Abstract
Many pharmaceuticals and personal care products (PPCPs) have been shown to be biotransformed in water treatment systems. However, little research exists on the effect of initial PPCP concentration on PPCP biotransformation or on the microbial communities treating impacted water. In this study, biological PPCP removal at various concentrations was assessed using laboratory columns inoculated with wastewater treatment plant effluent. Pyrosequencing was used to examine microbial communities in the columns and in soil from a soil aquifer treatment (SAT; a method of water treatment prior to reuse) site. Laboratory columns were supplied with different concentrations (0.25, 10, 100, or 1,000 μg liter−1) of each of 15 PPCPs. Five PPCPs (4-isopropyl-3-methylphenol [biosol], p-chloro-m-xylenol, gemfibrozil, ketoprofen, and phenytoin) were not removed at any tested concentrations. Two PPCPs (naproxen and triclosan) exhibited removals independent of PPCP concentration. PPCP removal efficiencies were dependent on initial concentrations for biphenylol, p-chloro-m-cresol, chlorophene, diclofenac, 5-fluorouracil, ibuprofen, and valproic acid, showing that PPCP concentration can affect biotransformation. Biofilms from sand samples collected from the 0.25- and 10-μg liter−1 PPCP columns were pyrosequenced along with SAT soil samples collected on three consecutive days of a wetting and drying cycle to enable comparison of these two communities exposed to PPCPs. SAT communities were similar to column communities in taxonomy and phylotype composition, and both were found to contain close relatives of known PPCP degraders. The efficiency of biological removal of PPCPs was found to be dependent on the concentration at which the contamination occurs for some, but not all, PPCPs.
INTRODUCTION
The world's expanding population and increasing demand for water necessitate establishment of sustainable water resources if severe shortages in the future are to be avoided. Various water recycling methods to increase potable and nonpotable water supplies have been developed and are in use around the world, but the public's acceptance for water recycling is diminished by reports of pathogens, toxic chemicals, disinfection by-products, and micropollutants, including pharmaceuticals and personal care products (PPCPs), persisting through treatment processes (1, 2). Concern over PPCPs in particular is engendered by studies describing their toxicological effects, such as one in which the growth of human embryonic kidney cells was inhibited after exposure to a low-concentration mixture of pharmaceuticals (3). Recent research has focused on the removal of these trace compounds from water as researchers aim to prevent potential negative effects on exposed ecosystems and nontarget organisms, and often biotransformation is found to be a major removal mechanism (4). Better understanding of the biological removal processes occurring during water recycling would provide insight into best management practices for optimal removal of micropollutants and raise public confidence in the treatment processes.
One water recycling method that is currently used in the southwestern United States and other arid locations around the world is a method of managed aquifer recharge known as soil aquifer treatment (SAT), in which wastewater treatment plant (WWTP) effluent is applied to a spreading basin and is allowed to infiltrate. It travels through less than 1 m of biologically active soil, percolates through 3 to 30 m of the vadose zone, and finally remains in the underlying aquifer for anywhere from 6 months to 10 years. Water can then be pumped from the aquifer for further use (5). Several factors are thought to influence the biotransformation of PPCPs during SAT, including effluent pretreatment, redox conditions, organic carbon concentration, wetting and drying cycles, and the composition of the microbial community (6).
PPCPs detected in aquatic environments during surface water and groundwater occurrence studies are often reported at ng liter−1concentrations (7–10), yet they have been detected in the μg liter−1 range in WWTP effluent. WWTP effluent is often the source of these micropollutants to the environment, and literature concentration values for the PPCPs included herein can vary significantly (Table 1). Many of the PPCP biotransformation studies in the literature were carried out at higher concentrations than are commonly observed in the environment due to analytical sensitivity limitations, provision of the PPCP as the sole carbon and energy sources, and adherence to published biodegradation study protocols (e.g., Organisation for Economic Co-operation and Development [OECD] [11]). PPCP initial experimental concentrations greater than or equal to 1 mg liter−1 (even as high as 1.86 g liter−1) are not uncommon (12–17). But with the wide range of concentrations found in the environment typically being less than 1 μg liter−1, it is unclear whether studies at higher concentrations can be extrapolated to predict the fate of PPCPs at environmentally relevant concentrations accurately. This study investigates the hypothesis that the starting concentration of a PPCP substrate affects its biotransformation. Increasing the concentration of a PPCP substrate supplied to a microorganism that uses the PPCP as a carbon and energy source would be expected to increase microbial growth, assuming the PPCP does not exert any toxic effect on the microorganism. However, a PPCP that is being degraded as a secondary substrate by cometabolism would not exhibit increased growth as a result of an increase in PPCP concentration.
TABLE 1.
Names, compound classifications, and WWTP effluent concentrations from the literature for the PPCPs included in this study
| PPCP | Classification | Literature WWTP effluent concentration(s) (ng liter−1) |
|---|---|---|
| Biosol | Antiseptic | 250a |
| Biphenylol | Antiseptic | 900a |
| p-Chloro-m-cresol | Antiseptic | 600a |
| p-Chloro-m-xylenol | Antiseptic | 400,a <84–3, 010b |
| Chlorophene | Antiseptic | 750,a <26–39b |
| Diclofenac (sodium) | NSAIDh | 110,a 6–496,b 211–486,c 120,d 80–290,e 599,f 187–855g |
| 5-Fluorouracil | Anticancer | NDa,i |
| Gabapentin | Anticonvulsant | 110,a 1,786–42,611,b 1,860–4,620e |
| Gemfibrozil | Antilipemic | 1,200,a 22–1,081,c 330,d 37–155e |
| 185–5,714g | ||
| Ibuprofen | NSAID | 1,900,a 65–491,b 18–219,c 150,d <10–161,e 4,201,f NDg |
| Ketoprofen | NSAID | 1,200,a <3–37,b 22–1,081,c 330d |
| Naproxen | NSAID | 3,200,a <2–703,b 42–289,c 250,d 100–587,e ND-560g |
| Phenytoin | Anticonvulsant | 450,a 90–373e |
| Triclosan | Antiseptic | 800,a 13–82,b 160,d <10e |
| Valproic acid | Anticonvulsant | NDa |
The microbial community is instrumental in determining the biological fate of PPCPs during water recycling (18), though little is known about the identity of key players in biotransformation of PPCPs in situ. The community can be affected by a number of environmental factors during SAT, including wetting and drying cycles of the soil during normal SAT operation. In this study, we employed pyrosequencing to investigate the microbial communities that developed in the laboratory columns under two different PPCP concentration conditions and in a full-scale SAT system over a wetting and drying period. We compared column communities with SAT communities to gain insights into the similarities and differences in microbial communities capable of PPCP biotransformation at the laboratory and field scales and to survey potential PPCP degraders.
MATERIALS AND METHODS
Column setup and operation.
Five glass columns (2.5-cm internal diameter [i.d.] and 30-cm length) were packed with sand and operated in an upflow direction. Details of the columns, fittings, tubing, and column packing can be found in our previous work (19). All columns were sterilized by autoclaving on three consecutive days and were then inoculated by pumping WWTP final effluent through the columns in a closed loop for 13 h using a multichannel peristaltic pump. WWTP final effluent was collected from the Back River Wastewater Treatment Plant (Baltimore, MD, USA), which includes tertiary treatment, chlorination, and dechlorination as part of its treatment process. After inoculation, the columns were supplied with medium continuously for 343 days, with periodic adjustments to pump settings made to maintain column flow rates close to 0.1 ml/min.
The medium compositions for each column were identical (8.5 mg liter−1 KH2PO4, 22 mg liter−1 K2HPO4, 33 mg liter−1 Na2HPO4, 28 mg liter−1 CaCl2, 0.25 mg liter−1 FeCl3, 2.7 mg liter−1 NH4Cl, and 23 mg liter−1 MgSO4 in ultrapure water) except for the concentration of PPCPs, which was the experimental variable. Acetate (100 μg liter−1) was provided as the primary substrate due to its ready biodegradability, environmental relevance, and demonstrated utilization in PPCP-degrading biofilters (19). Each of the 15 PPCPs in the suite was supplied to each column at the concentrations listed in Table 2. Also, the abiotic control column received mercuric chloride from a 17.6-g liter−1 stock as a bacteriostatic agent. The suite of PPCPs studied is listed in Table 1, and details of chemical purchases can be found elsewhere (20). Though gabapentin was included in the influent medium, its results are not discussed due to analytical sensitivity issues for this particular compound. The procedure for making the medium is provided in the supplemental materials. The complete medium was filter sterilized (superhydrophilic polyethersulfone membranes, 0.2 μm; Pall Life Sciences, Port Washington, NY) prior to being pumped into the columns. During continuous operation of the columns, influent medium flasks were replaced approximately every 6 days.
TABLE 2.
Composition of media supplied to each of the experimental biofiltration columnsa
| Column | [PPCP] (μg liter−1) | [Acetate] (μg liter−1) | [HgCl2] (mg liter−1) |
|---|---|---|---|
| Abiotic | 10 | 100 | 10 |
| Active | 0.25 | 100 | 0 |
| Active | 10 | 100 | 0 |
| Active | 100 | 100 | 0 |
| Active | 1000 | 100 | 0 |
The listed PPCP concentrations represent the concentration of each of the 15 PPCPs added to the medium.
Influent and effluent column samples were collected and compared to determine the extent of biological removal of PPCPs in the columns. Details of column sample collection and PPCP analysis by gas chromatography-mass spectrometry (GC-MS) can also be found in the supplemental materials. PPCP data are reported as the fraction of the PPCP applied to the column influent that remains in the column effluent, which is calculated as the mean of triplicate effluent sample concentrations divided by the mean of triplicate influent samples. Confidence intervals (95%) were calculated as described previously (19) and represent the error associated with the calibration curve, which was found to be larger than the error associated with triplicate analysis.
For each compound, a one-way analysis of variance (ANOVA) test was performed using Microsoft Excel 2003 at an α value of 0.05 to determine if the mean removals observed from day 138 to day 326 were plausibly the same across different supplied PPCP concentrations or if at least two or more of the columns were significantly different from each other, suggesting dependence on PPCP concentration (21). Samples were analyzed in this way with just the four active columns to determine if they were statistically different and, if not, with all five columns to determine if those four were statistically different from the abiotic control column. PPCPs that showed significant differences among the active columns were further analyzed with a Tukey analysis at the 95% confidence level using R statistical software (22).
Column acetate biodegradation.
Biodegradation of the acetate supplied to the columns was assessed using radiolabeled 14C-acetate on days 338 to 341. Details of the method are discussed elsewhere (19). Briefly, on day 338 of column operation, radiolabeled 14C-sodium acetate (1,2-14C) was applied as a step input to three of the columns: abiotic, 100 μg liter−1 PPCPs, and 1,000 μg liter−1 PPCPs. The step input was provided as a spike of 3.4 μCi of radioactivity (135 μl of 25 μCi/ml stock solution) to 500 ml of the regularly prepared medium for each column. Since this led to an increase of 3.6 μg liter−1 acetate over normal medium acetate concentrations, columns not receiving the radiolabeled spike received a spike of unlabeled acetate to yield the same final medium acetate concentration. Columns were sampled for biodegradation of 14C-acetate on days 340 and 341. Total 14C samples were collected for 10 min in 1 M NaOH, and 14C-acetate samples collected for 10 min were acidified with concentrated HCl and sparged with air to remove 14CO2. All samples were combined with 9.0 ml Opti-Fluor liquid scintillation cocktail (PerkinElmer, Waltham, MA) prior to liquid scintillation counting on a Beckman model LS 3801 liquid scintillation counter (Fullerton, CA). Results are reported as disintegrations per minute per gram of aqueous sample. As a previous study showed that acetate was degraded in similar column systems (19), positive confirmation in two active columns was considered a sufficient measure of whether or not acetate was being removed in the other two active columns.
Additionally, dissolved oxygen was measured in the influent and effluent of each column on days 335 and 336 using a dissolved oxygen probe, and the pH of the influent and effluent was measured on day 332 following methods outlined elsewhere (19).
Column biomass determination.
After 343 days of continuous operation, column flow was stopped, and the columns were sectioned into the following segments (distance from the inlet): 0 to 2, 2 to 3, 11 to 13, 18 to 20, 27 to 28, 28 to 30 cm. Biomass concentration method details are found elsewhere (19). Briefly, a Micro BCA (bicinchoninic acid) assay kit (Thermo Fisher Scientific, Waltham, MA) was employed to determine the protein concentrations associated with each column sand sample. Protein concentrations were used as a surrogate for biomass present in that segment. Column sand samples were combined with sterile medium, vortexed, sonicated, and centrifuged, after which the supernatant was decanted for BCA analysis and subsequent measurement with a spectrophotometer. Protein concentrations were normalized to the weight of the dry sand from which it had been extracted, and results are reported in units of micrograms of protein per gram of sand.
Column tracer tests.
A tracer test using a pulse input of 3H2O was performed on all five columns between days 291 and 293. The average pore water velocity and the dispersion coefficient were calculated by fitting the experimental tracer curves to the convection dispersion equation using CXTFIT software (23). The hydraulic retention time was calculated by dividing the average pore water velocity by the length of the column (30 cm). The tracer test method is detailed elsewhere (19).
Column sampling for microbial community analysis.
Two columns (0.25 μg liter−1 and 10 μg liter−1 PPCPs) were selected for microbial community analysis. On day 343, the columns were aseptically disassembled and sectioned. Subsamples of each section designated for biomass analysis were homogenized by vigorous mixing to provide a single, representative sample for each column.
SAT site description.
The field site for this study was the Sweetwater Underground Storage and Recovery Facility in Tucson, AZ. The site contains eight infiltration basins, but a single basin, RB-1, was selected for sampling. This basin, which abuts the west bank of the Santa Cruz River, has been in operation since 1990. The basin itself has a depth of about 3.7 m and is filled to a depth of about 0.5 m during the wetting phase of operation. Though this basin has previously been operated with wet and dry cycles as long as 5 days wetting and 7 days drying (5), it has been operating on a shorter cycle in recent years. Before and during the soil sampling for this study, RB-1 was on a regular wetting and drying cycle that consisted of WWTP effluent water application for 12 h on 3 days each week (Sunday, Tuesday, and Thursday) and drying at all other times. The water supplied to the basin was chlorinated secondary effluent from the Roger Road Wastewater Treatment Plant (B. Prior and T. Miley, personal communication). The topmost layer of soil, from which soil samples were obtained, consists of gravel, sand, and silt alluvium. Further details of the site characteristics have been described previously (5).
SAT site sampling.
The WWTP effluent being applied to RB-1 was sampled for water quality characterization as it was being pumped into the basin on 2 November 2010. Grab samples of water were collected near the inlet to the basin and analyzed for concentrations of bromide, chloride, fluoride, nitrate as N, nitrite as N, orthophosphate as P, and sulfate by ion chromatography according to EPA method 300.0. The total organic carbon concentration was measured according to method SM 5310, and total Kjeldahl nitrogen (i.e., sum of organic nitrogen and ammonia nitrogen) was measured by MWH Labs (Monrovia, CA) according to EPA method 351.2. Dissolved oxygen, temperature, conductivity, and pH were also measured using a Hydrolab Quanta multiparameter sonde (Hach Hydromet, Loveland, CO).
Three soil samples from discrete locations within RB-1 were collected on each of three consecutive days from 3 to 5 November 2010 and were designated days 1 through 3. The day 1 samples were collected through overlying water because the application of effluent water to the basin had ceased only approximately 12 h prior to sampling, which was not sufficient time for all of it to infiltrate the soil. All of the overlying water had drained by the sampling times on the second and third days, so that soil moisture decreased with time. The basin would have been refilled on 4 November under normal operation, but the process was postponed until after the 5 November soil collection to accommodate the sampling campaign.
Samples were collected using a bulb planter sterilized between sampling with alcohol-soaked wipes. Samples were collected to a depth of 15 cm, and the plug of soil was transferred to a sterile, plastic sampling bag via a lever-operated release mechanism. The sample was immediately sealed and then stored in a cooler on ice packs during transport back to a laboratory freezer. Following the final sampling and freezing on the third day, the samples were shipped overnight in a cooler with ice packs to the JHU laboratory, where they were transferred to a −80°C freezer until further processing. After the samples were thawed in a refrigerator, samples collected from the three locations on each day were homogenized by vigorous mixing to yield a single, representative sample for each day of sampling.
DNA extraction and PCR.
DNA was extracted from the homogenized SAT soil and column sand samples using a PowerSoil DNA isolation kit (MoBio, Carlsbad, CA) according to the manufacturer's instructions. The mass of sample applied to each spin filter tube was normalized to both the moisture content and protein concentration (data not shown) of the samples. Extracted quadruplicate samples were pooled and concentrated using ethanol precipitation, and the DNA concentration was determined by spectrophotometer (NanoDrop, Wilmington, DE).
The 530-to-1100 region of the 16S RNA gene was amplified from each sample using primers containing Roche GS FLX Titanium primer sequences, library key sequences, and an eight-base, unique multiplex identifier (MID) sequence in each direction (see Table S1 in the supplemental material). PCRs (25-μl volume) were conducted with 5 ng extracted DNA, 400 nM each primer, 200 nM each deoxynucleoside triphosphate (dNTP), 2.5 μl of 10× FastStart buffer number 2, and 1.25 U FastStart high-fidelity PCR system polymerase (Roche, Nutley, NJ) as specified in the Roche GS FLX Titanium amplicon library preparation method manual. The PCR program was 3 min at 94°C, 30 cycles of 15 s at 94°C, 45 s at 58°C, and 60 s at 72°C, a final 8 min at 72°C, and holding at 4°C. Amplicon libraries were purified using Agencourt AMPure XP beads (5-ml kit from Beckman Coulter, Brea, CA). After quantitation using a Qubit fluorometer (Life Technologies, Carlsbad, CA), samples were pooled to achieve equal numbers of molecules for sequencing.
Pyrosequencing and sequencing analyses.
Pooled amplicons were sequenced using Titanium chemistry on a 454 FLX genome sequencer (Roche) in a two-region picotiter plate. The 454 System Sequencing Software generated SFF files for each plate region through the shotgun data processing pipeline. Sequences were sorted by MID and then recombined into individual FASTA files containing all sequences for each sample with the MID sequences removed.
The FASTA files were corrected for pyrosequencing errors by using Acacia software with default settings (24). Forward and reverse reads were pooled and assigned to phylotypes with a 97% identity threshold using CD-HIT (25). The most abundant read was chosen as the representative for each cluster. Sequence reads were aligned using MOTHUR software (26). Chimera Slayer software was used to identify and remove sequences that were possibly chimeric (27). Alpha and beta diversity metrics Chao1, Good's coverage, Shannon index, Simpson index, rarefaction curves, and UniFrac distances (28) were calculated using QIIME (29). The analyses were performed with bootstrapping (n = 1,000 [sample size]) below the size of the smallest library (9,000 reads) to avoid library-size-dependent artifacts. This resulted in negligible loss of diversity for the column samples and sufficient remaining diversity for elucidating community differences among the SAT samples. The 2.5 and 97.5 percentiles of ranked observed numbers were utilized for establishing a 95% confidence interval. Reads were assigned taxonomic affiliations using the RDP naive Bayesian classifier (30). A Mega BLAST search of the NCBI nucleotide database (nr/nt) over the length of the 16S rRNA gene amplicon was performed. The most closely related cultured and named representative to each abundant phylotype was identified (31).
Nucleotide sequence accession number.
All pyrosequencing sequences are stored in the NCBI Sequence Read Archive under accession no. SRX181218.
RESULTS
PPCP removal at different concentrations.
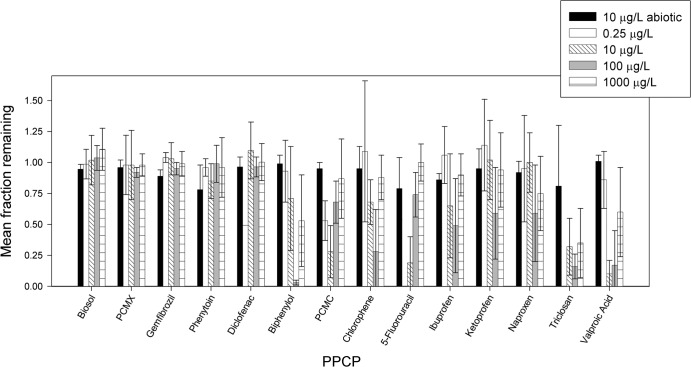
The mean removals for each PPCP supplied to the columns were calculated using column influent and effluent PPCP concentrations between days 138 and 326. Removals are shown for each sampling event (see Fig. S1 in the supplemental material) and as mean values between days 138 and 326 (Fig. 1). No data for the 0.25 μg liter−1 level of 5-fluorouracil or triclosan are included in Fig. 1 due to their nonattainment of the imposed quality control criterion of measured influent concentrations being within 50 to 150% of the supplied concentration (see the supplemental material). The minor losses of triclosan in the abiotic control column (Fig. 1; see also Fig. S1 in the supplemental material) were determined to be due to sorption to PharmMed ismaprene pump tubing (Ismatec, Switzerland), which was periodically replaced during the experiment as tubing became fatigued (data not shown). As a result, abiotic triclosan loss is assumed to have occurred to the same extent in the active columns.
FIG 1.
Mean removal values from day 138 through day 326 under each PPCP concentration condition. Error bars represent plus and minus one standard deviation.
ANOVA testing (α = 0.05) was performed to determine if there were any statistically significant differences in the mean PPCP removals among the different columns. P values (see Table S2 in the supplemental material) greater than 0.05 when comparing all five columns indicate that 4-isopropyl-3-methylphenol (biosol), p-chloro-m-xylenol, and phenytoin removals in the active columns neither varied in response to the concentration of the supplied PPCPs nor differed significantly from the removals demonstrated for the abiotic control column. The minor loss of gemfibrozil in the abiotic control column led to a P value of less than 0.05, but because the active column removals did not significantly differ from each other and were not less than that of the abiotic column, gemfibrozil is categorized with biosol, p-chloro-m-xylenol, and phenytoin. The variation of influent PPCP concentration from 0.25 to 1,000 μg liter−1 had no impact on the fraction of applied PPCP concentration that underwent biotransformation for these compounds. Furthermore, none of these compounds exhibited substantial removals in any of the columns. ANOVA testing revealed that triclosan and naproxen removals did not differ significantly among the active columns, but all of the active column removals were significantly greater than those in the abiotic control column. This suggests that these two PPCPs can be biotransformed, but the process is not appreciably influenced by the concentration at which the compounds are supplied. These six compounds (biosol, p-chloro-m-xylenol, phenytoin, gemfibrozil, triclosan, and naproxen) were not included in the subsequent Tukey analyses, as differences among the active columns were already shown not to be significant. Though the ANOVA results for ketoprofen suggested there was a significant difference in the active columns compared to the abiotic column, the subsequent Tukey analysis showed by individual comparisons that the removal in individual active columns was not significantly different than that found in the abiotic column (see Table S3 in the supplemental material).
Each of the other remaining PPCPs was well removed in at least one column across a majority of sample points, suggesting that microbes originating from the WWTP effluent can biotransform these compounds. P values (see Table S2 in the supplemental material) from ANOVA testing of the active columns suggest that the removals are not equivalent across all of the active columns for these PPCPs. For biphenylol, p-chloro-m-cresol, chlorophene, diclofenac, 5-fluorouracil, ibuprofen, and valproic acid, ANOVA testing revealed that all of the active columns had mean removals significantly greater than their abiotic control removal, but the active columns' mean removals were not all the same for each compound. A Tukey analysis was performed to test for differences among the active columns at the 95% confidence level (see Fig. S3 in the supplemental material), and resulting P values distinguished which differences in removals between columns were statistically significant. None of the PPCPs achieved its greatest removal in the 1,000-μg liter−1 column. Biphenylol, chlorophene, and ibuprofen were best removed in the 100-μg liter−1 column, though ibuprofen's removal in this column was not distinguishable from that in its 10-μg liter−1 column. 5-Fluorouracil was best removed at the 10-μg liter−1 level (though no data points were available for 0.25 μg liter−1). Valproic acid had its greatest removals in 10- and 100-μg liter−1 columns. p-Chloro-m-cresol had its greatest removals in the 10-μg liter−1 column, but this removal was not statistically different from that in the 0.25-μg liter−1 columns. Diclofenac had its greatest removal at 0.25 μg liter−1, though it should be noted that there was only one sample point for the 0.25-μg liter−1 concentration level that met the quality control requirements. Omitting that column from the ANOVA analyses shows that the mean removals of diclofenac are potentially equal among the abiotic and 10-, 100-, and 1,000-μg liter−1 columns. The mean PPCP concentrations remaining in the effluent of each column were compared to determine if, despite different fractions remaining in each column's effluent, the final concentrations were similar. For example, if the 1,000-μg liter−1 column had 5% of its PPCP influent concentration remaining and the 100-μg liter−1 column had 50% remaining, both columns would have 50 μg liter−1 of the PPCP in their effluent. This would suggest enzyme saturation or a threshold PPCP concentration below which microorganisms were not degrading the compounds. Also, the fraction of each PPCP remaining in each column's effluent was normalized to the mass of protein measured in each column (see below) and showed that differences in removal were not simply an effect of biomass concentrations (data not shown). Differences in PPCP removal in this study appear to be a function of the initial PPCP concentration.
Biomass results.
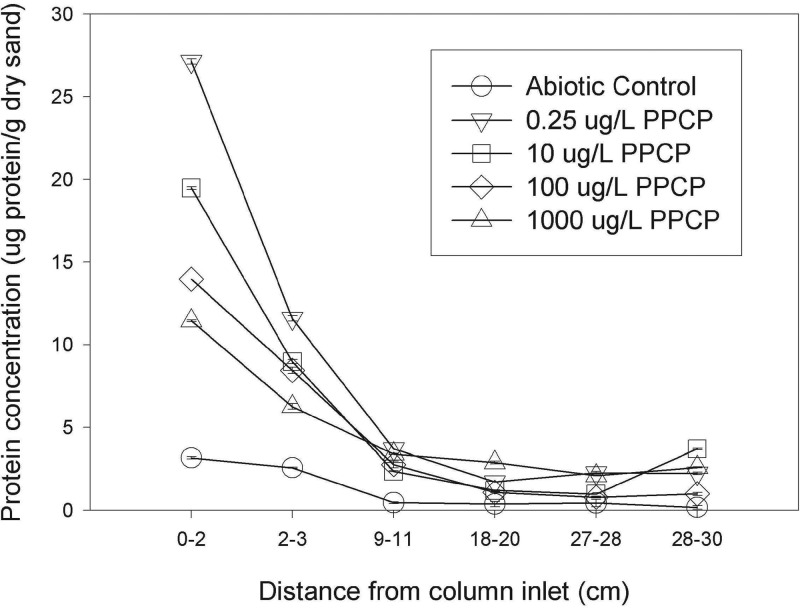
The protein concentrations associated with different segments of the columns were measured as a surrogate for biomass. Protein concentration measurements were made for six segments from each of the columns (Fig. 2). The abiotic control column shows a low-level protein concentration or minor medium interference with the assay. At every depth, the abiotic column's measured protein concentration was below those measured for the other columns, so its protein concentrations were considered to be a background level. Among the active columns, protein concentration was inversely related to the PPCP concentration supplied to the columns in the 3-cm column segments located closest to the inlets. This finding suggests that rather than serving as additional carbon and energy sources and thereby supporting more biomass growth, one or more of the PPCPs or a PPCP metabolite was suppressing biofilm growth.
FIG 2.
Protein concentrations in column segments. Columns were disassembled and subsampled at specified segment depths on day 343. Protein concentrations for the biomass associated with each segment are presented as micrograms of protein per gram of dry sand. Data shown are the means from duplicate assay measurements of the same column sample, and error bars represent the range of the two measurements. Nonvisible error bars mean that the bars are contained within the data marker.
Other measured indicators of biological growth were dissolved oxygen depletion and acetate biodegradation. Removal of dissolved oxygen during passage of the medium through the columns ranged from 47 to 69%, and biodegradation of 98 and 95% of supplied acetate was measured in the two active columns tested (100 and 1,000 μg liter−1, respectively) (see Table S4 and Results in the supplemental material). Measured pH values of influent (7.2 to 7.3) and effluent (6.6 to 6.8) were similar among the columns (see Results in the supplemental material). Results of the tritiated water tracer test can also be found in Table S5 and Results in the supplemental material.
SAT site influent water quality.
Concentrations of bromide, chloride, fluoride, nitrate as N, nitrite as N, orthophosphate as P, sulfate, total organic carbon, total Kjeldahl nitrogen (TKN), dissolved oxygen, temperature, pH, and conductivity measured in the water being pumped into the infiltration basin are shown in Table S6 in the supplemental material.
Microbial community analyses.
The numbers of sequences resulting from the pyrosequencing of the SAT soil samples and the laboratory column samples are shown in Table 3. A rarefaction curve was generated using an operational phylotype definition of 97% sequence similarity (see Fig. S2 in the supplemental material). Richness, evenness, and diversity were estimated for all column and SAT samples using library subsamples (9,000 sequences). Chao1, Good's coverage, Shannon index, and Simpson index estimates are summarized in Table 3 and are presented as 95% confidence interval ranges. Confidence intervals were based on rarefaction subsampling (1,000 resamples) below the size of the minimum library (9,000 reads).
TABLE 3.
Estimates of sample diversity, richness, and evennessa
| Sample | No. of sequences | No. of OTUs (full library) | No. of OTUs (subsample) | Chao1 richness | Good's coverage | Shannon index | Simpson index |
|---|---|---|---|---|---|---|---|
| SAT_day 1 | 9,911 | 837 | 792–813 | 1,163–1,297 | 0.9612–0.9613 | 6.459–6.500 | 0.947–0.949 |
| SAT_day 2 | 138,866 | 2,786 | 792–862 | 1,385–1,778 | 0.9512–0.9515 | 6.369–6.501 | 0.951–0.956 |
| SAT_day 3 | 158,532 | 3,019 | 812–881 | 1,420–1,815 | 0.9497–0.9500 | 6.200–6.337 | 0.942–0.948 |
| Column low PPCP | 30,848 | 285 | 175–198 | 225–347 | 0.9930–0.9931 | 4.494–4.577 | 0.915–0.919 |
| Column high PPCP | 147,470 | 469 | 196–222 | 246–367 | 0.9929–0.9930 | 5.209–5.299 | 0.944–0.948 |
The ranges shown are 95% confidence intervals. Libraries were subsampled below the size of the smallest library (9,000 reads) for diversity estimates in order to avoid sample-size-related biases. OTUs, operational taxonomic units.
A principal coordinates analysis (PCoA) was performed to assess the similarity of the communities in each sample based upon the weighted UniFrac distance metric (Fig. 3a). Confidence intervals (95%) for 9,000 sequence subsamples are represented by symbol size. The SAT site samples from all 3 days cluster closely, while there is greater distance between the two column samples. This suggests that the SAT site sample communities are more similar to each other than the two column sample communities are to each other. There is also significant UniFrac distance between the SAT samples and both column samples, suggesting that the two types of samples (column and SAT) are different from each other. This information is also shown by a plot (Fig. 3b) of the UniFrac distance metric values determined for the following groups of data: each SAT site compared to the other SAT sites; both columns compared to all SAT sites; columns compared to each other. The UniFrac distance metric is greatest for the column-SAT comparison (for both the samples from high- and low-PPCP-concentration reactors) and least for the SAT sample comparison.
FIG 3.
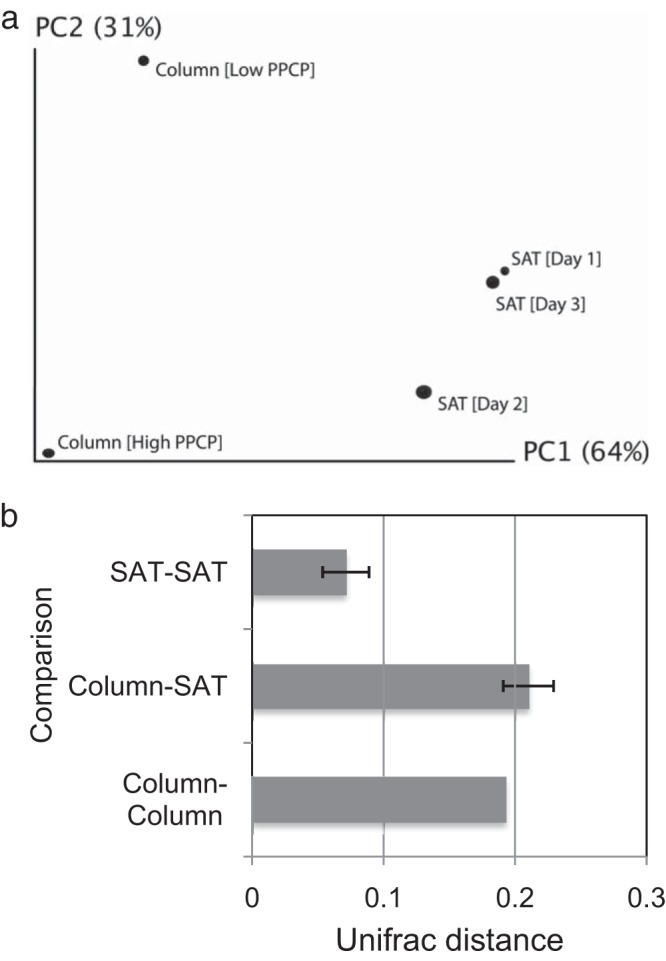
(a) Results of a principal coordinates analysis (PCoA) based on weighted UniFrac distances with 95% confidence intervals represented by the size of the symbol. (b) Community dissimilarity as represented by a plot of the UniFrac distance for three comparisons: SAT-SAT, column-SAT, and column-column. Error bars represent 95% confidence intervals.
Dominant taxa.
The microbial community composition of each of the samples was analyzed in terms of the relative abundance of taxa present (see Fig. S4 in the supplemental material). All samples were dominated by Proteobacteria, and this phylum was represented primarily by the genus Xanthobacter. Bacteriodetes were also well represented in all of the samples and were represented by Flavobacterium and genera belonging to the family Chitinophagaceae. The “Other” portion of each sample is composed of unclassified bacteria and the minute contributions (less than 0.1%) of the phyla OD1, WS3, SR1, Spirochaetes, OP11, Fusobacteria, BRC1, and Deinococcus-Thermus. The microbial communities were further analyzed at the 97% sequence similarity phylotype level, which is approximately species level and is the maximum level of resolution possible for the pyrosequencing method used (32). All sequences present in at least one sample at a relative abundance greater than or equal to 1% were evaluated using NCBI's GenBank to identify the most closely related named species listed in GenBank. These most abundant phylotypes are shown (see Table S7 in the supplemental material) with their consensus taxonomy, closest named species, percent identity to the closest named species, relative abundance (%) of the phylotype in each of the samples, and relative standard deviation (RSD) for the SAT samples. The RSD for the SAT samples across 3 days is shown graphically as well (see Fig. S4). Greater RSD values denote a larger variation in the population abundance over 3 days of SAT sampling.
DISCUSSION
Of the nine PPCPs that were biologically removed during passage through the laboratory columns, seven showed different removal efficiencies based upon the initial concentration at which the suite of PPCPs was supplied. With a mixture of PPCPs being supplied to the columns, it is not possible to determine whether a single compound, single metabolite, or a mixture is responsible for inhibiting biotransformation of the recalcitrant compounds. Another possibility is that the ratio of acetate to the combined concentration of PPCPs supplied to each column could have led to differences in the PPCP removal observed in each column. The highest concentration mixture tested did not favor the greatest removals for any of the compounds. This result suggests the possibility that poor PPCP biodegradability documented in the literature might be improved with lower initial PPCP concentrations. Cases where experiments using nonenvironmentally relevant, higher initial concentrations have predicted poor biodegradability in the environment might be worth reconsidering. As two of the compounds (triclosan and naproxen) were biotransformed in the active columns but were not removed to significantly different extents with various initial PPCP concentrations, the trend of increasing removal with decreasing substrate concentration would not be expected to apply to all PPCPs. A study of biodegradation of hormones estrone and 17-alpha-ethinylestradiol at environmentally relevant concentrations and at elevated concentrations showed that between initial concentrations of 0.03 to 10 μg liter−1, there was no significant difference in the biodegradation rates of the hormones, whereas an initial concentration of 100 μg liter−1 yielded a greater-than-3-fold decrease in biodegradation rate (33). Similar results were found in a continuous-flow system, in which the fraction of applied PPCP biodegraded decreased dramatically when the starting concentration was changed from 0.1 μg liter−1 to 100 μg liter−1 (33). Initial substrate concentrations have been shown to influence the fraction of compound biodegraded for non-PPCP compounds as well, such as pesticide isopropyl N-phenylcarbamate (34), pesticide benzonitrile (35), and herbicide intermediate and biocide 2,4,5-trichlorophenol (36).
The compounds most likely responsible for suppressing biofilm growth at higher concentrations as shown by the protein assay results are the antiseptics. Most data in the literature regarding MICs pertain to pure bacterial strains of medical concern, but these values can still provide an idea of the concentration range that would be toxic to the column bacteria. In Table S8 in the supplemental material, the MIC values for growth from the literature for triclosan, biphenylol, p-chloro-m-cresol, and p-chloro-m-xylenol are shown for four bacteria. For triclosan, the MICs ranged from 10 μg liter−1 to greater than 300,000 μg liter−1. Interestingly, the MIC of 10 μg liter−1 for Staphylococcus aureus is exceeded in two of our columns. Conversely, the ranges of reported MICs for biphenylol (1.0 × 105 to 1.0 × 106 μg liter−1), p-chloro-m-cresol (6.25 × 105 to 1.25 × 106), and p-chloro-m-xylenol (7.5 × 104 to 1.0 × 106) are all orders of magnitude higher than what was supplied to the columns. Yet, even if antiseptics are present below the MIC, they can still have deleterious effects. For instance, it has been found that triclosan can prevent cells from taking up necessary nutrients when supplied at a concentration less than the MIC (37), and subinhibitory concentrations have also been found to extend the lag phase and decrease overall growth rates (38). Synergistic toxicity involving multiple low-concentration antiseptics or even nonantiseptic compounds could also be responsible for inhibiting the biomass in the columns. For instance, exposure to 5 μg liter−1 of caffeine, acetaminophen, diclofenac, and their mixtures have been shown to cause a decrease in river biofilm biomass as well as disruptions to community structure (39). A study utilizing anonymous DNA microarrays demonstrated that biofilm microorganisms responded transcriptionally to exposure to environmentally relevant concentrations of erythromycin, gemfibrozil, sulfamethazine, and sulfamethoxazole (40).
In spite of the correlation between PPCP concentration and microbial biomass, even the highest concentration of PPCPs tested (1,000 μg liter−1) was insufficient to completely inhibit biotransformation. This may be due to intrinsic resistance of the microbes collected from the wastewater treatment plant, acquired resistance, or protection due to biofilm growth. In terms of tolerance to antiseptics, biofilms are advantageous for many reasons, including physical protection, favorable microenvironments, and the proximity of cells aiding in horizontal transfer of genes (41). Johnson et al. (42) found that six different bacterial strains evaluated for susceptibility to p-chloro-m-xylenol were more tolerant of the antiseptic when grown as biofilms than when grown and exposed in planktonic form.
In an earlier study, researchers assessed removals of selected PPCPs in the same SAT basin sampled for the microbial ecology portion of this study at different depths (1.5 m and 40 m below ground surface) and travel times (43). Six compounds (diclofenac, gemfibrozil, ibuprofen, naproxen, phenytoin, and triclosan) common to both studies were detected in the basin water at maximum concentrations of 200, 4,500, 1,075, 1,600, 810, and 433 ng liter−1, respectively. After 2 to 3 days of subsurface travel, triclosan was removed by more than 90%, while diclofenac, gemfibrozil, ibuprofen, and naproxen showed removals between 50 and 90%. Phenytoin was removed only by 25 to 50%. The PPCPs more effectively removed in the columns (e.g., ibuprofen and naproxen) also exhibited high removals at the SAT site. During 2 weeks of travel time, all of these compounds were removed by greater than 90%, except for phenytoin, which fell in the 50 to 90% removal category (43). The greater removals in the field are likely due to longer contact time that extends the opportunity for biodegradation and additional removal mechanisms, such as sorption.
Microbial diversity can play a major role in ecosystem function in soils (44); therefore, the richness and diversity of column and SAT communities were assessed with the estimates summarized in Table 3. The Chao1 richness estimate revealed that the number of phylotypes present in the columns is approximately four times lower than in the SAT site samples. The high-PPCP-concentration column has the slightly higher estimate of the two columns, but the 95% confidence intervals overlap. For the SAT samples, the estimated range for the day 1 sample is significantly less than the day 2 and 3 samples, suggesting that there is less richness on day 1 when soil is sampled through overlying water and richness increased as the water drained and the soil moisture content decreased, though future studies with more extensive sampling and replication are needed to determine if this trend is robust. Community evenness was estimated with the Shannon index. This estimate indicates that the low-PPCP-concentration column had greater evenness than the high-PPCP-concentration column. It also suggests that evenness increased over time in the SAT samples. Diversity was estimated using the Simpson index. Results show that the lowest-PPCP-concentration column had the least diversity, the day 2 SAT sample had the greatest diversity, and the others were not significantly different from each other. The columns were expected to have lower diversity because they had been operating in the laboratory for months with selective pressures exerted on the original WWTP effluent inoculum. The selective pressures for the laboratory column included supplying PPCPs and acetate at stable concentrations and operating the columns with continuous flow. The higher Simpson index in the day 2 SAT sample might be explained by more WWTP bacteria being included in the soil sample after a full day of water infiltration promoted their transfer to the soil. The decrease in the Simpson index between days 2 and 3 may be a result of the desiccation of the soil and the subsequent stress on the bacteria living in the soil. Further SAT studies across multiple wetting and drying cycles are necessary to confirm our hypotheses regarding temporal community shifts.
Overall, diverse phylotypes were found in both the laboratory columns and the top layer of the SAT infiltration basin (see Table S7 in the supplemental material). The sequences present in the highest abundances were most closely related to a range of microorganisms, including aerobes, anaerobes (facultative and obligate), xenobiotic degraders, and nitrifiers. Taxonomic composition of microbial communities in the columns and the SAT samples are remarkably similar considering that column inoculum was from the effluent of a geographically distant WWTP, sand (not soil) served as the biofilm substratum in the columns, and biogeochemical conditions in the laboratory columns cannot replicate the field environment perfectly. Consistent with this observation, recent work on managed aquifer recharge systems has also shown that lab scale systems can reproduce the microbial communities present in field systems at higher phylogenetic/taxonomic levels, such as phylum and class (45).
A large portion of all samples was composed of bacteria with Xanthobacter flavus (JN592464) as the closest named species, which suggests that this was the main component of Proteobacteria (see Fig. S3 in the supplemental material). The strain is identified in GenBank as having been isolated from a flooded rice field and being able to degrade the insecticide chlorpyrifos. It is interesting that a microbe isolated from a rice field that is likely operated under various wetting and drying cycles would thrive in an SAT system also subjected to similar moisture fluctuations. The second most abundantly found phylotype was most closely related to Mycobacterium sp. R5 (JN110434), which was isolated from a rice seed endosphere (46).
The greatest abundance of both Mycobacterium (phylum Actinobacteria) and Desulfosporosinus (phylum Firmicutes) was found in the low-PPCP-concentration column, suggesting that these taxa may be sensitive to higher concentrations of PPCPs or alternatively may have a selective advantage under lower total organic carbon conditions. Gemmatimonas (phylum Gemmatimonadetes) was not detected in the low-PPCP-concentration column at all, while phylum representatives were found in the high-PPCP-concentration column and in all the SAT samples (relative abundances between 2.5 and 2.7%). The columns were treated identically except for the concentrations of PPCPs supplied. As higher PPCP concentrations have been shown to suppress biofilm growth, it is possible that a PPCP metabolite being generated only in the low-concentration column was impeding Gemmatimonas growth. Another possibility is that Gemmatimonas was outcompeted in the low-concentration column by other microbes better able to utilize the trace concentrations of PPCPs. Similarly, Chloroflexi was not represented in the low-PPCP-concentration column, while it was found in all the other samples, albeit only at 0.0014% in the high-PPCP-concentration column and at 0.7 to 2.5% in the SAT samples. Conversely, OP10 was not detected in the high-PPCP-concentration column, while it was detected in the low-PPCP-concentration column at 0.0032% and in the SAT samples at 0.3% in all samples. TM7 was not found in either of the column samples, yet it ranged from 0.7 to 1.4% in the SAT samples. The absence of Cyanobacteria in column samples, while present ranging from 0.1 to 0.4% in the SAT samples, was anticipated, as laboratory columns were covered in aluminum foil when in operation to prevent algal growth and phototransformation of PPCPs.
There are very few isolated PPCP degraders in the literature, but one of the few is strain KCY1 (DQ983313), which was isolated from activated sludge and has been demonstrated to dechlorinate triclosan (47). Four sequences detected in our column and SAT samples at relative abundances as high as 0.071% had ≥97% similarity with strain KCY1 (see Table S9 in the supplemental material). Another identified triclosan degrader, Sphingomonas sp. PH-07 (DQ185574) (48), had at least 97% similarity with three detected sequences from the columns and SAT site at relative abundances as high as 3.503%. Strain VAL, a valproic acid degrader with 98% similarity to Sphingomonas aquatilis, was isolated from one of our previous laboratory experiments in which WWTP effluent was used to inoculate a sand column that was then exposed to PPCPs (49). A sequence with 97% similarity to VAL's sequence was detected once in the low-PPCP-concentration column. The presence of such close relatives of KCY1, PH-07, and VAL suggests a strong likelihood that there are triclosan and valproic acid degraders in our sampled systems. Several other isolated and characterized PPCP degraders were identified in the literature, but they were not close relatives of sequences found in the columns and SAT systems. They include ibuprofen degrader Sphingomonas sp. strain Ibu-2 (EF090268) (50) and triclosan degraders Sphingomonas sp. RD1 (AF292238) (51), Alcaligenes xylosoxidans subsp. denitrificans TR1 (51), Pseudomonas putida TriRY (52), Nitrosomonas europaea (53), and Pseudomonas sp. BDC1, -2, and -3 (GQ456128, GQ456129, and GQ456130, respectively) (54).
The greatest variation was observed for Trachelomonas volvocinopsis (RSD, 57%; FJ719709), Sphingobium yanoikuyae (RSD, 48%; JF681288), and Bosea thiooxidans (RSD, 43%; JQ659580) (see Fig. S4 in the supplemental material). T. volvocinopsis, if accurately identified as a photosynthesizer, could have varied in response to light infiltration through both the overlying water and soil, as well as in response to changes in moisture content. B. thiooxidans, a chemolithoheterotrophic soil microorganism (55), may have decreased abundance on consecutive days as a result of both soil moisture and the decrease in dissolved organic carbon (DOC) concentration over time. The RSD for S. yanoikuyae was calculated using two separate occurrences of sequences most closely related to that species. S. yanoikuyae, an aerobe which has a type strain isolated from a hospital sample and environmental strains isolated from plant root associations (55), increased in abundance over time when considering both of the two closely related but different phylotypes detected. This bacterium may be more resistant to desiccation or have simply benefited from the draining of the infiltrating water, allowing increased oxygen levels in the soil, leading to faster growth. However, making any statements about causation for these correlations would require more extensive sampling of the site over multiple wetting and drying cycles.
The SAT samples had the same phyla represented in the samples on all 3 days when considering those present at greater than 0.1% abundance. The relative abundance of these phyla shifted slightly across 3 days, possibly due to changes in soil moisture that were occurring, but again, more extensive sampling would be required to test this hypothesis. The literature shows that wetting and drying cycles can be responsible for shifts in microbial communities. For instance, a microbial community study using terminal restriction fragment length polymorphism to investigate the effect of the number of wetting and drying cycles (0 to 15) to which grassland and oak forest soils were exposed for a period of 2 months suggested that the grassland soil community composition was not affected by the wetting and drying cycles, but the oak canopy sample composition was. The authors pointed out that the grassland communities would have been previously exposed to such wetting and drying events in the field (56). With SAT sites constantly exposed to wetting and drying phases, soil communities in the top layers of a spreading basin may also be relatively insensitive to soil moisture changes like the grassland community. Other studies have shown that wetting and drying cycles can impact bacterial growth, biodegradation of xenobiotic compounds, respiration rates, and denitrification rates (57–61).
Understanding of the microbiological processes occurring during SAT and other water recycling processes would be greatly enhanced by future research that couples microbial community data with the removals of PPCPs at various depths in the SAT soil. Although community taxa were surveyed in the present study, further investigation to detect and identify genes within these communities involved in PPCP biodegradation as well as their level of transcription in the soil would allow more detailed information to be gathered regarding which microorganisms are actively degrading the micropollutants of interest. The dependence of PPCP biological removal on initial PPCP concentration in the laboratory column system suggests that determining PPCP concentration may be an important step in predicting PPCP fate during treatment. Overall, the demonstrated biological removal of many tested PPCPs in the laboratory column SAT simulations coupled with the diversity of the SAT microbes and their metabolic strategies suggest great potential for biotransformation of many contaminants of concern in SAT and other water reuse systems.
Supplementary Material
ACKNOWLEDGMENTS
We thank Bruce Prior and Terry Miley of Tucson Water for their collaboration on this project, Audrey Fischer Hesselbrock for her assistance with the sample homogenization and DNA extraction, and the Back River Wastewater Treatment Plant in Baltimore for the wastewater effluent sample. We also thank Jim Yu, Kevin Bisceglia, Mehmet Coelhan, and Lynn Roberts for their work on solid-phase extraction (SPE) and GC-MS method development and John Sivey, Lynn Roberts, Bill Ball, and anonymous reviewers for their helpful suggestions for manuscript improvement.
This material is based upon work supported by the National Science Foundation under grant no. 0606880 and a Graduate Research Fellowship (K.M.O.-B.), by the U.S. Environmental Protection Agency under STAR Research Assistance agreement no. FP-916857 (K.M.O.-B.), and by the Johns Hopkins University Global Water Program.
Any opinions, findings, and conclusions or recommendations expressed in this material are those of the authors and do not necessarily reflect the views of the National Science Foundation. It has not been formally reviewed by the EPA. The views expressed in this document are solely those of the authors, and the EPA does not endorse any products or commercial services mentioned in this publication.
Footnotes
Published ahead of print 7 February 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.03693-13.
REFERENCES
- 1.Asano T, Cotruvo JA. 2004. Groundwater recharge with reclaimed municipal wastewater: health and regulatory considerations. Water Res. 38:1941–1951. 10.1016/j.watres.2004.01.023 [DOI] [PubMed] [Google Scholar]
- 2.Jones OAH, Voulvoulis N, Lester JN. 2005. Human pharmaceuticals in wastewater treatment processes. Crit. Rev. Environ. Sci. Technol. 35:401–427. 10.1080/10643380590956966 [DOI] [Google Scholar]
- 3.Pomati F, Castiglioni S, Zuccato E, Fanelli R, Vigetti D, Rossetti C, Calamari D. 2006. Effects of a complex mixture of therapeutic drugs at environmental levels on human embryonic cells. Environ. Sci. Technol. 40:2442–2447. 10.1021/es051715a [DOI] [PubMed] [Google Scholar]
- 4.Onesios KM, Yu JT, Bouwer EJ. 2009. Biodegradation and removal of pharmaceuticals and personal care products in treatment systems: a review. Biodegradation 20:441–466. 10.1007/s10532-008-9237-8 [DOI] [PubMed] [Google Scholar]
- 5.Fox P, Houston S, Westerhoff P, Nellor M, Yanko W, Baird R, Rincon M, Gully J, Carr S, Arnold R, Lansey K, Quanrud D, Ela W, Amy G, Reinhard M, Drewes JE. 2006. Advances in soil aquifer treatment for sustainable water reuse. AWWA Research Foundation and American Water Works Association, Denver, CO [Google Scholar]
- 6.Fox P, Houston S, Westerhoff P. 2001. Soil aquifer treatment for sustainable water reuse. American Water Works Association, Denver, CO [Google Scholar]
- 7.Halling-Sørensen B, Nielsen SN, Lanzky PF, Ingerslev F, Holten Lützhøft Jørgensen. 1998. Occurrence, fate and effects of pharmaceutical substances in the environment—a review. Chemosphere 36:357–393. 10.1016/S0045-6535(97)00354-8 [DOI] [PubMed] [Google Scholar]
- 8.Kolpin DW, Furlong ET, Meyer MT, Thurman EM, Zaugg SD, Barber LB, Buxton HT. 2002. Pharmaceuticals, hormones, and other organic wastewater contaminants in U.S. streams, 1999–2000: a national reconnaissance. Environ. Sci. Technol. 36:1202–1211. 10.1021/es011055j [DOI] [PubMed] [Google Scholar]
- 9.Barnes KK, Kolpin DW, Furlong ET, Zaugg SD, Meyer MT, Barber LB. 2008. A national reconnaissance of pharmaceuticals and other organic wastewater contaminants in the United States—I. Groundwater. Sci. Total Environ. 402:192–200. 10.1016/j.scitotenv.2008.04.028 [DOI] [PubMed] [Google Scholar]
- 10.Focazio MJ, Kolpin DW, Barnes KK, Furlong ET, Meyer MT, Zaugg SD, Barber LB, Thurman ME. 2008. A national reconnaissance for pharmaceuticals and other organic wastewater contaminants in the United States—II. Untreated drinking water sources. Sci. Total Environ. 402:201–216. 10.1016/j.scitotenv.2008.02.021 [DOI] [PubMed] [Google Scholar]
- 11.OECD. 1992. Test no. 301: ready biodegradability, OECD guidelines for the testing of chemicals, section 3. OECD Publishing, Paris, France [Google Scholar]
- 12.Bergheim M, Gieré R, Kümmerer K. 2012. Biodegradability and ecotoxicity of tramadol, ranitidine, and their photoderivatives in the aquatic environment. Environ. Sci. Pollut. Res. 19:72–85. 10.1007/s11356-011-0536-y [DOI] [PubMed] [Google Scholar]
- 13.Carucci A, Cappai G, Piredda M. 2006. Biodegradability and toxicity of pharmaceuticals in biological wastewater treatment plants. J. Environ. Sci. Health A Tox. Hazard Subst. Environ. Eng. 41:1831–1842. 10.1080/10934520600779000 [DOI] [PubMed] [Google Scholar]
- 14.Haiss A, Kümmerer K. 2006. Biodegradability of the X-ray contrast compound diatrizoic acid, identification of aerobic degradation products and effects against sewage sludge micro-organisms. Chemosphere 62:294–302. 10.1016/j.chemosphere.2005.05.007 [DOI] [PubMed] [Google Scholar]
- 15.Kwon JW, Armbrust KL. 2006. Laboratory persistence and fate of fluoxetine in aquatic environments. Environ. Toxicol. Chem. 25:2561–2568. 10.1897/05-613R.1 [DOI] [PubMed] [Google Scholar]
- 16.Stasinakis AS, Petalas AV, Mamais D, Thomaidis NS, Gatidou G, Lekkas TD. 2007. Investigation of triclosan fate and toxicity in continuous-flow activated sludge systems. Chemosphere 68:375–381. 10.1016/j.chemosphere.2007.01.047 [DOI] [PubMed] [Google Scholar]
- 17.Trautwein C, Kümmerer K, Metzger JW. 2008. Aerobic biodegradability of the calcium channel antagonist verapamil and identification of a microbial dead-end transformation product studied by LC-MS/MS. Chemosphere 72:442–450. 10.1016/j.chemosphere.2008.02.022 [DOI] [PubMed] [Google Scholar]
- 18.Almeida B, Oehmen A, Marques R, Brito D, Carvalho G, Barreto Crespo M.T. 2013. Modelling the biodegradation of non-steroidal anti-inflammatory drugs (NSDAIDs) by activated sludge and a pure culture. Bioresour. Technol. 133:31–37. 10.1016/j.biortech.2013.01.035 [DOI] [PubMed] [Google Scholar]
- 19.Onesios KM, Bouwer EJ. 2012. Biological removal of pharmaceuticals and personal care products during laboratory soil aquifer treatment simulation with different primary substrate concentrations. Water Res. 46:2365–2375. 10.1016/j.watres.2012.02.001 [DOI] [PubMed] [Google Scholar]
- 20.Yu JT, Bouwer EJ, Coelhan M. 2006. Occurrence and biodegradability studies of selected pharmaceuticals and personal care products in sewage effluent. Agric. Water Management 86:72–80. 10.1016/j.agwat.2006.06.015 [DOI] [Google Scholar]
- 21.Navidi W. 2006. Statistics for engineers and scientists. McGraw Hill, New York, NY [Google Scholar]
- 22.R Core Development Team. 2008. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria [Google Scholar]
- 23.Toride N, Leij FK, van Genuchten MT. 1995. The CXTFIT code for estimating transport parameters from laboratory or fieldtracer experiments, version 2.1, research report 137. U.S. Salinity Laboratory, U.S. Department of Agriculture, Riverside, CA [Google Scholar]
- 24.Bragg L, Stone G, Imelfort M, Hugenholtz P, Tyson GW. 2012. Fast, accurate error-correction of amplicon pyrosequences using Acacia. Nat. Methods 9:425–426. 10.1038/nmeth.1990 [DOI] [PubMed] [Google Scholar]
- 25.Weizhong L, Jaroszewski L, Godzik A. 2001. Clustering of highly homologous sequences to reduce the size of large protein database. Bioinformatics 17:282–283. 10.1093/bioinformatics/17.3.282 [DOI] [PubMed] [Google Scholar]
- 26.Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ, Sahl JW, Stres B, Thallinger GG, Van Horn DJ, Weber CF. 2009. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 75:7537–7541. 10.1128/AEM.01541-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Haas BJ, Gevers D, Earl AM, Feldgarden M, Ward DV, Giannoukos G, Ciulla D, Tabbaa D, Highlander SK, Sodergren E, Methe B, DeSantis TZ, Petrosino JF, Knight R, Birren BW. 2011. Chimeric 16S rRNA sequence formation and detection in Sanger and 454-pyrosequenced PCR amplicons. Genome Res. 21:494–504. 10.1101/gr.112730.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lozupone C, Knight R. 2005. UniFrac: a new phylogenetic method for comparing microbial communities. Appl. Environ. Microbiol. 71:8228–8235. 10.1128/AEM.71.12.8228-8235.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello IK, Fierer N, Pena AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Tumbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R. 2010. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7:335–336. 10.1038/nmeth.f.303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang Q, Garrity GM, Tiedje JM, Cole JR. 2007. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 73:5261–5267. 10.1128/AEM.00062-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Morgulis A, Coulouris G, Raytselis Y, Madden TL, Agarwala R, Schaffer AA. 2008. Database indexing for production MegaBLAST searches. Bioinformatics 24:1757–1764. 10.1093/bioinformatics/btn322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kunin V, Engelbrektson A, Ochman H, Hugenholtz P. 2010. Wrinkles in the rare biosphere: pyrosequencing errors can lead to artificial inflation of diversity estimates. Environ. Microbiol. 12:118–123. 10.1111/j.1462-2920.2009.02051.x [DOI] [PubMed] [Google Scholar]
- 33.Xu N, Johnson AC, Jürgens Llewellyn MDNR, Hankins NP, Darton RC. 2009. Estrogen concentration affects its biodegradation rate in activated sludge. Environ. Toxicol. Chem. 28:2263–2270. 10.1897/08-577.1 [DOI] [PubMed] [Google Scholar]
- 34.Wang Y-S, Subba-Rao RV, Alexander M. 1984. Effect of substrate concentration and organic and inorganic compounds on the occurrence and rate of mineralization and cometabolism. Appl. Environ. Microbiol. 47:1195–1200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang P, Sheng G, Feng Y, Miller DM. 2006. Predominance of char sorption over substrate concentration and soil pH in influencing biodegradation of benzonitrile. Biodegradation 17:1–8. 10.1007/s10532-005-1919-x [DOI] [PubMed] [Google Scholar]
- 36.Marsolek M, Kirisits M, Rittmann B. 2007. Biodegradation of 2,4,5-trichlorophenol by aerobic microbial communities: biorecalcitrance, inhibition, and adaptation. Biodegradation 18:351–358. 10.1007/s10532-006-9069-3 [DOI] [PubMed] [Google Scholar]
- 37.Regos J, Hitz H. 1974. Investigations on the mode of action of triclosan, a broad spectrum antimicrobial agent. Zentralbl. Bakteriol. Orig. A 226:390–401 [PubMed] [Google Scholar]
- 38.Gomez Escalada M, Russell D, Maillard J-Y, Ochs D. 2005. Triclosan-bacteria interactions: single or multiple target sites? Lett. Appl. Microbiol. 41:476–481. 10.1111/j.1472-765X.2005.01790.x [DOI] [PubMed] [Google Scholar]
- 39.Lawrence JR, Zhu B, Swerhone GDW, Roy J, Tumber V, Waiser MJ, Topp E, Korber DR. 2012. Molecular and microxcopic assessment of the effects of caffeine, acetaminophen, diclofenac, and their mixtures on river biofilm communities. Environ. Tox. Chem. 31:508–517. 10.1002/etc.1723 [DOI] [PubMed] [Google Scholar]
- 40.Yergeau E, Lawrence JR, Waiser MJ, Korber DR, Greer CW. 2010. Metatranscriptomic analysis of the response of river biofilms to pharmaceutical products, using anonymous DNA microarrays. Appl. Environ. Microbiol. 76:5432–5439. 10.1128/AEM.00873-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McDonnell G, Russell D. 1999. Antiseptics and disinfectants: activity, action, and resistance. Clin. Microbiol. Rev. 12:147–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Johnson SA, Goddard PA, Iliffe C, Timmins B, Rickard AH, Robson G, Handley PS. 2002. Comparative susceptibility of resident and transient hand bacteria to para-chloro-meta-xylenol and triclosan. J. Appl. Microbiol. 93:336–344. 10.1046/j.1365-2672.2002.01691.x [DOI] [PubMed] [Google Scholar]
- 43.Drewes JE, Dickenson E, Snyder S. 2011. Development of surrogates to determine the efficacy of groundwater recharge systems for the removal of trace organic chemicals. WaterReuse Research Foundation, Alexandria, VA [Google Scholar]
- 44.Torsvik V, Ovreås L. 2002. Microbial diversity and function in soil: from genes to ecosystems. Curr. Opin. Microbiol. 5:240–245. 10.1016/S1369-5274(02)00324-7 [DOI] [PubMed] [Google Scholar]
- 45.Li D, Sharp JO, Saikaly PE, Ali S, Alidina M, Alarawi MS, Keller S, Hoppe-Jones C, Drewes JE. 2012. Dissolved organic carbon influences microbial community composition and diversity in managed aquifer recharge systems. Appl. Environ. Microbiol. 78:6819–6828. 10.1128/AEM.01223-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hardoim PR, Hardoim CCP, van Overbeek LS, van Elsas JD. 2012. Dynamics of seed-borne rice endophytes on early plant growth stages. PLoS One 7:e30438. 10.1371/journal.pone.0030438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lee DG, Zhao F, Rezenom YH, Russell DH, Chu K-H. 2012. Biodegradation of triclosan by a wastewater microorganism. Water Res. 46:4226–4234. 10.1016/j.watres.2012.05.025 [DOI] [PubMed] [Google Scholar]
- 48.Kim Y-M, Murugesan K, Schmidt S, Bokare V, Jeon J-R, Kim E-J, Chang Y-S. 2011. Triclosan susceptibility and co-metabolism: a comparison for three aerobic pollutant-degrading bacteria. Bioresour. Technol. 102:2206–2212. 10.1016/j.biortech.2010.10.009 [DOI] [PubMed] [Google Scholar]
- 49.Onesios KM. 2012. Biological removal of pharmaceuticals and personal care products during simulated water recycling. Ph.D. dissertation. Johns Hopkins University, Baltimore, MD [Google Scholar]
- 50.Murdoch RW, Hay AG. 2005. Formation of catechols via removal of acid side chains from ibuprofen and related aromatic acids. Appl. Environ. Microbiol. 71:6121–6125. 10.1128/AEM.71.10.6121-6125.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hay AG, Dees PM, Sayler GS. 2001. Growth of a bacterial consortium on triclosan. FEMS Microbiol. Ecol. 36:105–112. 10.1111/j.1574-6941.2001.tb00830.x [DOI] [PubMed] [Google Scholar]
- 52.Meade MJ, Waddell RL, Callahan TM. 2001. Soil bacteria Pseudomonas putida and Alcaligenes xylosoxidans subsp. denitrificans inactivate triclosan in liquid and solid substrates. FEMS Microbiol. Lett. 204:45–48. 10.1111/j.1574-6968.2001.tb10860.x [DOI] [PubMed] [Google Scholar]
- 53.Roh H, Subramanya N, Zhao F, Yu C-P, Sandt J, Chu K-H. 2009. Biodegradation potential of wastewater micropollutants by ammonia-oxidizing bacteria. Chemosphere 77:1084–1089. 10.1016/j.chemosphere.2009.08.049 [DOI] [PubMed] [Google Scholar]
- 54.Gangadharan Puthiya Veetil P, Vijaya Nadaraja A, Bhasi A, Khan S, Bhaskaran K. 2012. Degradation of triclosan under aerobic, anoxic, and anaerobic conditions. Appl. Biochem. Biotechnol. 167:1603–1612. 10.1007/s12010-012-9573-3 [DOI] [PubMed] [Google Scholar]
- 55.Staley JT, Brenner DJ, Krieg R. 2005. Bergey's manual of systematic bacteriology: the proteobacteria: the alpha-, beta-, delta-, and epsilonproteobacteria, vol 2 Springer, New York, NY [Google Scholar]
- 56.Fierer N, Schimel JP, Holden PA. 2003. Influence of drying-rewetting frequency on soil bacterial community structure. Microb. Ecol. 45:63–71. 10.1007/s00248-002-1007-2 [DOI] [PubMed] [Google Scholar]
- 57.Pesaro M, Nicollier G, Zeyer J, Widmer F. 2004. Impact of soil drying-rewetting stress on microbial communities and activities and on degradation of two crop protection products. Appl. Environ. Microbiol. 70:2577–2587. 10.1128/AEM.70.5.2577-2587.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bapiri A, Erland B, Rousk J. 2010. Drying-rewetting cycles affect fungal and bacterial growth differently in an arable soil. Microb. Ecol. 60:419–428. 10.1007/s00248-010-9723-5 [DOI] [PubMed] [Google Scholar]
- 59.Zhao B, Chen J, Zhang J, Qin S. 2010. Soil microbial biomass and activity response to repeated drying-rewetting cycles along a soil fertility gradient modified by long-term fertilization management practices. Geoderma 160:218–224. 10.1016/j.geoderma.2010.09.024 [DOI] [Google Scholar]
- 60.Song K, Lee S-H, Mitsch WJ, Kang H. 2010. Different responses of denitrification rates and denitrifying bacterial communities to hydrologic pulsing in created wetlands. Soil Biol. Biochem. 42:1721–1727. 10.1016/j.soilbio.2010.06.007 [DOI] [Google Scholar]
- 61.Manzoni S, Schimel JP, Porporato A. 2012. Responses of soil microbial communities to water stress: results from a meta-analysis. Ecology 93:930–938. 10.1890/11-0026.1 [DOI] [PubMed] [Google Scholar]
- 62.Kasprzyk-Hordern B, Dinsdale RM, Guwy AJ. 2009. The removal of pharmaceuticals, personal care products, endocrine disruptors and illicit drugs during wastewater treatment and its impact on the quality of receiving water. Water Res. 43:363–380. 10.1016/j.watres.2008.10.047 [DOI] [PubMed] [Google Scholar]
- 63.Rabiet M, Togola A, Brissaud F, Seidel J-L, Budzinski H, Elbaz-Poulichet F. 2006. Consequences of treated water recycling as regards pharmaceuticals and drugs in surface and ground waters of a medium-sized Mediterranean catchment. Environ. Sci. Technol. 40:5282–5288. 10.1021/es060528p [DOI] [PubMed] [Google Scholar]
- 64.Bendz D, Paxeus NA, Ginn TR, Loge FJ. 2005. Occurrence and fate of pharmaceutically active compounds in the environment, a case study: Hoje River in Sweden. J. Hazard. Mater. 122:195–204. 10.1016/j.jhazmat.2005.03.012 [DOI] [PubMed] [Google Scholar]
- 65.Reungoat J, Escher BI, Macova M, Keller J. 2011. Biofiltration of wastewater treatment plant effluent: effective removal of pharmaceuticals and personal care products and reduction of toxicity. Water Res. 45:2751–2762. 10.1016/j.watres.2011.02.013 [DOI] [PubMed] [Google Scholar]
- 66.Ashton D, Hilton M, Thomas KV. 2004. Investigating the environmental transport of human pharmaceuticals to streams in the United Kingdom. Sci. Total Environ. 333:167–184. 10.1016/j.scitotenv.2004.04.062 [DOI] [PubMed] [Google Scholar]
- 67.Teijon G, Candela L, Tamoh K, Molina-Díaz A, Fernández-Alba AR. 2010. Occurrence of emerging contaminants, priority substances (2008/105/CE) and heavy metals in treated wastewater and groundwater at Depurbaix facility (Barcelona, Spain). Sci. Total Environ. 408:3584–3595. 10.1016/j.scitotenv.2010.04.041 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.