Abstract
Silver nanoparticles (AgNPs) are considered to be a potentially useful tool for controlling various pathogens. However, there are concerns about the release of AgNPs into environmental media, as they may generate adverse human health and ecological effects. In this study, we developed and evaluated a novel micrometer-sized magnetic hybrid colloid (MHC) decorated with variously sized AgNPs (AgNP-MHCs). After being applied for disinfection, these particles can be easily recovered from environmental media using their magnetic properties and remain effective for inactivating viral pathogens. We evaluated the efficacy of AgNP-MHCs for inactivating bacteriophage ϕX174, murine norovirus (MNV), and adenovirus serotype 2 (AdV2). These target viruses were exposed to AgNP-MHCs for 1, 3, and 6 h at 25°C and then analyzed by plaque assay and real-time TaqMan PCR. The AgNP-MHCs were exposed to a wide range of pH levels and to tap and surface water to assess their antiviral effects under different environmental conditions. Among the three types of AgNP-MHCs tested, Ag30-MHCs displayed the highest efficacy for inactivating the viruses. The ϕX174 and MNV were reduced by more than 2 log10 after exposure to 4.6 × 109 Ag30-MHCs/ml for 1 h. These results indicated that the AgNP-MHCs could be used to inactivate viral pathogens with minimum chance of potential release into environment.
INTRODUCTION
With recent advances in nanotechnology, nanoparticles have been receiving increased attention worldwide in the fields of biotechnology, medicine, and public health (1, 2). Owing to their high surface-to-volume ratio, nano-sized materials, typically ranging from 10 to 500 nm, have unique physicochemical properties compared with those of larger materials (1). The shape and size of nanomaterials can be controlled, and specific functional groups can be conjugated on their surfaces to enable interactions with certain proteins or intracellular uptake (3–5).
Silver nanoparticles (AgNPs) have been widely studied as an antimicrobial agent (6). Silver is used in the creation of fine cutlery, for ornamentation, and in therapeutic agents. Silver compounds such as silver sulfadiazine and certain salts have been used as wound care products and as treatments for infectious diseases due to their antimicrobial properties (6, 7). Recent studies have revealed that AgNPs are very effective for inactivating various types of bacteria and viruses (8–11). AgNPs and Ag+ ions released from AgNPs interact directly with phosphorus- or sulfur-containing biomolecules, including DNA, RNA, and proteins (12–14). They have also been shown to generate reactive oxygen species (ROS), causing membrane damage in microorganisms (15). The size, shape, and concentration of AgNPs are also important factors that affect their antimicrobial capabilities (8, 10, 13, 16, 17).
Previous studies have also highlighted several problems when AgNPs are used for controlling pathogens in a water environment. First, existing studies on the effectiveness of AgNPs for inactivating viral pathogens in water are limited. In addition, monodispersed AgNPs are typically subject to particle-particle aggregation because of their small size and large surface area, and these aggregates reduce the effectiveness of AgNPs against microbial pathogens (7). Finally, AgNPs have been shown to have various cytotoxic effects (5, 18–20), and the release of AgNPs into a water environment could result in human health and ecological problems.
Recently, we developed a novel micrometer-sized magnetic hybrid colloid (MHC) decorated with AgNPs of various sizes (21, 22). The MHC core can be used to recover the AgNP composites from the environment. We evaluated the antiviral efficacy of these silver nanoparticles on MHCs (AgNP-MHCs) using bacteriophage ϕX174, murine norovirus (MNV), and adenovirus under different environmental conditions.
MATERIALS AND METHODS
Synthesis of Ag07-MHC, Ag15-MHC, and Ag30-MHC.
All of the magnetic hybrid colloids decorated with AgNPs of various sizes (AgNP-MHCs) were synthesized by the Molecular Recognition Research Center of the Korea Institute of Science and Technology (KIST), Seoul, South Korea. Materials were used as purchased and as reported in our previous study (21). Ag07-MHC, Ag15-MHC, and Ag30-MHC were synthesized using the same procedure as reported in our previous studies (21, 22). An aminopropyl-functionalized Fe3O4-SiO2 core-shell magnetic hybrid colloid (AP-MHC) stock solution containing 3.7 × 1010 AP-MHCs/ml was prepared and used as required.
Briefly, silver nanoparticles of 7, 15, and 30 nm were grown on the outermost surface of a MHC through a seeding, coalescing, and growing strategy (21, 22). Five milliliters of AP-MHC was mixed with 25 ml of Au seed solution (22). The Au-seeded MHC was collected by magnetic decantation and dispersed in 5 ml of deionized water (DW). To grow 7-nm (or 15-nm) AgNPs on the surface of the MHC, a mixture of 40 ml (or 80 ml) of AgNO3 (0.01% [wt/vol] in water) and 0.004 ml (or 0.008 ml) of NH4OH (30% in water) were prepared with stirring. After 5 min, 0.03 ml of formaldehyde (37% in water) was added. The mixture was stirred with a mechanical stirrer for 30 min and allowed to sit for 1.5 h. The solid was purified by washing with DW three times using magnetic decantation and dispersed in 20 ml of DW, resulting in a solution containing 9.2 × 109 Ag07-MHC (or Ag15-MHC) particles/ml of solution.
To synthesize Ag30-MHC, the pH-adjusted (pH ∼4) AP-MHC solution (5 ml) was slowly added to 25 ml of the Ag seed solution (21). The Ag-seeded MHC was collected by magnetic decantation and dispersed in 5 ml of DW. This solution was added to a mixture of AgNO3 (0.02 g) and NH4OH (30%, 0.04 ml) in 200 ml of DW and stirred for 10 min with a mechanical stirrer in an ice bath. After 0.05 ml of formaldehyde was added, the solution was stirred for 30 min and left for 1.5 h without perturbation. The resulting Ag30-MHC was purified by washing with DW three times using magnetic decantation and dispersed in 20 ml of DW (9.2 × 109 particles/ml). The completed AgNP-MHC solutions were stored at room temperature (25°C) in the dark.
The AgNP-MHCs were characterized by transmission electron microscopy (TEM) using a CM30 transmission electron microscope (Philips Inc., USA) equipped with an energy-dispersive spectrometer and by scanning electron microscopy (SEM) using an XL30 environmental scanning electron microscope (ESEM) (FEI Co., USA). Images were captured at the Advanced Analysis Center of KIST (Seoul, South Korea), as in our previous studies (21).
Preparation of target virus stock.
Bacteriophages MS2 (ATCC 15597-B1) and ϕX174 (ATCC 13706-B1) were propagated using the single-agar-layer technique (23–25). Initially, both bacteriophages were cultured overnight at 37°C with Escherichia coli C3000 (ATCC 15597) as the host bacteria. The phages were then washed using phosphate-buffered saline (PBS) and purified from E. coli lysates as described previously with some modifications (26). Briefly, an equal volume of chloroform was added to the lysates, followed by centrifugation at 5,000 × g for 20 min at 4°C. The supernatant was recovered as the phage stock and stored at −80°C until use.
MNV was propagated in RAW 264.7 cells as described previously with some modifications (26). RAW 264.7 cells were cultured in Dulbecco's modified Eagle's medium (DMEM) (Gibco, USA) containing 10% fetal bovine serum (Gibco), 10 mM HEPES (Gibco), 10 mM sodium bicarbonate (Gibco), 10 mM nonessential amino acids (Gibco), and 50 μg/μl gentamicin reagent (Gibco). MNV was then inoculated onto a monolayer of RAW 264.7 cells in a sterilized flask and cultivated for 3 to 4 days. Infected cells were subjected to three cycles of freezing and thawing to cause cell membrane damage, allowing the easy release of MNV inside the infected cells. An equal volume of chloroform was added to the cell lysates and mixed, followed by centrifugation at 5,000 × g for 20 min at 4°C. Centrifugal ultrafiltration using Amicon Ultra-15 tubes (Millipore, USA) was used to concentrate the titer of MNV, and the virus stock was stored at −80°C until use.
Adenovirus serotype 2 (AdV2) was propagated in A549 cells as described previously with some modifications (27). A549 cells were cultured in RPMI 1640 medium (Gibco) containing 10% fetal bovine serum, 10 mM HEPES, 10 mM sodium bicarbonate, 10 mM nonessential amino acids, and 50 μg/μl gentamicin reagent. AdV2 was then inoculated onto the cultivated A549 cells, and the infected cells were subjected to three cycles of freezing and thawing. An equal volume of chloroform was added to the cell lysates and mixed, followed by centrifugation at 5,000 × g for 20 min at 4°C. Centrifugal ultrafiltration using Amicon Ultra-15 tubes (Millipore, USA) was used to concentrate the titer of AdV2, and the virus stock was stored at −80°C until use.
Antiviral properties of AgNP-MHCs.
To assess the antiviral properties of AgNP-MHCs, each type of AgNP-MHC was used at three different concentrations (4.6 × 107, 4.6 × 108, and 4.6 × 109 particles/ml). Briefly, bacteriophage ϕX174, MNV, and AdV2 were treated with Ag07-MHCs, Ag15-MHCs, and Ag30-MHCs in a shaking incubator at 150 rpm for 1 h at 25°C. As a control, OH-MHCs with no AgNPs were used at 4.6 × 109 particles/ml. In addition, the viruses were exposed to each AgNP-MHC at 4.6 × 109 particles/ml for 1, 3, and 6 h at 25°C.
After exposure to the particles, plaque assays and real-time TaqMan PCR (RT-PCR) assays were used to measure the efficacy of AgNP-MHCs for inactivating the tested viruses. For the RT-PCR assays, the viral genomes of MNV and AdV2 were extracted using a QIAamp MinElute virus spin kit (Qiagen, USA). RT-PCR for MNV was performed using a AgPath-ID one-step RT-PCR kit (Ambion, USA) as described previously (28). Briefly, the nucleic acids from MNV were reverse transcribed at 48°C for 30 min and denatured initially at 95°C for 10 min, followed by 45 cycles of 95°C for 15 s and 60°C for 1 min using a 7300 real-time PCR system (Applied Biosystems, USA). For RT-PCR of AdV2, TaqMan universal PCR master mix (Applied Biosystems) was used. The AdV2 nucleic acids were reverse transcribed at 50°C for 2 min and denatured at 95°C for 10 min, followed by 45 cycles of 95°C for 15 s and 60°C for 1 min. The sequences of the primers and TaqMan probe used for RT-PCR are given in Table 1 (26, 27).
TABLE 1.
Oligonucleotide primer and probe sequences for TaqMan real-time RT-PCR assay of AdV2 and MNV
| Virus | Primer or probe | Name (position, 5′→3′) | Sequence | Reference |
|---|---|---|---|---|
| AdV2 | Forward primer | JHKXF (18891–19910) | 5′-GGA CGC CTC GGA GTA CCT GA-3′ | 27 |
| Reverse primer | JHKXR (19025–19007) | 5′-CGC TGI GAC CIG TCT GTG G-3′ | ||
| Probe | JHKXP (18939–18960) | 5′-FAM-CAC CGA TAC GTA CTT CAG CCT G-MGB-3′ | ||
| MNV | Forward primer | MNV1 F (5614–5630) | 5′-ACG CCA CTC CGC ACA AA-3′ | 26 |
| Reverse primer | MNV1 R (5649–5657) | 5′-GCG GCC AGA GAC CAC AAA-3′ | ||
| Probe | MNV1 P (5632–5646) | 5′-VIC-AGC CCG GGT GAT GAG-MGB-3′ |
Effects of pH on the efficacy of AgNP-MHCs.
To characterize the effects of pH on AgNP-MHCs, 4.6 × 109 particles/ml of Ag30-MHCs were exposed to acidic (pH 2.0 and 5.0) or alkaline (pH 9.0 and 12.0) DW solutions in a shaking incubator at 150 rpm for 10 min at 25°C as described previously (29). The pH values were measured using an Orion 3-Star benchtop pH meter (Thermo Fisher Scientific, USA). After acid or alkali treatment, the DW was neutralized to pH 7.6. The Ag30-MHCs were recovered from the solution using a strong magnet and suspended in DW at their initial concentration. The Ag30-MHCs exposed to pH 2.0, 7.0, and 12.0 were examined using a Libra energy-filtering transmission electron microscope (Carl Zeiss Co. Ltd., South Korea) at the National Instrumentation Center for Environmental Management (NICEM), Seoul National University. Approximately 1 × 106 PFU/ml of both bacteriophage MS2 and ϕX174 were mixed with Ag30-MHCs with and without previous exposure to acid or alkali. The mixtures were placed in a shaking incubator at 150 rpm for 1 h at 25°C, and the efficacy of virus inactivation was measured using the single-agar-layer technique described previously (24).
Antiviral activity of AgNP-MHCs in tap and surface water.
Surface water was collected from the Han River in Seoul, South Korea, in July 2011. All water samples were collected in 1-liter sterilized bottles and stored at 4°C until use. Physicochemical analyses of the sampled surface water and tap water were performed by a commercial company (Wendi-Bio Inc., Seongnam, South Korea), using the water analysis techniques applicable to the Korean standards for drinking water quality. Half of the collected surface water samples were filtered through a 0.22-μm-pore-size syringe filter (Millex; Millipore). The concentrations of microorganisms were measured by cultivation prior to an assay using the single-agar-layer technique (24).
Approximately 4.6 × 109 Ag30-MHCs/ml were added to the raw and filtered surface water samples and tap water samples in a shaking incubator at 150 rpm for 10 min at 25°C. Approximately 1 × 106 PFU/ml of MS2 and ϕX174 were then added to the water-treated Ag30-MHCs in a shaking incubator at 150 rpm for 1 h at 25°C, and virus inactivation was determined using the single-agar-layer technique.
Statistical analysis.
The data are expressed as means ± standard deviations (SD) from at least three independent experiments. When appropriate, the data were analyzed with one-way analysis of variance (ANOVA) or Kruskal-Wallis one-way ANOVA and Dunnett's test for multiple comparisons. P values of <0.05 were considered to indicate statistical significance. SPSS for Windows (ver. 19.0; IBM, USA) and Sigmaplot for Windows (ver. 12.0; Systat software Inc., USA) were used for statistical analyses.
RESULTS
Characterization of AgNP-MHCs.
AgNP-MHCs were coated with AgNPs of different sizes. The properties of the AgNP-MHCs are presented in Table 2. Given the concentration of AgNP-MHCs (9.2 × 109 particles/ml), the number of AgNPs per MHC and the silver concentration varied. As a result, AgNPs with the largest particle size (Ag30-MHCs) had the highest concentration of silver per ml (400 ppm) and the lowest number of AgNPs per MHC (290 particles) compared with Ag07-MHCs (57.5 ppm and 2,600 particles). Our data indicate that Ag15-MHCs had the highest total surface area of AgNPs per MHC (1.1 μm2) and the highest surface coverage by AgNPs per MHC (25%).
TABLE 2.
AgNP composite samples (AgNP-MHCs) used in this studya
| Type of AgNP | Silver concn (ppm)b | No. of AgNPs per MHC | Total surface area of AgNPs per MHC (μm2)c | Surface coverage by AgNPs per MHC (%) |
|---|---|---|---|---|
| Ag07-MHC | 57.5 | 2,600 | 0.41 | 8.8 |
| Ag15-MHC | 275 | 1,600 | 1.1 | 25 |
| Ag30-MHC | 400 | 290 | 0.81 | 18 |
Cited in our previous study (21). AgNP-MHCs were used at 9.2 × 109 particles/ml.
The Ag concentration was obtained from the Advanced Analysis Center of KIST using atomic absorption spectrometer (AAS) and inductively coupled plasma (ICP) analysis.
Calculated by the following formula: surface area of one AgNP (4πr2) × number of AgNPs/MHC.
Antiviral effects of AgNP-MHCs.
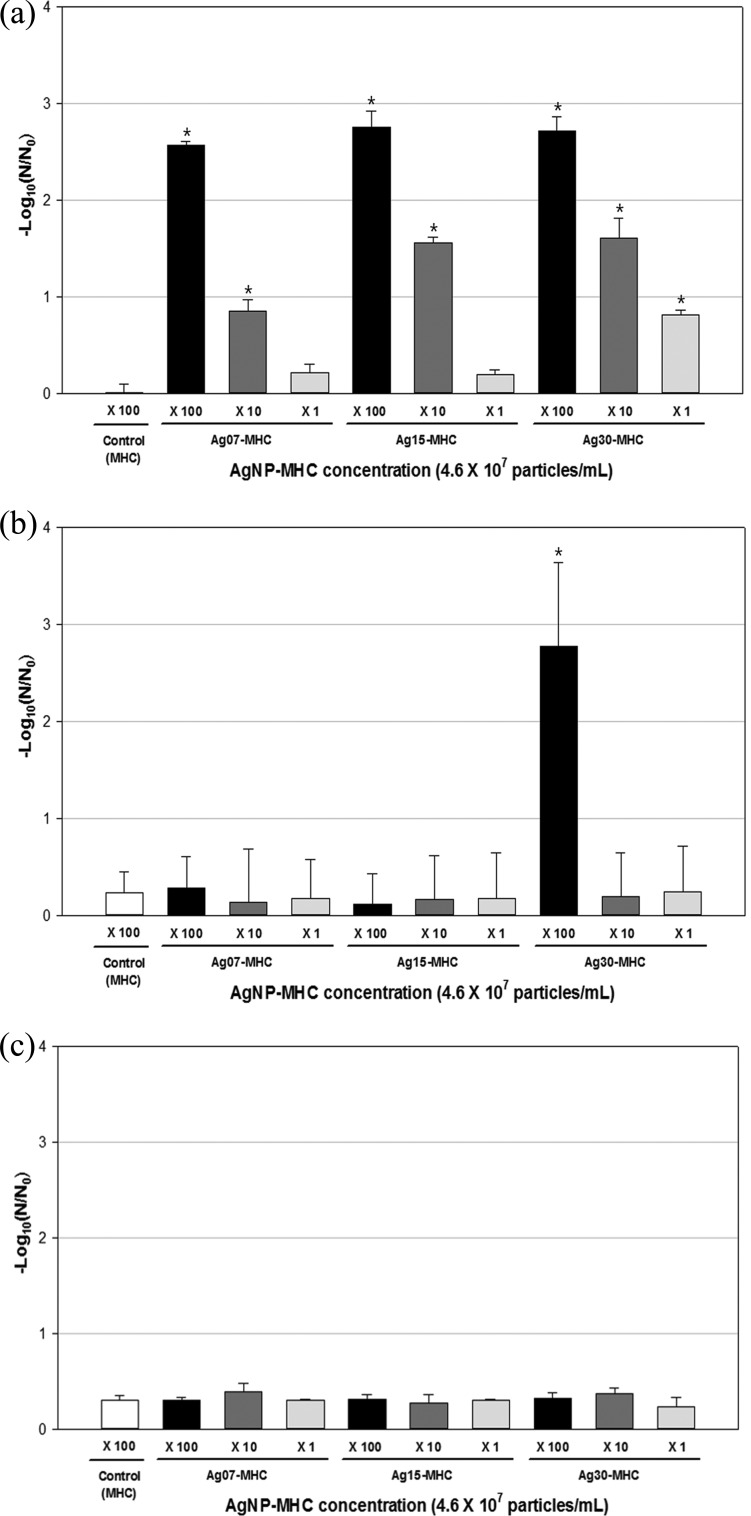
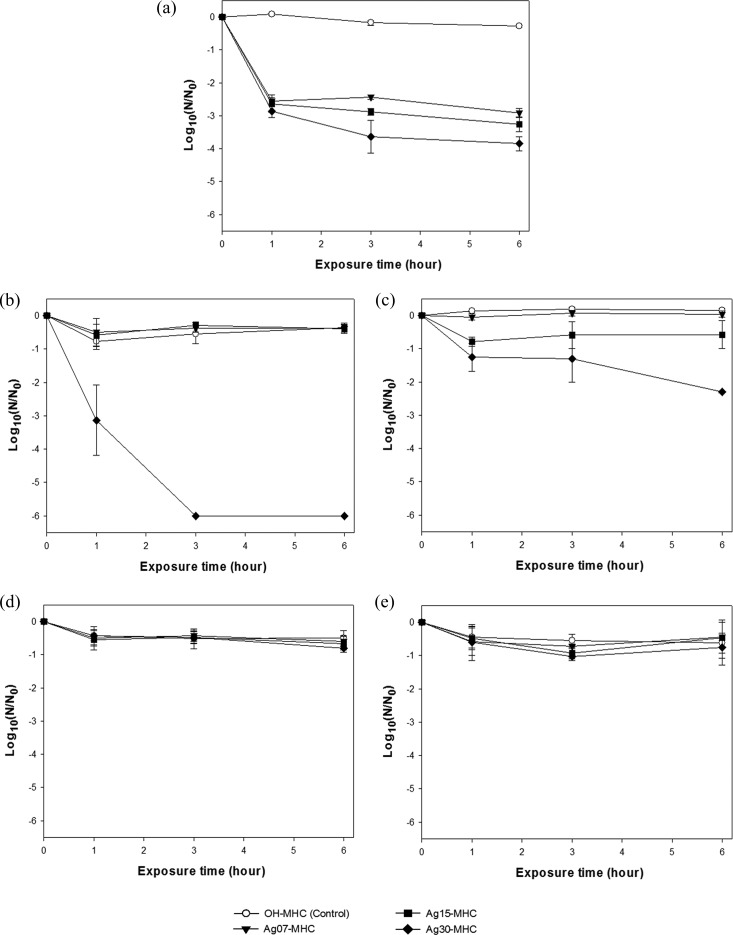
The antiviral effects of the AgNP-MHCs with various particle sizes and at different concentrations were measured by a plaque assay using bacteriophage ϕX174, MNV, and AdV2 (Fig. 1). The control treatment with OH-MHCs had no significant antiviral effect. In contrast, the AgNP-MHCs produced significant antiviral effects against bacteriophage ϕX174 and MNV but not against AdV2 (Fig. 1 and 2). A longer incubation time with the Ag30-MHCs produced a significantly greater reduction in bacteriophage ϕX174 and MNV. Compared with the other two AgNP-MHCs, Ag30-MHCs exhibited significantly greater antiviral effects for bacteriophage ϕX174 and MNV (P < 0.05). An approximate 6-log10 reduction of MNV and 4-log10 reduction of ϕX174 occurred after exposure to 4.6 × 109 Ag30-MHCs/ml for 6 h, as determined using a plaque assay. In contrast, when inactivation of MNV was analyzed by RT-PCR, only a 2-log10 reduction was observed. AdV2 had the highest resistance to AgNP-MHCs and did not display any significant inactivation regardless of the type of AgNP-MHCs used. The plaque assay and RT-PCR results for AdV2 were not significantly different (Fig. 2d and e).
FIG 1.
Antiviral effects of AgNP-MHCs at various concentrations against bacteriophage ϕX174 (a), MNV (b), and AdV2 (c). Target viruses were treated with different concentrations of AgNP-MHCs, and with OH-MHCs (4.6 × 109 particles/ml) as a control, in a shaking incubator (150 rpm, 1 h, 25°C). The plaque assay method was used to measure surviving viruses. Values are means ± standard deviations (SD) from three independent experiments. Asterisks indicate significantly different values (P < 0.05 by one-way ANOVA with Dunnett's test).
FIG 2.
Antiviral effects of AgNP-MHCs after various reaction times. (a) Bacteriophage ϕX174; (b) MNV, plaque assay; (c) MNV, RT-PCR; (d) AdV2, plaque assay; (e) AdV2, RT-PCR. Target viruses was treated with AgNP-MHCs or OH-MHCs (control) at 4.6 × 109 particles/ml in a shaking incubator (25°C, 150 rpm) for 1, 3, and 6 h. Surviving viruses were measured by plaque assay and RT-PCR. The results are expressed as means ± standard deviations (SD) from three independent experiments.
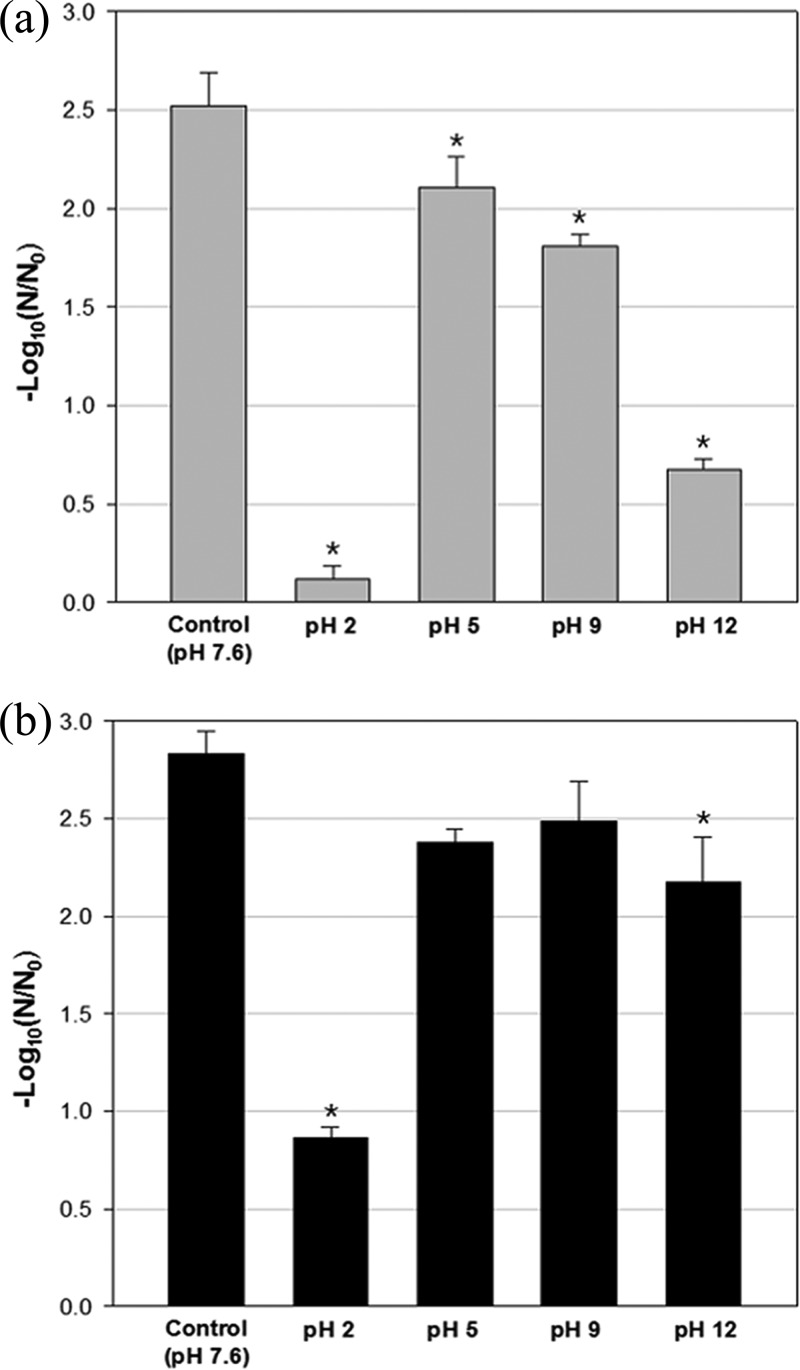
Effect of exposure of AgNP-MHCs to various pH conditions prior to use.
Figure 3 shows the antiviral efficacies of AgNP-MHCs that had been exposed to different pH conditions. Exposure to extremely acidic conditions (pH 2.0) significantly reduced the antiviral capabilities of Ag30-MHCs. After 10 min of exposure to pH 2.0 conditions, the Ag30-MHCs produced only a 0.1-log10 reduction of bacteriophage MS2 and a 0.8-log10 reduction of bacteriophage ϕX174, indicating antiviral efficacy significantly lower than that of untreated Ag30-MHCs (P < 0.05). Exposure to a strongly alkaline condition (pH 12.0) also decreased antiviral efficacy.
FIG 3.
Antiviral activities of Ag30-MHCs after exposure to various pH conditions. (a) Bacteriophage MS2; (b) bacteriophage ϕX174. First, Ag30-MHCs were exposed to acidic (pH 2.0 and 5.0) and alkaline (pH 9.0 and 12.0) conditions, created by adding 0.1 N HCl or 0.1 N NaOH to distilled water, in a shaking incubator (150 rpm, 10 min, 25°C). The acid or alkali was neutralized, and the target microorganisms were reacted with the treated and untreated (control, pH 7.6) Ag30-MHCs in a shaking incubator (150 rpm, 1 h, 25°C). Values are expressed as means ± standard deviations (SD) from three independent experiments. Asterisks indicate significantly different values (P < 0.05 by Kruskal-Wallis one-way ANOVA with Dunnett's test).
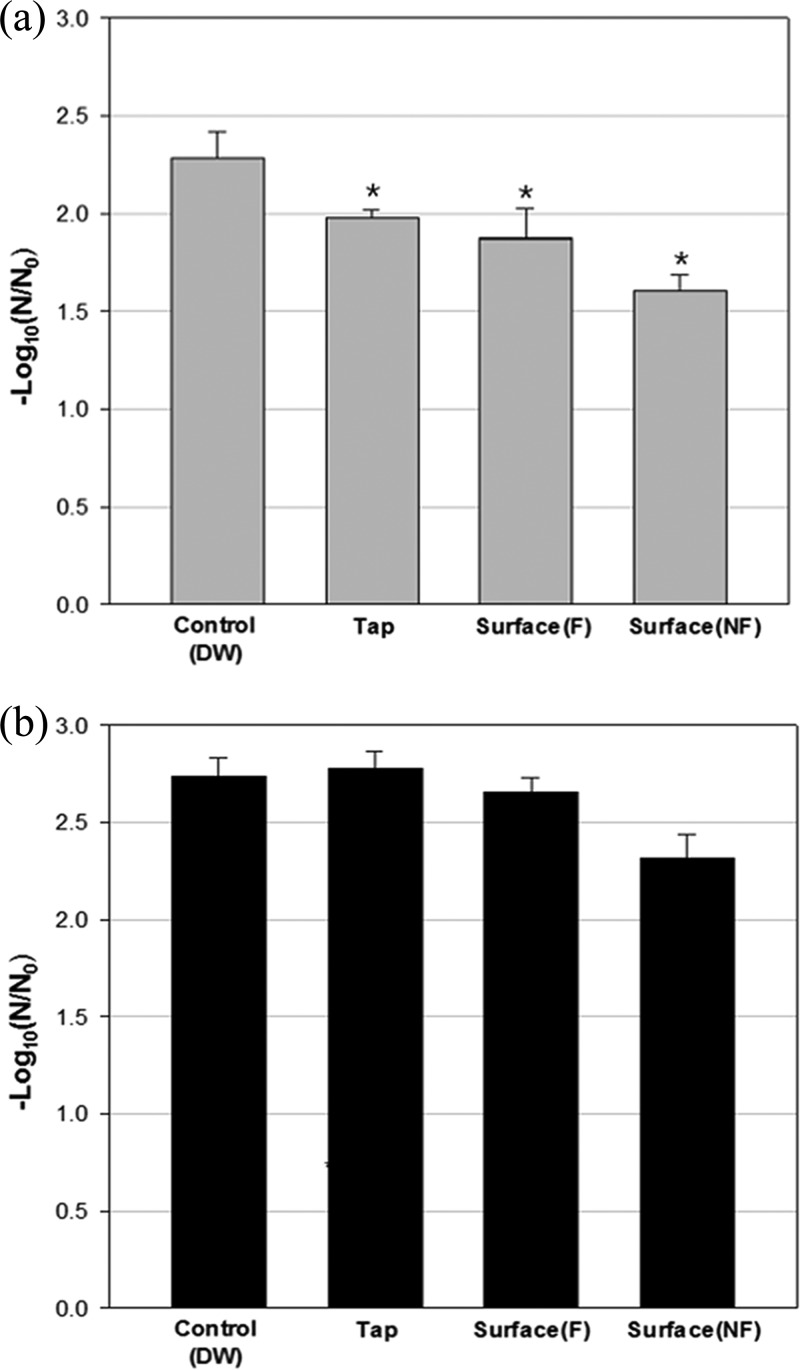
Antiviral effects of AgNP-MHCs in tap and surface water.
The antiviral efficacy of AgNP-MHCs against bacteriophages MS2 and ϕX174 in environmental water samples and in tap water samples was investigated (Fig. 4). Various physicochemical characteristics of the river water samples are summarized in Table 3. The efficacy of AgNP-MHCs against ϕX174 was maintained in the water samples, but the antiviral efficacy against MS2 was lower in the water samples than in DW.
FIG 4.
Inactivation of bacteriophages MS2 (a) and ϕX174 (b) by Ag30-MHCs exposed to tap and surface water. First, Ag30-MHCs were exposed to tap water or to surface water samples (F, filtered; NF, nonfiltered) from the Han River in a shaking incubator (150 rpm, 10 min, 25°C). Then, the bacteriophages were added to the water samples in a shaking incubator (150 rpm, 1 h, 25°C). Values are expressed as means ± standard deviations (SD) from three independent experiments. Asterisks indicate significantly different values (P < 0.05 by Kruskal-Wallis one-way ANOVA with Dunnett's test).
TABLE 3.
Results of water quality analysis of tap and surface water samples
| Parameter | Drinking water standarda | Value in: |
|
|---|---|---|---|
| Tap water | Surface water (Han River) | ||
| Total colony count | 100 CFU/ml | ND | 4,700c |
| Total coliforms | NDb/100 ml | ND | Detectedc |
| Escherichia coli/fecal coliforms | ND/100 ml | ND | Detectedc |
| Hardness | 300 mg/liter | 51 | 61.0 |
| pH | 5.8–8.5 | 7.3 | 7.4 |
| Turbidity | 0.5 NTUd | 0.15 | 6.37c |
| Total solids | 500 mg/liter | 94 | 87 |
| Chloride (Cl−) | 250 mg/liter | 11 | 7 |
| Chlorine residual concn | 4.0 mg/liter | ND | 0.48 |
| Sulfate (SO4−) | 200 mg/liter | 9 | 11 |
| Ammonium nitrogen (NH3-N) | 0.5 mg/liter | ND | 0.09 |
| Nitrate nitrogen (NO3-N) | 10 mg/liter | 2.3 | 2.0 |
| Fluoride (F) | 1.5 mg/liter | ND | ND |
| Lead (Pb) | 0.01 mg/liter | ND | ND |
| Mercury (Hg) | 0.001 mg/liter | ND | ND |
| Cadmium (Cd) | 0.005 mg/liter | ND | ND |
| Trihalomethanes | 0.1 mg/liter | 0.029 | ND |
| Toluene | 0.7 mg/liter | 0.006 | ND |
| Ethylbenzene | 0.3 mg/liter | 0.007 | ND |
| Xylene | 0.5 mg/liter | 0.011 | ND |
Standards for drinking water quality from the Korean EPA.
ND, not detected.
The level of the contaminant exceeded drinking water quality standards.
NTU, nephelometric turbidity units.
DISCUSSION
This study demonstrated that AgNP-MHCs are effective for inactivating bacteriophages and MNV, a surrogate for human norovirus, in water. In addition, AgNP-MHCs can be easily recovered with a magnet, effectively preventing the release of potentially toxic AgNPs into the environment. A number of previous studies have shown that the concentration and particle size of AgNPs are critical factors for inactivating targeted microorganism (8, 16, 17). The antimicrobial effects of AgNPs also depend on the type of microorganism. The efficacy of AgNP-MHCs for inactivating ϕX174 followed a dose-response relationship. Among the AgNP-MHCs tested, Ag30-MHCs had a higher efficacy for inactivating ϕX174 and MNV. For MNV, only Ag30-MHCs displayed antiviral activity, with the other AgNP-MHCs not generating any significant inactivation of MNV. None of the AgNP-MHCs had any significant antiviral activity against AdV2.
In addition to particle size, the concentration of silver in the AgNP-MHCs was also important. The concentration of silver appeared to determine the efficacy of the antiviral effects of AgNP-MHCs. The silver concentrations in solutions of Ag07-MHCs and Ag30-MHCs at 4.6 × 109 particles/ml were 28.75 ppm and 200 ppm, respectively, and correlated with the level of antiviral activity. Table 2 summarizes the silver concentrations and surface areas of the AgNP-MHCs tested. Ag07-MHCs displayed the lowest antiviral activity and had the lowest silver concentration and surface area, suggesting that these properties are related to the antiviral activity of AgNP-MHCs.
Our previous study indicated that the major antimicrobial mechanisms of AgNP-MHCs are the chemical abstraction of Mg2+ or Ca2+ ions from microbial membranes, the creation of complexes with thiol groups located at the membranes, and the generation of reactive oxygen species (ROS) (21). Because AgNP-MHCs have a relatively large particle size (∼500 nm), it is unlikely that they can penetrate a viral capsid. Instead, AgNP-MHCs appear to interact with viral surface proteins. AgNPs on the composites tend to bind thiol group-containing biomolecules embedded in the coat proteins of viruses. Therefore, the biochemical properties of viral capsid proteins are important for determining their susceptibility to AgNP-MHCs. Figure 1 shows the different susceptibilities of the viruses to the effects of AgNP-MHCs. The bacteriophages ϕX174 and MNV were susceptible to AgNP-MHCs, but AdV2 was resistant. The high resistance level of AdV2 is likely to be associated with its size and structure. Adenoviruses range in size from 70 to 100 nm (30), making them much larger than ϕX174 (27 to 33 nm) and MNV (28 to 35 nm) (31, 32). In addition to their large size, adenoviruses have double-stranded DNA, unlike other viruses, and are resistant to various environmental stresses such as heat and UV radiation (33, 34). Our previous study reported that almost a 3-log10 reduction of MS2 occurred with Ag30-MHCs within 6 h (21). MS2 and ϕX174 have similar sizes with different types of nucleic acid (RNA or DNA) but have similar rates of inactivation by Ag30-MHCs. Therefore, the nature of the nucleic acid does not appear to be the major factor for resistance to AgNP-MHCs. Instead, the size and shape of viral particle appeared to be more important, because adenovirus is a much larger virus. The Ag30-MHCs achieved almost a 2-log10 reduction of M13 within 6 h (our unpublished data). M13 is single-stranded DNA virus (35) and is ∼880 nm in length and 6.6 nm in diameter (36). The rate of inactivation of the filamentous bacteriophage M13 was intermediate between those of small, round-structured viruses (MNV, ϕX174, and MS2) and a large virus (AdV2).
In the present study, the inactivation kinetics of MNV were significantly different in the plaque assay and the RT-PCR assay (Fig. 2b and c). Molecular assays such as RT-PCR are known to significantly underestimate the inactivation rates of viruses (25, 28), as was found in our study. Because AgNP-MHCs interact primarily with the viral surface, they are more likely to damage viral coat proteins rather than viral nucleic acids. Therefore, an RT-PCR assay to measure viral nucleic acid may significantly underestimate the inactivation of viruses. The effect of Ag+ ions and the generation of reactive oxygen species (ROS) should be responsible for the inactivation of the tested viruses. However, many aspects of the antiviral mechanisms of AgNP-MHCs are still unclear, and further research using biotechnological approaches is required to elucidate the mechanism of the high resistance of AdV2.
Finally, we evaluated the robustness of the antiviral activity of Ag30-MHCs by exposing them to a wide range of pH values and to tap and surface water samples before measuring their antiviral activity (Fig. 3 and 4). Exposure to extremely low pH conditions resulted in the physical and/or functional loss of AgNPs from the MHC (unpublished data). In the presence of nonspecific particles, Ag30-MHCs consistently displayed antiviral activity, despite a decline in the antiviral activity against MS2. The antiviral activity was lowest in unfiltered surface water, as an interaction between Ag30-MHCs and nonspecific particles in the highly turbid surface water probably caused a reduction of antiviral activity (Table 3). Therefore, field evaluations of AgNP-MHCs in various types of water (e.g., with different salt concentrations or humic acid) should be performed in the future.
In conclusion, the new Ag composites, AgNP-MHCs, have excellent antiviral capabilities against several viruses, including ϕX174 and MNV. AgNP-MHCs maintain strong efficacy under different environmental conditions, and these particles can be easily recovered using a magnet, thus reducing their potential harmful effects on human health and the environment. This study showed that the AgNP composite can be an effective antiviral in various environmental settings, without significant ecological risks.
ACKNOWLEDGMENTS
This work was supported by National Research Foundation of Korea (NRF) grants funded by the Korean government (MEST) (no. 2012-0008692 and no. 2012-0009628) and by the BK21 Plus project (22A20130012682).
Footnotes
Published ahead of print 31 January 2014
REFERENCES
- 1.Mody VV, Siwale R, Singh A, Mody HR. 2010. Introduction to metallic nanoparticles. J. Pharm. Bioallied Sci. 2:282–289. 10.4103/0975-7406.72127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Salata O. 2004. Applications of nanoparticles in biology and medicine. J. Nanobiotechnol. 2:3. 10.1186/1477-3155-2-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sokolov K, Aaron J, Hsu B, Nida D, Gillenwater A, Follen M, MacAulay C, Adler-Storthz K, Korgel B, Descour M, Pasqualini R, Arap W, Lam W, Richards-Kortum R. 2003. Optical systems for in vivo molecular imaging of cancer. Technol. Cancer Res. Treat. 2:491–504 [DOI] [PubMed] [Google Scholar]
- 4.El-Sayed IH, Huang X, El-Sayed MA. 2005. Surface plasmon resonance scattering and absorption of anti-EGFR antibody conjugated gold nanoparticles in cancer diagnostics: applications in oral cancer. Nano Lett. 5:829–834. 10.1021/nl050074e [DOI] [PubMed] [Google Scholar]
- 5.Yen HJ, Hsu SH, Tsai CL. 2009. Cytotoxicity and immunological response of gold and silver nanoparticles of different sizes. Small 5:1553–1561. 10.1002/smll.200900126 [DOI] [PubMed] [Google Scholar]
- 6.Rai M, Yadav A, Gade A. 2009. Silver nanoparticles as a new generation of antimicrobials. Biotechnol. Adv. 27:76–83. 10.1016/j.biotechadv.2008.09.002 [DOI] [PubMed] [Google Scholar]
- 7.Lara HH, Garza-Trevino EN, Ixtepan-Turrent L, Singh DK. 2011. Silver nanoparticles are broad-spectrum bactericidal and virucidal compounds. J. Nanobiotechnol. 9:30. 10.1186/1477-3155-9-30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim JS, Kuk E, Yu KN, Kim JH, Park SJ, Lee HJ, Kim SH, Park YK, Park YH, Hwang CY, Kim YK, Lee YS, Jeong DH, Cho MH. 2007. Antimicrobial effects of silver nanoparticles. Nanomedicine 3:95–101. 10.1016/j.nano.2006.12.001 [DOI] [PubMed] [Google Scholar]
- 9.Lu L, Sun RW, Chen R, Hui CK, Ho CM, Luk JM, Lau GK, Che CM. 2008. Silver nanoparticles inhibit hepatitis B virus replication. Antivir. Ther. 13:253–262 [PubMed] [Google Scholar]
- 10.Lara HH, Ayala-Nunez NV, Ixtepan-Turrent L, Rodriguez-Padilla C. 2010. Mode of antiviral action of silver nanoparticles against HIV-1. J. Nanobiotechnol. 8:1. 10.1186/1477-3155-8-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Speshock JL, Murdock RC, Braydich-Stolle LK, Schrand AM, Hussain SM. 2010. Interaction of silver nanoparticles with Tacaribe virus. J. Nanobiotechnol. 8:19. 10.1186/1477-3155-8-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Panacek A, Kvítek L, Prucek R, Kolar M, Vecerova R, Pizúrova N, Sharma VK, Nevecna T, Zboril R. 2006. Silver colloid nanoparticles: synthesis, characterization, and their antibacterial activity. J. Phys. Chem. B 110:16248–16253. 10.1021/jp063826h [DOI] [PubMed] [Google Scholar]
- 13.Pal S, Tak YK, Song JM. 2007. Does the antibacterial activity of silver nanoparticles depend on the shape of the nanoparticle? A study of the gram-negative bacterium Escherichia coli. Appl. Environ. Microbiol. 73:1712–1720. 10.1128/AEM.02218-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sotiriou GA, Pratsinis SE. 2010. Antibacterial activity of nanosilver ions and particles. Environ. Sci. Technol. 44:5649–5654. 10.1021/es101072s [DOI] [PubMed] [Google Scholar]
- 15.He D, Jones AM, Garg S, Pham AN, Waite TD. 2011. Silver nanoparticle-reactive oxygen species interactions: application of a charging-discharging model. J. Phys. Chem. C 115:5461–5468. 10.1021/jp111275a [DOI] [Google Scholar]
- 16.Sondi I, Salopek-Sondi B. 2004. Silver nanoparticles as antimicrobial agent: a case study on E. coli as a model for Gram-negative bacteria. J. Colloid Interface Sci. 275:177–182. 10.1016/j.jcis.2004.02.012 [DOI] [PubMed] [Google Scholar]
- 17.Morones JR, Elechiguerra JL, Camacho A, Holt K, Kouri JB, Ramirez JT, Yacaman MJ. 2005. The bactericidal effect of silver nanoparticles. Nanotechnology 16:2346–2353. 10.1088/0957-4484/16/10/059 [DOI] [PubMed] [Google Scholar]
- 18.Hsin YH, Chen CF, Huang S, Shih TS, Lai PS, Chueh PJ. 2008. The apoptotic effect of nanosilver is mediated by a ROS- and JNK-dependent mechanism involving the mitochondrial pathway in NIH3T3 cells. Toxicol. Lett. 179:130–139. 10.1016/j.toxlet.2008.04.015 [DOI] [PubMed] [Google Scholar]
- 19.Park MV, Neigh AM, Vermeulen JP, de la Fonteyne LJ, Verharen HW, Briede JJ, van Loveren H, de Jong WH. 2011. The effect of particle size on the cytotoxicity, inflammation, developmental toxicity and genotoxicity of silver nanoparticles. Biomaterials 32:9810–9817. 10.1016/j.biomaterials.2011.08.085 [DOI] [PubMed] [Google Scholar]
- 20.Mukherjee SG, O'Claonadh N, Casey A, Chambers G. 2012. Comparative in vitro cytotoxicity study of silver nanoparticle on two mammalian cell lines. Toxicol. In Vitro 26:238–251. 10.1016/j.tiv.2011.12.004 [DOI] [PubMed] [Google Scholar]
- 21.Park HH, Park SJ, Ko G, Woo K. 2013. Magnetic hybrid colloid decorated with Ag nanoparticles bite away bacteria and chemisorb virus. J. Mater. Chem. B 21:2701–2709. 10.1039/C3TB20311E [DOI] [PubMed] [Google Scholar]
- 22.Park HH, Woo K, Ahn J. 2013. Core-shell bimetallic nanoparticles robustly fixed on the outermost surface of magnetic silica microspheres. Sci. Rep. 3:1497. 10.1038/srep01497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Battigelli DA, Sobsey MD, Lobe DC. 1993. The inactivation of hepatitis A virus and other model viruses by UV irradiation. Water Sci. Technol. 27:339–342 [Google Scholar]
- 24.U.S. Environmental Protection Agency. 2001. Manual of methods for virology, chapter 16 Procedures for detecting coliphages. http://www.epa.gov/microbes/about.html
- 25.Lim MY, Kim JM, Ko G. 2010. Disinfection kinetics of murine norovirus using chlorine and chlorine dioxide. Water Res. 44:3243–3251. 10.1016/j.watres.2010.03.003 [DOI] [PubMed] [Google Scholar]
- 26.Lee J, Zoh K, Ko G. 2008. Inactivation and UV disinfection of murine norovirus with TiO2 under various environmental conditions. Appl. Environ. Microbiol. 74:2111–2117. 10.1128/AEM.02442-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ko G, Jothikumar N, Hill VR, Sobsey MD. 2005. Rapid detection of infectious adenoviruses by mRNA real-time RT-PCR. J. Virol. Methods 127:148–153. 10.1016/j.jviromet.2005.02.017 [DOI] [PubMed] [Google Scholar]
- 28.Lim MY, Kim JM, Lee JE, Ko G. 2010. Characterization of ozone disinfection of murine norovirus. Appl. Environ. Microbiol. 76:1120–1124. 10.1128/AEM.01955-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xu H, Qu F, Xu H, Lai W, Wang YA, Aguilar ZP, Wei H. 2012. Role of reactive oxygen species in the antibacterial mechanism of silver nanoparticles on Escherichia coli O157:H7. Biometals 25:45–53. 10.1007/s10534-011-9482-x [DOI] [PubMed] [Google Scholar]
- 30.Kennedy MA, Parks RJ. 2009. Adenovirus virion stability and the viral genome: size matters. Mol. Ther. 17:1664–1666. 10.1038/mt.2009.202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bayer ME, DeBlois RW. 1974. Diffusion constant and dimension of bacteriophage ϕX174 as determined by self-beat laser light spectroscopy and electron microscopy. J. Virol. 14:975–980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wobus CE, Thackray LB, Virgin HW., IV 2006. Murine norovirus: a model system to study norovirus biology and pathogenesis. J. Virol. 80:5104–5112. 10.1128/JVI.02346-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Linden KG, Thurston J, Schaefer R, Malley JP., Jr 2007. Enhanced UV inactivation of adenoviruses under polychromatic UV lamps. Appl. Environ. Microbiol. 73:7571–7574. 10.1128/AEM.01587-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sauerbrei A, Wutzler P. 2009. Testing thermal resistance of viruses. Arch. Virol. 154:115–119. 10.1007/s00705-008-0264-x [DOI] [PubMed] [Google Scholar]
- 35.Henry TJ, Pratt D. 1969. The proteins of bacteriophage M13. Proc. Natl. Acad. Sci. U. S. A. 62:800–807. 10.1073/pnas.62.3.800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee BY, Zhang J, Zueger C, Chung WJ, Yoo SY, Wang E, Meyer J, Ramesh R, Lee SW. 2012. Virus-based piezoelectric energy generation. Nat. Nanotechnol. 7:351–356. 10.1038/nnano.2012.69 [DOI] [PubMed] [Google Scholar]