Abstract
Biopolymers are important substrates for heterotrophic bacteria in (ultra)oligotrophic freshwater environments, but information about their utilization at microgram-per-liter levels by attached freshwater bacteria is lacking. This study aimed at characterizing biopolymer utilization in drinking-water-related biofilms by exposing such biofilms to added carbohydrates or proteins at 10 μg C liter−1 in flowing tap water for up to 3 months. Individually added amylopectin was not utilized by the biofilms, whereas laminarin, gelatin, and caseinate were. Amylopectin was utilized during steady-state biofilm growth with simultaneously added maltose but not with simultaneously added acetate. Biofilm formation rates (BFR) at 10 μg C liter−1 per substrate were ranked as follows, from lowest to highest: blank or amylopectin (≤6 pg ATP cm−2 day−1), gelatin or caseinate, laminarin, maltose, acetate alone or acetate plus amylopectin, and maltose plus amylopectin (980 pg ATP cm−2 day−1). Terminal restriction fragment length polymorphism (T-RFLP) and 16S rRNA gene sequence analyses revealed that the predominant maltose-utilizing bacteria also dominated subsequent amylopectin utilization, indicating catabolic repression and (extracellular) enzyme induction. The accelerated BFR with amylopectin in the presence of maltose probably resulted from efficient amylopectin binding to and hydrolysis by inductive enzymes attached to the bacterial cells. Cytophagia, Flavobacteriia, Gammaproteobacteria, and Sphingobacteriia grew during polysaccharide addition, and Alpha-, Beta-, and Gammaproteobacteria, Cytophagia, Flavobacteriia, and Sphingobacteriia grew during protein addition. The succession of bacterial populations in the biofilms coincided with the decrease in the specific growth rate during biofilm formation. Biopolymers can clearly promote biofilm formation at microgram-per-liter levels in drinking water distribution systems and, depending on their concentrations, might impair the biological stability of distributed drinking water.
INTRODUCTION
Polysaccharides and proteins of phytoplanktonic and bacterial origin represent a significant fraction of the organic matter in natural aquatic environments (1, 2). Unlike low-molecular-weight (LMW) compounds, these biopolymers have to undergo extracellular enzymatic hydrolysis before bacteria can utilize them (3, 4). Nevertheless, biopolymers are important carbon and energy sources for heterotrophic aquatic bacteria, because the bacterial community composition in freshwater and marine environments changes when these compounds become abundant during phytoplankton blooms (5–8). Furthermore, various selected biopolymers were degraded when added individually to marine and estuarine water at >100 μg C liter−1 and to marine sediments at >10 mg C liter−1 (9–14). Marine bacterial communities have also been reported to degrade selected biopolymers at <10 μg C liter−1 (i.e., ultraoligotrophic) levels in seawater (15–18), but information on biopolymer degradation and utilization in (ultra)oligotrophic freshwater is scarce.
Planktonic members of the classes Cytophagia, Flavobacteriia, Betaproteobacteria, and Actinobacteria contribute significantly to biopolymer degradation in freshwater environments (13, 19–22). Certain planktonic freshwater representatives of the genus Flavobacterium are particularly adapted to growth with polysaccharides and proteins at a few μg C liter−1 in batch tests (23–25). However, under the turbulent flow conditions prevailing in drinking water distribution systems and in certain natural lotic freshwater systems (e.g., brooks and streams), surface-attached rather than planktonic microorganisms predominate (26). Biofilm formation in drinking water distribution systems can impair drinking water quality and safety by causing increased levels of coliform and heterotrophic bacteria, esthetic problems (e.g., unusual taste, odor, appearance, presence of invertebrates), and the growth of opportunistic pathogens such as Legionella pneumophila, nontuberculous mycobacteria, and Pseudomonas aeruginosa (27). LMW compounds in drinking water promote biofilm formation in unchlorinated distribution systems at only a few μg C per liter (28, 29). Various extracellular biopolymer-degrading enzymes have been detected in biofilms (30), but biopolymer degradation by biofilms in oligotrophic freshwater environments has, to our knowledge, not yet been quantified. Hence, it is not known whether biopolymers at microgram-per-liter levels can support the growth of attached heterotrophic bacteria under turbulent flow conditions in drinking water distribution systems and in natural (ultra)oligotrophic freshwater systems.
The objectives of our study were therefore (i) to assess the ability of attached heterotrophic bacteria to utilize biopolymers at microgram-per-liter levels in flowing ultraoligotrophic water by using biofilm monitors (28) supplemented with unchlorinated tap water and selected biopolymers and (ii) to assess the effect of polysaccharide or protein addition on the bacterial community composition of the biofilms formed in these monitors.
MATERIALS AND METHODS
Biofilm monitor.
Four different experiments (experiments A to D) were conducted to assess the biofilm-forming properties of tap water supplemented with microgram-per-liter levels of maltose and/or amylopectin (from corn), acetate and/or amylopectin, caseinate (from bovine milk), gelatin (type B, from bovine skin), or laminarin (from Laminaria digitata) (all compounds were from Sigma-Aldrich, Germany) in separate biofilm monitors (see Table S1 in the supplemental material). These biopolymers were selected because they are generally acknowledged model substrates for studies on biopolymer utilization in natural and engineered aquatic systems (9, 24, 31–33); maltose is a common amylopectin degradation product and was therefore important in elucidating the kinetics and mechanisms of amylopectin utilization. The biofilm monitor simulates the turbulent flow conditions prevailing during drinking water distribution in residential areas and has been described in detail elsewhere (28). In short, this test system consists of one or two vertical glass columns (diameter, 2.5 cm; height, 60 cm) placed in a series, each with approximately 40 cylindrical glass rings of 17.4 ± 0.2 cm2 that had been heated for 5 h at 550°C before use. Unchlorinated tap water, prepared from anaerobic groundwater by aeration and rapid sand filtration, continuously flowed downward through the column(s) at a rate of 275 liters h−1. The dissolved organic carbon (DOC) content in this feed water was 2.0 ± 0.1 mg C liter−1; the particulate organic carbon (POC) content was below the detection limit (0.2 mg C liter−1); the assimilable organic carbon (AOC) content was 3 to 5 μg acetate-C equivalents liter−1; and the phosphate and nitrate contents were 0.02 ± 0.02 and 0.12 ± 0.04 mg liter−1, respectively (34, 35). The feed water contained 6.4 (± 3.1) × 104 cells ml−1 (sample size [n] = 12) and 4.0 ± 1.3 ng ATP liter−1 and had an average temperature of 15.0 ± 2.4°C (n = 130) (see Table S2 in the supplemental material); other typical characteristics of the feed water have been reported previously (34, 35). During the first 2 to 4 weeks, only tap water without organic additives flowed through the system, and an initial biofilm developed on the glass rings. Subsequently, the addition of individual organic compounds or mixtures of organic compounds was started by dosing accurately prepared solutions containing 183 mg C liter−1 per organic compound at a flow rate of 15 ml h−1. Solutions of individual compounds were prepared in Milli-Q ultrapure water (Millipore, USA) and were autoclaved for 15 min at 103°C, except for the maltose solution, which was heated for 30 min at 60°C. The mixtures of amylopectin and maltose or acetate were prepared by aseptically mixing sterile solutions of the individual compounds. The water supply and organic compound addition were continuously monitored and controlled in order to ensure a constant inlet concentration of 3.0, 4.5, or 10 μg C liter−1 per added compound (± 5%) (n = 130).
Sampling.
One or two glass rings were removed from each column of the biofilm monitor two or three times per week. Each ring was transferred to a sterile capped glass tube (diameter, 1.9 cm; height, 25 cm) containing 10 ml of autoclaved tap water (15 min, 121°C) and was subjected to low-energy sonication (LES) using a Bransonic ultrasonic cleaner, model 5510 (Branson Ultrasonic Cooperation, USA) for 2 min at a constant 40-kHz frequency and 180-W power output to release the biofilm from the glass surface. The 10-ml biofilm suspension was transferred to a sterile, screw-cap Greiner tube (Greiner Bio One B.V., The Netherlands) and was placed on ice. The LES treatment was repeated twice, resulting in 30 ml of biofilm suspension per ring. Subsequently, the biofilm suspension was homogenized by applying high-energy sonication (HES) with a Branson Sonifier ultrasonic cell disruptor (model W-250D; Branson Ultrasonic Cooperation, USA) for 1 min at a constant 20-kHz frequency and an adjusted 90-W power output (45% amplitude). The biofilm suspension was kept on ice during and after HES treatment.
ATP analysis and bacterial counts.
The concentrations of ATP (as a measure of active biomass) in the biofilm suspensions obtained were measured using a bioluminescence assay as described previously (36). The growth rate (μ) per day during the exponential phase of biofilm formation with the organic additive(s) was calculated as (ln BFC2 − ln BFC1)/(t2 − t1), where BFC2 and BFC1 are the biofilm concentrations (expressed in picograms of ATP per square centimeter) at time points 2 and 1 (t2 and t1). The subsequent linear increase in the BFC as a function of time was used to determine the biofilm formation rate (BFR; expressed in picograms of ATP per square centimeter per day) (28). The BFR/BFC ratio (per day) is defined as the specific growth rate during the linear phases of biofilm formation. The minimum or critical specific growth rate (µmin) represents the BFR/BFC ratio in the steady-state phases of biofilm formation.
Heterotrophic plate counts (HPC) and total direct cell counts (TDC) of the biofilms were determined once to three times per 14 days, depending on the BFR. HPC values were determined using triplicate R2A agar spread plates, which were incubated for 10 days at 25°C (37). TDC values were determined using acridine orange and epifluorescence microscopy (38). The plating efficiency (expressed as a percentage) was calculated from the HPC/TDC ratio.
Identification of bacterial isolates.
When enhanced biofilm formation with the organic additive(s) was observed, the bacterial strains that formed predominant colonies on the R2A plates based on colony morphology were identified by 16S rRNA gene sequencing. Primers 8f (5′-AGAGTTTGATCMTGGCTCAG-3′) and 1392r (5′-ACGGGCGGTGTGTACA-3′) (Biolegio, The Netherlands) were used to amplify the 16S rRNA genes of the bacterial isolates. PCR mixtures (50 μl) contained 25 μl of PCR Master Mix (Promega, USA), 10 pmol of each primer, 5 μl of bovine serum albumin (BSA), and 5 μl of 100-times-diluted template DNA. PCR amplification was performed in the GeneAmp PCR system 9700 (Applied Biosystems, USA), according to the following program: 5 min at 95°C; 35 cycles of 45 s at 95°C, 60 s at 57°C, and 2 min at 72°C; and finally 7 min at 72°C. The size of the PCR product was confirmed on a 2% (wt/vol) agarose gel stained with SYBR Gold. For each bacterial isolate, approximately 700 bp of the 16S rRNA gene were sequenced by Macrogen (USA) using primer 8f. Close relatives of each 16S rRNA gene were obtained from the GenBank database by nucleotide-nucleotide BLAST homology searches.
DNA isolation.
One to 10 ml of a biofilm suspension was filtered through a 0.22-μm-pore-size, 50-mm-diameter polycarbonate track-etched membrane (type 23007; Sartorius, Germany). The filter was placed in a 2-ml tube containing Lysing Matrix E, sodium phosphate buffer, and the MT buffer of the FastDNA Spin kit for soil (MP Biomedicals, USA) and was stored at −20°C. DNA was isolated by using the FastDNA Spin kit for soil according to the manufacturer's protocol.
T-RFLP analysis.
16S rRNA genes were amplified using 6-carboxyfluorescein (FAM)-labeled primer 8f and primer 1392r, with the PCR amplification conditions described above. For each experiment (A1 to D4 [see Table S1 in the supplemental material]), terminal restriction fragment length polymorphism (T-RFLP) analysis was conducted on the biofilms sampled on the first day (day 0), on the final day, and on one to three intermediate days. Fluorescently labeled PCR products (45 μl) were purified using a DNA Clean & Concentrator-5 kit according to the manufacturer's instructions (Zymo Research, USA). Digestion mixtures (20 μl) contained 5 U of restriction enzyme HhaI (Promega, The Netherlands), 2 μl of buffer C (Promega), 0.2 μl of BSA, and 5 μl of the purified PCR product and were incubated for 6 h at 37°C. Subsequently, the digestion mixtures were purified as described above. Purified digestion products (5 μl) were mixed with 15 μl of the loading buffer Hi-Di formamide and 1 μl of the internal standard GeneScan 1000 ROX (Applied Biosystems, The Netherlands). After heating for 3 min at 95°C and subsequent cooling on ice, the fluorescently labeled terminal restriction fragments (T-RFs) were size separated on an ABI Prism genetic analyzer, model 310 (Applied Biosystems), in GeneScan analysis mode (Applied Biosystems). T-RFLP electropherograms were analyzed with BioNumerics software, version 6.0 (Applied Maths, Belgium). Cluster analysis using the unweighted-pair group method with arithmetic means (UPGMA) was based on the densitometric curve-based Pearson correlation coefficient between the T-RF band patterns of the biofilm samples.
16S rRNA gene cloning and sequencing.
Biofilm samples collected on the final days of experiments A2, A4, B2 phase 2, B3, B4 phase 3, and D1 through D4 (see Table 1; see also Table S1 in the supplemental material) were selected for 16S rRNA gene cloning and sequencing. The 16S rRNA genes were amplified using primers 8f and 1392r as described above. PCR products were cloned into Escherichia coli JM109 using the pGEM-T Easy vector system (Promega, The Netherlands). Inserts of randomly selected positive clones were amplified with pGEM T Easy vector-specific primers T7 and SP6, with the following PCR conditions: 94°C for 5 min; 30 cycles of 94°C for 20 s, 48°C for 45 s, and 72°C for 45 s; and finally 72°C for 7 min. About 700 bp of the 16S rRNA gene insert of each clone was sequenced by Macrogen (USA) using primer 8f. Approximately 96 clones were analyzed per biofilm sample. A distance matrix was generated in BioEdit (39) and was imported into DOTUR (40) in order to assign the 16S rRNA gene sequences obtained to operational taxonomical units (OTUs) on the basis of ≥99% similarity and to calculate the Shannon diversity index for each biofilm sample.
TABLE 1.
Biological parameters of biofilm formation when organic compounds with C at μg liter−1 levels are added to tap watera
| Expt | Period (days) | Added compound(s) (concn of C [μg liter−1]) | Avg BFR ± SE (pg ATP cm−2 day−1) | Estimated avg µmin ± SD (day−1) | Avg BFCmax ± SD (103 pg cm−2) | Avg TDCmax ± SD (107 cells cm−2) | HPC/TDC (%) | Avg amt of ATP cell−1 ± SD (fg cell−1) |
|---|---|---|---|---|---|---|---|---|
| A1 | 0–69 | Blank | 4.7 ± 0.3 | NAd | 0.35 | 0.5 ± 0.2 | ≤1 | 0.06 ± 0.01 |
| A2 | 0–32 | Maltose (10) | 267 ± 26 | 0.09 ± 0.001 | 3.0 ± 0.1 | 1.5 ± 0.2 | 25–50 | 0.18 ± 0.01 |
| A3 | 0–39 | Amylopectin (10) | 7.6 ± 0.6 | NA | 0.25 | 0.3 ± 0.05 | ≤1 | 0.09 ± 0.02 |
| 39–69 | Maltose (10), amylopectin (10) | 452 ± 177; 978 ± 137b | 0.10 ± 0.003; 0.07 ± 0.004c | 14 ± 0.8 | 7.6 ± 0.9 | 22–28 | 0.21 ± 0.05 | |
| A4 | 0–32 | Maltose (10), amylopectin (10) | 358 ± 56; 1,034 ± 70b | 0.15 ± 0.005; 0.08 ± 0.004c | 13 ± 0.3 | 6.5 ± 1.5 | 40–86 | 0.22 ± 0.01 |
| B1 | 0–86 | Blank | 4.0 ± 0.3 | NA | 0.42 | 0.5 ± 0.05 | ≤1 | 0.10 ± 0.01 |
| B2 | 0–57 | Acetate (3.0) | 40 ± 3 | 0.02 ± 0.002 | 2.5 | 1.1 ± 0.4 | 16–40e | 0.23 ± 0.03e |
| 57–86 | Acetate (4.5) | 158 ± 35 | 0.05 ± 0.002 | 4.9 | 2.8 ± 1.2 | |||
| B3 | 0–86 | Amylopectin (10) | 4.0 ± 0.3; 15 ± 2b | NA | 0.76 | 0.4 ± 0.2 | 2–3 | 0.12 ± 0.02 |
| B4 | 0–31 | Acetate (3.0) | 47 ± 12 | 0.03 ± 0.000 | NA | 0.5 ± 0.1 | 18–54e | 0.24 ± 0.04e |
| 31–57 | Acetate (3.0), amylopectin (10) | 46 ± 5 | 0.03 ± 0.003 | 2.3 | 1.1 ± 0.2 | |||
| 57–86 | Acetate (4.5), amylopectin (10) | 135 ± 21 | 0.04 ± 0.005 | 5.9 | 2.8 ± 0.8 | |||
| C1 | 0–52 | Blank | 5.5 ± 0.3 | NA | 0.24 | 0.1 ± 0.05 | ≤1 | 0.12 ± 0.02 |
| C2 | 0–52 | Acetate (10) | 613 ± 55; 737 ± 52b | 0.06 ± 0.009; 0.03 ± 0.001 | 23 | 3.3 ± 0.4 | 22–48 | 0.46 ± 0.17 |
| C3 | 0–52 | Amylopectin (10) | 9.2 ± 0.7 | NA | 0.32 | 0.1 ± 0.02 | 3–5 | 0.10 ± 0.02 |
| C4 | 0–20 | Acetate (10) | 823 ± 93 | 0.08 ± 0.008 | NA | 2.2 ± 0.7 | 18–37e | 0.48 ± 0.09e |
| 20–52 | Acetate (10), amylopectin (10) | 634 ± 52 | 0.03 ± 0.002 | 21 | 3.7 ± 1.2 | |||
| D1 | 0–88 | Blank | 5.3 ± 0.3 | NA | 0.49 | 0.6 ± 0.2 | ≤1 | 0.11 ± 0.02 |
| D2 | 0–82 | Caseinate (10) | 61 ± 4 | 0.013 ± 0.001 | 4.6 ± 0.3 | 2.5 ± 0.7 | 6–37 | 0.21 ± 0.02 |
| D3 | 0–88 | Gelatin (10) | 59 ± 4; 70 ± 10; 64 ± 9b | 0.057 ± 0.004; 0.036 ± 0.002; 0.017 ± 0.000c | 3.7 ± 0.01 | 1.7 ± 0.3 | 13–63 | 0.21 ± 0.01 |
| D4 | 0–82 | Laminarin (10) | 81 ± 10; 83 ± 13b | 0.058 ± 0.005; 0.030 ± 0.001c | 2.8 ± 0.1 | 1.2 ± 0.3 | 5–93 | 0.26 ± 0.05 |
BFR, biofilm formation rate; µmin, minimum or critical specific growth rate (calculated as BFR/BFC) in steady-state growth phases; BFCmax, maximum biofilm concentration on glass ring; HPC, heterotrophic plate count; TDCmax, maximum total direct cell count.
Where more than one BFR value is given for linear biofilm growth phases, the values were obtained at the following times: in experiment A3, before and after day 51; in experiment A4, before and after day 12; in experiment B3, from day 0 to 48 and day 51 to 86; in experiment C2, from day 8 to 20 and day 31 to 52; in experiment D3, from day 21 to 35, day 49 to 61, and day 73 to 84; and in experiment D4, from day 13 to 34 and day 59 to 68.
Critical growth rates determined in the steady-state growth phases following the different linear growth phases for which BFR values are given.
NA, not applicable.
Values apply to the entire experimental period.
Chemical analyses.
For determination of total iron (Fe) concentrations, tap water samples and selected biofilm suspensions were destructed with nitric acid at 103 to 175°C and were analyzed by inductively coupled plasma mass spectrometry (ICP-MS) using a Thermo Scientific XSeries 2 ICP-MS (Thermo Fisher Scientific Inc., USA). Tap water temperature was monitored with a calibrated digital thermometer (Testo B.V., The Netherlands).
Nucleotide sequence accession numbers.
The 16S rRNA gene sequences obtained in this study have been deposited in GenBank under accession numbers JQ791554 through JQ792006.
RESULTS
Biofilm formation with amylopectin in the absence or presence of maltose or acetate.
During the period of continuous supply of tap water without added organic compounds, biofilms were formed at a BFR ranging from 4.0 to 5.5 (± 0.3) pg ATP cm−2 day−1, and a maximum biofilm concentration (BFCmax) of approximately 240 to 490 pg ATP cm−2 was reached after 2 to 3 months (Table 1; see Fig. 1 to 3). When maltose was added to tap water at 10 μg C liter−1, biofilm growth was promoted within 4 days at an exponential rate (μ) of 1.0 ± 0.1 day−1 until day 8 (Fig. 1). Subsequently, the BFC increased linearly at a BFR of 267 ± 26 pg ATP cm−2 day−1 and reached a BFCmax of approximately 3,000 pg ATP cm−2 after 12 to 14 days (Fig. 1; Table 1). In contrast, amylopectin at 10 μg C liter−1 did not enhance biofilm formation over that with the blank for 39 days in the same experiment (Fig. 1). The addition of amylopectin at 10 μg C liter−1 for more than 50 days in another experiment eventually resulted in a BFR that was about four times higher than that of the blank (Fig. 2; Table 1).
FIG 1.
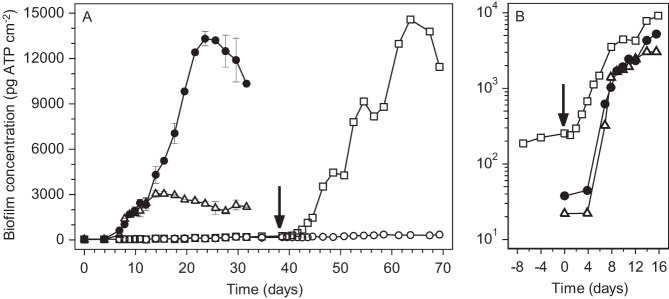
Biofilm formation on glass rings exposed to tap water with added maltose, amylopectin, or both, each at 10 μg C liter−1. (A) Linear scale; (B) log scale. Arrows indicate day 39 in panel A and its equivalent, day 0, in panel B. Symbols: ○, blank (no compounds added); △, maltose added; □, amylopectin added from day 0 to 39 in panel A (corresponding to times earlier than day 0 in panel B), and maltose and amylopectin added from day 39 on in panel A (corresponding to day 0 and later times in panel B); ●, maltose and amylopectin added from day 0 on in panels A and B. Error bars represent standard deviations of biofilm concentrations on duplicate glass rings with the addition of maltose only or with the addition of maltose and amylopectin from day 0 on.
FIG 3.
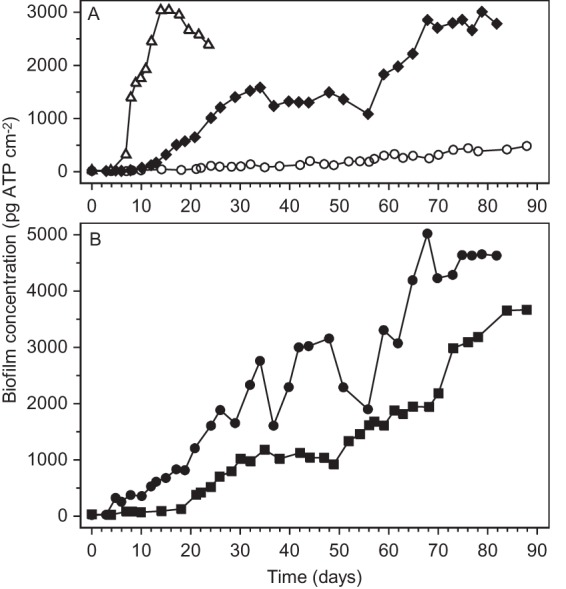
Biofilm formation on glass rings exposed to tap water with added laminarin (⧫), caseinate (●), or gelatin (■) at 10 μg C liter−1 compared with biofilm formation with tap water without additions (○) or with maltose at 10 μg C liter−1 (△).
FIG 2.
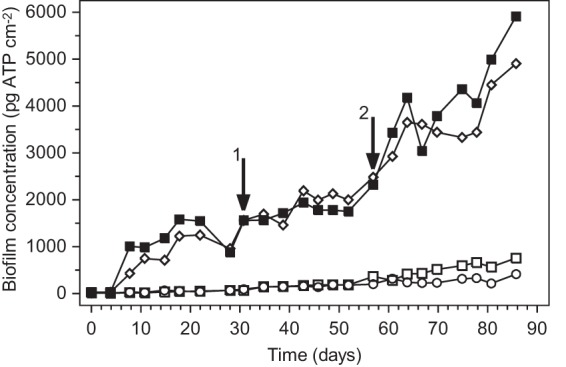
Biofilm formation on glass rings exposed to tap water to which acetate, amylopectin, or both were added with C at μg liter−1 levels. Symbols: ○, blank (no compounds added); □, amylopectin added at 10 μg C liter−1; ■, acetate added from day 0 to 57 at 3.0 μg C liter−1 and from day 57 on at 4.5 μg C liter−1; ■, acetate at 3.0 μg C liter−1 added from day 0 to 31, acetate at 3.0 μg C liter−1 and amylopectin at 10 μg C liter−1 added from day 31 to 57, and acetate at 4.5 μg C liter−1 and amylopectin at 10 μg C liter−1 added from day 57 on. Arrows 1 and 2 indicate the changes in compound supplementation on days 31 and 57, respectively.
During simultaneous addition of maltose and amylopectin, each at 10 μg C liter−1, the BFC increased exponentially at a rate of 0.81 ± 0.07 day−1 from day 4 until day 8 and then linearly at a BFR of 358 ± 56 pg ATP cm−2 day−1 until day 12. Hence, biofilm formation with maltose plus amylopectin was synchronic with biofilm formation with maltose alone during the first 12 days (Fig. 1; Table 1). Unlike biofilm formation with maltose alone, however, biofilm formation with maltose and amylopectin continued after day 12 at a BFR of 1,034 ± 70 pg ATP cm−2 day−1, and a BFCmax of about 13,000 pg cm−2 was attained (Fig. 1). Apparently, biofilm formation was promoted initially by maltose and subsequently by amylopectin in the presence of maltose. Similar findings were obtained during simultaneous addition of maltose and amylopectin, each at 10 μg C liter−1, to a biofilm monitor that had first been supplied only with amylopectin at 10 μg C liter−1 for 39 days (Fig. 1; Table 1). Thus, amylopectin strongly promoted biofilm formation in the presence of maltose at 10 μg C liter−1. Simultaneous addition of acetate at 3.0, 4.5, or 10 μg C liter−1 and amylopectin at 10 μg C liter−1, however, resulted in BFR and BFCmax values similar to those obtained with the addition of acetate alone (Fig. 2; Table 1). Hence, the biofilms formed with acetate at ≤10 μg C liter−1 did not utilize amylopectin.
Biofilm formation with laminarin, caseinate, or gelatin.
Laminarin, gelatin, and caseinate promoted biofilm formation individually at 10 μg C liter−1 in the biofilm monitor. During 82 days of laminarin addition, multiple phases in biofilm formation could be distinguished (Fig. 3A). The BFC increased exponentially at a rate of 0.34 ± 0.01 day−1 (days 5 to 15) and then linearly at a BFR of 81 ± 10 pg ATP cm−2 day−1 (days 16 to 34). The first steady-state growth phase (days 37 to 51) was followed by a second linear increase in BFC at a BFR of 83 ± 13 pg ATP cm−2 (days 56 to 68). Finally, a BFCmax of almost 3,000 pg ATP cm−2 was reached in the second steady-state phase (days 70 to 82) (Table 1).
The concentration of the biofilm formed with gelatin at 10 μg C liter−1 also increased discontinuously, i.e., before day 21 at an exponential growth rate of 0.30 ± 0.07 day−1 and from day 21 to day 35, day 49 to day 61, and day 73 to day 84 at BFRs of 59 ± 4, 70 ± 10, and 64 ± 9 pg ATP cm−2, respectively (Fig. 3B; Table 1). A BFCmax of 3,700 pg ATP cm−2 was attained after >84 days of gelatin addition. During the first week of caseinate addition at 10 μg C liter−1, exponential biofilm growth was promoted at an estimated rate of 0.85 ± 0.26 day−1, but insufficient data were available to accurately determine the growth rate (Fig. 3B). Subsequently, fluctuating but overall increasing BFCs at a BFR of 61 ± 4 pg ATP cm−2 day−1 were observed. After approximately 68 days of caseinate addition, a BFCmax of about 4,600 pg ATP cm−2 was reached.
ATP level cell−1 and plating efficiency.
The average ATP level cell−1 of the bacteria in the biofilms formed with either tap water (blank) or added amylopectin ranged from 0.06 ± 0.01 (experiment A1) to 0.12 ± 0.02 fg cell−1 (experiments B3 and C1), and only ≤1% of these bacteria could be cultivated (Table 1). The linear increase in the BFC upon addition of maltose alone or maltose and amylopectin was accompanied by an elevated average ATP level cell−1 of around 0.20 ± 0.02 fg cell−1 and a plating efficiency of >20 to 86% (Table 1). During linear biofilm growth with acetate alone or acetate and amylopectin, average ATP levels cell−1 also increased to ≥0.20 fg cell−1, and 16 to 54% of the biofilm bacteria were cultivated (Table 1). The bacteria that utilized laminarin, caseinate, or gelatin in the linear biofilm growth phases had average ATP levels of 0.26 ± 0.05, 0.21 ± 0.01, and 0.21 ± 0.02 fg cell−1, respectively. The plating efficiency observed in the first month of laminarin (93%) or caseinate (20 to 37%) addition decreased to <10% in the subsequent 2 months, whereas the plating efficiency in the first month of gelatin addition was >20 to 63% and decreased to 13% afterwards (Table 1). These results indicate a relation between plating efficiency and ATP level cell−1 in the linear biofilm growth phase, i.e., plating efficiency was ≤1% at ≤0.12 ± 0.02 fg ATP cell−1 in the absence of growth-promoting organic additives but ranged from ≥5 to 93% at ≥0.20 fg ATP cell−1 in the presence of growth-promoting organic additives.
T-RFLP profiles of attached bacterial communities.
The biofilms formed with tap water (blank) were ≥80% similar both to each other and to the biofilms formed with added amylopectin alone based on their T-RFLP profiles (Fig. 4). The high similarity between the bacterial communities of these biofilms is consistent with the observation that amylopectin did not enhance biofilm formation over that with the blank. In contrast, addition of the other tested compounds yielded biofilms with bacterial community compositions clearly different from those of the biofilms formed with tap water alone (Fig. 4). The bacterial communities in the biofilms formed with maltose (maltose-BF) or with maltose and amylopectin (maltose-amylopectin-BF) shared 74% similarity on day 16 (data not shown) and 87% similarity on day 24 (Fig. 4). Furthermore, these carbohydrate-specific biofilms clustered closer to the biofilms formed with laminarin (laminarin-BF) than to the biofilms formed with protein or acetate (Fig. 4). The biofilms formed after 32 to 82 days of caseinate addition (caseinate-BF) grouped together with the biofilms formed after 44 to 88 days of gelatin addition (gelatin-BF) in a protein-specific T-RFLP cluster (Fig. 4 [not all data shown]). The bacterial communities in the biofilms formed with acetate (acetate-BF) and with acetate and amylopectin (acetate-amylopectin-BF) were highly similar to each other (87 to 95% similarity) but largely dissimilar to the bacterial communities in the biofilms formed with other organic additives or with the blank (Fig. 4).
FIG 4.
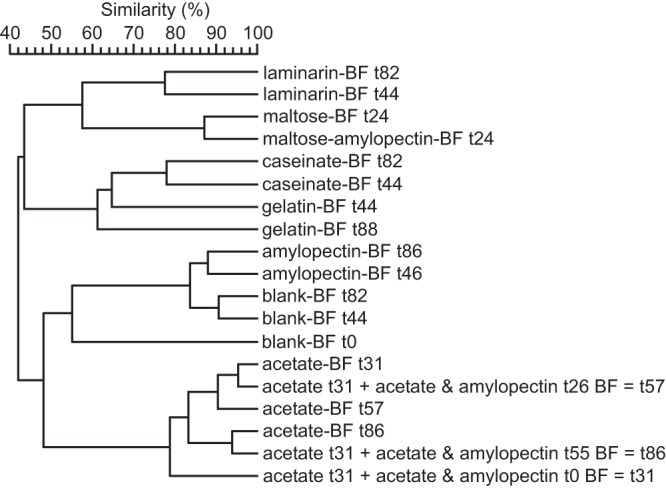
UPGMA cluster analysis based on the Pearson correlation coefficient measuring the degree of similarity between the T-RFLP patterns of biofilms formed with tap water (blank; from experiment D1) and those of biofilms formed with an organic compound(s) added to tap water at μg C liter−1 levels over 1 to 3 months (time [t] is given in days).
Identity and diversity of the predominating attached bacteria.
A total of 327 OTUs, including 263 biofilm type-specific OTUs, were obtained, revealing high bacterial diversities within and between the nine biofilm types (Table 2). The number of OTUs ranged from <30 for the maltose-BF (29 OTUs) and maltose-amylopectin-BF (25 OTUs) to >60 for the blank-BF (64 OTUs) and amylopectin-BF (62 OTUs); Shannon diversity indices ranged from 2.3 to 4.0 (Table 2). OTUs related to aquatic members of the family Nitrospiraceae (class Nitrospira) and the family Hyphomicrobiaceae (class Alphaproteobacteria) were found in nearly all nine biofilm types (see Table S3 in supplemental material). Ten of the 11 Nitrospiraceae OTUs had ≥97% similarity to Nitrospira spp., and five were shared among multiple biofilms (see Table S3). Together, the two largest shared Nitrospira OTUs included 1 to 12% of the clones from seven biofilm types. Most of the 21 Hyphomicrobiaceae OTUs were affiliated to Hyphomicrobium or Pedomicrobium spp., were biofilm type specific, and were singletons (see Table S3 [not all data shown]).
TABLE 2.
Classification of OTUs clustering with bacteria obtained from biofilms formed with tap water alone or with maltose, amylopectin, maltose and amylopectin, acetate, acetate and amylopectin, gelatin, caseinate, or laminarin added to tap water at μg C liter−1 levelsa
| Classification according to phylum and class | Blank-BF |
Maltose-BF |
Amylop-BF |
Mal-amp-BF |
Acetate-BF |
Ac-amp-BF |
Gelatin-BF |
Caseinate-BF |
Laminarin-BF |
Total of all biofilms |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Clonesb | OTUsb | Clones | OTUs | Clones | OTUs | Clones | OTUs | Clones | OTUs | Clones | OTUs | Clones | OTUs | Clones | OTUs | Clones | OTUs | Clones | OTUsc |
||
| Unique | Shared | ||||||||||||||||||||
| Actinobacteria | 4 (5) | 3 (5) | 2 (2) | 2 (7) | 2 (2) | 2 (3) | 2 (2) | 2 (8) | 1 (1) | 1 (2) | 4 (5) | 4 (8) | 4 (4) | 4 (8) | 19 (2.4) | 8 (2.4) | 5 (1.5) | ||||
| Bacteroidetes | |||||||||||||||||||||
| Cytophagia | 1 (1) | 1 (2) | 12 (14) | 3 (10) | 5 (6) | 1 (4) | 1 (1) | 1 (2) | 4 (5) | 3 (6) | 23 (2.9) | 6 (1.8) | 1 (0.3) | ||||||||
| Flavobacteriia | 3 (3) | 1 (3) | 11 (12) | 2 (8) | 2 (2) | 1 (2) | 5 (6) | 2 (4) | 6 (7) | 5 (9) | 1 (1) | 1 (2) | 28 (3.6) | 6 (1.8) | 3 (0.9) | ||||||
| Sphingobacteriia | 1 (1) | 1 (2) | 17 (20) | 3 (10) | 1 (1) | 1 (2) | 1 (1) | 1 (4) | 5 (6) | 4 (8) | 8 (9) | 3 (6) | 27 (29) | 6 (12) | 60 (7.7) | 9 (2.8) | 3 (0.9) | ||||
| Miscellaneous | 1 (1) | 1 (3) | 1 (1) | 1 (2) | 1 (1) | 1 (4) | 2 (2) | 1 (2) | 2 (2) | 2 (4) | 2 (2) | 1 (2) | 9 (1.2) | 7 (2.1) | |||||||
| Chloroflexi | 5 (6) | 5 (8) | 1 (1) | 1 (2) | 2 (2) | 2 (4) | 2 (2) | 1 (2) | 1 (1) | 1 (2) | 11 (1.4) | 9 (2.8) | 1 (0.3) | ||||||||
| Cyanobacteria | 1 (1) | 1 (2) | 1 (1) | 1 (2) | 4 (4) | 1 (2) | 6 (0.8) | 1 (0.3) | |||||||||||||
| Dictyoglomia | 1 (1) | 1 (2) | 1 (0.1) | 1 (0.3) | |||||||||||||||||
| Fibrobacteres and Acidobacteria | 7 (8) | 4 (6) | 1 (1) | 1 (3) | 5 (6) | 3 (5) | 2 (2) | 2 (5) | 4 (5) | 3 (6) | 8 (9) | 6 (11) | 5 (6) | 4 (7) | 3 (3) | 3 (6) | 35 (4.5) | 22 (6.7) | 2 (0.6) | ||
| Firmicutes | 3 (3) | 3 (5) | 8 (9) | 6 (10) | 2 (2) | 1 (2) | 3 (4) | 3 (6) | 1 (1) | 1 (2) | 2 (2) | 2 (4) | 19 (2.4) | 16 (4.9) | |||||||
| Gemmatimonadates | 1 (1) | 1 (2) | 1 (1) | 1 (2) | 2 (2) | 2 (4) | 4 (0.5) | 2 (0.6) | 1 (0.3) | ||||||||||||
| Nitrospirae | 10 (12) | 4 (6) | 2 (2) | 2 (7) | 16 (18) | 5 (8) | 1 (1) | 1 (4) | 7 (9) | 4 (9) | 4 (5) | 2 (4) | 17 (20) | 5 (9) | 12 (14) | 3 (6) | 11 (12) | 3 (6) | 80 (10.2) | 6 (1.8) | 5 (1.5) |
| Planctomycetes | 10 (12) | 7 (11) | 1 (1) | 1 (3) | 3 (3) | 3 (5) | 1 (1) | 1 (4) | 1 (1) | 1 (2) | 2 (2) | 2 (4) | 1 (1) | 1 (2) | 6 (7) | 6 (11) | 7 (8) | 6 (12) | 32 (4.1) | 21 (6.4) | 4 (1.2) |
| Proteobacteria | |||||||||||||||||||||
| Alphaproteobacteria | 24 (28) | 19 (30) | 11 (13) | 3 (10) | 18 (20) | 15 (24) | 3 (3) | 3 (12) | 11 (13) | 8 (19) | 10 (12) | 7 (15) | 19 (22) | 11 (21) | 17 (20) | 13 (24) | 5 (5) | 4 (8) | 118 (15.1) | 50 (15.3) | 12 (3.7) |
| Betaproteobacteria | 11 (13) | 7 (11) | 9 (10) | 6 (21) | 11 (12) | 9 (15) | 18 (20) | 8 (32) | 36 (44) | 16 (37) | 45 (54) | 23 (48) | 7 (8) | 6 (11) | 7 (8) | 6 (11) | 6 (7) | 5 (10) | 150 (19.2) | 44 (13.5) | 15 (4.6) |
| Gammaproteobacteria | 2 (2) | 2 (3) | 25 (29) | 4 (14) | 12 (13) | 8 (13) | 46 (51) | 4 (16) | 15 (18) | 4 (9) | 9 (11) | 3 (6) | 12 (14) | 6 (11) | 12 (14) | 5 (9) | 7 (8) | 6 (12) | 140 (17.9) | 22 (6.7) | 9 (2.8) |
| Deltaproteobacteria | 4 (5) | 4 (6) | 1 (1) | 1 (3) | 7 (8) | 6 (10) | 1 (1) | 1 (4) | 4 (5) | 4 (9) | 1 (1) | 1 (2) | 1 (1) | 1 (2) | 2 (2) | 2 (4) | 21 (2.7) | 18 (5.5) | 1 (0.3) | ||
| Verrumicrobia | 1 (1) | 1 (2) | 2 (2) | 2 (4) | 1 (1) | 1 (2) | 4 (0.5) | 4 (1.2) | |||||||||||||
| Unidentified bacteria | 1 (1) | 1 (2) | 1 (1) | 1 (3) | 6 (7) | 3 (5) | 1 (1) | 1 (2) | 3 (4) | 3 (6) | 9 (10) | 5 (10) | 21 (2.7) | 12 (3.7) | 1 (0.3) | ||||||
| Total | 86 (100) | 64 (100) | 86 (100) | 29 (100) | 90 (100) | 62 (100) | 90 (100) | 25 (100) | 82 (100) | 43 (100) | 83 (100) | 48 (100) | 86 (100) | 53 (100) | 86 (100) | 55 (100) | 92 (100) | 52 (100) | 781 (100) | 263 (80) | 64 (20) |
| Shannon diversity index | 4.0 | 2.8 | 3.9 | 2.3 | 3.3 | 3.4 | 3.7 | 3.7 | 3.6 | ||||||||||||
Blank-BF, biofilms formed with tap water alone (t, 82 days); maltose-BF, biofilms formed with maltose (t, 24 days); Amylop-BF, biofilms formed with amylopectin (t, 86 days); Mal-amp-BF, biofilms formed with maltose and amylopectin (t, 24 days); acetate-BF, biofilms formed with acetate (t, 86 days); Ac-amp-BF, biofilms formed with acetate and amylopectin (t, 86 days); gelatin-BF, biofilms formed with gelatin (t, 88 days); caseinate-BF, biofilms formed with caseinate (t, 88 days); laminarin-BF, biofilms formed with laminarin (t, 82 days).
All values are given as the number (percentage) of clones or OTUs.
The percentage of OTUs was calculated using the total number of 327 OTUs (100%).
Many of the clones from the acetate-BF and acetate-amylopectin-BF were affiliated to members of the family Comamonadaceae (33% and 39%, respectively; [data not shown]), e.g., Hydrogenophaga, Rhodoferax and Curvibacter spp. (see Table S3 in the supplemental material). In addition, a significant proportion of the clones (15% and 8%, respectively) were represented by an OTU with 99% similarity to Thiothrix fructosivorans (see Table S3).
The predominant OTU from the maltose-BF included 19% of the clones and showed 95% similarity to a Cellvibrio sp. (class Gammaproteobacteria), whereas an OTU with 96% similarity to another Cellvibrio sp. represented another 8% of the clones (see Table S3 in the supplemental material). These two Cellvibrio OTUs together also represented nearly 50% of the clones from the maltose-amylopectin-BF (see Table S3). Furthermore, bacterial strains with the same identity as these OTUs were isolated from both the maltose-BF and the maltose-amylopectin-BF (see Table S4 in the supplemental material). Apparently, bacteria affiliated to Cellvibrio spp. played an important role in the utilization of maltose and of amylopectin in the presence of maltose. In addition, two OTUs with 98 to 99% similarity to Flavobacterium spp. (class Flavobacteriia) contained 12% of the maltose-amylopectin-BF clones versus 3% of the maltose-BF clones (see Table S3), indicating that these bacteria probably also utilized amylopectin during simultaneous maltose and amylopectin addition. Members of the class Sphingobacteriia in the maltose-BF (19% of the clones) belonged primarily to two OTUs with either 97% or 98% similarity to Haliscomenobacter hydrossis, which were shared with the laminarin-BF (see Table S3). Of the nine biofilm types, the maltose-BF contained the highest percentage of Cytophagia representatives (14% of the clones), which were affiliated mainly to Emticicia spp. and were distributed over two unique OTUs and one OTU shared with the maltose-amylopectin-BF (Table 2; see also Table S3 in the supplemental material).
The clone library of the laminarin-BF contained the highest percentage of Sphingobacteriia members (29% of the clones), all of which belonged to the family Saprospiraceae (Table 2; see also Table S3 in the supplemental material). Besides the two aforementioned Haliscomenobacter hydrossis OTUs shared by the laminarin-BF (19% of the clones) and the maltose-BF (16% of the clones), three unique OTUs and another shared OTU were obtained (see Table S3). This third shared Saprospiraceae OTU comprised clones from biofilms formed with either laminarin (7%), caseinate (6%), gelatin (2%), maltose (1%), or amylopectin (1%) (see Table S3).
Alphaproteobacteria were relatively abundant in the clone libraries of the caseinate-BF and gelatin-BF (Table 2), but comparison with the clone libraries generated for the seven other biofilms in this study suggests that only some Sphingomonas or Sphingopyxis spp. and Parvularcula-related bacteria were specifically involved in caseinate and gelatin utilization (see Table S3 in the supplemental material). The family Xanthomonadaceae (class Gammaproteobacteria) occurred exclusively in the clone libraries of the caseinate-BF (13% of clones) and gelatin-BF (5% of clones) and was represented mainly by Lysobacter spp. (see Table S3 [singleton OTUs not shown]). The caseinate-BF also yielded four unique Flavobacterium OTUs (5% of clones) and one Flavobacterium OTU shared with the gelatin-BF, from which another Flavobacteriia-related OTU with 98% similarity to a Fluviicola sp. was derived (5% of clones) (see Table S3). Unique OTUs related to Cytophagia or Sphingobacteriia were distinguished in both the caseinate-BF (Pontibacter, Cytophaga, Chitinophaga, and Haliscomenobacter spp.) and the gelatin-BF (Haliscomenobacter spp.) (Table 2; see also Table S3 in the supplemental material).
Overall, the results of our study demonstrate that specific attached bacteria were involved in utilizing the added carbohydrates (e.g., members of the families Pseudomonadaceae, Saprospiraceae, Flavobacteriaceae, and Cytophagaceae) or proteins (e.g., members of the families Xanthomonadaceae, Saprospiraceae, Flavobacteriaceae, Sphingomonadaceae, Cytophagaceae, Cryomorphaceae, and Chitinophagaceae) (see Table S3 in the supplemental material).
DISCUSSION
Kinetics of attached growth under turbulent flow conditions.
The total bacterial counts in our biofilm samples are comparable to previously reported numbers of attached bacteria in drinking-water-related biofilms and thus confirm the suitability of the biofilm monitor as a model drinking water distribution system (41–43) (Table 1 [not all counts shown]). The data presented therefore prove that attached heterotrophic bacteria can utilize biopolymers at μg C liter−1 levels in flowing drinking water. Biofilm formation with individual biopolymers is, however, relatively slow compared with biofilm formation with LMW compounds, and specific LMW compounds may have to be present to accelerate biopolymer utilization, e.g., maltose for amylopectin utilization (Fig. 1). Additionally, the rates of exponential biofilm growth observed with acetate, maltose, laminarin, caseinate, or gelatin at 10 μg C liter−1 were approximately 2 to 10 times lower than the growth rates of bacterial strains with these compounds at 10 μg C liter−1 in batch tests (23, 24, 44, 45). These findings are consistent with previously observed differences in growth kinetics between biofouling and planktonic growth with acetate at μg C liter−1 levels (46). The average ATP levels cell−1 of the attached bacteria in the blank-BF (0.10 ± 0.03 fg cell−1) and the biofilms formed at 10 μg added C liter−1 (≥0.20 fg cell−1) (Table 1) exceeded the average ATP levels cell−1 reported previously for suspended bacteria in drinking water with no additions (0.017 ± 0.014 and 0.018 ± 0.095 fg cell−1) and with added carbohydrate or protein at 10 μg C liter−1 (0.061 ± 0.01 fg cell−1) (23, 36). Attached bacteria thus appear to be larger than suspended bacteria. In contrast to the relatively low growth rates observed in our study, the larger cell size suggests more-favorable growth conditions (47). Attachment may favor a larger cell size, and/or the growth rates observed may have been lower than the actual growth rates because of either (i) cell detachment under turbulent flow conditions or (ii) protozoan grazing on attached bacteria (48, 49). Furthermore, the lower growth rates with the added biopolymers than with acetate or maltose could indicate that adsorption of the biopolymers to the bacterial or biofilm surface and their subsequent extracellular hydrolysis before bacterial uptake were essential for biofilm growth with these high-molecular-weight (HMW) compounds and thus were potentially rate-determining (3, 18, 32, 50, 51).
Characteristics of biofilm formation with polysaccharides.
The 104-kDa to 5 × 105-kDa, heavily branched amylopectin molecule is a more complex homoglucan to degrade than the 5- to 6-kDa, mainly linear laminarin molecule (52, 53). These structural differences could explain why amylopectin promoted biofilm formation only in the presence of maltose, whereas laminarin promoted biofilm formation when added individually. Certain Gram-negative bacteria, such as Cellvibrio and Flavobacterium spp., can degrade polysaccharides, including starch, using free or cell-attached extracellular enzymes that contain noncatalytic carbohydrate-binding modules (CBM) (40, 54, 55). Extracellular enzymatic activities in aquatic biofilms are often cell associated, presumably to allow more-efficient substrate utilization (56, 57). It seems plausible, particularly considering the ultraoligotrophic and continuous turbulent flow conditions in our study, that the predominating Cellvibrio-like bacteria in the maltose-amylopectin-BFs employed mainly cell-attached, CBM-containing enzymes to bind amylopectin in a cell-specific manner and to hydrolyze it. The maltose-utilizing Cellvibrio-like bacteria, however, did not utilize amylopectin until the maltose-limited growth approached the steady-state growth phase at a µmin of 0.15 day−1 (i.e., BFR/BFC on day 11) (Fig. 1; Table 1). Hence, a minimum specific growth rate seems to be critical for inducing the synthesis of the extracellular amylopectin-binding and -degrading enzymes. This phenomenon resembles carbon catabolite repression, where the utilization of readily assimilable maltose (the preferred substrate) prevented the binding and utilization of amylopectin (the secondary substrate) until a minimum μ was attained with maltose, although maltose was probably still utilized during amylopectin utilization, because it is a typical amylopectin hydrolysis product (58–60). The finding that amylopectin was not utilized when the specific growth rates with simultaneously added acetate were at their minimum (0.02 to 0.06 day−1 [Table 1]) implies that a maltodextrin (e.g., maltose) or a certain level of maltodextrin-utilizing bacteria was required for CBM-containing enzyme induction. Furthermore, nonspecific adsorption of amylopectin to the acetate-amylopectin-BF or the amylopectin-BF either did not occur or did not induce enzyme synthesis. Hence, the accelerated biofilm growth rate with amylopectin in the presence of maltose may be attributed to highly efficient amylopectin binding to, and hydrolysis by, inductive CBM-containing enzymes.
The discontinuity in biofilm formation with laminarin and the large difference in plating efficiency between the two linear growth phases (viz., >90% and <10%) correspond to a significant shift in the bacterial community. In the first linear growth phase, bacterial strains BF-BP5 and BF-BP6, with ≥98% similarity to Flavobacterium spp., were repeatedly isolated and accounted for about 70% of the HPC based on colony morphology (see Table S4 in the supplemental material [HPC data not shown]). No clone library was generated in this phase, but the fact that Flavobacterium spp. represented >60% of the TDC demonstrates their dominance in laminarin utilization and is in line with the previously observed proficiency of Flavobacterium spp. in laminarin utilization at μg C liter−1 levels in batch tests (23, 61). In the second linear growth phase with laminarin, however, both the HPC and the Flavobacterium colony count decreased by a factor of 4, whereas the TDC increased by the same factor (data not shown). Moreover, according to the clone library of the 82-day-old laminarin-BF, Flavobacterium spp. were scarce (1% of clones) in the second linear growth phase, whereas Haliscomenobacter hydrossis and some other Saprospiraceae representatives prevailed (29% of clones). The HPC method used in this study clearly did not meet the specific cultivation conditions of the Haliscomenobacter strains (62, 63). The H. hydrossis genome encodes genes for degrading polysaccharides, but hitherto only starch utilization has been demonstrated experimentally at gram-per-liter levels (62, 64). Our study seems to be the first to report the growth of Haliscomenobacter spp. at microgram-per-liter levels and their involvement in laminarin utilization under turbulent flow conditions.
As the biofilm concentration increased at a constant BFR in the first linear growth phase with laminarin, the specific growth rate (μ, calculated as BFR/BFC) decreased, until eventually a value of 0.058 ± 0.005 day−1 was reached in the first steady-state growth phase (Table 1). Subsequently, the specific growth rate decreased further, to 0.030 ± 0.001 day−1, in the second linear growth phase. The succession of bacterial populations in the biofilms formed with laminarin appears to coincide with the decrease in the specific growth rate during biofilm formation, i.e., Haliscomenobacter spp. began to predominate at a specific growth rate that was minimum for steady-state growth of the Flavobacterium spp. that prevailed in the first linear growth phase. In terms of growth kinetics, the attached Haliscomenobacter spp. most likely had higher affinities for laminarin as a substrate than the Flavobacterium spp. and were therefore able to grow more efficiently with laminarin at lower growth rates. In addition, if Haliscomenobacter spp. were already present in the first linear growth phase with laminarin, their maximum growth rates must have been lower than those of the predominating Flavobacterium spp., which would be consistent with previous reports on the relatively slow growth of H. hydrossis (65, 66). Assessment of the growth kinetic parameters of attached, oligotrophic Flavobacterium and Haliscomenobacter spp. for laminarin was beyond the scope of our current study.
Characteristics of biofilm formation with proteins.
Intracellular proteolysis occurs continuously in all bacteria, and various broad-spectrum extracellular proteases capable of protein hydrolysis are generally produced constitutively by many bacteria (2, 67–69). Hence, extracellular protein degradation and incorporation of the degradation products into biomass seem relatively common among bacteria (69, 70), which could explain why (i) the 40- to 50-kDa gelatin molecules, as well as the 19- to 25-kDa casein molecules, promoted biofilm formation in the absence of a simultaneously added LMW substrate; (ii) a higher diversity of attached bacteria was involved in protein utilization (9 bacterial classes, 14 genera) than in polysaccharide utilization (4 bacterial classes, 4 genera) (see Tables S3 and S4 in the supplemental material); and (iii) most of the bacterial genera detected in the gelatin-BF or the caseinate-BF have been shown previously to include protein-degrading aquatic isolates (23, 62, 71–77).
Like extracellular polysaccharides, extracellular proteins in biofilms are primarily bound to and hydrolyzed by cell-attached enzymes (2, 56, 78). However, protein is degraded by a mixture of cell-attached proteases, and degradation products (i.e., peptides) may be repeatedly released and reattached for further hydrolysis until small enough (<700 Da) to enter the cell (12, 78, 79). Whether the same process occurred under the ultraoligotrophic conditions in our study is unknown, but the possibility that released intermediate peptides supported the growth of attached bacteria incapable of protein degradation or were further degraded to growth-promoting compounds of <700 Da by free extracellular enzymes retained in the biofilm matrix cannot be excluded (30).
The T-RFLP profile dissimilarities of 37 and 22% between the gelatin-BF of 88 days and the gelatin-BFs of 44 and 61 days, respectively, suggest that the discontinuity in biofilm formation with gelatin resulted from changes in the biofilm community composition as well. Also in this case, a relation seems to exist between the successional changes in the biofilm community and the decreasing specific growth rate, which was approximately 0.057, 0.036, and 0.017 day−1 in the first, second, and third steady-state growth phases, respectively (Table 1). By the end of the first or second steady-state growth phase, the bacterial population(s) that had predominated in the preceding linear growth phase was outgrown by other bacterial populations that could grow at lower specific growth rates and predominated in the subsequent linear growth phase (Table 1). Some of these changes were deduced from the predominant bacterial isolates and the plating efficiencies obtained during biofilm formation with gelatin (60 to 20%). Isolate BF-BP2, with 99% similarity to Flavobacterium sp. clone L-1, accounted for about 25% of the TDC during the first fortnight of biofilm formation, but in the following 3 weeks, isolates BF-BP7, BF-BP8, and BF-BP9, with 99% similarity to Brevundimonas, Caulobacter, and Undibacterium spp., respectively, became more dominant, and each represented 10 to 15% of the TDC (see Table S4 in the supplemental material [counts not shown]). In the last 6 weeks, the main isolates BF-BP12 and BF-BP14, with ≥98% similarity to Lysobacter spp., together constituted about 4% of the TDC, demonstrating that only a fraction of the predominant bacteria was isolated in this period. The clone library of the 88-day-old gelatin-BF confirmed the low levels (<3% of clones) of the five genera mentioned above in the final stage of biofilm formation. The T-RFLP profile dissimilarity of 30% between the caseinate-BFs of 32 and 82 days also indicates differences in biofilm community composition, which were partly revealed by shifts in the prevalences of bacterial strains isolated during caseinate addition. Strains BF-BP3, BF-BP4, BF-BP7, BF-BP13, and BF-BP14, with >98% similarity to Flavobacterium, Brevundimonas, Xanthomonas, or Lysobacter spp., each represented 4 to 8% of the TDC in the first 6 weeks and 0 to 2% in the last 6 weeks, during which strain BF-BP2, with 99% similarity to Flavobacterium spp., was also observed at approximately 2% of the TDC (see Table S4). Similarly, 0 to 2% of the clones in the clone library of the 82-day-old caseinate-BF were identical to these six strains (see Table S3). Generation of multiple clone libraries would have elucidated the succession of bacterial populations in the biofilms formed with gelatin and caseinate but was outside the scope of this study.
Biopolymer utilization by biofilms in drinking water distribution systems.
Drinking water in distribution systems and oligotrophic freshwater in natural environments contain complex mixtures of organic substrates for heterotrophic bacteria (61, 80, 81). LMW compounds are generally more readily utilized than HMW compounds, and aquatic bacterial communities adapted to LMW substrate utilization usually differ significantly from those adapted to HMW substrate utilization (13, 82). Indeed, the BFR at 10 μg C liter−1 in our study was 3 to 12 times higher with acetate or maltose added individually than with laminarin, gelatin, or caseinate added individually; amylopectin added individually was not utilized at all (Table 1). Furthermore, the biofilms grown with acetate, carbohydrate, or protein clearly differed in bacterial community composition (Fig. 2 and 4). The limited number of bacterial genera involved in utilizing maltose, amylopectin, and laminarin (e.g., a Cellvibrio-like genus, Haliscomenobacter, Flavobacterium) and the amylopectin utilization prerequisites (e.g., enzyme synthesis induction, minimum growth rate with inducing substrate, binding to enzymes) indicate that bacterial utilization of complex homopolysaccharides requires specialization and may not commonly occur in drinking water distribution systems. This hypothesis is supported by the fact that, to our knowledge, Cellvibrio-like bacteria and Haliscomenobacter spp. isolated from drinking water distribution systems have not been deposited in GenBank previously. Instead of homopolysaccharides, complex heteropolysaccharides included in extracellular polymeric substances (EPS) originating from freshwater phytoplankton and from biofilms in water treatment processes may be present in distributed drinking water (1, 83, 84). In the absence of enzyme-inducing oligosaccharides, utilization of polysaccharide EPS by relatively young (i.e., <1 year) biofilms in drinking water distribution systems may be severely limited. Studies on polysaccharide degradation under oligotrophic conditions in marine environments revealed rapid degradation of various homo- and heteropolysaccharides by bacterial communities at milligram-per-liter levels in (e.g., anoxic) sediments and aggregates, in contrast to slower or no degradation in seawater (33, 85–87). It is not clear to what extent and under what conditions sedimentary bacterial communities and mature biofilms in drinking water distribution systems could also be capable of utilizing complex polysaccharides. Nevertheless, the protein-adapted biofilm communities in our study were more diverse than the polysaccharide-adapted biofilm communities and had a relatively high content of heterotrophic bacteria typically found in drinking water, suggesting that proteins in distributed drinking water are more commonly utilized than polysaccharides and are therefore more likely to promote biofilm formation in drinking water distribution systems.
Taking all these data together, our study demonstrates that not only LMW compounds but also biopolymers can promote biofilm formation in drinking water distribution systems. Depending on their concentrations, biopolymers might therefore negatively impact the biological stability of distributed drinking water. The original AOC test, which is used to determine biological stability by quantifying assimilable organic carbon (i.e., LMW compounds) in drinking water, has recently been extended with Flavobacterium johnsoniae strain A3, which specializes in the utilization of carbohydrates and proteins at microgram-per-liter levels (23, 45, 61, 88). Additional research will focus on applying this extended AOC test to quantify changes in biopolymer concentrations during drinking-water production and distribution and to assess the effects of these changes on the biological stability of distributed drinking water.
Supplementary Material
ACKNOWLEDGMENTS
This study was supported by SenterNovem and the Joint Research Program of the Water Supply Companies in The Netherlands and was conducted within the framework of the NOM-project “Breakthrough in the biological stability of drinking water” (reference IS 054040).
Thanks are due to Lonneke Hensen, Anita van der Veen-Lugtenberg, Anke Brouwer-Hanzens, Marijan Uytewaal-Aarts, and Meindert de Graaf for skillful technical assistance and to Hauke Smidt and Marco Dignum for valuable discussions.
Footnotes
Published ahead of print 31 January 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.04105-13.
REFERENCES
- 1.Myklestad S. 2000. Dissolved organic carbon from phytoplankton, p 111–148 In Wangersky P. (ed), The handbook of environmental chemistry, vol 5, part D Springer-Verlag, Berlin, Germany [Google Scholar]
- 2.Janssen JMA. 1979. Ecological and kinetic aspects of amylolysis and proteolysis in activated sludge. Ph.D. thesis Landbouwhogeschool, Wageningen, The Netherlands [Google Scholar]
- 3.Salyers AA, Reeves A, D'Elia J. 1996. Solving the problem of how to eat something as big as yourself: diverse bacterial strategies for degrading polysaccharides. J. Ind. Microbiol. Biotechnol. 17:470–476 [Google Scholar]
- 4.Nikaido H. 2003. Molecular basis of bacterial outer membrane permeability revisited. Microbiol. Mol. Biol. Rev. 67:593–656. 10.1128/MMBR.67.4.593-656.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Eiler A, Bertilsson S. 2004. Composition of freshwater bacterial communities associated with cyanobacterial blooms in four Swedish lakes. Environ. Microbiol. 6:1228–1243. 10.1111/j.1462-2920.2004.00657.x [DOI] [PubMed] [Google Scholar]
- 6.Bruckner CG, Bahulikar R, Rahalkar M, Schick B, Kroth PG. 2008. Bacteria associated with benthic diatoms from Lake Constance: phylogeny and influences on diatom growth and secretion of extracellular polymeric substances. Appl. Environ. Microbiol. 74:7740–7749. 10.1128/AEM.01399-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Riemann L, Winding A. 2001. Community dynamics of free-living and particle-associated bacterial assemblages during a freshwater phytoplankton bloom. Microb. Ecol. 42:274–285. 10.1007/s00248-001-0018-8 [DOI] [PubMed] [Google Scholar]
- 8.Pinhassi J, Sala MM, Havskum H, Peters F, Guadayol Ò, Malits A, Marrasé C. 2004. Changes in bacterioplankton composition under different phytoplankton regimens. Appl. Environ. Microbiol. 70:6753–6766. 10.1128/AEM.70.11.6753-6766.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Arnosti C. 2008. Functional differences between Arctic seawater and sedimentary microbial communities: contrasts in microbial hydrolysis of complex substrates. FEMS Microbiol. Ecol. 66:343–351. 10.1111/j.1574-6941.2008.00587.x [DOI] [PubMed] [Google Scholar]
- 10.Taylor GT. 1995. Microbial degradation of sorbed and dissolved protein in seawater. Limnol. Oceanogr. 40:875–885 [Google Scholar]
- 11.Arnosti C, Ziervogel K, Ocampo L, Ghobrial S. 2009. Enzyme activities in the water column and in shallow permeable sediments from the northeastern Gulf of Mexico. Estuar. Coast. Shelf Sci. 84:202–208. 10.1016/j.ecss.2009.06.018 [DOI] [Google Scholar]
- 12.Nunn BL, Norbeck A, Keil RG. 2003. Hydrolysis patterns and the production of peptide intermediates during protein degradation in marine sediments. Mar. Chem. 83:59–73. 10.1016/S0304-4203(03)00096-3 [DOI] [Google Scholar]
- 13.Elifantz H, Malmstrom RR, Cottrell MT, Kirchman DL. 2005. Assimilation of polysaccharides and glucose by major bacterial groups in the Delaware estuary. Appl. Environ. Microbiol. 71:7799–7805. 10.1128/AEM.71.12.7799-7805.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Keith SC, Arnosti C. 2001. Extracellular enzyme activity in a river-bay-shelf transect: variations in polysaccharide hydrolysis rates with substrate and size class. Aquat. Microb. Ecol. 24:243–253. 10.3354/ame024243 [DOI] [Google Scholar]
- 15.Hollibaugh JT, Azam F. 1983. Microbial degradation of dissolved proteins in seawater. Limnol. Oceanogr. 28:1104–1116. 10.4319/lo.1983.28.6.1104 [DOI] [Google Scholar]
- 16.Keil RG, Kirchman DL. 1999. Utilization of dissolved protein and amino acids in the northern Sargasso Sea. Aquat. Microb. Ecol. 18:293–300. 10.3354/ame018293 [DOI] [Google Scholar]
- 17.Kirchman DL, Meon B, Ducklow HW, Carlson CA, Hansell DA, Steward GF. 2001. Glucose fluxes and concentrations of dissolved combined neutral sugars (polysaccharides) in the Ross Sea and Polar Front Zone, Antarctica. Deep Sea Res. II 48:4179–4197. 10.1016/S0967-0645(01)00085-6 [DOI] [Google Scholar]
- 18.BeMiller JN, Whistler RL. 1996. Carbohydrates, p 157–223 In Fennema OR. (ed), Food chemistry, 3rd ed. Marcel Dekker Inc, New York, NY [Google Scholar]
- 19.Zeder M, Peter S, Shabarova T, Pernthaler J. 2009. A small population of planktonic Flavobacteria with disproportionally high growth during the spring phytoplankton bloom in a prealpine lake. Environ. Microbiol. 11:2676–2686. 10.1111/j.1462-2920.2009.01994.x [DOI] [PubMed] [Google Scholar]
- 20.Eiler A, Bertilsson S. 2007. Flavobacteria blooms in four eutrophic lakes: linking population dynamics of freshwater bacterioplankton to resource availability. Appl. Environ. Microbiol. 73:3511–3518. 10.1128/AEM.02534-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kirchman DL. 2002. The ecology of Cytophaga-Flavobacteria in aquatic environments. FEMS Microbiol. Ecol. 39:91–100. 10.1111/j.1574-6941.2002.tb00910.x [DOI] [PubMed] [Google Scholar]
- 22.Glöckner FO, Fuchs BM, Amann R. 1999. Bacterioplankton compositions of lakes and oceans: a first comparison based on fluorescence in situ hybridization. Appl. Environ. Microbiol. 65:3721–3726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sack ELW, van der Wielen PWJJ, van der Kooij D. 2011. Flavobacterium johnsoniae as a model organism for characterizing biopolymer utilization in oligotrophic freshwater environments. Appl. Environ. Microbiol. 77:6931–6938. 10.1128/AEM.00372-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.van der Kooij D, Hijnen WAM. 1985. Determination of the concentration of maltose- and starch-like compounds in drinking water by growth measurements with a well-defined strain of a Flavobacterium species. Appl. Environ. Microbiol. 49:765–771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.van der Kooij D, Hijnen WAM. 1981. Utilization of low concentrations of starch by a Flavobacterium species isolated from tap water. Appl. Environ. Microbiol. 41:216–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Manz W, Szewzyk U, Lawrence JR. 2002. Biofilms in natural and drinking water systems, p 560–575 In Bitton G. (ed), Encyclopedia of environmental microbiology. Wiley, New York, NY [Google Scholar]
- 27.van der Wielen PWJJ, van der Kooij D. 2013. Nontuberculous mycobacteria, fungi, and opportunistic pathogens in unchlorinated drinking water in the Netherlands. Appl. Environ. Microbiol. 79:825–834. 10.1128/AEM.02748-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.van der Kooij D, Veenendaal HR, Baars-Lorist C, Van der Klift DW, Drost YC. 1995. Biofilm formation on surfaces of glass and Teflon exposed to treated water. Water Res. 29:1655–1662. 10.1016/0043-1354(94)00333-3 [DOI] [Google Scholar]
- 29.van der Kooij D, Vrouwenvelder HS, Veenendaal HR. 1995. Kinetic aspects of biofilm formation on surfaces exposed to drinking water. Water Sci. Technol. 32:61–65. 10.1016/0273-1223(96)00008-X [DOI] [Google Scholar]
- 30.Flemming HC, Wingender J. 2010. The biofilm matrix. Nat. Rev. Microbiol. 8:623–633. 10.1038/nrmicro2415 [DOI] [PubMed] [Google Scholar]
- 31.Cadoret A, Conrad A, Block J-C. 2002. Availability of low and high molecular weight substrates to extracellular enzymes in whole and dispersed activated sludges. Enzyme Microb. Technol. 31:179–186. 10.1016/S0141-0229(02)00097-2 [DOI] [Google Scholar]
- 32.Karahan O, Martins A, Orhon D, van Loosdrecht MCM. 2006. Experimental evaluation of starch utilization mechanism by activated sludge. Biotechnol. Bioeng. 93:964–970. 10.1002/bit.20795 [DOI] [PubMed] [Google Scholar]
- 33.Ziervogel K, Arnosti C. 2008. Polysaccharide hydrolysis in aggregates and free enzyme activity in aggregate-free seawater from the north-eastern Gulf of Mexico. Environ. Microbiol. 10:289–299. 10.1111/j.1462-2920.2007.01451.x [DOI] [PubMed] [Google Scholar]
- 34.Hijnen WAM, Cornelissen ER, Van der Kooij D. 2011. Threshold concentrations of biomass and iron for pressure drop increase in spiral-wound membrane elements. Water Res. 45:1607–1616. 10.1016/j.watres.2010.11.047 [DOI] [PubMed] [Google Scholar]
- 35.van der Kooij D, Veenendaal HR, Scheffer WJH. 2005. Biofilm formation and multiplication of Legionella in a model warm water system with pipes of copper, stainless steel and cross-linked polyethylene. Water Res. 39:2789–2798. 10.1016/j.watres.2005.04.075 [DOI] [PubMed] [Google Scholar]
- 36.van der Wielen PWJJ, van der Kooij D. 2010. Effect of water composition, distance and season on the adenosine triphosphate concentration in unchlorinated drinking water in the Netherlands. Water Res. 44:4860–4867. 10.1016/j.watres.2010.07.016 [DOI] [PubMed] [Google Scholar]
- 37.Reasoner DJ, Geldreich EE. 1985. A new medium for the enumeration and subculture of bacteria from potable water. Appl. Environ. Microbiol. 49:1–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hobbie JE, Daley RJ, Jasper S. 1977. Use of Nuclepore filters for counting bacteria by fluorescence microscopy. Appl. Environ. Microbiol. 33:1225–1228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Monod J. 1949. The growth of bacterial cultures. Annu. Rev. Microbiol. 9:371–394 [Google Scholar]
- 40.DeBoy RT, Mongodin EF, Fouts DE, Tailford LE, Khouri H, Emerson JB, Mohamoud Y, Watkins K, Henrissat B. 2008. Insights into plant cell wall degradation from the genome sequence of the soil bacterium Cellvibrio japonicus. J. Bacteriol. 190:5455–5463. 10.1128/JB.01701-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Boe-Hansen R, Martiny AC, Arvin E, Albrechtsen HJ. 2003. Monitoring biofilm formation and activity in drinking water distribution networks under oligotrophic conditions. Water Sci. Technol. 47(5):91–97 [PubMed] [Google Scholar]
- 42.Block JC, Haudidier K, Paquin JL, Miazga J, Levi Y. 1993. Biofilm accumulation in drinking water distribution systems. Biofouling 6:333–343. 10.1080/08927019309386235 [DOI] [Google Scholar]
- 43.Pedersen K. 1990. Biofilm development on stainless steel and PVC surfaces in drinking water. Water Res. 24:239–243. 10.1016/0043-1354(90)90109-J [DOI] [Google Scholar]
- 44.van der Kooij D, Visser A, Oranje JP. 1982. Multiplication of fluorescent pseudomonads at low substrate concentrations in tap water. Antonie Van Leeuwenhoek 48:229–243. 10.1007/BF00400383 [DOI] [PubMed] [Google Scholar]
- 45.van der Kooij D, Hijnen WAM. 1984. Substrate utilization by an oxalate-consuming Spirillum species in relation to its growth in ozonated water. Appl. Environ. Microbiol. 47:551–559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hijnen WA, Biraud D, Cornelissen ER, van der Kooij D. 2009. Threshold concentration of easily assimilable organic carton in feedwater for biofouling of spiral-wound membranes. Environ. Sci. Technol. 43:4890–4895. 10.1021/es900037x [DOI] [PubMed] [Google Scholar]
- 47.Koch AL. 1996. What size should a bacterium be? A question of scale. Annu. Rev. Microbiol. 50:317–348. 10.1146/annurev.micro.50.1.317 [DOI] [PubMed] [Google Scholar]
- 48.Parry JD. 2004. Protozoan grazing of freshwater biofilms. Adv. Appl. Microbiol. 54:167–196. 10.1016/S0065-2164(04)54007-8 [DOI] [PubMed] [Google Scholar]
- 49.Rickard AH, McBain AJ, Stead AT, Gilbert P. 2004. Shear rate moderates biofilm community in freshwater biofilms. Appl. Environ. Microbiol. 70:7426–7435. 10.1128/AEM.70.12.7426-7435.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mosquera-Corral A, Montras A, Heijnen JJ, van Loosdrecht MC. 2003. Degradation of polymers in a biofilm airlift suspension reactor. Water Res. 37:485–492. 10.1016/S0043-1354(02)00309-3 [DOI] [PubMed] [Google Scholar]
- 51.de Kreuk MK, Kishida N, Tsuneda S, van Loosdrecht MC. 2010. Behavior of polymeric substrates in an aerobic granular sludge system. Water Res. 44:5929–5938. 10.1016/j.watres.2010.07.033 [DOI] [PubMed] [Google Scholar]
- 52.BeMiller JN, Whistler RL. 1996. Carbohydrates, p 157–223 In Fennema OR. (ed), Food chemistry, 3rd ed. Marcel Dekker, Inc, New York, NY [Google Scholar]
- 53.Painter TJ. 1983. Structural evolution of glycans in algae. Pure Appl. Chem. 55:677–694 [Google Scholar]
- 54.Boraston AB, Bolam DN, Gilbert HJ, Davies GJ. 2004. Carbohydrate-binding modules: fine-tuning polysaccharide recognition. Biochem. J. 382:769–781. 10.1042/BJ20040892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.McBride MJ, Xie G, Martens EC, Lapidus A, Henrissat B, Rhodes RG, Goltsman E, Wang W, Xu J, Hunnicutt DW, Staroscik AM, Hoover TR, Cheng YQ, Stein JL. 2009. Novel features of the polysaccharide-digesting gliding bacterium Flavobacterium johnsoniae as revealed by genome sequence analysis. Appl. Environ. Microbiol. 75:6864–6875. 10.1128/AEM.01495-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wingender J, Jaeger K-E. 2002. Extracellular enzymes in biofilms, p 1207–1223 In Bitton G. (ed), Encyclopedia of environmental microbiology. Wiley, New York, NY [Google Scholar]
- 57.Chróst RJ, Rai H. 1993. Ectoenzyme activity and bacterial secondary production in nutrient-impoverished and nutrient-enriched freshwater mesocosms. Microb. Ecol. 25:131–150 [DOI] [PubMed] [Google Scholar]
- 58.Görke B, Stülke J. 2008. Carbon catabolite repression in bacteria: many ways to make the most out of nutrients. Nat. Rev. Microbiol. 6:613–624. 10.1038/nrmicro1932 [DOI] [PubMed] [Google Scholar]
- 59.Arnosti C. 2003. Microbial extracellular enzymes and their role in dissolved organic matter cycling, p 315–342 In Findley SEG, Sinsabaugh RL. (ed), Aquatic ecosystems: interactivity of dissolved organic matter. Academic Press, San Diego, CA [Google Scholar]
- 60.Chróst RJ. 1991. Environmental control of the synthesis and activity of aquatic microbial ectoenzymes, p 29–59 In Chróst RJ. (ed), Microbial enzymes in aquatic environments. Springer-Verlag, New York, NY [Google Scholar]
- 61.Sack ELW, van der Wielen PWJJ, van der Kooij D. 2010. Utilization of oligo- and polysaccharides at microgram-per-litre levels in freshwater by Flavobacterium johnsoniae. J. Appl. Microbiol. 108:1430–1440. 10.1111/j.1365-2672.2009.04546.x [DOI] [PubMed] [Google Scholar]
- 62.van Veen WL, van der Kooij D, Geuze EC, van der Vlies AW. 1973. Investigations on the sheathed bacterium Haliscomenobacter hydrossis gen.n., sp.n., isolated from activated sludge. Antonie Van Leeuwenhoek 39:207–216. 10.1007/BF02578853 [DOI] [PubMed] [Google Scholar]
- 63.Kämpfer P. 1997. Detection and cultivation of filamentous bacteria from activated sludge. FEMS Microbiol. Ecol. 23:169–181. 10.1111/j.1574-6941.1997.tb00400.x [DOI] [Google Scholar]
- 64.Daligault H, Lapidus A, Zeytun A, Nolan M, Lucas S, Del Rio TG, Tice H, Cheng JF, Tapia R, Han C, Goodwin L, Pitluck S, Liolios K, Pagani I, Ivanova N, Huntemann M, Mavromatis K, Mikhailova N, Pati A, Chen A, Palaniappan K, Land M, Hauser L, Brambilla EM, Rohde M, Verbarg S, Goker M, Bristow J, Eisen JA, Markowitz V, Hugenholtz P, Kyrpides NC, Klenk HP, Woyke T. 2011. Complete genome sequence of Haliscomenobacter hydrossis type strain (O). Stand. Genomic Sci. 4:352–360. 10.4056/sigs.1964579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Krul JM. 1977. Experiments with Haliscomenobacter hydrossis in continuous culture without and with Zoogloea ramigera. Water Res. 11:197–204. 10.1016/0043-1354(77)90126-9 [DOI] [Google Scholar]
- 66.Van Veen WL, Krul JM, Bulder CJEA. 1982. Some growth parameters of Haliscomenobacter hydrossis (syn. Streptothrix hyalina), a bacterium occurring in bulking activated sludge. Water Res. 16:531–534. 10.1016/0043-1354(82)90072-0 [DOI] [Google Scholar]
- 67.Gottesman S. 2003. Proteolysis in bacterial regulatory circuits. Annu. Rev. Cell Dev. Biol. 19:565–587. 10.1146/annurev.cellbio.19.110701.153228 [DOI] [PubMed] [Google Scholar]
- 68.Gupta R, Beg QK, Lorenz P. 2002. Bacterial alkaline proteases: molecular approaches and industrial applications. Appl. Microbiol. Biotechnol. 59:15–32. 10.1007/s00253-002-0975-y [DOI] [PubMed] [Google Scholar]
- 69.Chandra R, Rustgi R. 1998. Biodegradable polymers. Prog. Polymer Sci. 23:1273–1335. 10.1016/S0079-6700(97)00039-7 [DOI] [Google Scholar]
- 70.Leis A, Flemming H-C. 2002. Activity and carbon transformations in biofilms, p 81–92 In Bitton G. (ed), Encyclopedia of environmental microbiology. Wiley, New York, NY [Google Scholar]
- 71.Iuga M, Awram P, Nomellini JF, Smit J. 2004. Comparison of S-layer secretion genes in freshwater caulobacters. Can. J. Microbiol. 50:751–766. 10.1139/w04-046 [DOI] [PubMed] [Google Scholar]
- 72.Wei DQ, Yu TT, Yao JC, Zhou EM, Song ZQ, Yin YR, Ming H, Tang SK, Li WJ. 2012. Lysobacter thermophilus sp. nov., isolated from a geothermal soil sample in Tengchong, south-west China. Antonie Van Leeuwenhoek 102:643–651. 10.1007/s10482-012-9761-8 [DOI] [PubMed] [Google Scholar]
- 73.Xu M, Wang Y, Dai J, Jiang F, Rahman E, Peng F, Fang C. 2012. Pontibacter populi sp. nov., isolated from the soil of a Euphrates poplar (Populus euphratica) forest. Int. J. Syst. Evol. Microbiol. 62:665–670. 10.1099/ijs.0.030122-0 [DOI] [PubMed] [Google Scholar]
- 74.Eder W, Wanner G, Ludwig W, Busse HJ, Ziemke-Kageler F, Lang E. 2010. Description of Undibacterium oligocarboniphilum sp. nov., isolated from purified water, and Undibacterium pigrum strain CCUG 49012 as the type strain of Undibacterium parvum sp. nov., and emended descriptions of the genus Undibacterium and the species Undibacterium pigrum. Int. J. Syst. Evol. Microbiol. 61:384–391. 10.1099/ijs.0.018648-0 [DOI] [PubMed] [Google Scholar]
- 75.Donofrio RS, Bestervelt LL, Saha R, Bagley ST. 2010. Selective enumeration strategies for Brevundimonas diminuta from drinking water. J. Ind. Microbiol. Biotechnol. 37:407–417. 10.1007/s10295-010-0689-6 [DOI] [PubMed] [Google Scholar]
- 76.Woyke T, Chertkov O, Lapidus A, Nolan M, Lucas S, Del Rio TG, Tice H, Cheng JF, Tapia R, Han C, Goodwin L, Pitluck S, Liolios K, Pagani I, Ivanova N, Huntemann M, Mavromatis K, Mikhailova N, Pati A, Chen A, Palaniappan K, Land M, Hauser L, Brambilla EM, Rohde M, Mwirichia R, Sikorski J, Tindall BJ, Goker M, Bristow J, Eisen JA, Markowitz V, Hugenholtz P, Klenk HP, Kyrpides NC. 2011. Complete genome sequence of the gliding freshwater bacterium Fluviicola taffensis type strain (RW262). Stand. Genomic Sci. 5:21–29. 10.4056/sigs.2124912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Balkwill DL, Fredrickson JK, Romine MF. 2006. Sphingomonas and related genera, p 605–629 In Dworkin M, Falkow S, Rosenberg E, Schleifer K-H, Stackebrandt E. (ed), The prokaryotes. Springer, New York, NY [Google Scholar]
- 78.Confer DR, Logan BE. 1998. Location of protein and polysaccharide hydrolytic activity in suspended and biofilm wastewater cultures. Water Res. 32:31–38. 10.1016/S0043-1354(97)00194-2 [DOI] [Google Scholar]
- 79.Confer DR, Logan BE. 1997. Molecular weight distribution of hydrolysis products during biodegradation of model macromolecules in suspended and biofilm cultures. I. Bovine serum albumin. Water Res. 31:2127–2136 [Google Scholar]
- 80.Croué J-P, Korshin GV, Benjamin MM. 1999. Characterization of natural organic matter in drinking water. American Water Works Association and AWWA Research Foundation, Denver, CO [Google Scholar]
- 81.Volk CJ, Volk CB, Kaplan LA. 1997. Chemical composition of biodegradable dissolved organic matter in streamwater. Limnol. Oceanogr. 42:39–44. 10.4319/lo.1997.42.1.0039 [DOI] [Google Scholar]
- 82.Kujawinski EB. 2011. The impact of microbial metabolism on marine dissolved organic matter. Annu. Rev. Mar. Sci. 3:567–599. 10.1146/annurev-marine-120308-081003 [DOI] [PubMed] [Google Scholar]
- 83.Zhang X, Bishop PL. 2003. Biodegradability of biofilm extracellular polymeric substances. Chemosphere 50:63–69. 10.1016/S0045-6535(02)00319-3 [DOI] [PubMed] [Google Scholar]
- 84.Lazarova V, Manem J. 1995. Biofilm characterization and activity analysis in water and wastewater treatment. Water Res. 29:2227–2245. 10.1016/0043-1354(95)00054-O [DOI] [Google Scholar]
- 85.Arnosti C. 2000. Substrate specificity in polysaccharide hydrolysis: contrasts between bottom water and sediments. Limnol. Oceanogr. 45:1112–1119. 10.4319/lo.2000.45.5.1112 [DOI] [Google Scholar]
- 86.Teske A, Durbin A, Ziervogel K, Cox C, Arnosti C. 2011. Microbial community composition and function in permanently cold seawater and sediments from an Arctic fjord of Svalbard. Appl. Environ. Microbiol. 77:2008–2018. 10.1128/AEM.01507-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Arnosti C, Repeta DJ, Blough NV. 1994. Rapid bacterial degradation of polysaccharides in anoxic marine systems. Geochim. Cosmochim. Acta 58:2639–2652. 10.1016/0016-7037(94)90134-1 [DOI] [Google Scholar]
- 88.van der Kooij D. 1992. Assimilable organic carbon as an indicator of bacterial regrowth. J. Am. Water Works Assoc. 84:57–65 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


