Abstract
Water kefir is a sour, alcoholic, and fruity fermented beverage of which the fermentation is started with water kefir grains. These water kefir grains consist of polysaccharide and contain the microorganisms responsible for the water kefir fermentation. In this work, a water kefir fermentation process was followed as a function of time during 192 h to unravel the community dynamics, the species diversity, and the kinetics of substrate consumption and metabolite production. The majority of the water kefir ecosystem was found to be present on the water kefir grains. The most important microbial species present were Lactobacillus casei/paracasei, Lactobacillus harbinensis, Lactobacillus hilgardii, Bifidobacterium psychraerophilum/crudilactis, Saccharomyces cerevisiae, and Dekkera bruxellensis. The microbial species diversities in the water kefir liquor and on the water kefir grains were similar and remained stable during the whole fermentation process. The major substrate, sucrose, was completely converted after 24 h of fermentation, which coincided with the production of the major part of the water kefir grain polysaccharide. The main metabolites of the fermentation were ethanol and lactic acid. Glycerol, acetic acid, and mannitol were produced in low concentrations. The major part of these metabolites was produced during the first 72 h of fermentation, during which the pH decreased from 4.26 to 3.45. The most prevalent volatile aroma compounds were ethyl acetate, isoamyl acetate, ethyl hexanoate, ethyl octanoate, and ethyl decanoate, which might be of significance with respect to the aroma of the end product.
INTRODUCTION
Water kefir is a fermented beverage that is made by adding water kefir grains, which are polysaccharide grains that serve as the inoculum, to a mixture of water, sugar (sucrose), dried figs, and possibly other ingredients such as lemon, depending on the recipe (1–5). After 2 to 4 days of anaerobic incubation at room temperature, a sparkling, yellowish fermented beverage is obtained that has a fruity, acidic, slightly sweet, and slightly alcoholic taste and aroma. Water kefir is available worldwide, but it is still unknown what the real origin of the water kefir grains is. It has been postulated that the polysaccharide grains originate from the leaves of the Opuntia cactus fig plant (6). Besides the use of the name “water kefir grains” in western Europe, other names are also in use for this fermented beverage inoculum, depending on the geographic location, such as “ginger beer plants,” “Tibicos,” “Tibi grains,” “California bees,” “African bees,” “ale nuts,” “balm of Gilead,” “Bèbées,” “Japanese beer seeds,” and “sugary kefir grains” (2, 5–9).
Currently, research on water kefir is still very limited and most of the scientific information available deals with its species diversity (2, 4, 5, 8–16). Also, the chemical and structural composition of the water kefir grain polysaccharide has been studied (6, 7, 10–12, 15, 17–19). To date, it is known that the microbial species diversity of water kefir consists of a stable consortium of mainly lactic acid bacteria, yeasts, and acetic acid bacteria, as shown by both culture-dependent and culture-independent techniques (2, 4, 19, 20). Recently, Bifidobacterium psychraerophilum/crudilactis was also found in water kefir via culture-dependent and culture-independent techniques (2). It became clear, however, that different water kefirs display different species diversities. Hence, a systematic approach for the study of the microbiology of water kefir fermentation is necessary. Further, it is known that the water kefir grain polysaccharide consists of dextran, an α-(1→6)-linked glucose polymer, produced by certain Lactobacillus and/or Leuconostoc species (5, 7, 11, 17–19). However, until now no thorough metabolite analysis has been performed on a water kefir fermentation process.
The aim of the present study was to gain a deeper understanding of the water kefir fermentation process, in particular, its microbial species diversity, community dynamics, substrate consumption profile, and metabolite production course.
MATERIALS AND METHODS
Preparation of the inoculum.
To prepare an inoculum, approximately 100 g of water kefir grains was obtained from a private person, who cultivated water kefir at home (Ghent, Belgium). To obtain the necessary amount of kefir grains (± 600 g) to start an actual water kefir fermentation process, a series of consecutive prefermentations was performed in a common water kefir cultivation medium that was used at the household level. Therefore, for every 15 g of water kefir grains, 6 g of unrefined cane sugar (Candico Bio, Merksem, Belgium), 85 ml of tap water, and 5 g of dried figs (King Brand, Naziili, Turkey) were added. These prefermentations were consecutively performed in Schott bottles (1, 2, and 5 liters), each with a water lock composed of polytetrafluoroethylene (PTFE), that were incubated in a water bath at 21°C. A backslopping practice was applied every 72 h. Therefore, the water kefir grains were separated from the fermenting medium by sieving and recultivated in fresh medium under the same conditions. After each backslopping, the water kefir grain mass increased; this practice was applied until >600 g of water kefir grains was obtained.
Water kefir fermentation process.
The precultivated water kefir grains served as the inoculum for the water kefir fermentations (performed in triplicate using 12 bottles per fermentation) in 100-ml Schott bottles equipped with a water lock. Each bottle fermentation was started at the same time with 15 g of water kefir grains and 85 ml of sterile water kefir simulation medium. The water kefir simulation medium was prepared with 6 g of unrefined cane sugar (Candico Bio), 65 ml of tap water, and 20 ml of fig extract. Fig extract was prepared by adding 20 ml of distilled water to 5 g of dried figs, after which this suspension was mixed finely and centrifuged (7,200 × g, 20 min, 4°C). The supernatant was filtered through a coffee filter to obtain the final fig extract. The fermentation bottles were incubated in a water bath at 21°C. After closure of the bottles at the start of the fermentations, as well as before each sampling, the contents were homogenized by mildly turning the bottles. After 0, 3, 6, 12, 18, 24, 36, 48, 72, 96, 144, and 192 h of fermentation, the contents of three Schott bottles (representing the three independent fermentations) were analyzed. The results at each sampling point are presented as the mean ± standard deviation.
Water kefir grain mass and pH measurements.
After opening the bottles, their contents were sieved to separate the water kefir grains from the water kefir liquor. The water kefir grains were washed with 200 ml of sterile saline solution (8.5 g liter−1 of NaCl [Merck KGaA, Darmstadt, Germany]), and the total water kefir grain mass (wet mass) was weighed. Then, approximately 5 g of these water kefir grains was transferred into an aluminum recipient and dried at 105°C for 48 h to determine the dry mass of the water kefir grains. The pH of the water kefir liquor was measured with a SenTix 41 glass electrode (WTW GmbH, Weilheim, Germany).
Microbial enumerations.
To enumerate microorganisms in the water kefir liquor, the water kefir liquor obtained as described above was used to prepare appropriate decimal dilutions in sterile saline. For the enumerations of the microorganisms on the water kefir grains, 5.0 g of washed water kefir grains obtained as described above was brought into a sterile stomacher plastic bag. These grains were crushed by rolling a glass bottle over the outside of the bag, after which 45 ml of sterile maximum recovery diluent (8.5 g liter−1 of NaCl [Merck] and 1 g liter−1 of bacteriological peptone [Oxoid, Basingstoke, Hampshire, United Kingdom]) were added. This mixture was homogenized for 15 min at high speed in a Stomacher 400 instrument (Seward, Worthington, United Kingdom). The resulting water kefir grain suspension was used to prepare appropriate decimal dilutions in sterile saline solution.
The diluted suspensions were plated on de Man, Rogosa, and Sharpe (MRS) agar medium, modified deoxycholate-mannitol-sorbitol (mDMS) agar medium, and yeast extract-glucose (YG) agar medium to enumerate presumptive lactic acid bacteria, acetic acid bacteria, and yeasts, respectively (21, 22). In addition, cycloheximide (Sigma-Aldrich, Saint Louis, MO) (final concentration of 0.1 g liter−1) was added to the MRS and mDMS agar media to inhibit fungal growth, whereas chloramphenicol (Sigma-Aldrich) (final concentration of 0.1 g liter−1) was added to YG agar medium to inhibit bacterial growth. Further, the water kefir liquor and water kefir grain suspensions at 0 and 72 h of fermentation were plated on kanamycin-esculin-azide (KAA) agar medium (Oxoid) and violet-red-bile-glucose (VRBG) agar medium (Oxoid) to enumerate presumptive enterococci plus streptococci and to enumerate Enterobacteriaceae, respectively. MRS, mDMS, and YG agar media were incubated at 30°C for 2 to 4 days; KAA and VRBG agar media were incubated at 42°C for 24 h. Colony enumerations were expressed as log CFU per ml of water kefir liquor or per g of water kefir grains.
Culture-dependent species diversity analysis.
The culture-dependent species diversity analysis of lactic acid bacteria (based on MRS agar isolates), acetic acid bacteria (based on mDMS agar isolates), and yeasts (based on YG agar isolates) in the water kefir liquor and on the water kefir grains was assessed after 0, 24, 48, 72, and 192 h of fermentation. This was done by randomly picking up colonies (10% to 20% of the total colony count) from the agar plates with 30 to 300 colonies. The bacterial and yeast colonies were subcultivated in MRS medium (30°C, 24 h) and yeast extract-glucose-peptone (YGP) medium (30°C, 24 h), respectively. These cultures were supplemented with glycerol (final concentration of 25% [vol/vol]) and stored at −80°C.
To harvest cells for DNA extraction, 2 ml of overnight cultures was centrifuged (21,000 × g, 5 min, 4°C) and the supernatant was discarded. The DNA of the bacterial and yeast cultures was extracted and purified using a NucleoSpin 96 tissue kit (Macherey-Nagel GmbH, Düren, Germany), according to the instructions of the manufacturer. Bacterial cultures were treated with mutanolysin (Sigma-Aldrich) and lysozyme (VWR), and the yeast cultures were treated with lyticase (Sigma-Aldrich). Bacterial DNA was diluted to approximately 50 ng μl−1 and was subjected to (GTG)5-PCR fingerprinting as described before (23). Yeast DNA was diluted to approximately 20 ng μl−1 and was subjected to M13-PCR fingerprinting as described before (24). The fingerprint patterns obtained were clustered into similarity trees (based on the Pearson correlation coefficient and the unweighted-pair group method using average linkages [UPGMA] algorithm) with Bionumerics 5.10 software (Applied Maths, Sint-Martems-Latem, Belgium). Identification of the clusters was performed by the identification of several representatives within each cluster.
For identification of the bacteria, genomic DNA of the selected isolates was subjected to a PCR assay to amplify the 16S rRNA gene (1.5 kb) with primer pair pA and pH (25). For the identification of the yeast isolates, primer pair ITS1 and ITS4 (26) was used to amplify the internal transcribed spacer (ITS) region (variable length) and primer pair LR0R and LR3 (27) was used to amplify a part of the large subunit (LSU) rRNA gene (0.6 kb). These PCR amplicons were purified with a Wizard Plus SV Minipreps DNA purification system (Promega, Madison, WI) and sequenced in a commercial facility (Macrogen, Amsterdam, The Netherlands). The closest relatives of these sequenced fragments were identified with the BLAST algorithm (28) and the GenBank database (http://blast.ncbi.nlm.nih.gov/).
Culture-independent species diversity analysis.
For the culture-independent analyses of the microorganisms in the water kefir liquor and on the water kefir grains, 40 ml of sieved water kefir liquor and 10 ml of a water kefir grain suspension were centrifuged (7,200 × g, 20 min, 4°C). These pellets were resuspended in 2 ml of TES buffer (6.7% [mass/vol] sucrose [Merck], 50 mM Tris base [Merck], 1 mM EDTA [Sigma-Aldrich], pH 8.0) and centrifuged (21,000 × g, 20 min, 4°C). The resulting pellets were used for DNA extraction. Therefore, the pellets were treated with 200 U of lyticase (Sigma-Aldrich) in 600 μl of sorbitol buffer (30°C, 60 min), centrifuged (21,000 × g, 5 min, 4°C), and further processed as described previously (29). The DNA obtained was further purified with a NucleoSpin food kit (Macherey-Nagel), according to the instructions of the manufacturer. The purified DNA was diluted to approximately 50 ng μl−1 before PCR assays were performed. To assess the bacterial diversity, universal 16S rRNA primer pair 357f-GC and 518r (V3) was used (30). Group-specific primer pair LAC1 and LAC2-GC (LAC) was used for lactic acid bacteria (31), and genus-specific primer pair bif164f and bif662r-GC (Bif) was used for bifidobacteria (32). The yeast diversity was assessed with universal eukaryotic primer pair NL1-GC and LS2 (yeast) (33). A GC clamp was attached to one primer of each primer pair, as indicated. The PCR amplicons were separated in a 6% (vol/vol) polyacrylamide gel via denaturing gradient gel electrophoresis, as described before (21, 31). The denaturing gradients of the gels were, from top to bottom, 45% to 60% for the V3 and the yeast primer pairs, 40% to 55% for the LAC primer pair, and 45% to 55% for the Bif primer pair. Gel processing and DNA band sequencing were performed as described previously (21, 31). Identification of the DNA band sequences was performed as described above.
Substrate consumption and metabolite production.
For the measurement of the substrates, the metabolites, and the aroma compounds, sieved water kefir liquor was centrifuged (7,200 × g, 20 min, 4°C) to obtain cell-free supernatant. Carbohydrate concentrations were measured through high-performance anion-exchange chromatography with pulsed amperometric detection (HPAEC-PAD) as described before (34). Therefore, 50 μl of cell-free supernatant was added to 950 μl of ultrapure water. From this dilution, 50 μl was added to 950 μl of deproteinization solution (500 μl of acetonitrile [Sigma-Aldrich], 449.5 μl of ultrapure water, and 0.5 μl of 50 g liter−1 of rhamnose [internal standard; Sigma-Aldrich]).
Glycerol and mannitol concentrations were measured through HPAEC-PAD as described before (35). Therefore, 100 μl of cell-free supernatant was added to 400 μl of ultrapure water. From this dilution, 100 μl was added to 900 μl of deproteinization solution. Quantification was performed with an external standard curve with standards prepared in the same way as the samples.
Lactic acid and acetic acid concentrations were measured through high-performance liquid chromatography with refractive index detection, as described previously (36). Therefore, 800 μl of cell-free supernatant was added to 200 μl of 50% (mass/vol) trichloroacetic acid. Quantification was performed with an external calibration curve with standards prepared in the same way as the samples.
Ethanol concentrations were measured through gas chromatography (GC) with flame ionization detection, as described before (37). Therefore, 100 μl of cell-free supernatant was added to 1,100 μl of deproteinization solution (720 μl of acetonitrile [Sigma-Aldrich], 367.7 μl of ultrapure water, 12 μl of formate [VWR], and 0.3 μl of 1-butanol [internal standard; Merck]). Quantification was performed with an external standard curve with standards prepared in the same way as the samples.
After deproteinization, all samples were subjected to vortex mixing, centrifuged (21,000 × g, 20 min), filtered (0.2-μm-pore-size Whatman filters; GE Healthcare Life Sciences, Bucks, United Kingdom), and injected into the column.
Volatile aroma compounds in the water kefir liquor were measured through static headspace gas chromatography coupled to mass spectrometry (SH-GC-MS). Therefore, 5 ml of cell-free supernatant was brought into a 20-ml glass headspace vial (Gerstel GmbH & Co. KG, Mülheim-an-der-Ruhr, Germany) and closed with a magnetic screw cap (18-mm diameter) with a silicon/PTFE septum (Gerstel GmbH & Co. KG). Before analysis, 1.5 g of NaCl and 100 μl of internal standard solution (0.5 ml liter−1 of 4-methyl-2-pentanol [Sigma-Aldrich]) were added. The headspace vials were equilibrated at 40°C for 30 min at 400 rpm in a MPS2 Gerstel autosampler (Gerstel GmbH & Co. KG) and further analyzed as described before (23). The compounds were identified by comparison of the mass spectra with library data (NIST 08 database; http://www.nist.gov). For quantification, an external calibration curve was constructed to get an indication of the concentrations present. The standards were prepared in ultrapure water and analyzed in the same way as the samples. All volatile aroma compounds found in the water kefir fermentation samples taken after 72 h of fermentation were compared with their threshold values as reported in the literature.
Carbon recovery.
At each sampling time, the carbon recovery was calculated as the total amount of carbon at that sampling time point divided by the total amount of carbon at 0 h. The total amount of recovered carbon was calculated as the sum of the amount of carbon in the water kefir liquor plus that in the water kefir grains plus that produced as carbon dioxide. For these calculations, the measurements of the water kefir grain mass and water kefir grain dry mass and the measurements of the sucrose, glucose, fructose, ethanol, lactic acid, glycerol, acetic acid, and mannitol concentrations were used. It was assumed that the water kefir grain density was 1 g cm−3, that the dried water kefir grain mass consisted of pure polysaccharide, and that the ethanol and acetic acid present in the water kefir grain matrix were evaporated during the dry mass determinations. The ethanol and acetic acid concentrations in the water kefir grain matrix were assumed to be the same as those in the water kefir liquor.
RESULTS
Water kefir grain mass and pH measurements.
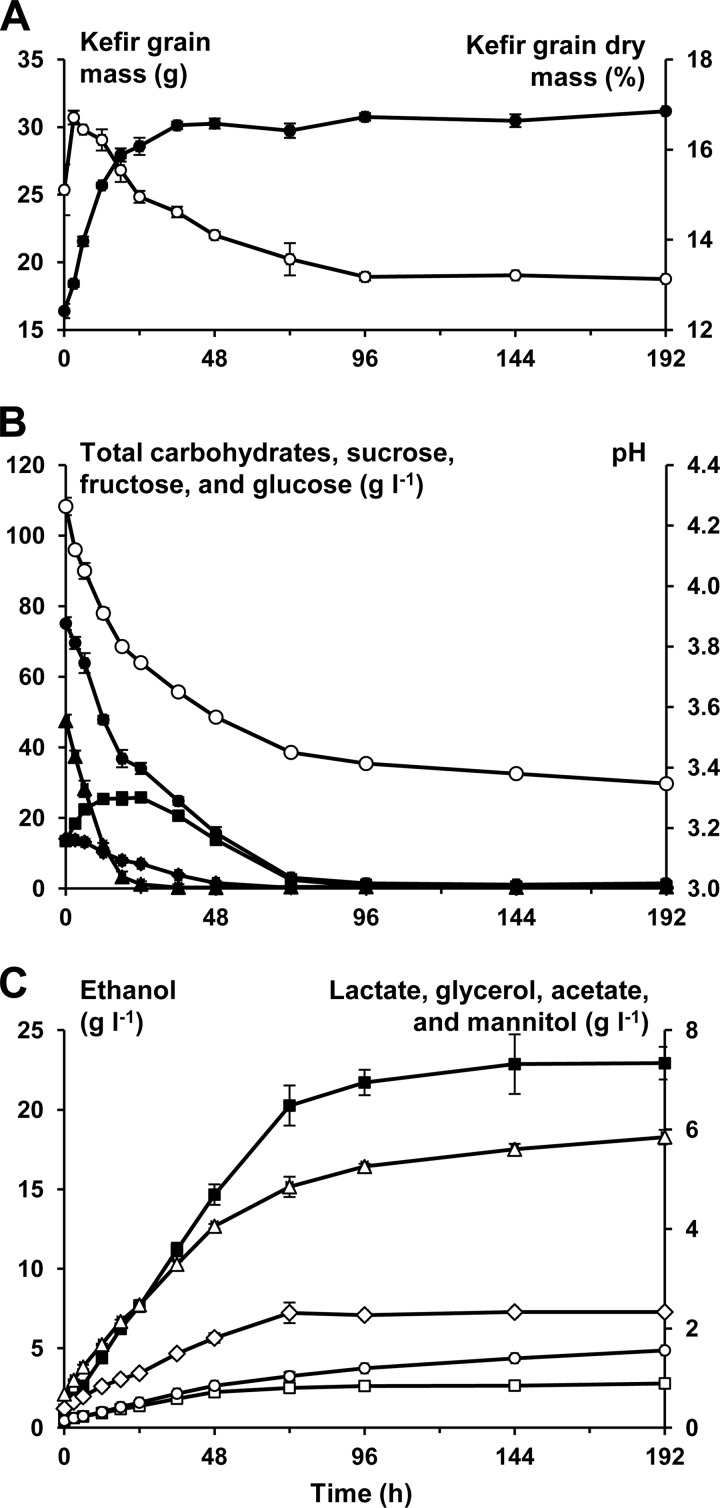
A water kefir fermentation process, using inoculation performed with water kefir grains that were obtained through a series of prefermentations, was carried out in triplicate. The water kefir grain mass increased from 16.4 ± 0.5 to 28.6 ± 0.6 g during the first 24 h of the fermentation; thereafter, the mass remained constant. The water kefir dry mass initially increased from 13.8% ± 0.1% (mass/mass) at 0 h (inoculum not yet added to the water kefir simulation medium) to 16.7% ± 0.2% (mass/mass) after 3 h of fermentation. Thereafter, the dry mass decreased until it remained stable at 13% to 14% (mass/mass) (Fig. 1A).
FIG 1.
(A) The production of water kefir grain mass (g, •) and of water kefir grain dry mass (%, ○) as a function of time (h). (B) The pH evolution and the consumption of carbohydrates (g liter−1) as a function of time (h). pH, ○; total carbohydrates, •; sucrose, ▲; fructose, ■; glucose, ⧫. (C) The production of metabolites (g liter−1) as a function of time (h). Ethanol, ■; lactate, Δ; glycerol, ♢; acetate, ○; mannitol, □.
The initial pH of the water kefir simulation medium was 4.85 ± 0.01. This value dropped to 4.26 ± 0.03 after the addition of the water kefir grains at 0 h. After 72 h of fermentation, the pH reached 3.45 ± 0.01, whereafter the pH decreased only slowly to reach 3.35 ± 0.01 after 192 h of fermentation (Fig. 1B).
Microbial enumerations.
The viable counts of the water kefir liquor and water kefir grains on the MRS and YG agar media remained constant during the whole fermentation process. Immediately after the water kefir grains were added to the water kefir simulation medium and the bottle was mildly turned, the viable counts of the lactic acid bacteria and the yeasts in the water kefir liquor plateaued at a certain level. The average viable counts of yeasts and lactic acid bacteria were 6.3 ± 0.2 and 6.9 ± 0.1 log CFU ml−1 of water kefir liquor, respectively, and 7.4 ± 0.1 and 8.2 ± 0.1 log CFU g−1 of water kefir grains, respectively. Quantifiable levels of acetic acid bacteria (>30 colonies on the agar medium with the lowest dilution) could be found only at 144 h and 192 h on the mDMS agar media from the water kefir liquor, namely, 3.8 ± 0.1 and 6.2 ± 0.1 log CFU ml−1, respectively. In the case of the water kefir grains, acetic acid bacteria could be quantified only after 192 h of fermentation (4.6 ± 0.1 log CFU g−1). No colonies were found on the KAA and the VRBG agar media, indicating the absence of enterococci plus streptococci and of Enterobacteriaceae, respectively.
The ratios of the viable counts of lactic acid bacteria to those of yeasts were also relatively constant during the whole fermentation, with averages of 4.4 ± 1.2 and 6.1 ± 2.4 in the water kefir liquor and on the water kefir grains, respectively, indicating that there were 2 to 10 lactic acid bacterial cells for each yeast cell, both in the water kefir liquor and on the water kefir grains. The ratios of the viable counts of lactic acid bacteria and yeasts on the water kefir grains (CFU g−1) to those in the water kefir liquor (CFU ml−1) were constant, too, with averages of 20.4 ± 8.4 and 14.7 ± 4.5, respectively, indicating that the cell density was 10 to 30 times higher on the water kefir grains than in the water kefir liquor. Taking the amounts of water kefir grains and of water kefir liquor into account, the ratios of the total amounts of cells on the water kefir grains (CFU) to those in the water kefir liquor (CFU) were again relatively constant, with averages of 8.8 ± 1.6 and 6.5 ± 1.5 for lactic acid bacteria and yeasts, respectively, indicating that there were 4 to 10 times more microorganisms on the water kefir grains than in the water kefir liquor. However, because the water kefir grain mass, with higher viable counts than the water kefir liquor, increased in mass as a function of time, there was an overall increase of the total cell count during the first 48 h of the fermentation.
Culture-dependent species diversity analysis and community dynamics.
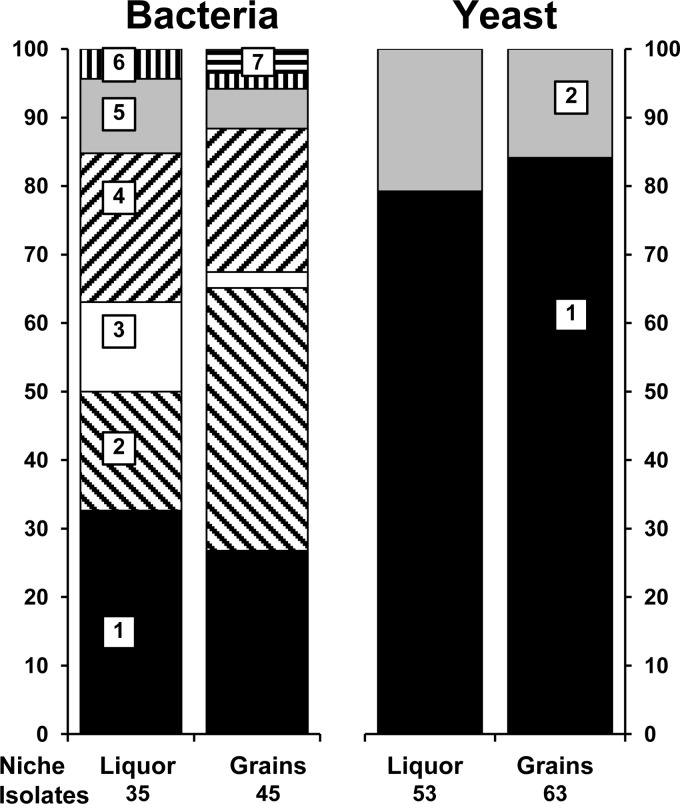
The culture-dependent bacterial species diversity analysis revealed the presence of Lactobacillus casei/paracasei, Lactobacillus hilgardii, Lactobacillus harbinensis, Lactobacillus nagelii, Acetobacter lovaniensis/fabarum, and Lactobacillus hordei/mali (in decreasing order), of which the first 3 were the most dominant (Fig. 2). The bacterial species diversities in the water kefir liquor and on the water kefir grains were similar and were more or less constant as a function of the fermentation time (data not shown). All mDMS agar isolates picked up after 192 h of fermentation were identified as Acetobacter lovaniensis/fabarum. The culture-dependent yeast species diversity analysis revealed the presence of two species, namely, Saccharomyces cerevisiae and Dekkera bruxellensis (Fig. 2). Also, the yeast species diversities in the water kefir liquor and on the water kefir grains were similar and were stable during the whole fermentation (data not shown).
FIG 2.
The microbial species diversity of the water kefir liquor and water kefir grains, represented by pooling all samplings. The closest relatives to the sequenced fragments are given. Left panel (bacterial species diversity): 1, Lactobacillus casei/paracasei (99% identity; accession no. KF500575/KF516078); 2, Lactobacillus hilgardii (99% identity; accession no. JX099894); 3, Lactobacillus nagelii (99% identity; accession no. AB370876); 4, Lactobacillus harbinensis (100% identity; accession no. KF418816); 5, Acetobacter lovaniensis/fabarum (99% identity; accession no. FJ157228/AB665084); 6, Lactobacillus mali (99% identity; accession no. AB326352); 7, unknown. Right panel (yeast species diversity): 1, Saccharomyces cerevisiae (LSU [99% identity; accession no. JQ914745] and ITS [99% identity; accession no. KC515374]); 2, Dekkera bruxellensis (LSU [100% identity; accession no.: JQ689028] and ITS [100% identity; accession no. FJ545249]).
Culture-independent species diversity analysis and community dynamics.
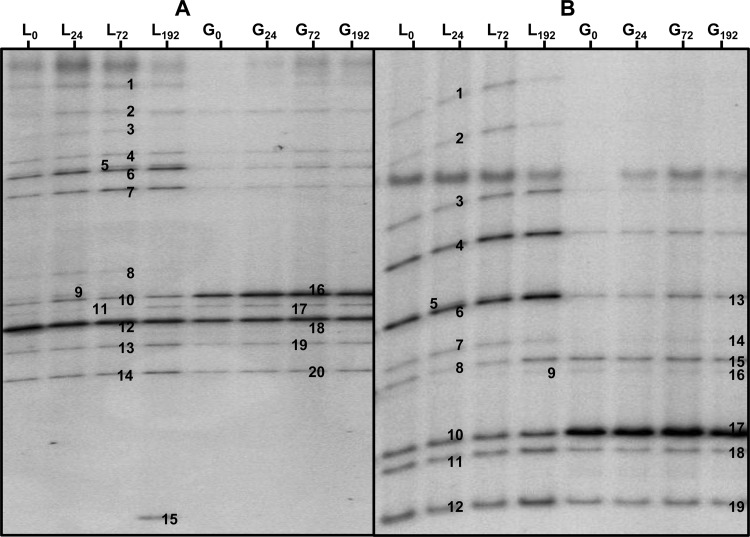
Via the culture-independent assessment of the microbial species diversity, it was confirmed that the three independent fermentations displayed identical PCR-denaturing gradient gel electrophoresis (PCR-DGGE) profiles with the primers used (V3, LAC, Bif, and yeast) at 0, 24, 72, and 192 h (data not shown). With the V3 primer pair, Lb. hordei/mali, Lb. harbinensis/brevis, Lb. casei/paracasei/rhamnosus, Lb. hilgardii/diolivorans, Lb. nagelii/ghanensis, B. psychraerophilum/crudilactis, and a species of Acetobacteraceae were found (Fig. 3). The latter appeared in the PCR-DGGE profile (V3 primer pair) of the water kefir liquor after 192 h of fermentation (for the three replicates), but this band was not present in the samples of the water kefir grains (Fig. 3).
FIG 3.
The PCR-DGGE profiles of the water kefir liquor (L) and the water kefir grains (G) after 0, 24, 72, and 192 h (indicated as subscripts) of fermentation. The closest relatives to the fragments sequenced are given. (A) Use of the universal V3 primer pair. 1, 2, 3, 5, and 7: Lactobacillus hordei/mali (98% identity; accession no. NR044394/AB326352); 4, unspecified bacterium; 6, Lactobacillus harbinensis/brevis (100% identity; accession no. KF418816/AY974809); 8, 9, 13, 14, 19, and 20, Lactobacillus casei/rhamnosus/paracasei (100% identity; accession no. KF500575/KF554252/KF516078); 10 and 16, Lactobacillus hilgardii/diolivorans (100% identity; accession no. KF418826/KF149643); 11 and 17, Lactobacillus ghanensis/nagelii (97% identity; accession no. AB690235/AB370876); 12 and 18, Bifidobacterium psychraerophilum (98% identity; accession no. AB437351); 15, Acetobacteraceae. (B) Use of the LAC primer pair. 5, 7, 11, 12, 14, 18, and 19, Lb. casei/paracasei (99% identity; accession no. KF500575/KF516078); 4, 8, 9, 15, and 16, Lb. nagelii (99% identity; accession no. AB370876); 1, 2, and 3, Lb. hordei/mali (99% identity; accession no. NR044394/AB326352); 6 and 13, Lb. harbinensis/brevis (99% identity; accession no. KF418816/AY974809); 10 and 17, Lb. hilgardii/diolivorans (99% identity; accession no. KF418826/KF149643).
The presence of the lactic acid bacteria detected with the universal V3 primers was confirmed with the use of the LAC primer pair. With this primer pair, Lb. hordei/mali, Lb. nagelii/ghanensis, Lb. casei/paracasei, Lb. hilgardii/diolivorans, and Lb. harbinensis/brevis were found. PCR-DGGE with the Bif primer pair was used to confirm the presence of bifidobacteria in water kefir fermentation. Only one band, identified as B. psychraerophilum (96% identity; GenBank accession no. AB437351), could be found in this gel (data not shown). However, this sequence was very similar to the sequence of an uncultivated Bifidobacterium species (100% identity; accession no. HE804184) found in water kefir grains. This band was present during the whole fermentation process in the water kefir liquor as well as on the water kefir grains. The PCR-DGGE analysis performed with the yeast primer pair confirmed the culture-dependent results. Both S. cerevisiae (100% identity; accession no. JX068683) and D. bruxellensis (100% identity; accession no. AY969049) were detected during the whole fermentation process. The PCR-DGGE profiles with the LAC, Bif, and yeast primer pairs showed no evolution of the species diversity as a function of time during the whole water kefir fermentation process.
Substrate consumption and metabolite production.
Sucrose was the main substrate present at the start of the fermentation (0 h). The concentration of sucrose decreased quickly from 47.5 ± 1.7 g liter−1 at 0 h to 1.2 ± 0.8 g liter−1 after 24 h of fermentation. This decrease in sucrose concentration gave rise to an increase in the fructose concentration, which reached a maximum after 24 h of fermentation. This was in contrast with the glucose concentration, which decreased continuously during the fermentation. After 72 h, most of the carbohydrates were consumed, with only 3.1 ± 1.0 g liter−1 of total carbohydrates left of the initial 75.1 ± 2.1 g liter−1 (Fig. 1B).
The ethanol concentration increased linearly from 1.1 ± 0.1 g liter−1 at 0 h to 20.3 ± 1.3 g liter−1 after 72 h. In this time frame, the lactic acid concentration increased from 0.7 ± 0.1 g liter−1 to 4.9 ± 0.2 g liter−1 and the acetic acid concentration increased from 0.1 ± 0.0 g liter−1 to 1.0 ± 0.1 g liter−1. The glycerol and mannitol concentrations reached 2.3 ± 0.2 g liter−1 and 0.8 ± 0.0 g liter−1 after 72 h of fermentation, respectively (Fig. 1C).
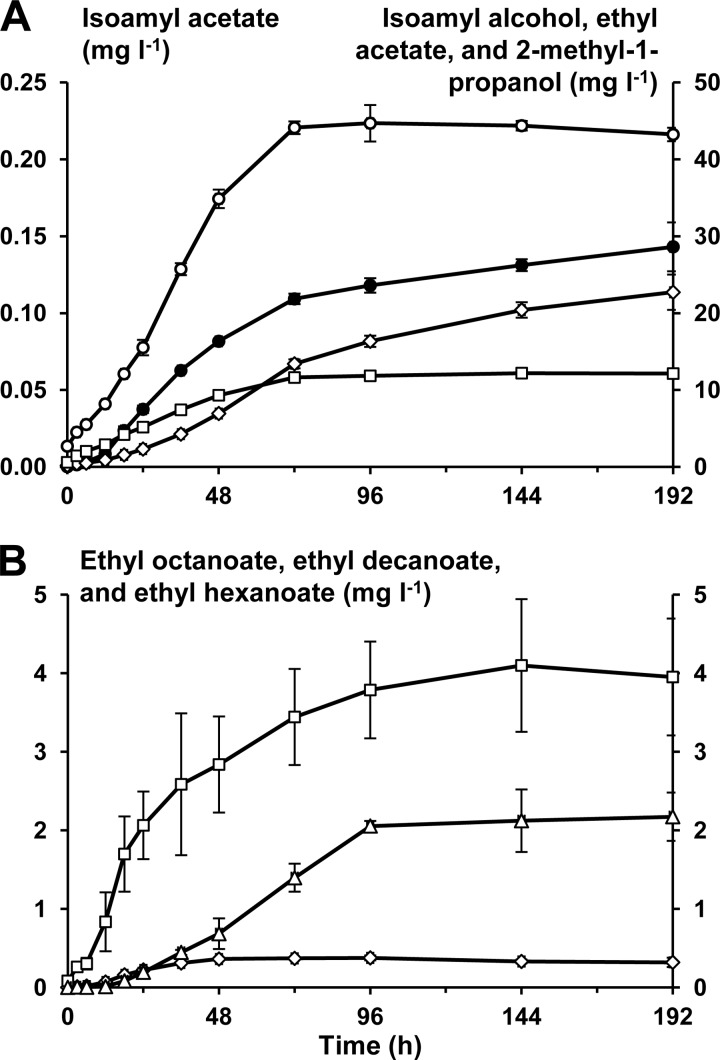
Only a limited amount of volatile aroma compounds (besides acetic acid and ethanol) could be found with the SH-GC-MS method used. These compounds were ethyl acetate, 2-methyl-propanol, isoamyl alcohol, isoamyl acetate, ethyl hexanoate, ethyl octanoate, and ethyl decanoate. Ethyl butanoate and ethyl 2-methyl-butanoate were also found, but these two compounds could not be quantified because their concentrations were too close to the quantification limit of the method used. Except for ethyl acetate and ethyl decanoate, the production of these compounds stopped as soon as the carbohydrates were exhausted (after 72 h of fermentation). The production of ethyl acetate and ethyl decanoate, however, continued until 192 h and 96 h of fermentation, respectively (Fig. 4). Compared to their threshold levels, the most dominant aroma components of this water kefir after 72 h were the esters (Table 1). In particular, ethyl octanoate was present at 688 times its threshold value.
FIG 4.
(A) The production of volatile short-chain aroma compounds (mg liter−1) as a function of time (h). Isoamyl acetate, •; isoamyl alcohol, ○; ethyl acetate, ♢; 2-methyl-1-propanol, □. (B) The production of volatile long-chain aroma compounds (mg liter−1) as a function of time (h). Ethyl octanoate, □; ethyl decanoate, Δ; ethyl hexanoate, ♢.
TABLE 1.
Concentrations of volatile aroma compounds found after 72 h of water kefir fermentationa
| Volatile aroma compound | Concn after 72 h (mg liter−1) | KI | Id | Threshold valuec (mg liter−1) | Aroma descriptord |
|---|---|---|---|---|---|
| 2-Methyl-1-propanol | 11.62 ± 0.05 | 1,097 | MS/RF | 40 | Spirituous, fuel |
| Isoamyl alcohol | 44.13 ± 0.82 | 1,222 | MS/RF | 30 | Harsh, nail polish remover |
| Ethyl acetate | 13.40 ± 0.58 | 831 | MS/RF | 7.5 | Fruity |
| >150 | Varnish, nail polish remover | ||||
| Isoamyl acetate | 0.11 ± 0.01 | 1,141 | MS/RF | 0.03 | Sweet, fruity, banana, pear |
| Ethyl hexanoate | 0.37 ± 0.01 | 1,250 | MS/RF | 0.014 | Fruity, apple, banana, violets |
| Ethyl octanoate | 3.44 ± 0.61 | 1,450 | MS/RF | 0.005 | Fruity, pineapple, pear |
| Ethyl decanoate | 1.40 ± 0.18 | 1,659 | MS/RF | 0.2 | Floral |
| Ethyl butanoate | DNQb | 1,043 | MS | 0.02 | Floral, fruity |
| Ethyl 2-methyl-butanoate | DNQb | 1,058 | MS | 0.001 | Fruity, strawberry, pineapple |
The Kovats index (KI) and the method of identification (Id) are given for every compound. Identification was via the mass spectrum (MS) and by comparison with the retention time of the reference compound (RF).
Detected but not quantified.
Carbon recovery.
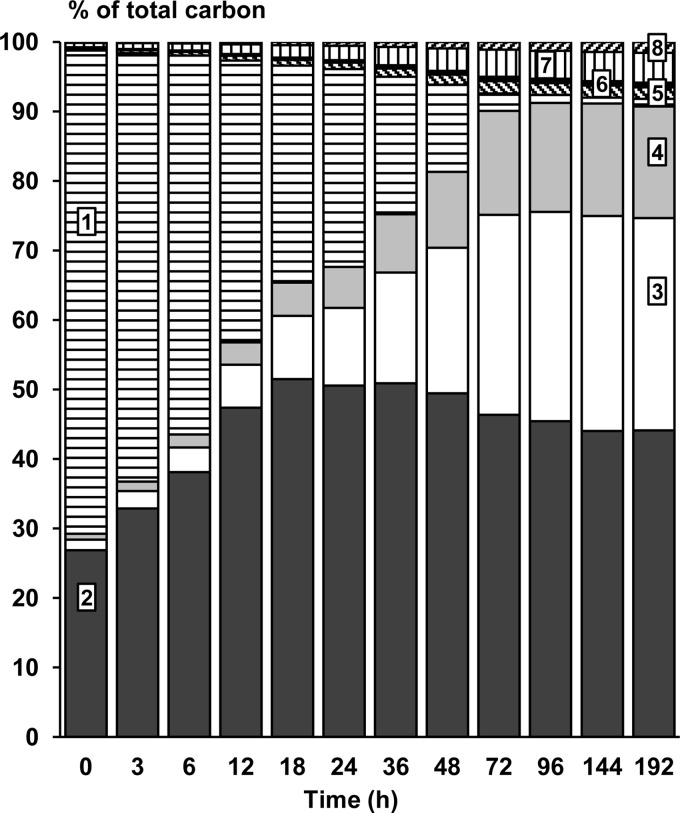
After 192 h of fermentation, a carbon recovery level of 100.6% was obtained, indicating that all major substrates and metabolites were recovered from this water kefir fermentation. After 72 h of fermentation, the majority of the metabolic activity had taken place. The major end products of the fermentation were ethanol, carbon dioxide, lactic acid, glycerol, mannitol, and acetic acid, besides the synthesis of water kefir grain mass (Fig. 5).
FIG 5.
Presence of different carbon-containing constituents of the water kefir fermentation process, as a function of time (h), expressed as a percentage (%) of the total amount of carbon recovered. 1, substrates; 2, kefir grain mass; 3, ethanol; 4, carbon dioxide; 5, glycerol; 6, mannitol; 7, lactate; 8, acetate.
DISCUSSION
The multiphasic microbial approach of the present study revealed lactic acid bacteria, yeasts, bifidobacteria, and acetic acid bacteria as the main microorganisms present during water kefir fermentation (2, 4, 19, 20). The species diversities in the water kefir liquor and on the water kefir grains were similar and remained stable in both phases during the whole fermentation process. However, the density of the microorganisms was higher on the water kefir grains than in the water kefir liquor, indicating that the water kefir grains harbored the microorganisms. This finding also explains the absence of an increasing microbial population during the water kefir fermentation process. Although the yeast metabolism predominated the fermentation, lactic acid bacteria were present in higher numbers than yeasts. As common fermentation times for water kefir are around 72 h, the acetic acid bacteria, appearing only after 144 h of fermentation, were not an important part of the water kefir ecosystem of the present study. Communities of acetic acid bacteria found in water kefir usually range from negligible (10) to >8 log CFU ml−1 (4). The absence of enterococci and Enterobacteriaceae was to be expected, considering the fast decrease of the pH during water kefir fermentation to a pH below 3.5.
The most dominant lactic acid bacteria (in decreasing order) were Lb. casei/paracasei, Lb. hilgardii, and Lb. harbinensis. The former two species have been associated with water kefir before (1, 2, 4, 5, 11, 13, 14, 18, 19, 38). To our knowledge, this is the first time that Lb. harbinensis was found in water kefir. This facultative heterofermentative lactic acid bacterium was first isolated from a Chinese vegetable fermentation (39) and was subsequently found in French cow milk (40), the oral ecosystem of healthy individuals (41), Parmigiano Reggiano cheese (42), and sorghum sourdough fermentation (43). It is worth notice that Lb. harbinensis produces antifungal compounds that inhibit yeasts (40, 44). Lactobacillus casei, also a facultative heterofermentative lactic acid bacterium, is also associated with the oral ecosystem of healthy individuals, the human intestinal tract, and raw and fermented dairy and vegetable products (45). Some strains of Lb. casei show probiotic potential (46), which makes the water kefir ecosystem a possible source of novel probiotic Lb. casei strains. Lactobacillus hilgardii, an obligate heterofermentative lactic acid bacterium, occurs also in wine and cocoa fermentations (47, 48). This bacterium is believed to be the main exopolysaccharide (EPS) producer in the water kefir ecosystem (5, 18, 19, 38). However, not all Lb. hilgardii strains from water kefir produce EPS (4). Other EPS-producing lactic acid bacteria isolated from water kefir include Leuconostoc mesenteroides, Lb. brevis, Lb. casei, Lb. nagelii, and Lb. hordei (4, 7). The water kefir of the present study harbored Lb. nagelii, a homofermentative lactic acid bacterium frequently found in water kefir (1, 2, 4). For homopolysaccharide production, sucrose is the necessary substrate (49). In the water kefir of the present study, the water kefir grain mass was produced only in the early stages of the fermentation, as long as sucrose was present. The accumulation of fructose in the water kefir liquor indicates that the water kefir grains were composed of glucan. The homopolysaccharide produced did not serve as a reserve polymer, as the kefir grain (dry) mass did not decrease upon prolonged fermentation.
The detection of B. psychraerophilum/crudilactis confirms a recent finding that water kefir harbors bifidobacteria (2, 3, 20). This species was first isolated from a porcine cecum (50). Bifidobacteria are obligate anaerobic bacteria that produce more acetate than lactate. Because of the low acetate concentrations of the water kefir of the present study, their metabolic activity was limited.
The most dominant yeast species was S. cerevisiae. This yeast species is frequently associated with water kefir (3, 4, 10, 13, 14, 19, 20, 51). Dekkera bruxellensis (anamorph Brettanomyces bruxellensis) was shown to be associated with water kefir only recently (3, 20). This yeast plays a key role in the spontaneous fermentation of typical Belgian acid ales (52), although it is usually associated with spoilage of beer and wine (53). Whether the presence of D. bruxellensis during water kefir fermentation has a positive or negative influence on the end product is unclear at this moment.
The wide meta-metabolomics approach of the present study elucidated the substrate consumption and metabolite production profiles of the microbial consortium described above. The major metabolites were ethanol, carbon dioxide, and lactic acid. Lactic acid, responsible for the fresh sour taste of water kefir, was the main metabolite of the lactic acid bacterial species, although smaller amounts of ethanol, acetate, and mannitol were produced, too. Although there were high concentrations of fructose present at the initial stage of the fermentation process, the production of mannitol from fructose by the heterofermentative lactic acid bacterial species was limited but could explain part of the acetate production. Mannitol has a sweet taste and possesses antioxidant activity (54); both properties might be desirable in water kefir. Ethanol and carbon dioxide were the main metabolites produced by the yeasts, although smaller amounts of glycerol and acetic acid were produced too. Glycerol is a slightly sweet molecule that may slightly increase the viscosity of a fermented beverage but does not seem to have a direct influence on the taste and aroma of fermented beverages (55).
All esters and higher alcohols found in the water kefir liquor are associated with yeast metabolism (56). For instance, hexanoic acid, octanoic acid, and decanoic acid, necessary for the production of the corresponding ethyl esters, originate from the fatty acid biosynthesis pathway in yeasts. All these volatile aroma compounds are also found in wine and beer, but a direct comparison of the water kefir liquor with beer or wine is difficult because of the multitude of interactions among all the chemical components in each fermented beverage. This also makes it difficult to estimate the impact of individual aroma compounds on the overall flavor. However, with regard to the threshold values of the different aroma compounds, the esters isoamyl acetate, ethyl hexanoate, ethyl octanoate, and ethyl decanoate exerted the greatest influence on the aroma of the water kefir of the present study, contributing to its fruity and floral notes.
In conclusion, a sound water kefir fermentation with water kefir grains that grew well was obtained during the present study, which can be used as reference for other water kefirs. This water kefir fermentation was dominated by the lactic acid bacterial species Lb. casei/paracasei, Lb. harbinensis, and Lb. hilgardii and by the yeasts S. cerevisiae and D. bruxellensis. More lactic acid bacteria were present than yeasts, although the metabolism of the yeasts prevailed. The majority of the microorganisms were present on the water kefir grains. The water kefir grain mass increased as long as sucrose was present, and the main metabolites produced during the fermentation were ethanol, carbon dioxide, lactic acid, glycerol, and acetic acid. Isoamyl acetate, ethyl hexanoate, ethyl octanoate, and ethyl decanoate dominated the aroma of the water kefir of the present study.
ACKNOWLEDGMENTS
We acknowledge the financial support of the Research Council of the Vrije Universiteit Brussel (SRP, IRP, and IOF projects), the Hercules Foundation, and the Research Foundation-Flanders (FWO-Vlaanderen). D.L. is the recipient of a Ph.D. fellowship of the Vrije Universiteit Brussel.
Footnotes
Published ahead of print 14 February 2014
REFERENCES
- 1.Stadie J, Gulitz A, Ehrmann MA, Vogel RF. 2013. Metabolic activity and symbiotic interactions of lactic acid bacteria and yeasts isolated from water kefir. Food Microbiol. 35:92–98. 10.1016/j.fm.2013.03.009 [DOI] [PubMed] [Google Scholar]
- 2.Gulitz A, Stadie J, Ehrmann MA, Ludwig W, Vogel RF. 2013. Comparative phylobiomic analysis of the bacterial community of water kefir by 16S rRNA gene amplicon sequencing and ARDRA analysis. J. Appl. Microbiol. 114:1082–1091. 10.1111/jam.12124 [DOI] [PubMed] [Google Scholar]
- 3.Hsieh HH, Wang SY, Chen TL, Huang YL, Chen MJ. 2012. Effects of cow's and goat's milk as fermentation media on the microbial ecology of sugary kefir grains. Int. J. Food Microbiol. 157:73–81. 10.1016/j.ijfoodmicro.2012.04.014 [DOI] [PubMed] [Google Scholar]
- 4.Gulitz A, Stadie J, Wenning M, Ehrmann MA, Vogel RF. 2011. The microbial diversity of water kefir. Int. J. Food Microbiol. 151:284–288. 10.1016/j.ijfoodmicro.2011.09.016 [DOI] [PubMed] [Google Scholar]
- 5.Pidoux M. 1989. The microbial flora of sugary kefir grain (the gingerbeer plant) - biosynthesis of the grain from Lactobacillus hilgardii producing a polysaccharide gel. J. Appl. Microbiol. 5:223–238. 10.1007/Bf01741847 [DOI] [Google Scholar]
- 6.Lutz ML. 1899. Recherches biologiques sur la constitution du Tibi. Bull. Soc. Mycol. France 15:68–72 [Google Scholar]
- 7.Pidoux M, Brillouet JM, Quemener B. 1988. Characterization of the polysaccharides from a Lactobacillus brevis and from sugary kefir grains. Biotechnol. Lett. 10:415–420. 10.1007/BF01087442 [DOI] [Google Scholar]
- 8.Kebler LF. 1921. California bees. J. Am. Pharm. Assoc. 10:939–943. 10.1002/jps.3080101206 [DOI] [Google Scholar]
- 9.Ward HM. 1892. The ginger-beer plant and the organisms composing it; a contribution to the study of fermentation-yeasts and bacteria. Philos. Trans. R. Soc. London 183:125–197. 10.1098/rstb.1892.0006 [DOI] [Google Scholar]
- 10.Franzetti L, Galli A, Pagani MA, De Noni I. 1998. Microbiological and chemical investigations on “sugar kefir” drink. Ann. Microbiol. Enzimol. 48:67–80 [Google Scholar]
- 11.Galli A, Fiori E, Franzetti L, Pagani MA, Ottogalli G. 1995. Microbiological and chemical composition of sugar kefir grains. Ann. Microbiol. Enzimol. 45:85–95 [Google Scholar]
- 12.Magalhães KT, Pereira GVD, Campos CR, Dragone G, Schwan RF. 2011. Brazilian kefir: structure, microbial communities and chemical composition. Braz. J. Microbiol. 42:693–702. 10.1590/S1517-83822011000200034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Magalhães KT, Pereira GVD, Dias DR, Schwan RF. 2010. Microbial communities and chemical changes during fermentation of sugary Brazilian kefir. World J. Microbiol. Biotechnol. 26:1241–1250. 10.1007/s11274-009-0294-x [DOI] [PubMed] [Google Scholar]
- 14.Miguel MGDP, Cardoso PG, Magalhaes KT, Schwan RF. 2011. Profile of microbial communities present in Tibico (sugary kefir) grains from different Brazilian states. World J. Microbiol. Biotechnol. 27:1875–1884. 10.1007/s11274-010-0646-6 [DOI] [Google Scholar]
- 15.Moinas M, Horisberger M, Bauer H. 1980. The structural organization of the Tibi grain as revealed by light, scanning and transmission microscopy. Arch. Microbiol. 128:157–161. 10.1007/BF00406153 [DOI] [Google Scholar]
- 16.Neve H, Heller KJ. 2002. The microflora of water kefir: a glance by scanning electron microscopy. Kieler Milchw. Forsch. 54:337–349 [Google Scholar]
- 17.Horisberger M. 1969. Structure of the dextran of the Tibi grain. Carbohydr. Res. 10:379–385. 10.1016/S0008-6215(00)80897-6 [DOI] [Google Scholar]
- 18.Pidoux M, Deruiter GA, Brooker BE, Colquhoun IJ, Morris VJ. 1990. Microscopic and chemical studies of a gelling polysaccharide from Lactobacillus hilgardii. Carbohyd. Polym. 13:351–362. 10.1016/0144-8617(90)90035-Q [DOI] [Google Scholar]
- 19.Waldherr FW, Doll VM, Meissner D, Vogel RF. 2010. Identification and characterization of a glucan-producing enzyme from Lactobacillus hilgardii TMW 1.828 involved in granule formation of water kefir. Food Microbiol. 27:672–678. 10.1016/j.fm.2010.03.013 [DOI] [PubMed] [Google Scholar]
- 20.Marsh AJ, O'Sullivan O, Hill C, Ross RP, Cotter PD. 2013. Sequence-based analysis of the microbial composition of water kefir from multiple sources. FEMS Microbiol. Lett. 348:79–85. 10.1111/1574-6968.12248 [DOI] [PubMed] [Google Scholar]
- 21.Papalexandratou Z, Falony G, Romanens E, Jimenez JC, Amores F, Daniel HM, De Vuyst L. 2011. Species diversity, community dynamics, and metabolite kinetics of the microbiota associated with traditional Ecuadorian spontaneous cocoa bean fermentations. Appl. Environ. Microbiol. 77:7698–7714. 10.1128/AEM.05523-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Papalexandratou Z, Lefeber T, Bahrim B, Lee OS, Daniel HM, De Vuyst L. 2013. Hanseniaspora opuntiae, Saccharomyces cerevisiae, Lactobacillus fermentum, and Acetobacter pasteurianus predominate during well-performed Malaysian cocoa bean box fermentations, underlining the importance of these microbial species for a successful cocoa bean fermentation process. Food Microbiol. 35:73–85. 10.1016/j.fm.2013.02.015 [DOI] [PubMed] [Google Scholar]
- 23.Wouters D, Grosu-Tudor S, Zamfir M, De Vuyst L. 2013. Bacterial community dynamics, lactic acid bacteria species diversity and metabolite kinetics of traditional Romanian vegetable fermentations. J. Sci. Food Agric. 93:749–760. 10.1002/jsfa.5788 [DOI] [PubMed] [Google Scholar]
- 24.Daniel HM, Vrancken G, Takrama JF, Camu N, De Vos P, De Vuyst L. 2009. Yeast diversity of Ghanaian cocoa bean heap fermentations. FEMS Yeast Res. 9:774–783. 10.1111/j.1567-1364.2009.00520.x [DOI] [PubMed] [Google Scholar]
- 25.Edwards U, Rogall T, Blocker H, Emde M, Bottger EC. 1989. Isolation and direct complete nucleotide determination of entire genes - characterization of a gene coding for 16S-ribosomal RNA. Nucleic Acids Res. 17:7843–7853. 10.1093/nar/17.19.7843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.White T, Buns T, Lee S, Taylor J. 1990. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics, p 681–694 In Innis N, Gelfand D, Snisky J, White T. (ed), PCR protocols: a guide to methods and applications. Academic Press, New York, NY [Google Scholar]
- 27.Vilgalys R, Hester M. 1990. Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. J. Bacteriol. 172:4238–4246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403–410 [DOI] [PubMed] [Google Scholar]
- 29.Camu N, De Winter T, Verbrugghe K, Cleenwerck I, Vandamme P, Takrama JS, Vancanneyt M, De Vuyst L. 2007. Dynamics and biodiversity of populations of lactic acid bacteria and acetic acid bacteria involved in spontaneous heap fermentation of cocoa beans in Ghana. Appl. Environ. Microbiol. 73:1809–1824. 10.1128/AEM.02189-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ercolini D, Moschetti G, Blaiotta G, Coppola S. 2001. Behavior of variable V3 region from 16S rDNA of lactic acid bacteria in denaturing gradient gel electrophoresis. Curr. Microbiol. 42:199–202. 10.1007/s002840010204 [DOI] [PubMed] [Google Scholar]
- 31.Garcia-Armisen T, Papalexandratou Z, Hendryckx H, Camu N, Vrancken G, De Vuyst L, Cornelis P. 2010. Diversity of the total bacterial community associated with Ghanaian and Brazilian cocoa bean fermentation samples as revealed by a 16S rRNA gene clone library. Appl. Microbiol. Biotechnol. 87:2281–2292. 10.1007/s00253-010-2698-9 [DOI] [PubMed] [Google Scholar]
- 32.Satokari RM, Vaughan EE, Akkermans ADL, Saarela M, de Vos WM. 2001. Bifidobacterial diversity in human feces detected by genus-specific PCR and denaturing gradient gel electrophoresis. Appl. Environ. Microbiol. 67:504–513. 10.1128/AEM.67.2.504-513.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cocolin L, Bisson LF, Mills DA. 2000. Direct profiling of the yeast dynamics in wine fermentations. FEMS Microbiol. Lett. 189:81–87. 10.1111/j.1574-6968.2000.tb09210.x [DOI] [PubMed] [Google Scholar]
- 34.Janssens M, Myter N, De Vuyst L, Leroy F. 2012. Species diversity and metabolic impact of the microbiota are low in spontaneously acidified Belgian sausages with an added starter culture of Staphylococcus carnosus. Food Microbiol. 29:167–177. 10.1016/j.fm.2011.07.005 [DOI] [PubMed] [Google Scholar]
- 35.Wouters D, Bernaert N, Conjaerts W, Van Droogenbroeck B, De Loose M, De Vuyst L. 2013. Species diversity, community dynamics, and metabolite kinetics of spontaneous leek fermentations. Food Microbiol. 33:185–196. 10.1016/j.fm.2012.09.016 [DOI] [PubMed] [Google Scholar]
- 36.Makras L, Van Acker G, De Vuyst L. 2005. Lactobacillus paracasei subsp. paracasei 8700:2 degrades inulin-type fructans exhibiting different degrees of polymerization. Appl. Environ. Microbiol. 71:6531–6537. 10.1128/AEM.71.11.6531-6537.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rimaux T, Vrancken G, Vuylsteke B, De Vuyst L, Leroy F. 2011. The pentose moiety of adenosine and inosine is an important energy source for the fermented-meat starter culture Lactobacillus sakei CTC 494. Appl. Environ. Microbiol. 77:6539–6550. 10.1128/AEM.00498-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Leroi F, Pidoux M. 1993. Characterization of interactions between Lactobacillus hilgardii and Saccharomyces florentinus isolated from sugary kefir grains. J. Appl. Bacteriol. 74:54–60 [DOI] [PubMed] [Google Scholar]
- 39.Miyamoto M, Seto Y, Hao DH, Teshima T, Sun YB, Kabuki T, Yao LB, Nakajima H. 2005. Lactobacillus harbinensis sp. nov., consisted of strains isolated from traditional fermented vegetables ‘Suan cai' in Harbin, northeastern China and Lactobacillus perolens DSM 12745. Syst. Appl. Microbiol. 28:688–694. 10.1016/j.syapm.2005.04.001 [DOI] [PubMed] [Google Scholar]
- 40.Delavenne E, Ismail R, Pawtowski A, Mounier J, Barbier G, Le Blay G. 2013. Assessment of lactobacilli strains as yogurt bioprotective cultures. Food Control 30:206–213. 10.1016/j.foodcont.2012.06.043 [DOI] [Google Scholar]
- 41.Lönnermark E, Nowrouzinan F, Adlerberth I, Ahrne S, Wold A, Friman V. 2012. Oral and faecal lactobacilli and their expression of mannose-specific adhesins in individuals with and without IgA deficiency. Int. J. Med. Microbiol. 302:53–60. 10.1016/j.ijmm.2011.08.004 [DOI] [PubMed] [Google Scholar]
- 42.Solieri L, Bianchi A, Giudici P. 2012. Inventory of non starter lactic acid bacteria from ripened Parmigiano Reggiano cheese as assessed by a culture dependent multiphasic approach. Syst. Appl. Microbiol. 35:270–277. 10.1016/j.syapm.2012.04.002 [DOI] [PubMed] [Google Scholar]
- 43.Sekwati-Monang B, Valcheva R, Ganzle MG. 2012. Microbial ecology of sorghum sourdoughs: effect of substrate supply and phenolic compounds on composition of fermentation microbiota. Int. J. Food Microbiol. 159:240–246. 10.1016/j.ijfoodmicro.2012.09.013 [DOI] [PubMed] [Google Scholar]
- 44.Belguesmia Y, Rabesona H, Mounier J, Pawtowsky A, Le Blay G, Barbier G, Haertlé T, Chobert J-M. 2014. Characterization of antifungal organic acids produced by Lactobacillus harbinensis K.V9.3.1Np immobilized in gellan–xanthan beads during batch fermentation. Food Control 36:205–211. 10.1016/j.foodcont.2013.08.028 [DOI] [Google Scholar]
- 45.Cai H, Rodriguez BT, Zhang W, Broadbent JR, Steele JL. 2007. Genotypic and phenotypic characterization of Lactobacillus casei strains isolated from different ecological niches suggests frequent recombination and niche specificity. Soc. Gen. Microbiol. 153:2655–2665. 10.1099/mic.0.2007/006452-0 [DOI] [PubMed] [Google Scholar]
- 46.Galdeano CM, Perdigon G. 2006. The probiotic bacterium Lactobacillus casei induces activation of the gut mucosal immune system through innate immunity. Clin. Vaccine Immunol. 13:219–226. 10.1128/CVI.13.2.219-226.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rodriguez AV, Denadra MCM. 1995. Mixed culture of Lactobacillus hilgardii and Leuconostoc oenos isolated from Argentine wine. J. Appl. Bacteriol. 78:521–525. 10.1111/j.1365-2672.1995.tb03094.x [DOI] [Google Scholar]
- 48.Ardhana MM, Fleet GH. 2003. The microbial ecology of cocoa bean fermentations in Indonesia. Int. J. Food Microbiol. 86:87–99. 10.1016/S0168-1605(03)00081-3 [DOI] [PubMed] [Google Scholar]
- 49.Monsan P, Bozonnet S, Albenne C, Joucla G, Willemot RM, Remaud-Simeon M. 2001. Homopolysaccharides from lactic acid bacteria. Int. Dairy J. 11:675–685. 10.1016/S0958-6946(01)00113-3 [DOI] [Google Scholar]
- 50.Simpson PJ, Ross RP, Fitzgerald GF, Stanton C. 2004. Bifidobacterium psychraerophilum sp. nov. and Aeriscardovia aeriphila gen. nov., sp. nov., isolated from a porcine caecum. Int. J. Syst. Evol. Microbiol. 54:401–406. 10.1099/ijs.0.02667-0 [DOI] [PubMed] [Google Scholar]
- 51.Diosma G, Romanin DE, Rey-Burusco MF, Londero A, Garrote GL. 2014. Yeasts from kefir grains: isolation, identification, and probiotic characterization. World J. Microbiol. Biotechnol. 30:43–53. 10.1007/s11274-013-1419-9 [DOI] [PubMed] [Google Scholar]
- 52.Martens H, Iserentant D, Verachtert H. 1997. Microbiological aspects of a mixed yeast-bacterial fermentation in the production of a special Belgian acidic ale. J. Inst. Brew. 103:85–91. 10.1002/j.2050-0416.1997.tb00939.x [DOI] [Google Scholar]
- 53.Wedral D, Shewfelt R, Frank J. 2010. The challenge of Brettanomyces in wine. Food Sci. Technol. 43:1474–1479. 10.1016/j.lwt.2010.06.010 [DOI] [Google Scholar]
- 54.Shen B, Jensen RG, Bohnert HJ. 1997. Mannitol protects against oxidation by hydroxyl radicals. Plant Physiol. 115:527–532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Picinelli A, Suarez B, Moreno J, Rodriguez R, Caso-Garcia LM, Mangas JJ. 2000. Chemical characterization of Asturian cider. J. Agric. Food Chem. 48:3997–4002. 10.1021/jf991284d [DOI] [PubMed] [Google Scholar]
- 56.Lambrechts MG, Pretorius IS. 2000. Yeast and its importance to wine aroma - a review. S. Afr. J. Enol. Vitic. 21:97–129 [Google Scholar]
- 57.Ferreira V, Lopez R, Cacho JF. 2000. Quantitative determination of the odorants of young red wines from different grape varieties. J. Sci. Food Agric. 80:1659–1667. [DOI] [Google Scholar]
- 58.Guth H. 1997. Quantitation and sensory studies of character impact odorants of different white wine varieties. J. Agric. Food Chem. 45:3027–3032. 10.1021/jf970280a [DOI] [Google Scholar]
- 59.Molina AM, Guadalupe V, Varela C, Swiegers JH, Pretorius IS, Agosin E. 2009. Differential synthesis of fermentative aroma compounds of two related commercial wine yeast strains. Food Chem. 117:189–195. 10.1016/j.foodchem.2009.03.116 [DOI] [Google Scholar]
- 60.Mamede MEO, Cardello HMAB, Pastore GM. 2005. Evaluation of an aroma similar to that of sparkling wine: sensory and gas chromatography analyses of fermented grape musts. Food Chem. 89:63–68. 10.1016/j.foodchem.2004.02.012 [DOI] [Google Scholar]
- 61.Corison CA, Ough CS, Berg HW, Nelson KE. 1979. Must acetic acid and ethyl acetate as mold and rot indicators in grapes. Am. J. Enol. Vitic. 30:130–134 [Google Scholar]