Abstract
Increasing evidence suggests that perturbations in the intestinal microbiota composition of infants are implicated in the pathogenesis of food allergy (FA), while the actual structure and composition of the intestinal microbiota in human beings with FA remain unclear. Microbial diversity and composition were analyzed with parallel barcoded 454 pyrosequencing targeting the 16S rRNA gene hypervariable V1-V3 regions in the feces of 34 infants with FA (17 IgE mediated and 17 non-IgE mediated) and 45 healthy controls. Here, we showed that several key FA-associated bacterial phylotypes, but not the overall microbiota diversity, significantly changed in infancy fecal microbiota with FA and were associated with the development of FA. The proportion of abundant Bacteroidetes, Proteobacteria, and Actinobacteria phyla were significantly reduced, while the Firmicutes phylum was highly enriched in the FA group (P < 0.05). Abundant Clostridiaceae 1 organisms were prevalent in infants with FA at the family level (P = 0.016). FA-enriched phylotypes negatively correlated with interleukin-10, for example, the genera Enterococcus and Staphylococcus. Despite profound interindividual variability, levels of 20 predominant genera were significantly different between the FA and healthy control groups (P < 0.05). Infants with IgE-mediated FA had increased levels of Clostridium sensu stricto and Anaerobacter and decreased levels of Bacteroides and Clostridium XVIII (P < 0.05). A positive correlation was observed between Clostridium sensu stricto and serum-specific IgE (R = 0.655, P < 0.001). The specific microbiota signature could distinguish infants with IgE-mediated FA from non-IgE-mediated ones. Detailed microbiota analysis of a well-characterized cohort of infants with FA showed that dysbiosis of fecal microbiota with several FA-associated key phylotypes may play a pathogenic role in FA.
INTRODUCTION
Food allergy (FA) is a common allergic disorder characterized by adverse immune and hypersensitivity reactions to food proteins (1). The prevalence of FA has been increasing worldwide in recent decades, reaching 6% among children <3 years of age and approximately 4% of adults in the United States, resulting in a considerable public health and economic burden (2). The key feature of FA is a T-helper type 2 (Th2)-predominant allergen-specific immune response, with the production of IgE antibodies specific for the food allergen (3). Except for IgE-mediated FA, many non-IgE reactions, which are poorly defined clinically and scientifically, are believed to be T-cell mediated. However, the etiology of FA remains unclear; a failure to establish or a breakdown in the maintenance of oral tolerance may be responsible. Generally, oral tolerance can be profoundly influenced by genetic predisposition, the route of allergic sensitization (placental, skin, and breast milk), timing and dose of allergen exposure, environment (diet, pollutants, gastric pH, and microbiota), and tissue milieu, in particular a bias toward Th2- versus tolerogenic Th1-mediated responses. No single unifying cause has been identified, but there is recent evidence showing that lack of microbial exposure during infancy is one factor responsible for FA (4–7). Clinical epidemiologic studies have postulated that a reduced microbial pressure in westernized countries underlies the increase in development of allergies. Indeed, alterations in the early gut microbiota may precede the development of allergies (7–11). Experimental evidence also demonstrates the importance of microbiota in shaping the development of the immune system (12), and disease-associated microbiota may play a pathogenic role in FA (5). Stimulation of the immune system by the commensal gut microbiota in the early stage of childhood might prevent the development of FA. Establishment of oral tolerance to dietary antigens is, at least in part, dependent on the presence of commensal microbes (13). However, a detailed assessment of the human gut microbiota for FA in infants has not been performed. Moreover, a good definition of gut microbiota and an understanding of the relationship to FA are essential in preventing and combating FA.
To date, the gut microbiota can be viewed as a forgotten “organ,” exquisitely tuned to our physiology, which performs functions that we have not had to evolve on our own. A growing body of evidence suggests that gut microbiota plays an essential role in host health by processing energy from food, protecting intestinal epithelial cells from injury, and promoting local and systemic immunity (12, 14), which has been extensively studied in recent years using culture-independent molecular methods. The next-generation DNA sequencing technologies, including high-throughput 454 pyrosequencing, provide a large number of sequencing reads in a single run, resulting in a large sampling depth and the detection of low-abundance taxa. Thus, the results of studies using high-throughput sequencing technologies have revolutionized our understanding of the gut microbiota in healthy and disease conditions.
The aim of this study was to identify differences in gut microbiota between healthy and FA subjects in Chinese infants using massively parallel barcoded 454 pyrosequencing targeting the 16S rRNA gene V1-V3 hypervariable regions to verify our hypothesis that altered human gut microbiota was associated with the development of FA in infants. Using feces as a proxy for gut microbiota, we sought to identify specific microbiota signatures for FA and their relationships to clinical patterns and physiologic measures.
MATERIALS AND METHODS
Recruitment of subjects.
Seventy-nine infants were enrolled (34 infants with FA and 45 healthy controls) (Table 1). FA was diagnosed and assessed according to the National Institute for Health and Clinical Excellence (NICE) guidelines (1, 15). Infants with suspected FA who were brought to the Department of Pediatrics of Peking University Third Hospital for evaluation between June 2012 and June 2013 had their FA established by means of clinical history or symptoms and confirmed with skin prick tests (SPTs) and oral food challenges with successive doses of milk, eggs, wheat, nut, peanuts, fish, shrimp, and soy beans. Measurement of serum-specific IgE (sIgE) to the different food allergens mentioned above (FEIA; Pharmacia Diagnostics, Uppsala, Sweden) was performed on all subjects according to the protocol of the manufacturer. Only infants with suspected FA undergoing all three tests required for this study (oral food challenge, determination of sIgE, and SPTs) were included in the study. Control infants were from the same cohorts and did not have allergic manifestations or increased total or specific IgE levels. Serum was also analyzed for interleukin-10 (IL-10) using the human IL-10 immunoassay kit (eBioscience, San Diego, CA, USA). The following exclusion criteria were established: allergic rhinitis; atopic eczema; asthma; use of antibiotics, probiotics, prebiotics, or synbiotics in the previous month; and known active bacterial, fungal, and/or viral infection(s). The study protocol was approved by the Ethics Committee of Peking University Third Hospital (Beijing, China). Informed written consent was obtained from the parents or guardians of all participants prior to enrollment.
TABLE 1.
Descriptive data of infants in the study
| Parameter | Value for infant group: |
||
|---|---|---|---|
| Healthy control (n = 45) | IgE mediated (n = 17) | Non-IgE mediated (n = 17) | |
| Age of infants (mo; means ± SD) (range) | 5.60 ± 2.33 (2–11) | 5.12 ± 1.83 (2–8) | 5.00 ± 1.37 (3–8) |
| Weight at birth (g; means ± SD) | 3,671 ± 605 | 3,536 ± 584 | 3,579 ± 608 |
| Proportion of males, no. (%) | 21 (46.7) | 9 (52.9) | 9 (52.9) |
| Vaginal delivery, no. (%) | 42 (93.3) | 15 (88.2) | 14 (82.3) |
| Breast-fed, no. (%) | 38 (84.4) | 16 (94.1) | 15 (88.2) |
| Vomiting, no. (%) | 0 (0) | 16 (94.1) | 8 (47.1) |
| Diarrhea (no. of times; means ± SD) | 2.09 ± 0.73 | 5.82 ± 2.16 | 5.53 ± 1.87 |
| Stool characteristics, no. | |||
| Normal | 45 | 0 | 0 |
| Watery | 0 | 1 | 5 |
| Mucous | 0 | 4 | 1 |
| Bloody | 0 | 7 | 7 |
| Mucous bloody | 0 | 5 | 4 |
| Skin prick test (>6 mm; no. positive) | |||
| Cow's milk | NDa | 4 | 4 |
| Egg | ND | 4 | 6 |
| Wheat | ND | 1 | 4 |
| Nuts | ND | 3 | 2 |
| Peanut | ND | 1 | 1 |
| Fish | ND | 1 | 1 |
| Shrimp | ND | 0 | 1 |
| Soy bean | ND | 0 | 2 |
| Specific-IgE of >0.35 KU/liter (no. of patients)b | |||
| Cow's milk | 0 | 12 | 0 |
| Egg | 0 | 11 | 0 |
| Wheat | 0 | 1 | 0 |
| Nuts | 0 | 0 | 0 |
| Peanut | 0 | 3 | 0 |
| Fish | 0 | 0 | 0 |
| Shrimp | 0 | 2 | 0 |
| Soy bean | 0 | 0 | 0 |
| IL-10 (pg/ml; means ± SD) | 313.4 ± 58.3 | 209.5 ± 62.0 | 189.0 ± 62.0 |
| Diagnostic food challenge (positive/negative) | 0/45 | 17/0 | 17/0 |
| Eczema, no. | 0 | 10 | 10 |
ND, not detected.
KU, kilo-international units.
Sample collection and DNA extraction.
Approximately 2 g of a fresh fecal sample was collected in a sterile plastic cup when the infant underwent initial examination at the hospital and was kept in an ice box. Samples for bacterial genomic DNA extraction were transferred immediately to the laboratory and stored at −80°C after preparation within 15 min until use. DNA was extracted from 300 mg of feces using a QIAamp DNA stool minikit (Qiagen, Hilden, Germany) according to the manufacturer's instructions, with additional glass bead-beating steps on a mini-beadbeater (FastPrep; Thermo Electron Corporation, Boston, MA, USA). The amount of DNA was determined using a NanoDrop ND-1000 spectrophotometer (Thermo Electron Corporation); the integrity and size were checked by 1.0% agarose gel electrophoresis containing 0.5 mg/ml ethidium bromide. All DNA was stored at −20°C before further analysis.
PCR and pyrosequencing.
The bacterial genomic DNA was amplified with the 27F (5′-AGAGTTTGATCCTGGCTCAG-3′) and 533R (5′-TTACCGCGGCTGCTGGCAC-3′) primers specific for the V1-V3 hypervariable regions of the 16S rRNA gene (16). Each forward primer incorporated FLX Titanium adapters and a sample barcode at the 5′ end of the reverse primer to allow all samples to be included in the single 454 FLX sequencing run (see Table S1 in the supplemental material). All PCRs were performed in 50-μl triplicates and combined after PCR. The products were extracted with the QIAquick gel extraction kit (Qiagen) and quantified on a NanoDrop ND-1000 spectrophotometer, QuantiFluor-ST fluorometer (Promega, USA), and Agilent 2100 bioanalyzer (Agilent Technologies, Palo Alto, CA, USA). Equimolar concentrations of 79 samples were pooled and sequenced on a 454 Life Sciences genome sequencer FLX system (Roche, Basel, Switzerland) according to the manufacturer's recommendations.
Bioinformatics and statistical analysis.
Raw pyrosequencing reads obtained from the sequencer were denoised using Titanium PyroNoise software (17–19). The resulting pyrosequencing reads were filtered according to barcode and primer sequences using a combination of tools from Mothur (version 1.25.0; http://www.mothur.org) and custom Perl scripts. Preliminary quality control steps included the removal of sequences shorter than 150 nucleotides with homopolymers longer than 8 nucleotides, those with an average quality score of <25, and all reads containing ambiguous base calls or >2 incorrect primer sequences. Using the Mothur implementation of the ChimeraSlayer algorithm (20), chimera sequences arising from the PCR amplification were detected and excluded from the denoised sequences. The high-quality sequences were assigned to samples according to barcodes. The high-quality reads were clustered into operational taxonomic units (OTUs) using Mothur (21). The OTUs that reached a 97% nucleotide similarity level were used for alpha diversity (Shannon, Simpson, and evenness indices), richness (ACE and Chao1), Good's coverage, Venn diagram, and rarefaction curve analysis using Mothur. A heatmap was generated on the basis of the relative abundance of OTUs using R (version 2.15; The R Project for Statistical Computing; http://www.R-project.org). Phylogenetic beta diversity measures, such as unweighted UniFrac distance metrics analysis, was performed using OTUs for each sample using the Mothur program. Principal coordinate analysis (PCoA) was conducted according to the distance matrices created by Mothur, and three-dimensional graphical outputs were drawn using SigmaPlot (version 12.0; Systat Software Inc., USA) (22).
Taxonomy-based analyses were performed by classifying each sequence using the Naive Bayesian Classifier program of the Michigan State University Center for Microbial Ecology Ribosomal Database Project (RDP) database (http://rdp.cme.msu.edu/) with a 50% bootstrap score (23). The Metastats program from Mothur was used to identify statistically different phylotypes among groups (21). Microbiome features of healthy controls were compared to those of patients with FA with Metastats using the P value and the false discovery rate (Q value) for nonnormal distributions. Only taxa with average abundances of >1%, P < 0.05, and low Q values (i.e., low risk of false discovery) were considered significant (24). The characterization of microorganismal features differentiating the fecal microbiota specific to FA was performed using the linear discriminant analysis (LDA) effect size (LEfSe) method (http://huttenhower.sph.harvard.edu/lefse/) for biomarker discovery, which emphasizes both statistical significance and biological relevance (25). With a normalized relative abundance matrix, LEfSe uses the Kruskal-Wallis rank sum test to detect features with significantly different abundances between assigned taxa and performs LDA to estimate the effect size of each feature. A significance alpha of 0.05 and an effect size threshold of 2 were used for all biomarkers discussed in this study.
The correlation between variables was computed using the Spearman rank correlation. Statistical analyses were performed using the SPSS data analysis program (version 16.0; SPSS Inc., Chicago, IL, USA). All tests for significance were two sided, and P < 0.05 was considered statistically significant.
Sequence read accession number.
The sequence data from this study have been deposited in the GenBank Sequence Read Archive under accession number SRP028835.
RESULTS
Overall structural changes of FA-associated fecal microbiota.
Of 770,480 high-quality sequences produced from 79 samples, accounting for 61.5% of valid sequences (1,251,846 reads in total) according to barcode and primer sequence filtering, an average of 9,752 (range, 4,031 to 15,596) sequences per barcoded sample was recovered for downstream analysis. Thus, a total of 433,554 sequences were obtained from healthy infants for phylogenetic analysis, while 336,926 sequences were obtained from infants with FA. The total number of unique sequences from the 2 groups was 11,501 and represented all phylotypes. Specifically, 7,268 species-level OTUs in healthy control infants and 6,495 OTUs in infants with FA were delineated at 97% similarity level. The summary information is shown in Table 2, and detailed characteristics of each sample are shown in Table S2 in the supplemental material. Good coverage was nearly 99.0% for all sequences in the 2 groups, indicating a greater sequencing depth for FA-associated fecal microbiota investigation in infants. Analysis of alpha diversity, such as Shannon and Simpson indices, showed that the infants with FA, as a whole, have average phylogenetic diversity of fecal microbiota similar to that of healthy controls (see Fig. S1). Although the Shannon index was lower in infants with FA and the Simpson index was higher, these differences failed to reach statistical significance (P > 0.05). By rarefaction analysis estimates, the trend of species richness in infants with FA was also similar to that of healthy controls (see Fig. S2A and S3). Based on the observed OTU analysis, a long tail in the rank abundance curves was observed, indicating that the majority of OTUs were present at low abundance (see Fig. S2B). Beta diversity analysis indicates the extent of similarity between microbial communities by measuring the degree to which membership or structure is shared between communities. Due to significant interindividual variations, the fecal microbiota from two groups could not be divided into different clusters according to community composition using unweighted UniFrac metrics (see Fig. S4) and could not be separated clearly by principal coordinates analysis (see Fig. S5); however, the clustering was complemented by an analysis of bacterial richness using the number of shared and unique OTUs in the two groups by a Venn diagram, which was generated to compare OTUs between two groups. At the 97% similarity level, 2,262 OTUs were shared between the 2 groups, which accounted for 34.8% and 31.1% of the total OTUs in infants with and without FA, respectively (see Fig. S6). These results indicated that infants shared a core set of bacteria in fecal microbiota regardless of health status. In contrast to previous studies, which focused only on predominant bacteria (4, 6), the present study demonstrated an unaltered overall diversity of FA-associated fecal microbiota in infants but fewer OTUs and phylotypes, which could result from the greater depth of sequencing coverage.
TABLE 2.
Comparison of phylotype coverage and diversity estimation of the 16S rRNA gene libraries at 97% similarity from the pyrosequencing analysis
| Group | No. of reads | No. of OTUsa | Good'sb (%) | Richness estimator |
Diversity index |
Evennessc | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| ACE | 95% CI | Chao1 | 95% CI | Shannon | Simpson | |||||
| Healthy control | 433,554 | 7,268 | 99.09 | 21,913 | 21,323–22,527 | 14,853 | 14,201–15,567 | 5.003 | 0.0178 | 0.0077 |
| Food allergy | 336,926 | 6,495 | 98.89 | 21,671 | 21,044–22,325 | 14,531 | 13,802–15,332 | 4.775 | 0.0235 | 0.0066 |
The operational taxonomic units (OTUs) were defined with 97% similarity level.
The coverage percentage (Good's) and richness estimators (ACE and Chao1) were calculated using Good's method, and diversity indices (Shannon and Simpson) were calculated using the Mothur program. CI, confidence interval.
The Shannon index of evenness was calculated with the formula E = H/ln(S), where E is evenness, H is the Shannon diversity index, and S is the total number of sequences in that group.
Associations between fecal microbiota and infancy FA.
A taxon-dependent analysis using the RDP classifier was conducted to describe the composition of infancy FA-associated fecal microbiota. Eleven phyla and two candidate divisions (TM7 and OD1) were found. Three phyla (Firmicutes, Bacteroidetes, and Proteobacteria) were the most predominant in infancy fecal samples and comprised >97% of all sequences, which showed a higher proportion of Proteobacteria in infants than in adults (26). In total, sequences from fecal microbiota could be classified as 270 genera, with 237 genera in healthy infants and 183 genera in infants with FA. Of the total number of genera identified in fecal microbiota, 17 predominant genera were detected in healthy infants and 18 predominant genera were detected in infants with FA, with 11 predominant genera being shared by both groups. The predominant genera were defined as having >1% of the total DNA sequences (27). These predominant genera accounted for 89.89% and 93.33% of the total sequences from healthy controls and infants with FA, respectively. The two most predominant genera were Bacteroides and Veillonella in healthy controls and Clostridium sensu stricto and Bacteroides in infants with FA. Among the shared predominant genera in both groups, Bacteroides was a member of the phylum Bacteroidetes, Escherichia/Shigella and Klebsiella belonged to the phylum Proteobacteria, and the other 8 genera were all classified as Firmicutes.
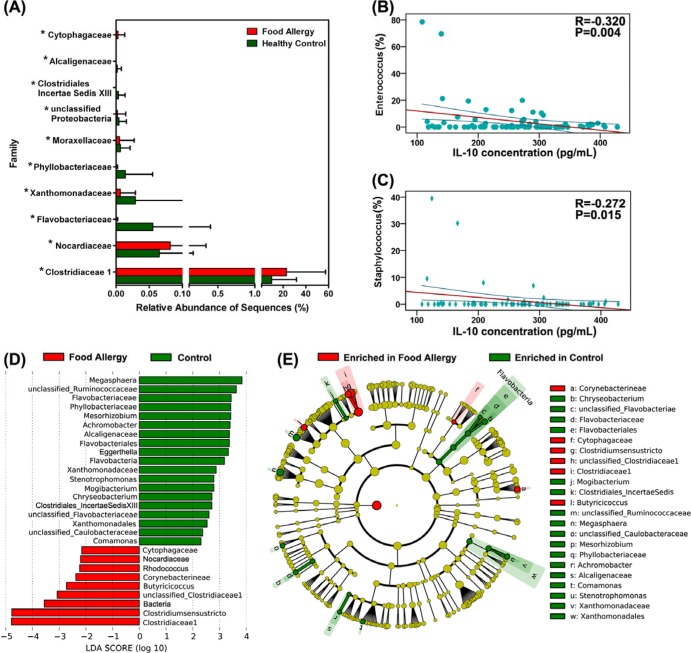
To investigate the associations between fecal microbiota and FA and to identify specific phylotypes responding to FA, we examined the composition of infancy fecal microbiota using Metastats. At the phylum level, the proportion of Firmicutes and Fusobacteria increased in FA cases, whereas the proportion of Bacteroidetes, Proteobacteria, Actinobacteria, and Verrucomicrobia decreased (P < 0.05) (see Table S3 in the supplemental material). At the family level, Clostridiaceae 1, Cytophagaceae and Nocardiaceae were prevalent in infants with FA, while Alcaligenaceae, Clostridiales incertae sedis XIII, Phyllobacteriaceae, Xanthomonadaceae, Flavobacteriaceae, and Moraxellaceae were enriched in healthy controls (Fig. 1A). At the genus level, 74 genera were identified differentially between the 2 groups, including 20 predominant (>1% of the total sequences in either group) and 54 less predominant genera (see Table S4). Among the predominant differential genera, 11 genera, such as Clostridium sensu stricto, Enterococcus, Escherichia/Shigella, Lactobacillus, Staphylococcus, Faecalibacterium, Clostridium XIVa, Anaerostipes, Prevotella, Clostridium XVIII, and Flavonifractor, showed increases in FA, whereas 9 genera, including Bacteroides, Streptococcus, Veillonella, Klebsiella, Blautia, Clostridium XI, Lachnospiracea incertae sedis, Megamonas, and Megasphaera, showed decreases (P < 0.05). Intriguingly, FA-enriched differential phylotypes, such as the genera Enterococcus and Staphylococcus, were negatively correlated with anti-inflammatory cytokines, such as IL-10 (P < 0.05) (Fig. 1B and C). Sixteen of the 20 differentially predominant genera belonged to the phylum Firmicutes, and 2 each of the remaining 4 belonged to Bacteroidetes and Proteobacteria. Interestingly, well-known probiotic bacteria, such as Lactobacillus and Bifidobacterium (28), were found to be significantly increased in infants with FA.
FIG 1.
Comparison of relative abundance at the bacterial family level between infants with food allergy and healthy controls. (A) Asterisks indicate P < 0.05. Correlation between interleukin-10 and the relative abundance of the genera Enterococcus (B) and Staphylococcus (C). The Spearman rank correlation (R) and probability (P) were used to evaluate statistical importance. (D) Linear discriminant analysis (LDA) coupled with effect size measurements identifies the most differentially abundant taxons between infants with food allergy and healthy controls. Healthy control-enriched taxa are indicated with a positive LDA score (green), and taxa enriched in food allergy have a negative score (red). Only taxa meeting an LDA significant threshold of >2 are shown. (E) A cladogram representation of data shown in panel A. Red, food allergy-enriched taxa; green, taxa enriched in healthy controls. The brightness of each dot is proportional to its effect size.
To identify bacterial taxa that had sequences that were more abundant in infants with FA than in healthy controls, a metagenomic biomarker discovery approach (LEfSe) was used to assess the size of effect of each differentially abundant taxon. Using LEfSe, we found that Clostridium sensu stricto and Butyricicoccus sequences were significantly enriched in infants with FA, as were sequences from the families Clostridiaceae 1 and Ruminococcaceae (Fig. 1D and E). Other taxa were enriched in healthy controls, but the LDA enrichment scores were lower by one order of magnitude or more. The high abundance of Clostridium sensu stricto sequences, with an average relative abundance of >20% of the total bacterial sequences, was a feature of some, but not all, infants with FA. Our present data showed that the aberrant compositions of fecal microbiota were associated with the development of infancy FA.
Phylotype signatures for IgE-mediated and non-IgE-mediated FA.
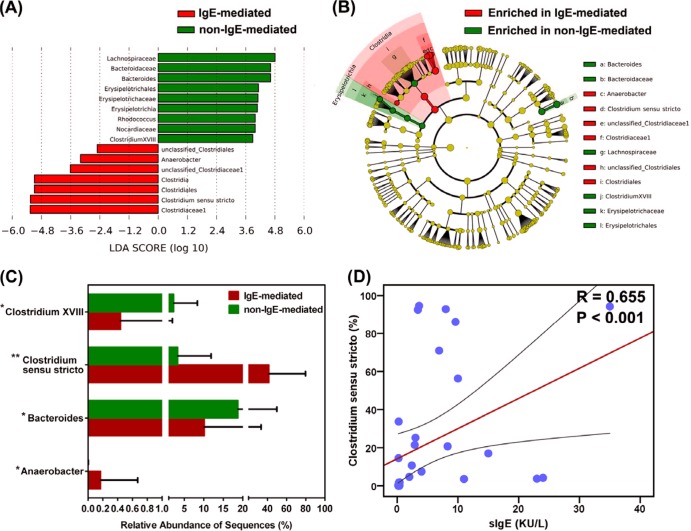
Consistent with our comparisons of fecal microbiota between healthy controls and infants with FA, no significant differences were detected in the overall microbial diversity between IgE-mediated and non-IgE-mediated FA (P > 0.05). However, further analysis suggested that several key phylotypes affected the development of IgE-mediated FA. Using LEfSe, we compared the infancy FA-associated fecal microbiota between IgE-mediated and non-IgE-mediated FA to identify specific phylotypes with OTU levels. We showed that Clostridium sensu stricto and Anaerobacter sequences were significantly enriched in the IgE-mediated FA, while Lachnospiraceae, Bacteroides, Erysipelotrichia, Rhodococcus, Nocardiaceae, and Clostridium XVIII sequences were significantly enriched in the non-IgE-mediated FA, which could be used as a potential biomarker for IgE-mediated FA (Fig. 2A and B). Statistical analysis identified four genera that were differentially abundant between the two groups (P < 0.05) (Fig. 2C). The genera of Clostridium sensu stricto, Anaerobacter, and Clostridium XVIII belonged to the phylum Firmicutes, and Bacteroides belonged to the phylum Bacteroidetes. The increased abundance of Clostridium sensu stricto and Anaerobacter and the decreased abundance of Bacteroides and Clostridium XVIII could be used to discriminate different types of FA. We then used the Kendall tau rank correlation coefficient to directly measure the correlation between sIgE and the relative abundance of the key phylotypes in fecal microbiota separately for IgE-mediated and non-IgE-mediated FA. With significant interindividual variability, we identified that only one genus, Clostridium sensu stricto, significantly correlated with sIgE in infants with FA (R = 0.655, P < 0.001), while the other key phylotypes showed no strong correlations with sIgE (Fig. 2D). The genus Clostridium sensu stricto was proportionally associated with sIgE, indicating that infants with IgE-mediated FA carried a greater load of this bacterium (see Fig. S7). Interestingly, the non-IgE-mediated FA infants carried significantly less of this bacterium (P < 0.05).
FIG 2.
LEfSe identifies the most differentially abundant taxons between IgE-mediated food allergy infants and non-IgE-mediated food allergy ones. Non-IgE-mediated food allergy-enriched taxa are indicated with a positive LDA score (green), and taxa enriched in IgE-mediated food allergy have a negative score (red). (A) Only taxa meeting an LDA significant threshold of >2 are shown. (B) A cladogram representation of data shown in panel A. Red, IgE-mediated food allergy-enriched taxa; green, taxa enriched in non-IgE-mediated food allergy infants. The brightness of each dot is proportional to its effect size. (C) The relative abundance of different genera obtained in fecal microbiota from IgE-mediated and non-IgE-mediated food allergy infants. The Mann-Whitney test was used to evaluate the two groups. *, P < 0.05; **, P < 0.01. (D) Correlation between specific serum IgE and the relative abundance of Clostridium sensu stricto. The Spearman rank correlation (R) and probability (P) were used to evaluate statistical importance. KU/L, kilo-international units per liter.
DISCUSSION
We used barcoded multiplexed 454 pyrosequencing to examine the relationship between fecal microbiota and FA in Chinese infants. This approach allowed a relatively comprehensive description of fecal microbiota associated with FA. This study also provided an interindividual comparison of fecal microbiota to identify the key phylotypes that might be associated with the development of infancy FA. Alterations in gut microbiota have been reported in patients with allergic diseases, although it is also possible that these changes precede the development of allergic manifestations and are a cause rather than (or as well as) a consequence. Unlike previous studies, which only focused on predominant bacterial phylotypes in allergic diseases, the work presented here was designed to better understand the alterations of overall bacterial diversity and composition in infants with FA at a much deeper level (4, 6, 29). Previous studies have shown that reduced diversity of the early intestinal microbiota during infancy is associated with an increased risk of allergic disease such as FA using DNA fingerprinting techniques, which can only detect bacteria with a relative abundance of >1% (4, 6, 30). Fewer OTUs and genera were found in infants with FA; however, the current study demonstrated an unaltered overall bacterial diversity of fecal microbiota in infants with FA (5). This discrepancy might be ascribed to a different depth of coverage for fecal microbiota with different methods. Our study showed that microbial diversity in the feces of infants was much greater than that of previous estimates, which were based on conventional molecular techniques. Good's coverage was >99.0% for all sequences in the FA and healthy control groups, indicating that less than one additional phylotype would be expected for every 100 additional sequenced reads. This level of coverage indicated that the 16S rRNA sequences identified in these groups represent the majority of bacterial sequences present in the samples under study, which could approach to the real world of infancy fecal microbiota. With an average of nearly 10,000 reads per sample, a large number of rare taxa present at relatively low abundances were detected, which influenced the overall bacterial diversity of fecal microbiota (31). The major strength of the 454 pyrosequencing method used in the current study is allowing a much deeper and more comprehensive analysis of changes in the gut microbiota responding to FA than in previous reports.
Consistent with previous studies using molecular methods, we sampled the fecal microbiota from infants at a single time point. In contrast to those well-controlled experimental FA animal models (5), profound interindividual variations were found in the composition of the fecal microbiota of infants with FA. In fact, the establishment of infancy gut microbiota was influenced by a number of factors, including delivery and feeding modes, birth order, degree of social exposure, and environmental bacterial content, which subsequently influenced the risk of atopic manifestations (11, 32–35). The interindividual variation implied that the same phylotypes occupy somewhat different niches in different individuals and have different linkages to other taxa, displaying different responses to FA. Generally, interindividual variability was greater than differences within the same subjects over time (36). The relative long-term stability of gut microbiota within individuals could be used to monitor the microbiota of patients with FA (37). Therefore, our data at a single time point could be used to identify the associations between specific phylotypes and FA with the relatively large cohort of subjects enrolled in our study.
Although the overall bacterial diversity of the fecal microbiota was unaltered in the present study, statistical analysis showed that some bacterial phylotypes were associated with the development of FA in infants. Most of these phylotypes were classified at the genus level but not into the higher taxonomic levels, such as bacterial families and orders. Using a high-density 16S rRNA gene oligonucleotide microarray (PhyloChip), Noval et al. exhibited a specific microbiota signature characterized by coordinate changes in the abundance of taxa of several bacterial families, including the Lachnospiraceae, Lactobacillaceae, Rikenellaceae, and Porphyromonadaceae, in experimental FA mouse models (5). The present fecal microbiota analysis also revealed a dysbiosis in the feces of infants with FA. In fact, the dysbiotic microbiota alone is sufficient to drive the intestinal immune disorders (38). Recent studies in animal models, however, have reinforced the notion that commensal microbiota contribute to systemic autoimmune and allergic diseases at sites distal to the intestinal mucosa (14). In the cross-talk between intestinal microbiota and host immune homeostasis, IL-10 plays a central role in the regulation of immune responses against commensal bacteria (39). Our present study found that FA-enriched phylotypes, such as the genera Enterococcus and Staphylococcus, were negatively correlated with IL-10. In contrast to previous studies, we identified several FA-associated specific phylotypes at the genus level, not just the changing patterns of fecal microbiota (4, 6). A total of 20 differentially abundant genera were observed in infancy fecal microbiota. Clostridium sensu stricto, Enterococcus, Escherichia/Shigella, Lactobacillus, Staphylococcus, Faecalibacterium, Clostridium XIVa, Anaerostipes, Prevotella, Clostridium XVIII, and Flavonifractor were detected more frequently in the FA group than in the healthy control group, and the others were present more frequently in the healthy control group than in the FA group. These abundant genera could be used as potential biomarkers for diagnosing FA and selecting treatment. The other 54 less abundant genera were also differentially present in the two groups, while the roles were difficult to accurately predict. All of these features contributed to the dysbiosis of fecal microbiota in infants with FA.
The major component of infancy fecal microbiota, Clostridium sensu stricto (clostridial cluster I), the core genus of the family Clostridiaceae, was strongly associated with infancy FA. Recently, Penders et al. reported that the prevalence of Clostridium cluster I (Clostridium sensu stricto) is positively associated with atopic dermatitis at 5 and 13 weeks of age (11); however, there was still no consensus of opinion about the role of Clostridium cluster I (Clostridium sensu stricto) in allergic diseases (11, 40). The present study is the first to identify the associations between Clostridium sensu stricto and infancy FA. Unlike other bacteria in Clostridium cluster I, such as Clostridium butyricum (41), our data indicated that Clostridium sensu stricto could not suppress immune disorders but directly affect the pathogenesis of infancy FA as the most predominant bacteria. One intriguing observation was an increased abundance of Clostridium cluster XIVa, Clostridium cluster XVIII, Lactobacillus, Faecalibacterium, and Bifidobacterium in FA, which behaved as potent inducers of regulatory T cell (Treg) or anti-inflammatory commensal bacteria (42–45). Atarashi et al. demonstrated that a rationally selected mixture of Clostridia strains, such as clusters IV, XIVa, and XVIII from the human microbiota, rather than a single Clostridia strain can induce the production of Treg cells for immune tolerance (45). The present study seems to contradict recent studies involving the roles of the bacteria mentioned above; however, the immunomodulatory capacities of these bacteria were strain or species specific (46). Some strains in the same genus have a proinflammatory effect, whereas others are more anti-inflammatory. A previous study also demonstrated that the presence of Bifidobacterium pseudocatenulatum, not B. bifidum, in feces was associated with atopic eczema (29). This might be the reason for an increased relative abundance of well-known probiotic bacteria, such as the genera Lactobacillus and Bifidobacterium, in infancy FA (9). The three most predominant genera, Escherichia/Shigella, Enterococcus, and Staphylococcus, were increased significantly in infants with FA. A previous study also showed that these genera are differentially associated with eczema and respiratory problems in infants (35, 47). Another two FA-associated genera, Anaerostipes and Prevotella, were shown to be the predominant bacteria in infancy fecal microbiota. However, Abrahamsson et al. reported that there was no significant correlation between Anaerostipes, Prevotella, and atopic eczema in infants (30). The genus of Flavonifractor, prevailing in minimal hepatic encephalopathy with cirrhosis (48), was also shown to have an increased relative abundance in infants with FA. Our current study indicated that the prevalence of those genera mentioned above would increase the risk for infancy FA.
The present study demonstrated the relationship between infancy fecal microbiota and IgE-mediated FA for the first time. FA can be classified into IgE-mediated and non-IgE-mediated reactions. IgE-mediated FA usually manifests as immediate and acute allergic reactions within 2 h of exposure, while many non-IgE-mediated reactions are relatively mild and poorly defined both clinically and scientifically. In the current study, an increased relative abundance of Clostridium sensu stricto and Anaerobacter and a decreased relative abundance of Bacteroides and Clostridium XVIII might disturb immune homeostasis in infants and induce food protein sIgE production. Interestingly, the prevalence of Clostridium sensu stricto was positively associated with IgE-mediated FA in infants but not non-IgE-mediated FA (P < 0.05). The sIgE level, in combination with the relatively high abundance of Clostridium sensu stricto, could be used to discriminate IgE-mediated FA from non-IgE-mediated FA more specifically; however, the mechanisms for IgE induction in infants are still unknown. Identifying the mechanisms for IgE induction by Clostridium sensu stricto was of great value, because such an understanding could help to develop novel tools or approaches to prevent and treat infancy FA. Further studies focused on the host-microbe interactions might be helpful in unraveling the mystery of Clostridium sensu stricto in allergic diseases.
The present study also had several limitations that should be acknowledged. First, feces was used as a proxy for gut microbiota in the current study, which was the only realistic sample for large, noninvasive epidemiologic studies. However, fecal microbiota only represents the composition of gut microbiota in the lumen, not on the mucosal surfaces, and mucosa-associated microbiota may affected infancy FA risk primarily through direct interaction with the host. Second, several key phylotypes were detected in infancy FA; however, the exact mechanisms for FA development remain to be identified. The restoration of infancy fecal microbiota after successful treatment also should be performed to verify the role of these key phylotypes in future studies. Third, the establishment of infancy fecal microbiota with different delivery and feeding modes was not considered in the current study, as almost all infants were delivered vaginally and breastfed for >6 months, while different delivery and feeding modes would influence the composition of infancy fecal microbiota and the risk of FA. Fourth, there were no microbiome data on the fecal samples after the infants had been placed on an allergen-free diet, which would strengthen the relationships between fecal microbiota and FA. We conclude from our results that the altered infancy fecal microbiota composition was correlated with immunologic alterations and the risk of developing FA in infants. This study used a comparatively large number of well-characterized FA infants and deep-level pyrosequencing to identify key phylotypes that may associate with the pathology of FA. This is the first study to show that the high relative abundance of Clostridium sensu stricto is associated with an increased risk of IgE-mediated FA in infants. We have extended previous studies and provided new key bacterial phylotypes and associations; some will require further investigation to fully define their role in FA.
Supplementary Material
ACKNOWLEDGMENTS
We thank Chung Owyang from the University of Michigan Health System for his critical reading of our manuscript.
The present work was funded by grants of the National Basic Research Program of China (973 Program), grant 2013CB531404.
We have no conflicts of interest to declare.
Footnotes
Published ahead of print 14 February 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.00003-14.
REFERENCES
- 1.Sackeyfio A, Senthinathan A, Kandaswamy P, Barry PW, Shaw B, Baker M. 2011. Diagnosis and assessment of food allergy in children and young people: summary of NICE guidance. BMJ 342:d747. 10.1136/bmj.d747 [DOI] [PubMed] [Google Scholar]
- 2.Sicherer SH, Sampson HA. 2010. Food allergy. J. Allergy Clin. Immunol. 125:S116–S125. 10.1016/j.jaci.2009.08.028 [DOI] [PubMed] [Google Scholar]
- 3.Noti M, Wojno ED, Kim BS, Siracusa MC, Giacomin PR, Nair MG, Benitez AJ, Ruymann KR, Muir AB, Hill DA, Chikwava KR, Moghaddam AE, Sattentau QJ, Alex A, Zhou C, Yearley JH, Menard-Katcher P, Kubo M, Obata-Ninomiya K, Karasuyama H, Comeau MR, Brown-Whitehorn T, de Waal Malefyt R, Sleiman PM, Hakonarson H, Cianferoni A, Falk GW, Wang ML, Spergel JM, Artis D. 2013. Thymic stromal lymphopoietin-elicited basophil responses promote eosinophilic esophagitis. Nat. Med. 19:1005–1013. 10.1038/nm.3281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang M, Karlsson C, Olsson C, Adlerberth I, Wold AE, Strachan DP, Martricardi PM, Aberg N, Perkin MR, Tripodi S, Coates AR, Hesselmar B, Saalman R, Molin G, Ahrne S. 2008. Reduced diversity in the early fecal microbiota of infants with atopic eczema. J. Allergy Clin. Immunol. 121:129–134. 10.1016/j.jaci.2007.09.011 [DOI] [PubMed] [Google Scholar]
- 5.Noval Rivas M, Burton OT, Wise P, Zhang YQ, Hobson SA, Garcia Lloret M, Chehoud C, Kuczynski J, DeSantis T, Warrington J, Hyde ER, Petrosino JF, Gerber GK, Bry L, Oettgen HC, Mazmanian SK, Chatila TA. 2013. A microbiota signature associated with experimental food allergy promotes allergic sensitization and anaphylaxis. J. Allergy Clin. Immunol. 131:201–212. 10.1016/j.jaci.2012.10.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bisgaard H, Li N, Bonnelykke K, Chawes BL, Skov T, Paludan-Muller G, Stokholm J, Smith B, Krogfelt KA. 2011. Reduced diversity of the intestinal microbiota during infancy is associated with increased risk of allergic disease at school age. J. Allergy Clin. Immunol. 128:646–652. 10.1016/j.jaci.2011.04.060 [DOI] [PubMed] [Google Scholar]
- 7.Russell SL, Gold MJ, Hartmann M, Willing BP, Thorson L, Wlodarska M, Gill N, Blanchet MR, Mohn WW, McNagny KM, Finlay BB. 2012. Early life antibiotic-driven changes in microbiota enhance susceptibility to allergic asthma. EMBO Rep. 13:440–447. 10.1038/embor.2012.32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bjorksten B, Sepp E, Julge K, Voor T, Mikelsaar M. 2001. Allergy development and the intestinal microflora during the first year of life. J. Allergy Clin. Immunol. 108:516–520. 10.1067/mai.2001.118130 [DOI] [PubMed] [Google Scholar]
- 9.Penders J, Thijs C, van den Brandt PA, Kummeling I, Snijders B, Stelma F, Adams H, van Ree R, Stobberingh EE. 2007. Gut microbiota composition and development of atopic manifestations in infancy: the KOALA birth cohort study. Gut 56:661–667. 10.1136/gut.2006.100164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thompson-Chagoyan OC, Vieites JM, Maldonado J, Edwards C, Gil A. 2010. Changes in faecal microbiota of infants with cow's milk protein allergy–a Spanish prospective case-control 6-month follow-up study. Pediatr. Allergy Immunol. 21:e394–e400. 10.1111/j.1399-3038.2009.00961.x [DOI] [PubMed] [Google Scholar]
- 11.Penders J, Gerhold K, Stobberingh EE, Thijs C, Zimmermann K, Lau S, Hamelmann E. 2013. Establishment of the intestinal microbiota and its role for atopic dermatitis in early childhood. J. Allergy Clin. Immunol. 132:601–607. 10.1016/j.jaci.2013.05.043 [DOI] [PubMed] [Google Scholar]
- 12.Mazmanian SK, Liu CH, Tzianabos AO, Kasper DL. 2005. An immunomodulatory molecule of symbiotic bacteria directs maturation of the host immune system. Cell 122:107–118. 10.1016/j.cell.2005.05.007 [DOI] [PubMed] [Google Scholar]
- 13.Rolinck-Werninghaus C, Staden U, Mehl A, Hamelmann E, Beyer K, Niggemann B. 2005. Specific oral tolerance induction with food in children: transient or persistent effect on food allergy? Allergy 60:1320–1322. 10.1111/j.1398-9995.2005.00882.x [DOI] [PubMed] [Google Scholar]
- 14.Hooper LV, Littman DR, Macpherson AJ. 2012. Interactions between the microbiota and the immune system. Science 336:1268–1273. 10.1126/science.1223490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Walsh J, O'Flynn N. 2011. Diagnosis and assessment of food allergy in children and young people in primary care and community settings: NICE clinical guideline. Br. J. Gen. Pract. 61:473–475. 10.3399/bjgp11X583498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen W, Liu F, Ling Z, Tong X, Xiang C. 2012. Human intestinal lumen and mucosa-associated microbiota in patients with colorectal cancer. PLoS One 7:e39743. 10.1371/journal.pone.0039743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Quince C, Lanzen A, Davenport RJ, Turnbaugh PJ. 2011. Removing noise from pyrosequenced amplicons. BMC Bioinformatics 12:38. 10.1186/1471-2105-12-38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Quince C, Lanzen A, Curtis TP, Davenport RJ, Hall N, Head IM, Read LF, Sloan WT. 2009. Accurate determination of microbial diversity from 454 pyrosequencing data. Nat. Methods 6:639–641. 10.1038/nmeth.1361 [DOI] [PubMed] [Google Scholar]
- 19.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Pena AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R. 2010. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7:335–336. 10.1038/nmeth.f.303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Haas BJ, Gevers D, Earl AM, Feldgarden M, Ward DV, Giannoukos G, Ciulla D, Tabbaa D, Highlander SK, Sodergren E, Methe B, DeSantis TZ, Petrosino JF, Knight R, Birren BW. 2011. Chimeric 16S rRNA sequence formation and detection in Sanger and 454-pyrosequenced PCR amplicons. Genome Res. 21:494–504. 10.1101/gr.112730.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ, Sahl JW, Stres B, Thallinger GG, Van Horn DJ, Weber CF. 2009. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 75:7537–7541. 10.1128/AEM.01541-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Charland MB. 1995. Sigma Plot for scientists. McGraw-Hill Professional, Brown Communications, Inc., Dubuque, IA [Google Scholar]
- 23.Wang Q, Garrity GM, Tiedje JM, Cole JR. 2007. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 73:5261–5267. 10.1128/AEM.00062-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.White JR, Nagarajan N, Pop M. 2009. Statistical methods for detecting differentially abundant features in clinical metagenomic samples. PLoS Comput. Biol. 5:e1000352. 10.1371/journal.pcbi.1000352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Segata N, Izard J, Waldron L, Gevers D, Miropolsky L, Garrett WS, Huttenhower C. 2011. Metagenomic biomarker discovery and explanation. Genome Biol. 12:R60. 10.1186/gb-2011-12-6-r60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ling Z, Liu X, Luo Y, Yuan L, Nelson KE, Wang Y, Xiang C, Li L. 2013. Pyrosequencing analysis of the human microbiota of healthy Chinese undergraduates. BMC Genomics 14:390. 10.1186/1471-2164-14-390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim HB, Borewicz K, White BA, Singer RS, Sreevatsan S, Tu ZJ, Isaacson RE. 2012. Microbial shifts in the swine distal gut in response to the treatment with antimicrobial growth promoter, tylosin. Proc. Natl. Acad. Sci. U. S. A. 109:15485–15490. 10.1073/pnas.1205147109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Veiga P, Gallini CA, Beal C, Michaud M, Delaney ML, DuBois A, Khlebnikov A, van Hylckama Vlieg JE, Punit S, Glickman JN, Onderdonk A, Glimcher LH, Garrett WS. 2010. Bifidobacterium animalis subsp. lactis fermented milk product reduces inflammation by altering a niche for colitogenic microbes. Proc. Natl. Acad. Sci. U. S. A. 107:18132–18137. 10.1073/pnas.1011737107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gore C, Munro K, Lay C, Bibiloni R, Morris J, Woodcock A, Custovic A, Tannock GW. 2008. Bifidobacterium pseudocatenulatum is associated with atopic eczema: a nested case-control study investigating the fecal microbiota of infants. J. Allergy Clin. Immunol. 121:135–140. 10.1016/j.jaci.2007.07.061 [DOI] [PubMed] [Google Scholar]
- 30.Abrahamsson TR, Jakobsson HE, Andersson AF, Bjorksten B, Engstrand L, Jenmalm MC. 2012. Low diversity of the gut microbiota in infants with atopic eczema. J. Allergy Clin. Immunol. 129:434–440. 10.1016/j.jaci.2011.10.025 [DOI] [PubMed] [Google Scholar]
- 31.Sogin ML, Morrison HG, Huber JA, Mark Welch D, Huse SM, Neal PR, Arrieta JM, Herndl GJ. 2006. Microbial diversity in the deep sea and the underexplored “rare biosphere.” Proc. Natl. Acad. Sci. U. S. A. 103:12115–12120. 10.1073/pnas.0605127103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dominguez-Bello MG, Costello EK, Contreras M, Magris M, Hidalgo G, Fierer N, Knight R. 2010. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc. Natl. Acad. Sci. U. S. A. 107:11971–11975. 10.1073/pnas.1002601107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.van Nimwegen FA, Penders J, Stobberingh EE, Postma DS, Koppelman GH, Kerkhof M, Reijmerink NE, Dompeling E, van den Brandt PA, Ferreira I, Mommers M, Thijs C. 2011. Mode and place of delivery, gastrointestinal microbiota, and their influence on asthma and atopy. J. Allergy Clin. Immunol. 128:948–971. 10.1016/j.jaci.2011.07.027 [DOI] [PubMed] [Google Scholar]
- 34.Hanski I, von Hertzen L, Fyhrquist N, Koskinen K, Torppa K, Laatikainen T, Karisola P, Auvinen P, Paulin L, Makela MJ, Vartiainen E, Kosunen TU, Alenius H, Haahtela T. 2012. Environmental biodiversity, human microbiota, and allergy are interrelated. Proc. Natl. Acad. Sci. U. S. A. 109:8334–8339. 10.1073/pnas.1205624109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jakobsson HE, Abrahamsson TR, Jenmalm MC, Harris K, Quince C, Jernberg C, Bjorksten B, Engstrand L, Andersson AF. 7 August 2013. Decreased gut microbiota diversity, delayed Bacteroidetes colonisation and reduced Th1 responses in infants delivered by Caesarean section. Gut 10.1136/gutjnl-2012-303249 [DOI] [PubMed] [Google Scholar]
- 36.Human Microbiome Project Consortium. 2012. Structure, function and diversity of the healthy human microbiome. Nature 486:207–214. 10.1038/nature11234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Faith JJ, Guruge JL, Charbonneau M, Subramanian S, Seedorf H, Goodman AL, Clemente JC, Knight R, Heath AC, Leibel RL, Rosenbaum M, Gordon JI. 2013. The long-term stability of the human gut microbiota. Science 341:1237439. 10.1126/science.1237439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Elinav E, Strowig T, Kau AL, Henao-Mejia J, Thaiss CA, Booth CJ, Peaper DR, Bertin J, Eisenbarth SC, Gordon JI, Flavell RA. 2011. NLRP6 inflammasome regulates colonic microbial ecology and risk for colitis. Cell 145:745–757. 10.1016/j.cell.2011.04.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu B, Tonkonogy SL, Sartor RB. 2011. Antigen-presenting cell production of IL-10 inhibits T-helper 1 and 17 cell responses and suppresses colitis in mice. Gastroenterology 141:653–662. 10.1053/j.gastro.2011.04.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nakayama J, Kobayashi T, Tanaka S, Korenori Y, Tateyama A, Sakamoto N, Kiyohara C, Shirakawa T, Sonomoto K. 2011. Aberrant structures of fecal bacterial community in allergic infants profiled by 16S rRNA gene pyrosequencing. FEMS Immunol. Med. Microbiol. 63:397–406. 10.1111/j.1574-695X.2011.00872.x [DOI] [PubMed] [Google Scholar]
- 41.Hayashi A, Sato T, Kamada N, Mikami Y, Matsuoka K, Hisamatsu T, Hibi T, Roers A, Yagita H, Ohteki T, Yoshimura A, Kanai T. 2013. A single strain of Clostridium butyricum induces intestinal IL-10-producing macrophages to suppress acute experimental colitis in mice. Cell Host Microbe 13:711–722. 10.1016/j.chom.2013.05.013 [DOI] [PubMed] [Google Scholar]
- 42.Sokol H, Pigneur B, Watterlot L, Lakhdari O, Bermudez-Humaran LG, Gratadoux JJ, Blugeon S, Bridonneau C, Furet JP, Corthier G, Grangette C, Vasquez N, Pochart P, Trugnan G, Thomas G, Blottiere HM, Dore J, Marteau P, Seksik P, Langella P. 2008. Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proc. Natl. Acad. Sci. U. S. A. 105:16731–16736. 10.1073/pnas.0804812105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Atarashi K, Tanoue T, Shima T, Imaoka A, Kuwahara T, Momose Y, Cheng G, Yamasaki S, Saito T, Ohba Y, Taniguchi T, Takeda K, Hori S, Ivanov II, Umesaki Y, Itoh K, Honda K. 2011. Induction of colonic regulatory T cells by indigenous Clostridium species. Science 331:337–341. 10.1126/science.1198469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chiba T, Seno H. 2011. Indigenous clostridium species regulate systemic immune responses by induction of colonic regulatory T cells. Gastroenterology 141:1114–1116. 10.1053/j.gastro.2011.07.013 [DOI] [PubMed] [Google Scholar]
- 45.Atarashi K, Tanoue T, Oshima K, Suda W, Nagano Y, Nishikawa H, Fukuda S, Saito T, Narushima S, Hase K, Kim S, Fritz JV, Wilmes P, Ueha S, Matsushima K, Ohno H, Olle B, Sakaguchi S, Taniguchi T, Morita H, Hattori M, Honda K. 2013. Treg induction by a rationally selected mixture of clostridia strains from the human microbiota. Nature 500:232–236. 10.1038/nature12331 [DOI] [PubMed] [Google Scholar]
- 46.Klaenhammer TR, Kleerebezem M, Kopp MV, Rescigno M. 2012. The impact of probiotics and prebiotics on the immune system. Nat. Rev. Immunol. 12:728–734. 10.1038/nri3312 [DOI] [PubMed] [Google Scholar]
- 47.Gosalbes MJ, Llop S, Valles Y, Moya A, Ballester F, Francino MP. 2013. Meconium microbiota types dominated by lactic acid or enteric bacteria are differentially associated with maternal eczema and respiratory problems in infants. Clin. Exp. Allergy 43:198–211. 10.1111/cea.12063 [DOI] [PubMed] [Google Scholar]
- 48.Zhang Z, Zhai H, Geng J, Yu R, Ren H, Fan H, Shi P. 2013. Large-scale survey of gut microbiota associated with MHE via 16S rRNA-based pyrosequencing. Am. J. Gastroenterol. 108:1601–1611. 10.1038/ajg.2013.221 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.