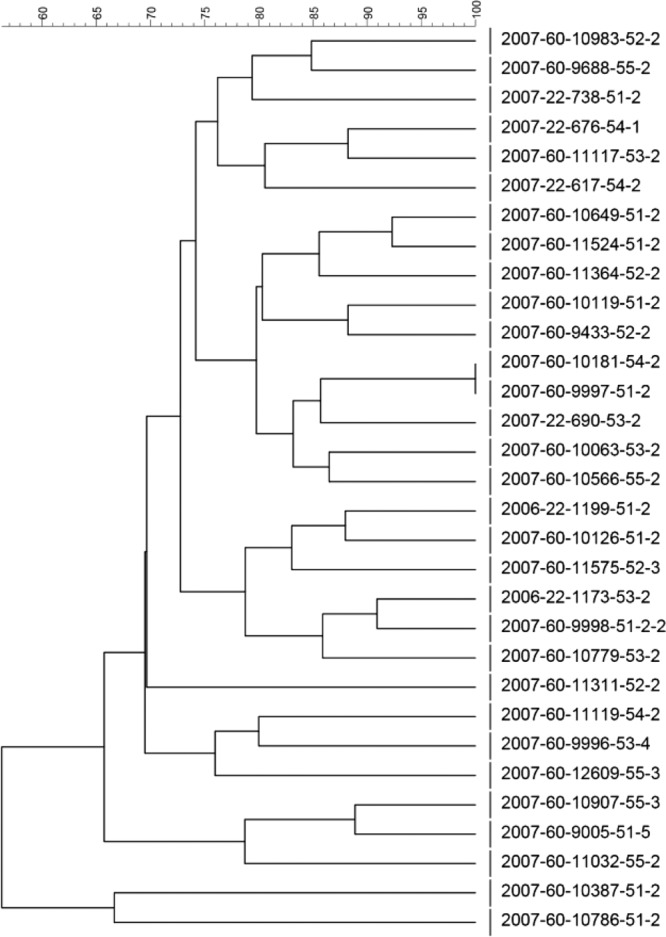
Dendrogram with percent similarity of PFGE profiles of E. coli O103:H25 isolates (isolate numbers are given for all isolates).
FIG 3.
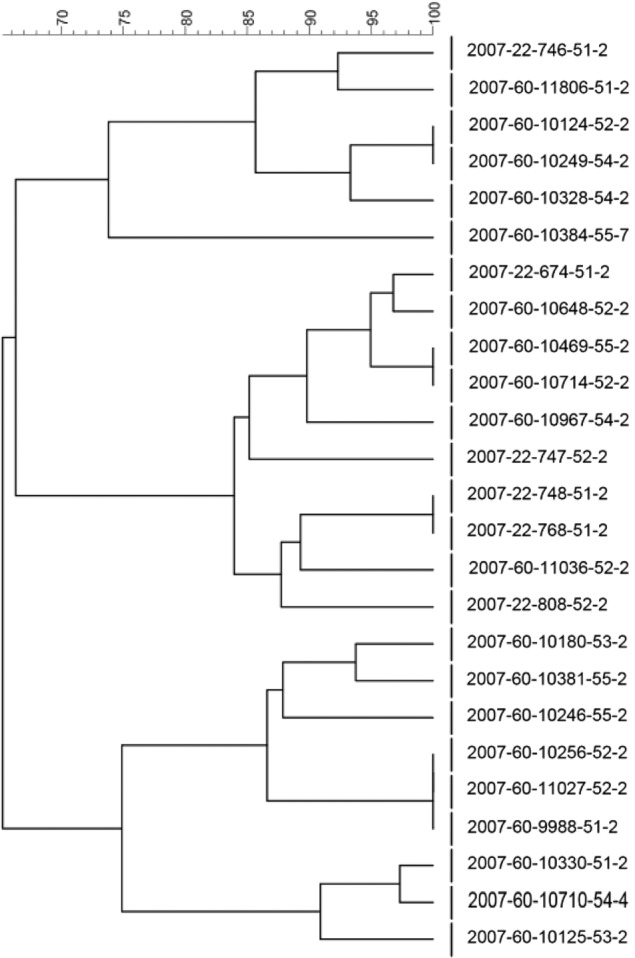
Dendrogram with percent similarity of PFGE profiles of E. coli O26:H11 isolates (isolate numbers are given for all isolates).
Biofilm production on polystyrene (microtiter plate assay).
When biofilm production by all isolates was tested in the microtiter plate assay, the results displayed significant differences between the serotypes (Table 1). E. coli O103:H2 isolates produced significantly more biofilm at all three temperatures than isolates of the other two serotypes (ANOVA, P < 0.0001). E. coli O26:H11 produced more biofilm than E. coli O103:H25 at 37°C (P = 0.003). Biofilm production was also influenced to some extent by temperature. E. coli O103:H2 produced significantly less biofilm at 37°C than at 12°C and 20°C (ANOVA, P < 0.0001), whereas the biofilm production of E. coli O26:H11 isolates was higher at 20 and 37°C than at 12°C (Wilcoxon rank test, P = 0.04 and 0.0002, respectively).
TABLE 1.
Biofilm production in the microtiter plate assay at 12, 20, and 37°C
| Serotype | No. of isolates tested | Mean OD595a ± SD (range) at temp (°C) of: |
||
|---|---|---|---|---|
| 12 | 20 | 37 | ||
| O103:H2 | 42 | 1.35 ± 0.60 (0.05–2.51) a | 1.38 ± 0.45 (0.07–2.01) a | 0.58 ± 0.64 (0.04–2.15) b |
| O103:H25 | 32 | 0.03 ± 0.01 (0.00–0.06) c | 0.04 ± 0.03 (0.00–0.10) cd | 0.04 ± 0.01 (0.02–0.08) c |
| O26:H11 | 25 | 0.06 ± 0.12 (0.00–0.49) c | 0.09 ± 0.11 (0.00–0.43) de | 0.15 ± 0.18 (0.02–0.66) e |
| O103:H2 eae positive | 19 | 1.34 ± 0.52 (0.05–2.18) a | 1.38 ± 0.51 (0.07–1.99) a | 0.12 ± 0.09 (0.04–0.34) f |
| O103:H2 eae negative | 23 | 1.37 ± 0.68 (0.16–2.51) a | 1.37 ± 0.41 (0.72–2.01) a | 0.95 ± 0.65 (0.04–2.15) g |
Different letters signify that the means are statistically different at a P value of <0.05.
Two of the E. coli O26:H11 isolates (2007-60-10125-53-2 and 2007-60-10710-54-4) produced more biofilm than the other isolates of this serotype both at 12 and 20°C. At 12°C, the OD595 values of the two isolates were 0.388 and 0.485, respectively, whereas the rest of the isolates had a mean of 0.020 (range, 0 to 0.136). The corresponding results at 20°C were 0.301 and 0.431 for the two isolates, while the rest of the isolates had a mean of 0.048 (range, 0.002 to 0.224). The two isolates were from two farms located in different counties, but they clustered together when genotyped by PFGE (Fig. 3). The third isolate in this cluster (2007-60-10330-51-2) also had relatively high OD595 values at these temperatures (0.101 and 0.224), but they were not as high as the two others.
Serotype O103:H2 was the only serotype that encompassed both eae-positive and eae-negative isolates. Within this serotype, the mean biofilm production at 37°C (as indicated by the OD595 in the microtiter plate assay) was significantly higher in eae-negative isolates than in eae-positive isolates (ANOVA, P < 0.0001), whereas no differences in mean OD595 were observed between eae-negative and eae-positive isolates at 12°C and 20°C (Table 1).
As there were no stx-positive isolates in our material, we infected three isolates with a Stx bacteriophage and compared their biofilm production with and without this bacteriophage. Two transductants were produced from each of the isolates 2006-22-1153-55-2 (serotype O103:H2) and 2006-22-1199-51-2 (serotype O103:H25) and one from the laboratory strain C600. When comparing the biofilm-forming abilities of isolates with and without the Stx bacteriophage in the microtiter plate assay, only minor variations in the total biofilm production (as indicated by the OD595 in the microtiter plate assay) were observed between mother strains and transductants (Table 2).
TABLE 2.
Biofilm production in the microtiter plate assay of three E. coli strains before and after the Stx bacteriophage Φ731 was inserted
| Isolatea | Serotype | Mean OD595 ± SD at temp (°C) of: |
||
|---|---|---|---|---|
| 12 | 20 | 37 | ||
| 2006-22-1153-55-2 | O103:H2 | 1.14 ± 0.08 | 1.67 ± 0.06 | 0.15 ± 0.03 |
| 2006-22-1153-55-2:Φ731 A | O103:H2 | 1.47 ± 0.11 | 1.70 ± 0.08 | 0.17 ± 0.13 |
| 2006-22-1153-55-2:Φ731 B | O103:H2 | 1.27 ± 0.08 | 1.52 ± 0.06 | 0.17 ± 0.05 |
| 2006-22-1199-51-2 | O103:H25 | 0.08 ± 0.01 | 0.01 ± 0.01 | 0.02 ± 0.01 |
| 2006-22-1199-51-2:Φ731 A | O103:H25 | 0.06 ± 0.04 | 0.02 ± 0.01 | 0.03 ± 0.03 |
| 2006-22-1199-51-2:Φ731 B | O103:H25 | 0.06 ± 0.01 | 0.03 ± 0.04 | 0.03 ± 0.27 |
| C600 | 0.06 ± 0.01 | 1.78 ± 0.01 | 0.10 ± 0.06 | |
| C600:Φ731 | 0.16 ± 0.02 | 2.03 ± 0.02 | 0.24 ± 0.24 | |
Two different transductant strains (A and B) were produced from each of the strains in serogroup O103, whereas one was produced from the laboratory strain C600.
Biofilm production on glass and stainless steel.
A subset of 12 isolates, including all three serotypes, different levels of biofilm production, and both eae-positive and -negative isolates, was tested on glass and stainless steel coupons with the semiquantitative method for biofilm production (Table 3). The isolates displayed various degrees of biofilm formation, ranging from no visible biofilm (scored as 0) to a thick biofilm at the liquid-air interface, which was scored on a scale from 1 to 3. In general, there was a good correlation between the amount of biofilm formed in microtiter plates and on glass and stainless steel, i.e., isolates with high OD595 values in the microtiter plates achieved high scores on glass and stainless steel and vice versa (ANOVA, P < 0.0001) (Fig. 4). There was also a relatively good correlation between the scores obtained on glass and stainless steel, although a tendency to higher scores on steel than on glass was observed (Table 3). This was particularly pronounced at 12°C, where all of the E. coli O103:H2 isolates displayed low scores (1) for biofilm production on glass and yet achieved higher scores (2 or 3) for biofilm production on steel.
TABLE 3.
Biofilm formation in a microtiter plate assay and on glass and stainless steel coupons and hydrophobicity of a subset of 12 ovine E. coli strains at different temperatures
| Isolate | Serotype | eae type | Median OD595 in microtiter plate assay (n = 3) at temp (°C) of: |
Mean scorea at indicated temp (°C) on: |
Mean hydrophobicity (%) ± SD (n = 3) at temp (°C) of: |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Glass coupons |
Steel coupons |
|||||||||||||
| 12 | 20 | 37 | 12 | 20 | 37 | 12 | 20 | 37 | 12 | 20 | 37 | |||
| 2006-22-1242-54-2 | O103:H2 | Negativeb | 2.194 | 1.886 | 0.479 | 1 | 3 | 2 | 3 | 3 | 1 | 51 ± 19 | 52 ± 16 | 93 ± 2 |
| 2006-22-1246-51-2 | O103:H2 | Negative | 1.048 | 1.744 | 1.364 | 1 | 3 | 1 | 3 | 3 | 2 | 47 ± 3 | 66 ± 5 | 90 ± 1 |
| 902006-22-1271-55-2 | O103:H2 | Negative | 1.089 | 1.722 | 0.644 | 1 | 3 | 1 | 3 | 3 | 2 | 62 ± 1 | 57 ± 7 | 93 ± 0 |
| 2006-22-1153-55-2 | O103:H2 | eaeε | 1.579 | 1.478 | 0.217 | 1 | 3 | 1 | 2 | 2 | 1 | 24 ± 3 | 34 ± 3 | 63 ± 1 |
| 2007-60-10651-51-2 | O103:H2 | eaeε | 1.284 | 1.804 | 0.344 | 1 | 3 | 1 | 3 | 3 | 2 | 36 ± 2 | 55 ± 7 | 67 ± 3 |
| 2007-60-10709-51-51 | O103:H2 | eaeε | 1.292 | 1.558 | 0.083 | 1 | 3 | 1 | 2 | 2 | 1 | 42 ± 8 | 63 ± 6 | 65 ± 8 |
| 2006-22-1173-53-2 | O103:H25 | eaeγ2/θ | 0.018 | 0.000 | 0.030 | 0 | 1 | 0 | 0 | 0 | 1 | 23 ± 7 | 29 ± 0 | 24 ± 10 |
| 2006-22-1199-51-2 | O103:H25 | eaeγ2/θ | 0.020 | 0.097 | 0.022 | 0 | 1 | 1 | 0 | 1 | 1 | 27 ± 12 | 25 ± 14 | 20 ± 3 |
| 2007-60-10125-53-2 | O26:H11 | eaeβ | 0.388 | 0.301 | 0.061 | 3 | 3 | 2 | 3 | 3 | 2 | 37 ± 6 | 38 ± 7 | 33 ± 5 |
| 2007-60-10710-54-4 | O26:H11 | eaeβ | 0.485 | 0.431 | 0.658 | 3 | 3 | 2 | 3 | 3 | 3 | 39 ± 5 | 39 ± 6 | 38 ± 0 |
| 2007-60-10180-53-2 | O26:H11 | eaeβ | 0.002 | 0.012 | 0.025 | 0 | 0 | 0 | 0 | 0 | 1 | 22 ± 4 | 25 ± 9 | 32 ± 6 |
| 2007-60-10246-55-2 | O26:H11 | eaeβ | 0.000 | 0.019 | 0.017 | 0 | 0 | 0 | 0 | 0 | 1 | 46 ± 3 | 43 ± 1 | 54 ± 3 |
There was no variation between parallel assays in the coupon test, i.e., the standard deviation was 0.
Negative, isolate did not have the eae gene.
FIG 4.
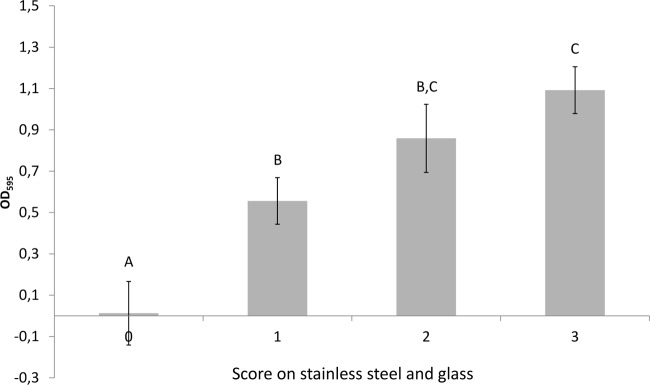
Mean OD595 values in microtiter plate assay of subset of 12 isolates with different biofilm formation scores on glass slides and steel coupons. Results from all temperatures are included. Bars represent standard deviations (SD). Different letters signify that the means are statistically different at a P value of <0.05.
Hydrophobicity.
The subset of 12 isolates tested for hydrophobicity showed differences both between and within serotypes at all three temperatures tested (Table 3; Fig. 5). The O103:H2 isolates displayed significantly higher hydrophobicities at both 20 and 37°C than the other two serotypes (ANOVA, P value range of 0.001 to 0.04). Within the serotype O103:H2, the hydrophobicity was higher at 37°C than at 12 and 20°C (P = 0.0001 and 0.005). Similar differences in hydrophobicities could not be seen between or within E. coli O103:H25 and O26:H11. Furthermore, the eae-negative O103:H2 isolates had higher hydrophobicities at 37°C than the eae-positive isolates (P = 0.0001) (Table 3).
FIG 5.
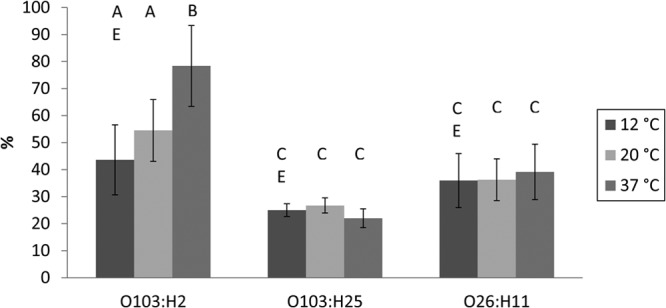
Mean hydrophobicities of different serotypes in subset of 12 isolates at 12, 20, and 37°C. Bars represent standard deviation (SD). Different letters signify that the means are statistically different at a P value of <0.05.
Isolates with no visible biofilm on the glass and stainless steel coupons (score of 0) displayed a lower mean hydrophobicity than isolates with biofilm scores of 1 to 3 (mean of 32% and SD of 11% versus mean of 50% and SD of 20%, respectively; ANOVA, P = 0.001). These results were the same on both glass and steel coupons. Similarly, isolates with very low biofilm production in the microtiter plate assay, i.e., OD595 of <0.1, displayed a significantly lower mean hydrophobicity than isolates with OD595 values of 0.1 and higher (mean of 33% and SD of 13% versus mean of 54% and SD of 19%, respectively; ANOVA, P = 0.002).
DISCUSSION
The results from the present study show that potentially pathogenic E. coli isolates of serotypes O103:H2, O103:H25, and O26:H11 can produce biofilm on stainless steel, glass, and polystyrene at 12, 20, and 37°C. There was a good general correlation between the results obtained on different surfaces. The main exception was found at 12°C, where isolates of E. coli O103:H2 produced large amounts of biofilm on polystyrene and stainless steel but little on glass. The microtiter plate assay is well suited for screening a large number of strains, and our results indicate that this assay can provide relevant information on biofilm-forming abilities on stainless steel and glass as well. This has previously been observed in Salmonella enterica subsp. enterica, where correlations between biofilm formation in microtiter plates, on stainless steel, and at the liquid-air interface were found (18).
Furthermore, we observed significant differences in the biofilm-forming abilities of the serotypes in all assays used. E. coli O103:H2 isolates produced much more biofilm than those of the other two serotypes at all three temperatures, and E. coli O26:H11 produced more biofilm than E. coli O103:H25 at 37°C. In particular, three isolates of E. coli O26:H11 belonging to the same PFGE subcluster produced more biofilm than the rest of the isolates of this serotype, although not as much as the E. coli O103:H2 isolates.
Different biofilm-forming abilities between serotypes of Listeria monocytogenes (27, 28) and between serovars of Salmonella (18, 29) have also been reported. Little has previously been reported on possible differences in biofilm production between serotypes or serogroups of E. coli (30), although differences between strains have been published by several (31–34). When comparing serotypes in our study, we aimed at reducing bias by having relatively large groups of isolates with as low an epidemiological relationship as possible. To achieve this, isolates from as many different counties as possible were included. This made it more likely that a large number of different clones were represented within each serotype, as moving sheep from one county to another is prohibited in Norway. This assumption was supported by the results from the PFGE analyses. Consequently, the results of the present study indicate that biofilm-forming ability in general is more closely correlated to serotype than to strain or clone.
All of the E. coli O103:H25 isolates displayed very low biofilm formation at all temperatures in all assays. However, we showed previously that the total number of CFU in biofilm produced on glass slides by one of these isolates (2006-22-1199-51-2) at 20°C was as high as 106 (20). This indicates that even isolates of the serotypes that were poor biofilm producers in our test systems still can form some biofilm under conditions relevant for the food production industry.
In the present study, significant differences in hydrophobicity were observed between the serotypes, with E. coli O103:H2 displaying the highest hydrophobicity. In addition, the hydrophobicity of these isolates increased with increasing temperature, whereas the isolates of the other two serotypes displayed low hydrophobicity at all temperatures. The hydrophobicity of a bacterial cell is largely influenced by the residues and structures on the surface of the cell, and it varies between species and strains and even within the same strain depending on the mode and stage of growth and the composition of the growth medium (35). It is not surprising that the surface structures and, thereby, the surface hydrophobicity may vary between isolates of different serotypes. Variations due to adaptations to different temperatures could also be expected, as this has been reported in other species, e.g., Listeria monocytogenes (27) and Serratia marcescens (36).
We did not observe any general correlation between hydrophobicity and the amount of biofilm formed in any of our test systems. However, we did find that isolates which did not form a visible biofilm on stainless steel and glass displayed lower hydrophobicity than isolates that did form a visible biofilm. Similarly, isolates with very poor biofilm production in the microtiter plate assay had lower hydrophobicity than those with better biofilm production. These results strongly indicate that the hydrophobicity of the isolates influenced the attachment of the bacteria rather than their ability to form biofilm once attached. A positive correlation between the hydrophobic properties of E. coli isolates and their attachment abilities has been reported in some studies (37–40), whereas others have failed to find such a relationship (41–43). It has been suggested that the differences between these studies could be explained, e.g., by the number and variety of strains included in the study and/or a lack of sensitivity within methods (35).
One aim of the study was to investigate whether the two main virulence factors of STEC, i.e., Stx bacteriophages and the eae gene encoding intimin, could influence the biofilm-producing abilities of E. coli. A number of cell surface structures are involved in the attachment of E. coli to host cells and/or abiotic surfaces (44, 45). To the best of our knowledge, a possible role of intimin in biofilm production is unknown. In our material, E. coli O103:H2 was the only serotype where we had both eae-positive and eae-negative isolates. Interestingly, the eae-negative isolates were significantly better biofilm producers than the eae-positive isolates at 37°C within this serotype. The eae-negative isolates also displayed higher hydrophobicity than the eae-positive isolates at this temperature. Isolates of the other two serotypes, which were all eae positive, displayed both low hydrophobicity and biofilm formation at 37°C. Consequently, one could speculate that intimin contributed to the lower hydrophobicity and biofilm production at this temperature. On the other hand, eae-positive O103:H2 isolates were very good biofilm producers at 12 and 20°C, whereas the eae-positive isolates of the other serotypes produced the same amount of biofilm at these temperatures as at 37°C. One explanation could be that intimin was only expressed at 37°C and that other factors were more important for biofilm production at the other temperatures. It has been reported previously that the LEE operons are repressed at 27°C and expressed at 37°C (46). The differences might also be attributed to the differences in intimin type found between the serotypes in our study. However, these are speculations only, as our investigations of eae were performed at the gene level, and we do not know whether intimin was expressed at the cell surface in any of our assays. An alternative explanation for the different biofilm-forming abilities of eae-positive and -negative E. coli O103:H2 isolates may be that eae was a confounding variable in our material. If so, the observed difference in biofilm formation would have been due to other factors that also differed between the two groups. In the PFGE analyses, the majority of the eae-negative E. coli O103:H2 isolates clustered together, but this was not as obvious for the eae-positive isolates. Thus, the results from the PFGE analyses are only partly supportive of this theory. Consequently, the observed difference in biofilm formation between eae-positive and -negative isolates remains interesting but unexplained. On the other hand, our results from testing strains before and after infection with a Stx bacteriophage strongly suggest that the presence of the bacteriophage does not influence biofilm-forming abilities.
In conclusion, our results show that potentially human-pathogenic E. coli from the ovine reservoir can form biofilm on various surfaces and at several temperatures relevant for food production and handling, although variation could be seen between and within serotypes. In food-producing facilities, the biofilm may serve as a reservoir for cross contamination of the products and as an environment for the dissemination of virulence genes and emergence of new pathogenic E. coli isolates.
ACKNOWLEDGMENTS
This study was supported by grant no. 178161/I10 from the Research Council of Norway. Part of this work was supported by the COST ACTION FA1202 BacFoodNet.
We are grateful to Gunvor Ruud for excellent technical assistance.
Footnotes
Published ahead of print 20 December 2013
REFERENCES
- 1.Paton JC, Paton AW. 1998. Pathogenesis and diagnosis of Shiga toxin-producing Escherichia coli infections. Clin. Microbiol. Rev. 11:450–479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Karmali MA. 2004. Infection by Shiga toxin-producing Escherichia coli: an overview. Mol. Biotechnol. 26:117–122. 10.1385/MB:26:2:117 [DOI] [PubMed] [Google Scholar]
- 3.Girard F, Batisson I, Frankel GM, Harel J, Fairbrother JM. 2005. Interaction of enteropathogenic and Shiga toxin-producing Escherichia coli and porcine intestinal mucosa: role of intimin and Tir in adherence. Infect. Immun. 73:6005–6016. 10.1128/IAI.73.9.6005-6016.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tzipori S, Gunzer F, Donnenberg MS, de Montigny L, Kaper JB, Donohue-Rolfe A. 1995. The role of the eaeA gene in diarrhea and neurological complications in a gnotobiotic piglet model of enterohemorrhagic Escherichia coli infection. Infect. Immun. 63:3621–3627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fitzhenry RJ, Pickard DJ, Hartland EL, Reece S, Dougan G, Phillips AD, Frankel G. 2002. Intimin type influences the site of human intestinal mucosal colonisation by enterohaemorrhagic Escherichia coli O157:H7. Gut 50:180–185. 10.1136/gut.50.2.180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mundy R, Schuller S, Girard F, Fairbrother JM, Phillips AD, Frankel G. 2007. Functional studies of intimin in vivo and ex vivo: implications for host specificity and tissue tropism. Microbiology 153:959–967. 10.1099/mic.0.2006/003467-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shaikh N, Tarr PI. 2003. Escherichia coli O157:H7 Shiga toxin-encoding bacteriophages: integrations, excisions, truncations, and evolutionary implications. J. Bacteriol. 185:3596–3605. 10.1128/JB.185.12.3596-3605.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brooks JT, Sowers EG, Wells JG, Greene KD, Griffin PM, Hoekstra RM, Strockbine NA. 2005. Non-O157 Shiga toxin-producing Escherichia coli infections in the United States, 1983–2002. J. Infect. Dis. 192:1422–1429. 10.1086/466536 [DOI] [PubMed] [Google Scholar]
- 9.Bielaszewska M, Kock R, Friedrich AW, von Eiff C, Zimmerhackl LB, Karch H, Mellmann A. 2007. Shiga toxin-mediated hemolytic uremic syndrome: time to change the diagnostic paradigm? PLoS One 2:e1024. 10.1371/journal.pone.0001024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hofshagen M, Lange H, Hauge K. 2011. Trends and source of zoonoses and zoonotic agents in humans foodstuffs, animals and feedingstuffs. Norwegian Veterinary Institute, Oslo, Norway: http://www.vetinst.no/eng/Publications/Zoonosis-Reports/Zoonosis-report-2011 [Google Scholar]
- 11.Schimmer B, Nygard K, Eriksen HM, Lassen J, Lindstedt BA, Brandal LT, Kapperud G, Aavitsland P. 2008. Outbreak of haemolytic uraemic syndrome in Norway caused by stx2-positive Escherichia coli O103:H25 traced to cured mutton sausages. BMC Infect. Dis. 8:41. 10.1186/1471-2334-8-41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sekse C, O'Sullivan K, Granum PE, Rorvik LM, Wasteson Y, Jorgensen HJ. 2009. An outbreak of Escherichia coli O103:H25—bacteriological investigations and genotyping of isolates from food. Int. J. Food Microbiol. 133:259–264. 10.1016/j.ijfoodmicro.2009.05.026 [DOI] [PubMed] [Google Scholar]
- 13.Chmielewski RAN, Frank JF. 2003. Biofilm formation and control in food processing facilities. Comp. Rev. Food Sci. Technol. 2:22–32. 10.1111/j.1541-4337.2003.tb00012.x [DOI] [PubMed] [Google Scholar]
- 14.Costerton JW, Lewandowski Z, Caldwell DE, Korber DR, Lappin-Scott HM. 1995. Microbial biofilms. Annu. Rev. Microbiol. 49:711–745. 10.1146/annurev.mi.49.100195.003431 [DOI] [PubMed] [Google Scholar]
- 15.Zogaj X, Nimtz M, Rohde M, Bokranz W, Romling U. 2001. The multicellular morphotypes of Salmonella typhimurium and Escherichia coli produce cellulose as the second component of the extracellular matrix. Mol. Microbiol. 39:1452–1463. 10.1046/j.1365-2958.2001.02337.x [DOI] [PubMed] [Google Scholar]
- 16.Vestby LK, Moretro T, Ballance S, Langsrud S, Nesse LL. 2009. Survival potential of wild type cellulose deficient Salmonella from the feed industry. BMC Vet. Res. 5:43. 10.1186/1746-6148-5-43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Moretro T, Vestby LK, Nesse LL, Storheim S, Kotlarz K, Langsrud S. 2009. Evaluation of efficacy of disinfectants against Salmonella from the feed industry. J. Appl. Microbiol. 106:1005–1012. 10.1111/j.1365-2672.2008.04067.x [DOI] [PubMed] [Google Scholar]
- 18.Vestby LK, Moretro T, Langsrud S, Heir E, Nesse LL. 2009. Biofilm forming abilities of Salmonella are correlated with persistence in fish meal and feed factories. BMC Vet. Res. 5:20. 10.1186/1746-6148-5-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Møretrø T, Langsrud S. 2004. Listeria monocytogenes: biofilm formation and persistence in food-processing environments. Biofilms 1:107–121. 10.1017/S1479050504001322 [DOI] [Google Scholar]
- 20.Solheim HT, Sekse C, Urdahl AM, Wasteson Y, Nesse LL. 2013. Biofilm as an environment for dissemination of stx genes by transduction. Appl. Environ. Microbiol. 79:896–900. 10.1128/AEM.03512-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sekse C, Sunde M, Lindstedt BA, Hopp P, Bruheim T, Cudjoe KS, Kvitle B, Urdahl AM. 2011. Potentially human-pathogenic Escherichia coli O26 in Norwegian sheep flocks. Appl. Environ. Microbiol. 77:4949–4958. 10.1128/AEM.00189-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sekse C, Sunde M, Hopp P, Bruheim T, Cudjoe KS, Kvitle B, Urdahl AM. 2013. Low prevalence of Shiga toxin-producing Escherichia coli O103 in Norwegian sheep flocks. Appl. Environ. Microbiol. 79:7502–7509. 10.1128/AEM.01825-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang WL, Kohler B, Oswald E, Beutin L, Karch H, Morabito S, Caprioli A, Suerbaum S, Schmidt H. 2002. Genetic diversity of intimin genes of attaching and effacing Escherichia coli strains. J. Clin. Microbiol. 40:4486–4492. 10.1128/JCM.40.12.4486-4492.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Blanco M, Blanco JE, Mora A, Dahbi G, Alonso MP, Gonzalez EA, Bernardez MI, Blanco J. 2004. Serotypes, virulence genes, and intimin types of Shiga toxin (verotoxin)-producing Escherichia coli isolates from cattle in Spain and identification of a new intimin variant gene (eae-ξ). J. Clin. Microbiol. 42:645–651. 10.1128/JCM.42.2.645-651.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sekse C, Muniesa M, Wasteson Y. 2008. Conserved Stx2 phages from Escherichia coli O103:H25 isolated from patients suffering from hemolytic uremic syndrome. Foodborne Pathog. Dis. 5:801–810. 10.1089/fpd.2008.0130 [DOI] [PubMed] [Google Scholar]
- 26.Mattos-Guaraldi AL, Formiga LC, Andrade AF. 1999. Cell surface hydrophobicity of sucrose fermenting and nonfermenting Corynebacterium diphtheriae strains evaluated by different methods. Curr. Microbiol. 38:37–42. 10.1007/PL00006769 [DOI] [PubMed] [Google Scholar]
- 27.Di Bonaventura G, Piccolomini R, Paludi D, D'Orio V, Vergara A, Conter M, Ianieri A. 2008. Influence of temperature on biofilm formation by Listeria monocytogenes on various food-contact surfaces: relationship with motility and cell surface hydrophobicity. J. Appl. Microbiol. 104:1552–1561. 10.1111/j.1365-2672.2007.03688.x [DOI] [PubMed] [Google Scholar]
- 28.Norwood DE, Gilmour A. 1999. Adherence of Listeria monocytogenes strains to stainless steel coupons. J. Appl. Microbiol. 86:576–582. 10.1046/j.1365-2672.1999.00694.x [DOI] [PubMed] [Google Scholar]
- 29.Schonewille E, Nesse LL, Hauck R, Windhorst D, Hafez HM, Vestby LK. 2012. Biofilm building capacity of Salmonella enterica strains from the poultry farm environment. FEMS Immunol. Med. Microbiol. 65:360–365. 10.1111/j.1574-695X.2012.00966.x [DOI] [PubMed] [Google Scholar]
- 30.Martinez-Medina M, Naves P, Blanco J, Aldeguer X, Blanco JE, Blanco M, Ponte C, Soriano F, Darfeuille-Michaud A, Garcia-Gil LJ. 2009. Biofilm formation as a novel phenotypic feature of adherent-invasive Escherichia coli (AIEC). BMC Microbiol. 9:202. 10.1186/1471-2180-9-202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang R, Bono JL, Kalchayanand N, Shackelford S, Harhay DM. 2012. Biofilm formation by Shiga toxin-producing Escherichia coli O157:H7 and non-O157 strains and their tolerance to sanitizers commonly used in the food processing environment. J. Food Prot. 75:1418–1428. 10.4315/0362-028X.JFP-11-427 [DOI] [PubMed] [Google Scholar]
- 32.Cremet L, Corvec S, Bemer P, Bret L, Lebrun C, Lesimple B, Miegeville AF, Reynaud A, Lepelletier D, Caroff N. 2012. Orthopaedic-implant infections by Escherichia coli: molecular and phenotypic analysis of the causative strains. J. Infect. 64:169–175. 10.1016/j.jinf.2011.11.010 [DOI] [PubMed] [Google Scholar]
- 33.Salo J, Sevander JJ, Tapiainen T, Ikaheimo I, Pokka T, Koskela M, Uhari M. 2009. Biofilm formation by Escherichia coli isolated from patients with urinary tract infections. Clin. Nephrol. 71:501–507. 10.5414/CNP71501 [DOI] [PubMed] [Google Scholar]
- 34.Reisner A, Krogfelt KA, Klein BM, Zechner EL, Molin S. 2006. In vitro biofilm formation of commensal and pathogenic Escherichia coli strains: impact of environmental and genetic factors. J. Bacteriol. 188:3572–3581. 10.1128/JB.188.10.3572-3581.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Goulter RM, Gentle IR, Dykes GA. 2009. Issues in determining factors influencing bacterial attachment: a review using the attachment of Escherichia coli to abiotic surfaces as an example. Lett. Appl. Microbiol. 49:1–7. 10.1111/j.1472-765X.2009.02591.x [DOI] [PubMed] [Google Scholar]
- 36.Rosenberg M, Blumberger Y, Judes H, Bar-Ness R, Rubinstein E, Mazor Y. 1986. Cell surface hydrophobicity of pigmented and nonpigmented clinical Serratia marcescens strains. Infect. Immun. 51:932–935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yousefi Rad AY, Ayhan H, Piskin E. 1998. Adhesion of different bacterial strains to low-temperature plasma-treated sutures. J. Biomed. Mater. Res. 41:349–358. [DOI] [PubMed] [Google Scholar]
- 38.Liu Y, Yang SF, Li Y, Xu H, Qin L, Tay JH. 2004. The influence of cell and substratum surface hydrophobicities on microbial attachment. J. Biotechnol. 110:251–256. 10.1016/j.jbiotec.2004.02.012 [DOI] [PubMed] [Google Scholar]
- 39.Abban S, Jakobsen M, Jespersen L. 2012. Attachment behaviour of Escherichia coli K12 and Salmonella Typhimurium P6 on food contact surfaces for food transportation. Food Microbiol. 31:139–147. 10.1016/j.fm.2012.04.003 [DOI] [PubMed] [Google Scholar]
- 40.Zita A, Hermansson M. 1997. Effects of bacterial cell surface structures and hydrophobicity on attachment to activated sludge flocs. Appl. Environ. Microbiol. 63:1168–1170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li J, McLandsborough LA. 1999. The effects of the surface charge and hydrophobicity of Escherichia coli on its adhesion to beef muscle. Int. J. Food Microbiol. 53:185–193. 10.1016/S0168-1605(99)00159-2 [DOI] [PubMed] [Google Scholar]
- 42.Hassan AN, Frank JF. 2004. Attachment of Escherichia coli O157:H7 grown in tryptic soy broth and nutrient broth to apple and lettuce surfaces as related to cell hydrophobicity, surface charge, and capsule production. Int. J. Food Microbiol. 96:103–109. 10.1016/S0168-1605(03)00160-0 [DOI] [PubMed] [Google Scholar]
- 43.Rivas L, Fegan N, Dykes GA. 2007. Attachment of Shiga toxigenic Escherichia coli to stainless steel. Int. J. Food Microbiol. 115:89–94. 10.1016/j.ijfoodmicro.2006.10.027 [DOI] [PubMed] [Google Scholar]
- 44.Beloin C, Roux A, Ghigo JM. 2008. Escherichia coli biofilms. Curr. Top. Microbiol. Immunol. 322:249–289 http://europepmc.org/articles/PMC2864707;jsessionid=AHnDJOcAmVOgt5A9ugrr.0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Farfan MJ, Torres AG. 2012. Molecular mechanisms that mediate colonization of Shiga toxin-producing Escherichia coli strains. Infect. Immun. 80:903–913. 10.1128/IAI.05907-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Umanski T, Rosenshine I, Friedberg D. 2002. Thermoregulated expression of virulence genes in enteropathogenic Escherichia coli. Microbiology 148:2735–2744 http://mic.sgmjournals.org/content/148/9/2735.long [DOI] [PubMed] [Google Scholar]