Abstract
Methane is an important greenhouse gas and the most abundant hydrocarbon in the Earth's atmosphere. Methanotrophic microorganisms can use methane as their sole energy source and play a crucial role in the mitigation of methane emissions in the environment. “Candidatus Methylomirabilis oxyfera” is a recently described intra-aerobic methanotroph that is assumed to use nitric oxide to generate internal oxygen to oxidize methane via the conventional aerobic pathway, including the monooxygenase reaction. Previous genome analysis has suggested that, like the verrucomicrobial methanotrophs, “Ca. Methylomirabilis oxyfera” encodes and transcribes genes for the Calvin-Benson-Bassham (CBB) cycle for carbon assimilation. Here we provide multiple independent lines of evidence for autotrophic carbon dioxide fixation by “Ca. Methylomirabilis oxyfera” via the CBB cycle. The activity of ribulose-1,5-bisphosphate carboxylase/oxygenase (RubisCO), a key enzyme of the CBB cycle, in cell extracts from an “Ca. Methylomirabilis oxyfera” enrichment culture was shown to account for up to 10% of the total methane oxidation activity. Labeling studies with whole cells in batch incubations supplied with either 13CH4 or [13C]bicarbonate revealed that “Ca. Methylomirabilis oxyfera” biomass and lipids became significantly more enriched in 13C after incubation with 13C-labeled bicarbonate (and unlabeled methane) than after incubation with 13C-labeled methane (and unlabeled bicarbonate), providing evidence for autotrophic carbon dioxide fixation. Besides this experimental approach, detailed genomic and transcriptomic analysis demonstrated an operational CBB cycle in “Ca. Methylomirabilis oxyfera.” Altogether, these results show that the CBB cycle is active and plays a major role in carbon assimilation by “Ca. Methylomirabilis oxyfera” bacteria. Our results suggest that autotrophy might be more widespread among methanotrophs than was previously assumed and implies that a methanotrophic community in the environment is not necessarily revealed by 13C-depleted lipids.
INTRODUCTION
Methane is an important volatile product of the anaerobic degradation of organic matter and is the most abundant hydrocarbon in the Earth's atmosphere (1). It is the most reduced form of carbon, but while bearing a vast amount of energy, it is thermodynamically one of the most difficult organic compounds to activate. The biological oxidation of methane occurs under both oxic and anoxic conditions and is performed by specialized groups of Bacteria or Archaea. Aerobic methanotrophs belong to the Bacteria, and all oxidize methane in a similar manner, using oxygen for the first step of oxidation of methane to methanol by a monooxygenase. In the following reactions, catalyzed by dehydrogenases, methanol is oxidized to carbon dioxide, with formaldehyde and formate being intermediates (2).
The aerobic methanotrophs belonging to the Proteobacteria were divided into two types on the basis of their morphology and physiological properties, including the route of C1 assimilation (3). Type I methanotrophs mostly utilize the ribulose monophosphate (RuMP) pathway, in which all cellular carbon is derived from methane and enters the pathway at the level of formaldehyde (2–4). Type II methanotrophs assimilate carbon via a combination of the serine and ethylmalonyl coenzyme A (ethylmalonyl-CoA) pathways, in which approximately one half of the cellular carbon is derived from methane via formaldehyde and the other half originates from multiple carboxylation reactions in both pathways (5–7). Both serine and RuMP pathways represent chemoorganoheterotrophic modes of metabolism and were considered to be universal among aerobic methanotrophs. However, some proteobacterial methanotrophs do possess complete gene sets for autotrophic CO2 fixation by the reductive pentose phosphate cycle, commonly known as the Calvin-Benson-Bassham (CBB) cycle (8–13): the type I methanotrophs Methylococcus capsulatus and Methylocaldum szegediense O-12 and the type II methanotrophs Methyloferula stellata AR4, Methylocella silvestris BL2, and Methylocapsa acidiphila B2. It still remains to be experimentally validated which role the CBB cycle plays in these organisms.
For a century after their first discovery in 1906 by Söhngen, methanotrophic bacteria were believed to be restricted to the Alphaproteobacteria and Gammaproteobacteria (14). However, since 2007 several independent studies have shown that bacterial methanotrophs are phylogenetically much more diverse and are also found within the verrucomicrobial and NC10 phyla (15–18). The discovery of the (acidophilic) verrucomicrobial methanotrophs not only revealed a wider environmental and phylogenetic spectrum for aerobic methanotrophy but also demonstrated that these methanotrophs lack essential genes of both the RuMP and the serine pathways (19). Instead, they were shown to utilize the CBB cycle for carbon dioxide fixation, challenging the paradigm that methanotrophs are heterotrophs that derive a large part of their biomass from methane (20). Similarly, genome analysis suggested that the first described member of the NC10 phylum, the nitrite-dependent methane oxidizer “Candidatus Methylomirabilis oxyfera,” may also employ the CBB cycle for carbon assimilation (16, 21). “Ca. Methylomirabilis oxyfera” oxidizes methane via a sequence of reactions similar to those employed by aerobic methanotrophs; however, it does so in the complete absence of external oxygen. Instead, nitrite is reduced to nitric oxide, and the latter is hypothesized to be dismutated to molecular nitrogen and oxygen (16, 22). The internally produced oxygen can then be used for methane oxidation by a methane monooxygenase.
Before the recent discovery of widespread autotrophy among verrucomicrobial methanotrophs (20, 23), all methanotrophs were believed to derive at least half of their cellular carbon from methane. This fact was used for the characterization of methanotrophic communities by stable carbon isotope probing (SIP) (24–29). The detection of aerobic methanotrophy in culture-independent environmental studies was also based on labeling of biomass and lipids with the strongly depleted 13C signature of biogenic methane (30–33). On the basis of these observations, autotrophy was generally dismissed as a dominant mode of carbon fixation in methanotrophic organisms. However, a recent study by Sharp et al. (23) has demonstrated that a modified SIP method with [13C]bicarbonate is necessary to reveal autotrophic methanotrophic communities in a geothermal environment.
The current study aimed to investigate the mode of C1 assimilation in “Ca. Methylomirabilis oxyfera” enrichment cultures through a tiered approach. We carried out detailed genome and transcriptome analysis focusing on the potential for autotrophic CO2 fixation via the CBB cycle and employed enzyme activity assays to detect the activity of ribulose-1,5-bisphosphate carboxylase/oxygenase (RubisCO) in cell extracts. In whole-cell batch incubations, we performed 13C-labeling experiments using labeled methane and/or bicarbonate to identify the effective source of C assimilation in “Ca. Methylomirabilis oxyfera”-specific lipid biomarkers and total enrichment culture biomass.
MATERIALS AND METHODS
“Ca. Methylomirabilis oxyfera” enrichment culture.
The culture of “Ca. Methylomirabilis oxyfera” strain Ooij was enriched in an anoxic sequencing batch bioreactor as previously described (34). The enrichment culture was dominated by “Ca. Methylomirabilis oxyfera,” making up approximately 80% of the total community, as estimated from previous fluorescence in situ hybridization (FISH) analysis.
Transcriptomic and genomic analysis.
Phylogenetic sequence analysis of the large-chain RubisCO genes was performed by comparison of cbbL gene sequences obtained from GenBank using the ClustalW algorithm within MEGA (version 5.0) software (35, 36). The resulting alignment was manually checked. The phylogenetic tree was calculated using the neighbor-joining method (37) and a bootstrap test of 1,000 replicates. The Dayhoff matrix was used for reconstruction of evolutionary distances. The transcriptome data used in this study were obtained from reference 38. Mapping of the total RubisCO reads was performed with CLC Bio Genomics Workbench (version 5.0) software using a sequence data set downloaded from GenBank as a mapping reference. The genome analysis was performed on the basis of published data with additional BLAST analysis.
Cell extract preparations.
Biomass was obtained from the enrichment culture and centrifuged (under oxic conditions) for 30 min at 16,500 × g (Sorvall RC5B Plus centrifuge; DuPont, Bad Homburg, Germany) at 4°C. The supernatant was discarded, and the pellet was resuspended in 20 mM Tris-HCl (pH 8.0) containing 50 mM sodium pyrophosphate (PPi). The final volume of resuspended pellet was 7 ml, to which 1 tablet of a protease inhibitor cocktail (Boehringer, Mannheim, Germany) and 1 mg DNase (Roche Diagnostics, Mannheim, Germany) were added. The disruption of cells was performed on ice by ultrasonication at 110 MPa 15 times for 10 s each time with pause intervals of 10 s between each ultrasonication. The sonicated biomass was centrifuged for 10 min at 12,000 × g and 4°C. The pellet was discarded, and the supernatant was further centrifuged for 10 min under identical conditions. Finally, the dark reddish-brownish supernatant was stored on ice for enzyme assays.
RubisCO activity assays.
The method for the RubisCO activity assay was adapted from reference 20. It is based on the determination of 13CO2 liberation after the destruction of labeled 3-phosphoglycerate, which is formed after the RubisCO-specific carboxylation of ribulose-1,5-bisphosphate (RuBP) by CO2 originating from labeled bicarbonate. The RubisCO activity can then be deduced from the increase in 13CO2 during a time series of samples.
The assay was performed in 2-ml septum vials (Labco, United Kingdom) with a liquid volume of 250 μl. Depending on the amount of cell extract used, either 175 or 200 μl of 20 mM potassium phosphate buffer (pH 6.9) containing 10 mM MgCl2 was used. Cell extract (50 or 25 μl) and 20 μl of 100 mM NaH13CO3 were added with a syringe to closed vials containing buffer, and the vials were vortexed for 2 s and incubated for 10 min at 30°C. Then, 5 μl of 25 mM RuBP was added and the vials were vortexed for 2 s. Further incubation was performed at 30°C, and samples of 50 μl were withdrawn by a syringe and injected into closed 3-ml Exetainer vials (Labco Limited, High Wycombe, United Kingdom) at intervals of 5 or 10 min over a total period of 20 min. Each Exetainer vial was amended with 20 μl of 0.5 M HCl, and the contents were dried under vacuum at 50°C overnight. The dried samples were amended with 0.5 ml ice-cold 0.1% KMnO4 in 0.1 M H3PO4 in closed Exetainer vials, and the vials were vortexed for 5 s and incubated for 25 min at 50°C. After incubation, the samples were kept at room temperature for 1 h for equilibration. The final measurement of headspace CO2 was performed by gas chromatography (GC) with a gas chromatograph coupled to a mass spectrometer (MS; 5975C; Agilent, Santa Clara, CA). Each measurement was performed in duplicate by injection of 100 μl of headspace gas with a gas-tight syringe.
Carbon isotope tracing experiments.
The effective assimilation of carbon from methane or bicarbonate/CO2 into the biomass and lipids of “Ca. Methylomirabilis oxyfera” was investigated with SIP experiments. Various treatments were performed in which “Ca. Methylomirabilis oxyfera” biomass was incubated with either [13C]methane or [13C]bicarbonate. The [13C]methane treatment also contained unlabeled bicarbonate, and the treatment with [13C]bicarbonate also received methane as the energy source. A control with [13C]bicarbonate without methane was included, and as a reference, biomass was also incubated with unlabeled methane and unlabeled bicarbonate. Additional treatments were included to test the use of H2 and/or formate as alternative electron donors. All treatments, summarized in Table 1, received nitrite as the electron acceptor.
TABLE 1.
Summary of treatments of the “Ca. Methylomirabilis oxyfera” enrichment culture in labeling experiments
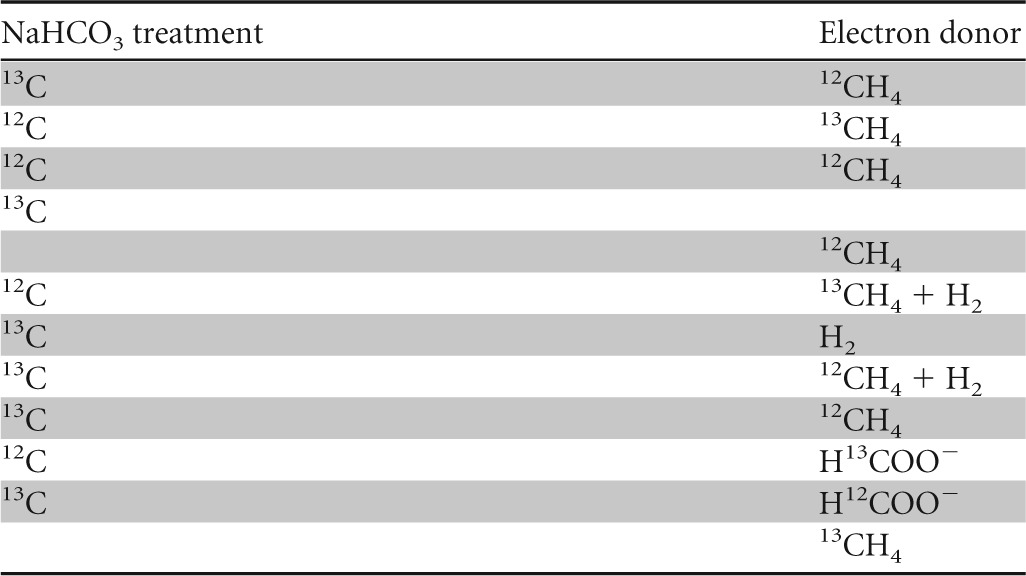
Incubation setup.
The reactor biomass was washed in 10 mM 3-(N-morpholino)propanesulfonic acid (MOPS) buffer (pH 7.4) and resuspended in nitrate-free mineral salt medium described previously (31). MOPS was used as a buffering agent (pH 7.4) at a final concentration of 5 mM. Duplicate incubations were performed in 60-ml glass serum bottles amended with different combinations of 13C-labeled or unlabeled sodium bicarbonate (2.5 mM), methane (∼4% in headspace), or formate (2.5 mM). All incubation mixtures were amended with sodium nitrite (0.3 mM) at the start of incubation and spiked with additional nitrite when the concentration in the bottles was close to 0, as estimated by Merckoquant test strips. The bottles were sealed with red butyl rubber stoppers and crimped with aluminum rings. Immediately after sealing, the batch cultures were made anoxic by 5 cycles of successive vacuuming and gassing with argon and a final flushing with argon for at least 10 min. In each bottle, an overpressure of 0.4 bar was applied. Thereafter, the incubation mixtures were amended with 13C-labeled (99 atom%; Isotec Inc., Matheson Trigas Products Division) or unlabeled methane (4%; Air Liquide, Eindhoven, The Netherlands), and some treatments were amended with hydrogen (16% headspace gas). Additionally, 2 ml unlabeled CO2 was added to each bottle to achieve bicarbonate/CO2 equilibrium. The cultures were incubated horizontally on a shaker (170 rpm; Innova40; New Brunswick Scientific) at 30°C. The pH was measured with a pH meter (Metrohm 691; Herisau, Switzerland) at the start and the end of incubation. The headspace pressure was monitored several times during the experiment with a needle pressure meter (GMH 3111; Greisinger, Germany).
Analysis of methane, nitrite, formate, and protein concentrations.
The methane concentration in the headspace was measured by gas chromatography with a flame ionization detector (HP 5890 series II; Agilent Technologies, Santa Clara, CA). Each measurement was performed in duplicate by injection of 100 μl headspace gas with a gas-tight syringe. Measurements of carbon dioxide, oxygen, and hydrogen were performed by GC-MS (see above). At the same time, liquid samples of 200 μl each were taken for nitrite and formate determination. After each activity experiment, 0.5 ml of biomass was taken from each incubation bottle for determination of the total protein content. The remaining biomass was centrifuged in 50-ml tubes at 4,000 × g for 20 min, the supernatant was discarded, and the pellets were kept at −80°C until subsequent freeze-drying and following lipid and isotope analysis. Nitrite and protein concentrations were determined colorimetrically as described in reference 34. Formate determination was performed with a method slightly modified from one described previously (39). A sample of 0.1 ml was mixed with 0.2 ml freshly prepared working solution (0.5 g citric acid and 10 g acetamide in 100 ml 2-propanol), 0.01 ml sodium acetate (30% [wt/vol] in Milli-Q water), and 0.7 ml acetic anhydride. All samples (in triplicate) were incubated for 2 h at room temperature in the dark, and the extinction was measured at 510 nm with a spectrophotometer (Ultrospec K; LKB Biochrom Ltd., Cambridge, United Kingdom).
During the experiment, the enrichment culture exhibited methanotrophic activity of 1.3 to 1.6 nmol CH4 min−1 mg−1 protein. Associated nitrite consumption closely followed the theoretical stoichiometry of 3:8 (CH4/NO2−). In the treatments where we checked for the use of alternative electron donors, we confirmed that neither formate nor hydrogen was used as the electron donor by the “Ca. Methylomirabilis oxyfera” enrichment culture (data not shown). The presence of hydrogen did not have any effect on the methanotrophic activity. Interestingly, the nitrite- and methane-oxidizing activity was considerably lower in incubations without added bicarbonate, although the difference in pH from that in cultures with added bicarbonate was not significant.
Bulk and compound-specific isotope analysis.
The freeze-dried biomass was separated into subsamples for bulk and compound-specific isotope analysis. For bulk isotope analysis of the biomass, the subsamples were acidified with 2 N HCl in silver foil cups to remove all inorganic carbon and dried at 60°C overnight. Isotope values were determined by elemental analysis, followed by isotope ratio mass spectrometry (with a Thermo Flash 2000 elemental analyzer coupled to a Thermo DeltaV information request management system). Lipid extraction of the subsample for compound-specific analysis was carried out as described previously (40). In short, after saponification the obtained extracts were methylated with boron trifluoride (BF3) in methanol and subsequently separated into apolar and polar fractions by column chromatography over activated alumina (Al2O3). Previous analysis had revealed that the apolar fraction comprised the majority of the total lipids (40). The apolar fractions were subsequently analyzed by GC (Agilent 6890) and GC-MS (with a Thermo Trace GC Ultra GC coupled to a Thermo Trace DSQ MS). The GC was equipped with a fused-silica column (25 m by 0.32 mm) coated with CP Sil-5 (film thickness = 0.12 μm), and He was the carrier gas. The relative abundance of the fatty acid methyl esters was derived from the integrated GC profile. Compounds were identified by GC-MS following methods described previously (40). For stable carbon isotope analysis, fractions were analyzed by GC coupled to isotope ratio mass spectrometry as described previously (41). Raw data were corrected for the δ13C of the carbon derived from BF3-methanol that was added during preparation of the methyl esters of the fatty acids.
RESULTS
Genome and transcriptome analysis of C1 assimilation potential by “Ca. Methylomirabilis oxyfera.”
Detailed analysis of the previously published genome data (16) showed that neither the serine nor the RuMP pathway was complete in “Ca. Methylomirabilis oxyfera.” For both pathways, important key genes could not be annotated. In the RuMP pathway, 3-hexulose-6-phosphate synthase (HPS) and 6-phospho-3-hexuloisomerase (PHI) were missing. Malyl-CoA lyase, malate thiokinase, and glycerate 2-kinase were absent in the serine pathway.
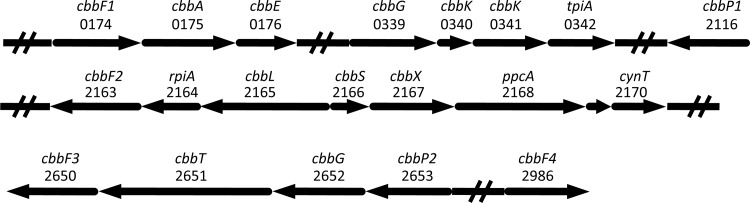
When we consider the six known autotrophic pathways, only the CBB cycle gene set was complete. Analysis of the previously published transcriptomic and proteomic data (16, 38) revealed that all CBB cycle genes were also significantly expressed in “Ca. Methylomirabilis oxyfera” bacteria. The relative expression of CBB cycle-associated genes and, for comparison, key catabolic genes is shown in Table 2. The CBB genes were arranged in four separate clusters (Fig. 1), with additional copies of a phosphoribulokinase (PRK) and a fructose-1,6-bisphosphatase (FBPase) not being located within a cbb operon.
TABLE 2.
Relative expression of CBB cycle-associated genes and (for comparison) key catabolic genes in “Ca. Methylomirabilis oxyfera”a
| Open reading frame identifier | Gene name | Encoded protein | ECb no. | Relative expressionc |
|---|---|---|---|---|
| damo_0174 | cbbF1 | Fructose-1,6-bisphosphatase II | 3.1.3.11 | 8.3 |
| damo_0175 | cbbA | Fructose-bisphosphate aldolase II, CBB cycle subtype | 4.1.2.13 | 9.8 |
| damo_0176 | cbbE | Ribulose-phosphate 3-epimerase | 5.1.3.1 | 8.8 |
| damo_0339 | cbbG1 | Glyceraldehyde 3-phosphate dehydrogenase | 1.2.1.12 | 9.4 |
| damo_0340 | cbbK | Phosphoglycerate kinase, 5′ end | 2.7.2.3 | 9.1 |
| damo_0341 | cbbK | Phosphoglycerate kinase, 3′ end | 2.7.2.3 | 8.3 |
| damo_0342 | tpiA | Triosephosphate isomerase | 5.3.1.1 | 9.0 |
| damo_0804 | oxyR | LysR-type transcriptional activator | 7.5 | |
| damo_0861 | oxyR | LysR-type transcriptional activator | 8.7 | |
| damo_2116 | cbbP1 | Phosphoribulokinase | 2.7.1.19 | 3.9 |
| damo_2163 | cbbF2 | Fructose-1,6-bisphosphatase II | 3.1.3.11 | 9.6 |
| damo_2164 | rpiA | Ribulose-5-phosphate isomerase A | 5.3.1.6 | 8.1 |
| damo_2165 | cbbL | Ribulose-1,6-bisphosphate carboxylase/oxygenase, large chain | 4.1.1.39 | 9.3 |
| damo_2166 | cbbS | Ribulose-1,6-bisphosphate carboxylase/oxygenase, small chain | 4.1.1.39 | 10.1 |
| damo_2167 | cbbX | Ribulose-1,6-bisphosphate carboxylase/oxygenase activase, AAA ATPase | 9.9 | |
| damo_2168 | ppcA | Phosphoenolpyruvate carboxylase | 4.1.1.31 | 9.3 |
| damo_2170 | cynT | Carbonic anhydrase | 4.2.1.1 | 10.3 |
| damo_2650 | cbbF3 | Fructose-1,6-bisphosphatase II | 3.1.3.11 | 9.6 |
| damo_2651 | cbbT | Transketolase | 2.2.1.1 | 7.9 |
| damo_2652 | cbbG2 | Glyceraldehyde-3-phosphate-dehydrogenase | 1.2.1.12 | 7.9 |
| damo_2653 | cbbP2 | Phosphoribulokinase | 2.7.1.19 | 9.9 |
| damo_2986 | cbbF4 | Fructose-1,6-bisphosphatase II | 3.1.3.11 | 9.1 |
| damo_0134 | mxaF3 | Methanol dehydrogenase, large subunit | 1.1.99.8 | 10.3 |
| damo_0136 | mxaJ3 | Hypothetical protein involved in methanol dehydrogenase | 1.1.99.8 | 8.7 |
| damo_0138 | mxaG3 | Cytochrome c1 involved in methanol dehydrogenase | 1.1.99.8 | 9.7 |
| damo_0454 | fae | Formaldehyde-activating enzyme | 4.3.−.− | 14.6 |
| damo_0455 | mtdB | Methylenetetrahydromethanopterine dehydrogenase | 1.5.1.− | 10.4 |
| damo_0456 | fwdD | Formylmethanofuran dehydrogenase | 1.2.99.5 | 10.6 |
| damo_0457 | fhcB | Formyltransferase/hydrolase complex, subunit β | 9.7 | |
| damo_0458 | fhcA | Formyltransferase/hydrolase complex, subunit α | 10.0 | |
| damo_0459 | fhcD | Formyltransferase/hydrolase complex, subunit ε | 10.0 | |
| damo_0460 | fhcC | Formyltransferase/hydrolase complex, subunit γ | 10.6 | |
| damo_0461 | mch | Methenyltetrahydromethanopterin cyclohydrolase | 3.5.4.27 | 8.3 |
| damo_0853 | fdhA2 | Formate dehydrogenase, subunit α | 1.2.1.2 | 8.1 |
| damo_0854 | fdhB2/C2 | Formate dehydrogenase, subunits β and γ | 1.6.99.5 | 8.0 |
| damo_1134 | fwdD2 | Formylmethanofuran dehydrogenase | 1.2.99.5 | 8.3 |
| damo_1135 | fhcB2 | Formyltransferase/hydrolase complex, subunit β, 5′ end | 7.5 | |
| damo_1136 | fhcB2 | Formyltransferase/hydrolase complex, subunit β, 3′ end | 8.3 | |
| damo_1852 | folD | 5,10-Methylene-tetrahydrofolate dehydrogenase; 5,10-methenyl-tetrahydrofolate cyclohydrolase | 1.5.1.5; 3.5.4.9 | 8.3 |
| damo_2415 | nirS | Nitrite reductase, cytochrome cd1 type | 1.7.2.1 | 12.0 |
| damo_2434 | norZ2 | Putative nitric oxide dismutase | 11.7 | |
| damo_2437 | norZ3 | Putative nitric oxide dismutase | 13.6 | |
| damo_2448 | pmoB | Particulate methane monooxygenase, subunit α | 1.14.13.25 | 11.4 |
| damo_2450 | pmoA | Particulate methane monooxygenase, subunit β | 1.14.13.25 | 12.3 |
| damo_2451 | pmoC | Particulate methane monooxygenase, subunit γ | 1.14.13.25 | 0.0 |
The transcriptome analysis was performed on the basis of data published previously (38). The presented data refer to the anoxic period.
EC, enzyme nomenclature designation.
Relative expression was determined from log2(RPKM + 1), where RPKM is reads per kilobase per million mapped reads.
FIG 1.
Gene clusters and single genes involved in or associated with the CBB cycle in “Ca. Methylomirabilis oxyfera.” The direction of transcription is indicated with arrows, and open reading frame identifiers are shown. The encoding genes were as follows: cbbF, fructose-1,6-bisphosphatase II; cbbA, fructose-bisphosphate aldolase II; cbbE, ribulose-phosphate 3-epimerase; cbbG, glyceraldehyde-3-phosphate dehydrogenase; cbbK, phosphoglycerate kinase; tpiA, triosephosphate isomerase; cbbP, phosphoribulokinase; rpiA, ribulose-5-phosphate isomerase A; cbbL, ribulose-1,6-bisphosphate carboxylase/oxygenase, large chain; cbbS, ribulose-1,6-bisphosphate carboxylase/oxygenase, small chain; cbbX, putative ribulose-1,6-bisphosphate carboxylase/oxygenase activase; ppcA, phosphoenolpyruvate carboxylase; cynT, carbonic anhydrase; cbbT, transketolase.
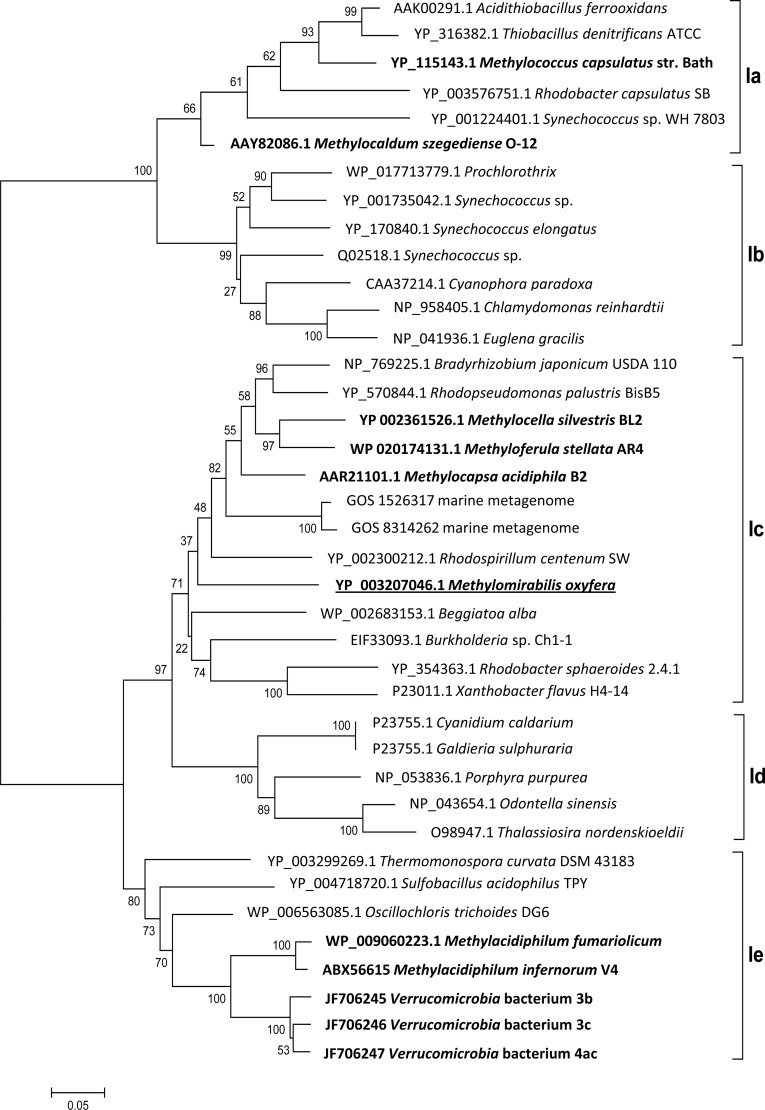
Two enzymes are exclusive to the CBB cycle: RubisCO and PRK (42). The large subunit of the “Ca. Methylomirabilis oxyfera” RubisCO belongs to the red-like type 1c (Fig. 2), indicating its proteobacterial origin and no close relatedness to verrucomicrobial sequences.
FIG 2.
Bootstrap consensus tree of selected RubisCO form I sequences calculated from 1,000 replicates. The evolutionary history was inferred using the neighbor-joining method, and the evolutionary distances were determined using the Dayhoff matrix. The scale bar represents the number of amino acid changes per site. The alignment gaps were eliminated in a pairwise comparison. RubisCO sequences of methanotrophs are highlighted in bold, and the sequence of “Ca. Methylomirabilis oxyfera” is underlined. The classification is based on a previous study (55), with types Ia and Ib belonging to the green-like RubisCOs and types Ic, Id, and Ie belonging to the red-like RubisCOs.
Additionally, two isoforms of an LysR-type transcriptional activator were identified in the genome of “Ca. Methylomirabilis oxyfera.” Many autotrophs regulate CBB cycle gene expression via the LysR-type transcriptional regulator designated CbbR (43). It was shown previously that CbbR is essential for the induction of autotrophy in Xanthobacter flavus (44–46). However, the cbbR-like genes of “Ca. Methylomirabilis oxyfera” are not located in the vicinity of any cbb cluster, and it remains to be shown whether the expressed LysR-type proteins are specifically involved in the regulation of the CBB cycle or other metabolic processes.
Two accessory elements are often associated with an active CBB cycle: the presence of CO2-concentrating microcompartments, commonly known as carboxysomes, and a photorespiration pathway employed to detoxify the 2-phosphoglycolate formed due to the oxygenase activity of RubisCO (47, 48). However, neither carboxysome-encoding genes nor carboxysome-like structures could be observed in “Ca. Methylomirabilis oxyfera” cells using electron microscopy (49), indicating a cytoplasmic CO2 fixation. Besides CO2, O2 is a competing substrate of RubisCO, which leads to a reduced CO2 fixation efficiency and the production of 2-phosphoglycolate (47), a toxic compound. The detoxification of phosphoglycolate eventually leads to the formation of 3-phosphoglycerate, which can reenter the CBB cycle. So far, three routes of the photorespiratory pathway are known: the C2 cycle, the glycerate pathway, and decarboxylation (50, 51). In each pathway, glycerate is the key intermediate, and steps leading to its formation are common to all. Glycerate is formed via glycolate from 2-phosphoglycolate, being catalyzed by a phosphoglycolate phosphatase and glycolate oxidase. No genes encoding a phosphoglycolate phosphatase were identified in “Ca. Methylomirabilis oxyfera”; however, the glycolate oxidase-encoding gene glcD is present and transcribed. Furthermore, except for a glycerate kinase catalyzing the last step, all other proteins involved in the C2 cycle are present in “Ca. Methylomirabilis oxyfera.”
RubisCO activity.
Cell extract preparations of a “Ca. Methylomirabilis oxyfera” enrichment culture exhibited an average RubisCO-specific activity of 0.2 nmol 13CO2 min−1 mg protein−1 (Fig. 3). This activity corresponded to approximately 10% of the methanotrophic activity of the culture at the time of the experiments. No activity was observed in incubations without addition of either RuBP or cell extract (data not shown). Interestingly, only freshly prepared cell extract exhibited the CO2-fixing activity; freezing or storing on ice for longer than 1 day resulted in a complete loss of activity.
FIG 3.
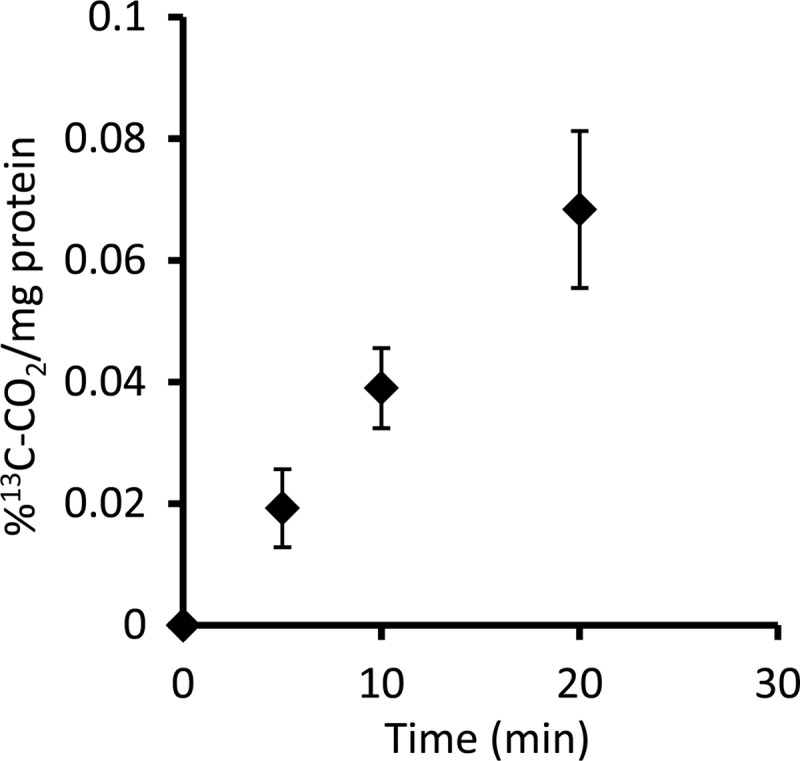
Increase of 13CO2 over time as an indication of specific RubisCO activity. The evolution of 13CO2 results from the destruction of labeled 3-phosphoglycerate. The activity test was performed with a whole-cell extract. The data presented originated from five replicates, and error bars show standard deviations.
Since the cell extract was prepared from an enrichment culture with approximately 20% other community members, we also investigated the transcriptome for reads mapping to a comprehensive RubisCO data set obtained from GenBank. Over 90% of the total RubisCO reads mapped to the “Ca. Methylomirabilis oxyfera” gene sequence, suggesting that the measured specific RubisCO activity could be attributed mainly to “Ca. Methylomirabilis oxyfera.”
13C labeling of bulk biomass and specific lipids.
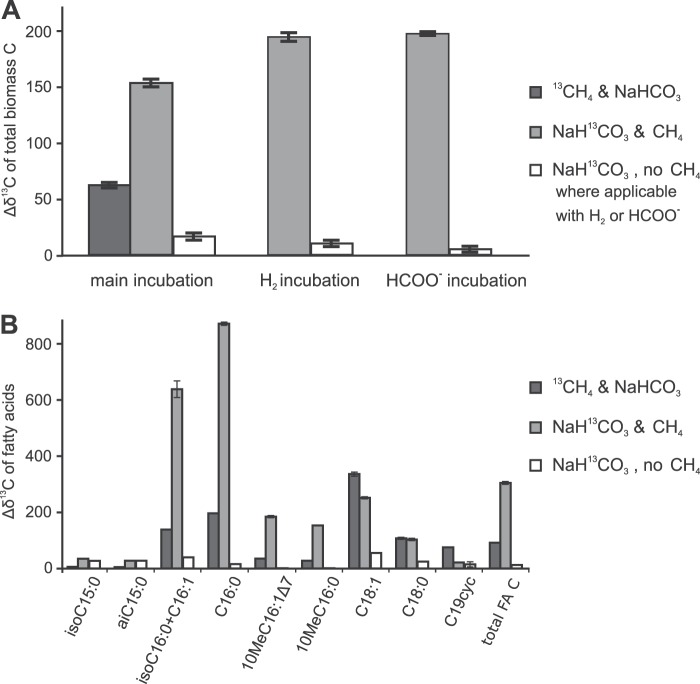
Bulk stable carbon isotope data showed that clearly more 13C was incorporated into the total biomass in the treatment with [13C]bicarbonate than in the treatment with [13C]methane (Fig. 4A). When CH4 was replaced by formate or H2 as an additional electron donor, the incorporation of 13C from [13C]bicarbonate was minimal. The absolute amount of 13C incorporation into biomass was estimated from the bulk isotope enrichment and the total organic carbon data, together with the overall amount of methane oxidation (1.3 to 1.6 nmol CH4 min−1 mg−1 protein), amounting to an average of 422 μg C oxidized per total batch incubated. By the end of the incubation, the [13C]bicarbonate treatment contained approximately 27 μg more 13C than the control treatment without any label (absolute amount per batch incubated). In the [13C]methane treatment, about 11 μg 13C was incorporated, which approximates the amount of C indirectly assimilated from methane via CO2. In total, approximately 38 μg of 13C was thus assimilated into the biomass of each batch over the course of the experiment, which amounts to about 9% of the amount of methane carbon oxidized. This correlates well with the observed specific RubisCO activity.
FIG 4.
Changes in δ13C (Δδ13C) in total biomass and fatty acids of “Ca. Methylomirabilis oxyfera” after incubation with 13C-labeled substrates relative to that in a control incubation without 13C labeling. (A) Changes in δ13C for the total biomass in the main incubations next to those for the additional treatments that were included to test the use of H2 and/or formate as additional electron donors; (B) changes in δ13C for the individual and total fatty acids for the main incubations. cyc, a cyclopropyl moiety; total FA C, weighted average of the change in δ13C in all fatty acids. The δ13C data for the various treatments are all averages of two experimental replicates, and the δ13C data for each experimental replicate are averages of multiple analytical replicates.
The compound-specific stable carbon isotopic analysis of fatty acids showed the same trend seen with the bulk carbon isotope enrichment, with the highest 13C enrichment seen when bicarbonate was 13C labeled in the presence of unlabeled methane (Fig. 4B). The three fatty acids C16:0, 10-methyl-C16:0 (10MeC16:0) and 10MeC16:1Δ7, which are of main interest because of their relative abundance and specificity (40), all followed the same trend seen for total 13C uptake. Some minor fatty acids showed a different response to the labeling: the fatty acids iso-C15 and ai-C15 became similarly enriched in 13C from [13C]bicarbonate, regardless of the presence of methane. For the fatty acids C18:1, C18:0, and C19cyc a higher degree of labeling was obtained from [13C]methane than from [13C]bicarbonate.
DISCUSSION
Based on the genome information, we followed the hypothesis that the CBB cycle would be the major route of C assimilation in “Ca. Methylomirabilis oxyfera.” Our labeling experiments (Fig. 4) now provide strong indications that “Ca. Methylomirabilis oxyfera” is an autotrophic organism, which was also recently shown for verrucomicrobial methanotrophs and which is in contrast to the findings for characterized proteobacterial methanotrophs. In the absence of CH4, the cultures did not incorporate significant amounts of 13C label, showing that CH4 is essential as an energy source, but the bulk stable carbon isotope analysis suggested that bicarbonate/CO2 is the carbon source for “Ca. Methylomirabilis oxyfera” cells (Fig. 4A). However, as the “Ca. Methylomirabilis oxyfera” biomass is available only in enrichment culture, the possibility remains that other community members may have contributed to the observed 13C incorporation. The compound-specific stable carbon isotope analysis provided the decisive data: “Ca. Methylomirabilis oxyfera”-specific fatty acids 10MeC16:0 and 10MeC16:1Δ7 (40) showed a clearly higher 13C content after incubation with 13C-labeled bicarbonate (Fig. 4) than after incubation with 13C-labeled methane, confirming that bicarbonate/CO2 was indeed the carbon source for cell material of “Ca. Methylomirabilis oxyfera.” Although some labeling was observed in the incubations with 13CH4 and unlabeled bicarbonate/CO2, its extent was clearly lower than that in the incubations with [13C]bicarbonate and unlabeled methane. This labeling signal was most probably caused by the indirect incorporation of labeled CO2 originating from labeled methane oxidation (i.e., scrambling). Additionally, the labeling results for a minor fatty acid fraction also indicated the presence of other CO2-fixing community members (iso-C15 and ai-C15 fatty acids became similarly enriched by [13C]bicarbonate with and without methane) and chemoorganoheterotrophic methanotrophs (C18:1, C18:0, and C19cyc fatty acids became more enriched in 13C by [13C]methane than by [13C]bicarbonate).
At the time of the earliest nitrite-dependent methane-oxidizing enrichment culture, a labeling experiment with [13C]methane showed that after 3 to 6 days of incubation the 13C label was indeed incorporated into bacterial lipids (52). However, the anticipated biomarker lipid 10MeC16:0 fatty acid did not become significantly enriched and the 13C content of the 10MeC16:1Δ7 fatty acid could not be determined due to its low abundance and coelution (52). This could be caused, at least in part, by the slow growth of “Ca. Methylomirabilis oxyfera.” However, a carbon source other than methane can also explain this observation. This was not tested at that time; our data now provide strong indications that this alternative carbon source is CO2.
The test for the specific activity of RubisCO with cell extracts of “Ca. Methylomirabilis oxyfera” confirmed that “Ca. Methylomirabilis oxyfera” indeed exhibited CO2-fixing activity. The measured activity rate may seem low, even when it is compared to the 2-week doubling time of “Ca. Methylomirabilis oxyfera” reported previously (53). However, this doubling time was observed during the initial enrichment period and represents exponential growth of “Ca. Methylomirabilis oxyfera.” In the current study, the culture exhibited constant steady-state activity, indicating that it was probably close to stationary phase, during which enzyme expression and specific activity are expected to be lower than they are during exponential growth phase. Moreover, the observed specific RubisCO activity compared well with the estimated C assimilation activity determined on the basis of our 13C labeling experiments.
The genomic information and the absence of carboxysome-like structures (46) suggest that the CO2 fixation in “Ca. Methylomirabilis oxyfera” is not carboxysomal but, rather, is cytoplasmic. Carboxysomal CO2 fixation probably evolved in order to increase the rates in the presence of low ambient CO2 concentrations and to minimize photorespiration (51). As in the environmental niche of “Ca. Methylomirabilis oxyfera” CO2 concentrations are unlikely to be limiting and external oxygen is not present, carboxysomal CO2 fixation might not offer an advantage in comparison with cytoplasmic CO2 fixation. The incomplete photorespiration pathway also implies that photorespiration might not be relevant in “Ca. Methylomirabilis oxyfera”; however, the mechanism of the hypothesized internal oxygen metabolism is not yet known, and the possibility of internal RubisCO oxygen exposure cannot be ruled out.
The first methanotroph to be shown to possess the functional CBB cycle was M. capsulatus; however, the main route of carbon assimilation in this methanotroph was shown to be the RuMP pathway (9, 54). The autotrophic growth of M. capsulatus could not be achieved in liquid and could be achieved only poorly on solid medium with CO2 and H2 or formate (8). Some other proteobacterial methanotrophs were also shown to possess the complete machinery for an operational CBB cycle (10–12), but so far it remains unclear what role it might play in those methanotrophs. In contrast, it was recently shown that autotrophy is widespread among the newly discovered verrucomicrobial methanotrophs (20, 23). The finding that autotrophy might be a more common mode of C1 metabolism among methanotrophs has implications for the detection of methanotrophy and assessment of its significance in the environment. It implies that the stable carbon isotopic signature of these bacteria would not identify them as part of the methanotrophic community in situ and in conventional stable isotope studies. A recent report on methanotrophy in a geothermal soil demonstrated that labeling with [13C]methane, as commonly used in SIP studies targeting methanotrophs, failed in detecting the active methanotrophic community (23). Instead, a modified SIP method including [13C]bicarbonate/CO2 was necessary to successfully detect autotrophic methanotrophs (23). The same would apply to the detection of “Ca. Methylomirabilis oxyfera”-like methanotrophs in mesophilic, anoxic, and suboxic environments. Thus, “Ca. Methylomirabilis oxyfera” might have escaped detection by means of stable isotope analysis, and, more generally, the contribution of autotrophic methanotrophic bacteria to methane cycling might have been hitherto overlooked.
ACKNOWLEDGMENTS
We thank Mingliang Wu for help with cell extract and SDS-PAGE techniques, Huub Op den Camp for providing a cbbL core alignment, Claudia Lüke for access to methanotroph genome data, and Margaret Butler for the initial genome annotation.
O.R. and M.S.M.J. were supported by ERC advanced grants (232937 and 339880), and D.M.K. and K.F.E. were supported by the Darwin Center for Biogeosciences (project 142.16.3071). M.S.M.J. and J.S.S.D. were supported by gravitation grant SIAM (24002002).
Footnotes
Published ahead of print 7 February 2014
REFERENCES
- 1.Cicerone RJ, Oremland RS. 1988. Biogeochemical aspects of atmospheric methane. Global Biogeochem. Cycles 2:299–327. 10.1029/GB002i004p00299 [DOI] [Google Scholar]
- 2.Hanson RS, Hanson TE. 1996. Methanotrophic bacteria. Microbiol. Rev. 60:439–471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Whittenbury R, Phillips KC, Wilkinson JF. 1970. Enrichment, isolation and some properties of methane-utilizing bacteria. J. Gen. Microbiol. 61:205–218. 10.1099/00221287-61-2-205 [DOI] [PubMed] [Google Scholar]
- 4.Anthony C. 1986. Bacterial oxidation of methane and methanol. Adv. Microb. Physiol. 27:113–210. 10.1016/S0065-2911(08)60305-7 [DOI] [PubMed] [Google Scholar]
- 5.Peyraud R, Kiefer P, Christen P, Massou S, Portais J-C, Vorholt JA. 2008. Demonstration of the ethylmalonyl-CoA pathway by using 13C metabolomics. Proc. Natl. Acad. Sci. U. S. A. 106:4846–4851. 10.1073/pnas.0810932106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Matsen JB, Yang S, Stein LY, Beck D, Kalyuzhnaya MG. 2013. Global molecular analyses of methane metabolism in methanotrophic alphaproteobacterium, Methylosinus trichosporium OB3b. Part I. Transcriptomic study. Front. Microbiol. Chem. 4:1–16. 10.3389/fmicb.2013.00040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yang S, Matsen JB, Konopka M, Green-Saxena A, Clubb J, Sadilek M, Orphan VJ, Beck D, Kalyuzhnaya MG. 2013. Global molecular analyses of methane metabolism in methanotrophic Alphaproteobacterium, Methylosinus trichosporium OB3b. Part II. Metabolomics and 13C-labeling study. Front. Microbiol. Chem. 4:1–13. 10.3389/fmicb.2013.00070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Baxter NJ, Hirt RP, Bodrossy L, Kovacs KL, Embley MT, Prosser JI, Murrell CJ. 2002. The ribulose-1,5-bisphosphate carboxylase/oxygenase gene cluster of Methylococcus capsulatus (Bath). Arch. Microbiol. 177:279–289. 10.1007/s00203-001-0387-x [DOI] [PubMed] [Google Scholar]
- 9.Taylor SC, Dalton H, Dow CS. 1981. Ribulose-1,5-bisphosphate carboxylase/oxygenase and carbon assimilation in Methylococcus capsulatus (Bath). J. Gen. Microbiol. 122:89–94 [Google Scholar]
- 10.Chen Y, Crombie A, Rahman MT, Dedysh SN, Liesack W, Stott MB, Alam M, Theisen AR, Murrell JC, Dunfield PF. 2010. Complete genome sequence of the aerobic facultative methanotroph Methylocella silvestris BL2. J. Bacteriol. 192:3840–3841. 10.1128/JB.00506-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dedysh SN, Khmelenina VN, Suzina NE, Trotsenko YA, Semrau JD, Liesack W, Tiedje JM. 2002. Methylocapsa acidiphila gen. nov., sp. nov., a novel methane-oxidizing and dinitrogen-fixing acidophilic bacterium from Sphagnum bog. Int. J. Syst. Evol. Microbiol. 52(Pt 1):251–261 [DOI] [PubMed] [Google Scholar]
- 12.Vorobev AV, Baani M, Doronina NV, Brady AL, Liesack W, Dunfield PF, Dedysh SN. 2011. Methyloferula stellata gen. nov., sp. nov., an acidophilic, obligately methanotrophic bacterium that possesses only a soluble methane monooxygenase. Int. J. Syst. Evol. Microbiol. 61:2456–2463. 10.1099/ijs.0.028118-0 [DOI] [PubMed] [Google Scholar]
- 13.Ward N, Larsen Ø, Sakwa J, Bruseth L, Khouri H, Durkin AS, Dimitrov G, Jiang L, Scanlan D, Kang KH, Lewis M, Nelson KE, Methé B, Wu M, Heidelberg JF, Paulsen IT, Fouts D, Ravel J, Tettelin H, Ren Q, Read T, DeBoy RT, Seshadri R, Salzberg SL, Jensen HB, Birkeland NK, Nelson WC, Dodson RJ, Grindhaug SH, Holt I, Eidhammer I, Jonasen I, Vanaken S, Utterback T, Feldblyum TV, Fraser CM, Lillehaug JR, Eisen JA. 2004. Genomic insights into methanotrophy: the complete genome sequence of Methylococcus capsulatus (Bath). PLoS Biol. 2:e303. 10.1371/journal.pbio.0020303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Trotsenko YA, Murrell JC. 2008. Metabolic aspects of aerobic obligate methanotrophy. Adv. Appl. Microbiol. 63:183–229. 10.1016/S0065-2164(07)00005-6 [DOI] [PubMed] [Google Scholar]
- 15.Dunfield PF, Yuryev A, Senin P, Smirnova AV, Stott MB, Hou SB, Ly B, Saw JH, Zhou ZM, Ren Y, Wang JM, Mountain BW, Crowe MA, Weatherby TM, Bodelier PLE, Liesack W, Feng L, Wang L, Alam M. 2007. Methane oxidation by an extremely acidophilic bacterium of the phylum Verrucomicrobia. Nature 450:879–882. 10.1038/nature06411 [DOI] [PubMed] [Google Scholar]
- 16.Ettwig KF, Butler MK, Le Paslier D, Pelletier E, Mangenot S, Kuypers MMM, Schreiber F, Dutilh BE, Zedelius J, de Beer D, Gloerich J, Wessels H, van Alen T, Luesken F, Wu ML, van de Pas-Schoonen KT, Op den Camp HJM, Janssen-Megens EM, Francoijs KJ, Stunnenberg H, Weissenbach J, Jetten MSM, Strous M. 2010. Nitrite-driven anaerobic methane oxidation by oxygenic bacteria. Nature 464:543–548. 10.1038/nature08883 [DOI] [PubMed] [Google Scholar]
- 17.Islam T, Jensen S, Reigstad LJ, Larsen O, Birkeland NK. 2008. Methane oxidation at 55°C and pH 2 by a thermoacidophilic bacterium belonging to the Verrucomicrobia phylum. Proc. Natl. Acad. Sci. U. S. A. 105:300–304. 10.1073/pnas.0704162105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pol A, Heijmans K, Harhangi HR, Tedesco D, Jetten MSM, Op den Camp HJM. 2007. Methanotrophy below pH 1 by a new Verrucomicrobia species. Nature 450:874–878. 10.1038/nature06222 [DOI] [PubMed] [Google Scholar]
- 19.Hou S, Makarova K, Saw J, Senin P, Ly B, Zhou Z, Ren Y, Wang J, Galperin M, Omelchenko M, Wolf Y, Yutin N, Koonin E, Stott M, Mountain B, Crowe M, Smirnova A, Dunfield P, Feng L, Wang L, Alam M. 2008. Complete genome sequence of the extremely acidophilic methanotroph isolate V4, Methylacidiphilum infernorum, a representative of the bacterial phylum Verrucomicrobia. Biol. Direct 3:26. 10.1186/1745-6150-3-26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Khadem AF, Pol A, Wieczorek A, Mohammadi SS, Francoijs KJ, Stunnenberg HG, Jetten MSM, Op den Camp HJM. 2011. Autotrophic methanotrophy in Verrucomicrobia: Methylacidiphilum fumariolicum SolV uses the Calvin-Benson-Bassham cycle for carbon dioxide fixation. J. Bacteriol. 193:4438–4446. 10.1128/JB.00407-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wu ML, Ettwig KF, Jetten MSM, Strous M, Keltjens JT, van Niftrik L. 2011. A new intra-aerobic metabolism in the nitrite-dependent anaerobic methane-oxidizing bacterium Candidatus ‘Methylomirabilis oxyfera.' Biochem. Soc. Trans. 39:243–248. 10.1042/BST0390243 [DOI] [PubMed] [Google Scholar]
- 22.Ettwig KF, Speth DR, Reimann J, Wu ML, Jetten MSM, Keltjens JT. 2012. Bacterial oxygen production in the dark. Front. Microbiol. 3:273. 10.3389/fmicb.2012.00273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sharp C, Stott M, Dunfield P. 2012. Detection of autotrophic verrucomicrobial methanotrophs in a geothermal environment using stable isotope probing. Front. Microbiol. 3:303. 10.3389/fmicb.2012.00303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Blumenberg M, Seifert R, Reitner J, Pape T, Michaelis W. 2004. Membrane lipid patterns typify distinct anaerobic methanotrophic consortia. Proc. Natl. Acad. Sci. U. S. A. 101:11111–11116. 10.1073/pnas.0401188101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Coolen MJL, Hopmans EC, Rijpstra WIC, Muyzer G, Schouten S, Volkman JK, Sinninghe Damsté JS. 2004. Evolution of the methane cycle in Ace Lake (Antarctica) during the Holocene: response of methanogens and methanotrophs to environmental change. Org. Geochem. 35:1151–1167. 10.1016/j.orggeochem.2004.06.009 [DOI] [Google Scholar]
- 26.Deines P, Bodelier PL, Eller G. 2007. Methane-derived carbon flows through methane-oxidizing bacteria to higher trophic levels in aquatic systems. Environ. Microbiol. 9:1126–1134. 10.1111/j.1462-2920.2006.01235.x [DOI] [PubMed] [Google Scholar]
- 27.Freeman KH, Hayes JM, Trendel J-M, Albrecht P. 1990. Evidence from carbon isotope measurements for diverse origins of sedimentary hydrocarbons. Nature 343:254–256. 10.1038/343254a0 [DOI] [PubMed] [Google Scholar]
- 28.Hinrichs K-U, Summons RE, Orphan V, Sylva SP, Hayes JM. 2000. Molecular and isotopic analysis of anaerobic methane-oxidizing communities in marine sediments. Org. Geochem. 31:1685–1701. 10.1016/S0146-6380(00)00106-6 [DOI] [Google Scholar]
- 29.Orphan VJ, House CH, Hinrichs K-U, McKeegan KD, DeLong EF. 2001. Methane-consuming archaea revealed by directly coupled isotopic and phylogenetic analysis. Science 293:484–487. 10.1126/science.1061338 [DOI] [PubMed] [Google Scholar]
- 30.Cébron A, Bodrossy L, Chen Y, Singer AC, Thompson IP, Prosser JI, Murrell JC. 2007. Identity of active methanotrophs in landfill cover soil as revealed by DNA-stable isotope probing. FEMS Microbiol. Ecol. 62:12–23. 10.1111/j.1574-6941.2007.00368.x [DOI] [PubMed] [Google Scholar]
- 31.Dumont MG, Pommerenke B, Casper P, Conrad R. 2011. DNA-, rRNA- and mRNA-based stable isotope probing of aerobic methanotrophs in lake sediment. Environ. Microbiol. 13:1153–1167. 10.1111/j.1462-2920.2010.02415.x [DOI] [PubMed] [Google Scholar]
- 32.Hutchens E, Radajewski S, Dumont MG, McDonald IR, Murrell JC. 2004. Analysis of methanotrophic bacteria in Movile Cave by stable isotope probing. Environ. Microbiol. 6:111–120. 10.1046/j.1462-2920.2003.00543.x [DOI] [PubMed] [Google Scholar]
- 33.Qiu Q, Noll M, Abraham W-R, Lu Y, Conrad R. 2008. Applying stable isotope probing of phospholipid fatty acids and rRNA in a Chinese rice field to study activity and composition of the methanotrophic bacterial communities in situ. ISME J. 2:602–614. 10.1038/ismej.2008.34 [DOI] [PubMed] [Google Scholar]
- 34.Ettwig KF, Shima S, Van De Pas-Schoonen KT, Kahnt J, Medema MH, Op Den Camp HJM, Jetten MSM, Strous M. 2008. Denitrifying bacteria anaerobically oxidize methane in the absence of Archaea. Environ. Microbiol. 10:3164–3173. 10.1111/j.1462-2920.2008.01724.x [DOI] [PubMed] [Google Scholar]
- 35.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28:2731–2739. 10.1093/molbev/msr121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thompson JD, Higgins DG, Gibson TJ. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673–4680. 10.1093/nar/22.22.4673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Saitou N, Nei M. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406–425 [DOI] [PubMed] [Google Scholar]
- 38.Luesken FA, Wu ML, Op den Camp HJM, Keltjens JT, Stunnenberg H, Francoijs KJ, Strous M, Jetten MSM. 2012. Effect of oxygen on the anaerobic methanotroph ‘Candidatus Methylomirabilis oxyfera': kinetic and transcriptional analysis. Environ. Microbiol. 14:1024–1034. 10.1111/j.1462-2920.2011.02682.x [DOI] [PubMed] [Google Scholar]
- 39.Sleat R, Mah RA. 1984. Quantitative method for colorimetric determination of formate in fermentation media. Appl. Environ. Microbiol. 47:884–885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kool DM, Zhu BL, Rijpstra WIC, Jetten MSM, Ettwig KF, Sinninghe Damsté JS. 2012. Rare branched fatty acids characterize the lipid composition of the intra-aerobic methane oxidizer “Candidatus Methylomirabilis oxyfera.” Appl. Environ. Microbiol. 78:8650–8656. 10.1128/AEM.02099-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schouten S, Klein Breteler WCM, Blokker P, Schogt N, Rijpstra WIC, Grice K, Baas M, Sinninghe Damsté JS. 1998. Biosynthetic effects on the stable carbon isotopic compositions of algal lipids: implications for deciphering the carbon isotopic biomarker record. Geochim. Cosmochim. Acta 62:1397–1406. 10.1016/S0016-7037(98)00076-3 [DOI] [Google Scholar]
- 42.Tabita FR. 1988. Molecular and cellular regulation of autotrophic carbon dioxide fixation in microorganisms. Microbiol. Rev. 52:155–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gibson JL, Tabita FR. 1993. Nucleotide sequence and functional analysis of cbbR, a positive regulator of the Calvin cycle operons of Rhodobacter sphaeroides. J. Bacteriol. 175:5778–5784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Meijer WG, van den Bergh ER, Smith LM. 1996. Induction of the gap-pgk operon encoding glyceraldehyde-3-phosphate dehydrogenase and 3-phosphoglycerate kinase of Xanthobacter flavus requires the LysR-type transcriptional activator CbbR. J. Bacteriol. 178:881–887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.van den Bergh ER, Dijkhuizen L, Meijer WG. 1993. CbbR, a LysR-type transcriptional activator, is required for expression of the autotrophic CO2 fixation enzymes of Xanthobacter flavus. J. Bacteriol. 175:6097–6104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.van Keulen G, Ridder ANJA, Dijkhuizen L, Meijer WG. 2003. Analysis of DNA binding and transcriptional activation by the LysR-type transcriptional regulator CbbR of Xanthobacter flavus. J. Bacteriol. 185:1245–1252. 10.1128/JB.185.4.1245-1252.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bowes G, Ogren WL, Hageman RH. 1971. Phosphoglycolate production catalyzed by ribulose diphosphate carboxylase. Biochem. Biophys. Res. Commun. 45:716–722. 10.1016/0006-291X(71)90475-X [DOI] [PubMed] [Google Scholar]
- 48.Yeates TO, Kerfeld CA, Heinhorst S, Cannon GC, Shively JM. 2008. Protein-based organelles in bacteria: carboxysomes and related microcompartments. Nat. Rev. Microbiol. 6:681–691. 10.1038/nrmicro1913 [DOI] [PubMed] [Google Scholar]
- 49.Wu ML, van Teeseling MCF, Willems MJR, van Donselaar EG, Klingl A, Rachel R, Geerts WJC, Jetten MSM, Strous M, van Niftrik L. 2012. Ultrastructure of the denitrifying methanotroph “Candidatus Methylomirabilis oxyfera,” a novel polygon-shaped bacterium. J. Bacteriol. 194:284–291. 10.1128/JB.05816-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Eisenhut M, Ruth W, Haimovich M, Bauwe H, Kaplan A, Hagemann M. 2008. The photorespiratory glycolate metabolism is essential for cyanobacteria and might have been conveyed endosymbiontically to plants. Proc. Natl. Acad. Sci. U. S. A. 105:17199–17204. 10.1073/pnas.0807043105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zarzycki J, Axen SD, Kinney JN, Kerfeld CA. 2013. Cyanobacterial-based approaches to improving photosynthesis in plants. J. Exp. Bot. 64:787–798. 10.1093/jxb/ers294 [DOI] [PubMed] [Google Scholar]
- 52.Raghoebarsing AA, Pol A, van de Pas-Schoonen KT, Smolders AJP, Ettwig KF, Rijpstra WIC, Schouten S, Sinninghe Damsté JS, Op den Camp, HJM. Jetten MSM, Strous M. 2006. A microbial consortium couples anaerobic methane oxidation to denitrification. Nature 440:918–921. 10.1038/nature04617 [DOI] [PubMed] [Google Scholar]
- 53.Ettwig KF, van Alen T, van de Pas-Schoonen KT, Jetten MSM, Strous M. 2009. Enrichment and molecular detection of denitrifying methanotrophic bacteria of the NC10 phylum. Appl. Environ. Microbiol. 75:3656–3662. 10.1128/AEM.00067-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Taylor SC. 1977. Evidence for the presence of ribulose-1,5-bisphosphate carboxylase and phosphoribulokinase in Methylococcus capsulatus (Bath). FEMS Microbiol. Lett. 2:305–307. 10.1111/j.1574-6968.1977.tb00966.x [DOI] [Google Scholar]
- 55.Tabita FR. 1995. The biochemistry and metabolic regulation of carbon metabolism and CO2 fixation in purple bacteria, p 885–914 In Blankenship RE, Madigan MT, Bauer CE. (ed), Anoxygenic photosynthetic bacteria, vol 2 Springer, Dordrecht, The Netherlands [Google Scholar]