Abstract
The alphaproteobacterium Magnetospirillum gryphiswaldense biomineralizes magnetosomes, which consist of monocrystalline magnetite cores enveloped by a phospholipid bilayer containing specific proteins. Magnetosomes represent magnetic nanoparticles with unprecedented magnetic and physicochemical characteristics. These make them potentially useful in a number of biotechnological and biomedical applications. Further functionalization can be achieved by expression of foreign proteins via genetic fusion to magnetosome anchor peptides. However, the available genetic tool set for strong and controlled protein expression in magnetotactic bacteria is very limited. Here, we describe versatile vectors for either inducible or high-level constitutive expression of proteins in M. gryphiswaldense. The combination of an engineered native PmamDC promoter with a codon-optimized egfp gene (Mag-egfp) resulted in an 8-fold increase in constitutive expression and in brighter fluorescence. We further demonstrate that the widely used Ptet promoter is functional and tunable in M. gryphiswaldense. Stable and uniform expression of the EGFP and β-glucuronidase (GusA) reporters was achieved by single-copy chromosomal insertion via Tn5-mediated transposition. In addition, gene duplication by Mag-EGFP–EGFP fusions to MamC resulted in further increased magnetosome expression and fluorescence. Between 80 and 210 (for single MamC–Mag-EGFP) and 200 and 520 (for MamC–Mag-EGFP–EGFP) GFP copies were estimated to be expressed per individual magnetosome particle.
INTRODUCTION
For magnetic orientation, magnetotactic bacteria (MTB) biomineralize bacterial (ferri)magnetic nanoparticles. In the model organism Magnetospirillum gryphiswaldense and other MTB, these organelles consist of magnetite (Fe3O4) cores enveloped by the magnetosome membrane (MM) (1, 2). Because of their unprecedented material properties, such as high crystallinity, strong magnetization, uniform shapes and sizes, and biocompatibility, the use of isolated magnetosome particles has been suggested for a number of biotechnological and biomedical applications, such as using them as nanocarriers in magnetic drug targeting, magnetosome-based immunoassays and as reporters for magnetic resonance imaging (MRI) (3). Many of these applications require further functionalization, for instance, by displaying additional functional moieties on the magnetosome surface such as antibodies, oligonucleotides, fluorophores, or enzymes (3, 4). It was shown that in M. gryphiswaldense, in addition to chemical functionalization of isolated particles in vitro (5, 6), magnetosomes can also be engineered in vivo by expression of foreign proteins via genetic fusion to native magnetosome anchors. For example, the small (12.5-kDa), highly abundant MamC protein was shown to provide tight and stable attachment of foreign proteins to the MM. This was first demonstrated by a MamC-green fluorescent protein (GFP) fusion, which displayed stable fluorescence in vivo and also after purification of magnetosomes (7). In different studies, a red fluorescent protein (RFP)-binding nanobody (RBP) and the endogenous RNA subunit C5 of the multisubunit chimeric bacterial RNase P enzyme were functionally expressed on magnetosomes by translational fusion with MamC (8, 9).
However, previous approaches were hampered by the unavailability of appropriate systems for controlled protein expression in M. gryphiswaldense. For example, so far only a few promoters have been identified as being functional for transcription in M. gryphiswaldense. The native PmamDC, which drives transcription of the mamGFDC operon, yielded the highest constitutive expression of the reporter EGFP, while weaker expression was found with other promoters like PmamAB (10, 11). Known inducible promoters yielded only weak (Plac [10]) or no expression (e.g., Pure [our unpublished data]). However, inducible expression systems are prerequisite for display of proteins that may interfere with magnetosome biomineralization or cellular processes. In the related strain Magnetospirillum magneticum AMB-1, the strong native msp13 and mms16 promoters were employed for magnetosome display of fusion proteins (12, 13). A tetracycline (Tet)-inducible expression system was described based on a hybrid promoter consisting of the combined msp1 promoter and tetO sequences (14). However, the transcriptional strength of the hybrid promoter compared to the strong constitutive msp13 and mms16 has not been reported. In addition, all expression studies so far were based on multicopy replicative plasmids, which have the disadvantage of segregational instability and nonuniform expression (10).
Here, we describe two versatile vectors for either inducible or high-level constitutive chromosomal expression. We demonstrate their use for cytoplasmic and magnetosome expression of foreign proteins. Furthermore, we show that codon optimization and multicopy expression are powerful approaches to enhance heterologous expression of proteins in M. gryphiswaldense. We also provide an estimation of protein copies expressed per magnetosome particle.
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture conditions.
Plasmids and bacterial strains used in this study are listed in Tables S1 and S2 in the supplemental material. M. gryphiswaldense strains were grown microaerobically in modified flask standard medium (FSM) at 30°C (15) with moderate agitation (120 rpm). Escherichia coli strains were cultivated as previously described (16) for growth of E. coli BW29427 (K. Datsenko and B. L. Wanner, unpublished data) and WM3064 (W. Metcalf, unpublished data); 1 mM d,l-α,ε-diaminopimelic acid (DAP) was added to lysogeny broth (LB) medium. Strains were routinely cultured on plates solidified with 1.5% (wt/vol) agar. For strains carrying recombinant plasmids, media were supplemented with 25 μg ml−1 kanamycin (Km) and 50 μg ml−1 ampicillin (Amp) for E. coli strains and 5 μg ml−1 Km for M. gryphiswaldense strains. For induction experiments, media were supplemented with various concentrations of anhydrotetracycline (Atet).
Molecular and genetic techniques.
Oligonucleotides (see Table S3 in the supplemental material) were purchased from Sigma-Aldrich (Steinheim, Germany). Chromosomal DNA of M. gryphiswaldense was isolated using a genomic DNA isolation kit (Zymo Research, USA). Plasmids were constructed by standard recombinant techniques as described in detail below. All constructs were sequenced on an ABI 3730 capillary sequencer (Applied Biosystems, Darmstadt, Germany), utilizing BigDye Terminator v3.1. Sequence data were analyzed with Vector NTI Advance 11.5 software (Invitrogen, Darmstadt, Germany). Magnetospirillum-optimized EGFP (Mag-EGFP) was optimized by proprietary algorithms for increased mRNA stability and avoidance of sequence repetitions and secondary structures and was purchased from GeneArt (Invitrogen, Darmstadt, Germany).
Construction of insertional expression plasmids.
For the construction of pSB6, first Mag-egfp was amplified from p11AAGJZC using the primers oEGFP BamHI Rev and oEGFP HindIII Fw. The resulting PCR fragment was subcloned into pJET1.2/blunt and after restriction with BamHI and HindIII inserted into pAP150 and pAP160 to replace egfp with Mag-egfp. Afterwards, the PmamDC45 promoter, the spacing-optimized ribosomal binding site (oRBS) and Mag-egfp were amplified from modified pAP150 (pSB1) using pBam_pAP150 Fw and pBam_DC w/o Term Rev, adding EcoRI and SanDI restriction sites for insertion into pBAM1, generating pSB6. For the generation of pSB7, the whole expression cassette from modified pAP160, containing the optimized Mag-egfp version plus Ptet and TetR, was amplified using the primers pBam_pAP160 Fw and pBam_Tet w/o Term Rev and inserted into pBAM1. For the generation of the expression plasmids containing gusA as a transcriptional reporter, gusA was amplified from pLYJ97 using the primers GusA BamHI Fw and GusA NdeI Rev and cloned into pSB7, replacing Mag-egfp with gusA, thereby generating pSB8. Transposition of the expression cassettes resulted in single-copy genomic insertion into phenotypical neutral sites as verified by arbitrary PCR (17) and sequencing.
Construction of MamC fusion proteins.
mamC and Mag-egfp were amplified and fused via overlap PCR (18) using the primers described in Table S3 in the supplemental material, resulting in C-terminal fusions of Mag-egfp to mamC; afterwards, the fusion gene was inserted into the NdeI and BamHI restriction sites of pSB6 and pSB7, resulting in pJH1 and pJH2. Additionally a tandem fusion of mamC with Mag-egfp and egfp was generated, following the same strategy, yielding plasmid pJH3. M. gryphiswaldense strains were conjugated with pJH1, pJH2, and pJH3 and were grown in 3 liters of FSM medium to stationary phase at 30°C and 120 rpm. Cells were harvested by centrifugation at 4°C and 6,500 rpm and used for magnetosome isolation.
Analytical methods.
Magnetic reaction of cells was detected by light microscopy applying a bar magnet. Optical density (OD) and magnetic response (Cmag) of exponentially growing cells were measured photometrically at 565 nm as previously reported (19). Iron concentrations of the isolated magnetosomes were determined by a modified ferrozine assay (7); 10 μl of magnetosome suspension was used for determination of iron concentration.
GFP expression in M. gryphiswaldense was assayed by fluorometry, as described previously (10).
GusA activity assay.
Cells were grown to exponential phase, collected via centrifugation, resuspended in phosphate-buffered saline (PBS), and disrupted using a sonifier. Cell debris was spun down, and the supernatant was used to determine protein concentration using the bicinchoninic acid (BCA) kit from Thermo Scientific. β-Glucuronidase (GusA) activity in the supernatant was also measured, and the assay was carried out at 37°C as described by Wilson et al. (20). Units were nanomoles of product formed per minute per milligram of protein. Duplicate assays were performed, and reported values were averaged from at least three independent cultures.
Biochemical methods.
Magnetosome isolation from M. gryphiswaldense strains was performed as described previously (7). Polyacrylamide gels were prepared according to the method of Laemmli (21). Magnetosome membranes were dissolved by incubation in 1% SDS at 65°C for 25 min. Protein concentrations were determined using the BCA protein kit (Thermo Scientific), and 100 ng to 1 μg of magnetosome membrane protein was loaded onto 10% (wt/vol) SDS gels and analyzed via quantitative immunoblotting to quantify the expression level of the MamC-GFP fusion protein.
Proteins were electroblotted onto polyvinylidene difluoride (PVDF) membranes (Roth, Germany). Membranes were blocked overnight at 4°C. Primary rabbit anti-GFP IgG antibody (1:500 dilution [Santa Cruz, USA]) was added to the blocking solution and incubated 1 h at room temperature. Membranes were washed 4 times with blocking solution (2.5% [wt/vol] milk powder in Tris-buffered saline [TBS] [50 mM Tris-HCl, pH 7.6; 150 mM NaCl]) for 10 min and incubated with a secondary horseradish peroxidase-labeled goat anti-rabbit IgG antibody (1:2,000 dilution [Promega, USA]) for 1 h. Membranes were washed 4 times with blocking solution for 10 min and finally 5 min in TBS, and immunoreactive proteins were visualized by using an Ace Glow substrate (Peqlab, Erlangen) and detected with the LumiImager (Peqlab, Erlangen).
RESULTS
Engineering of a cassette for high constitutive gene expression.
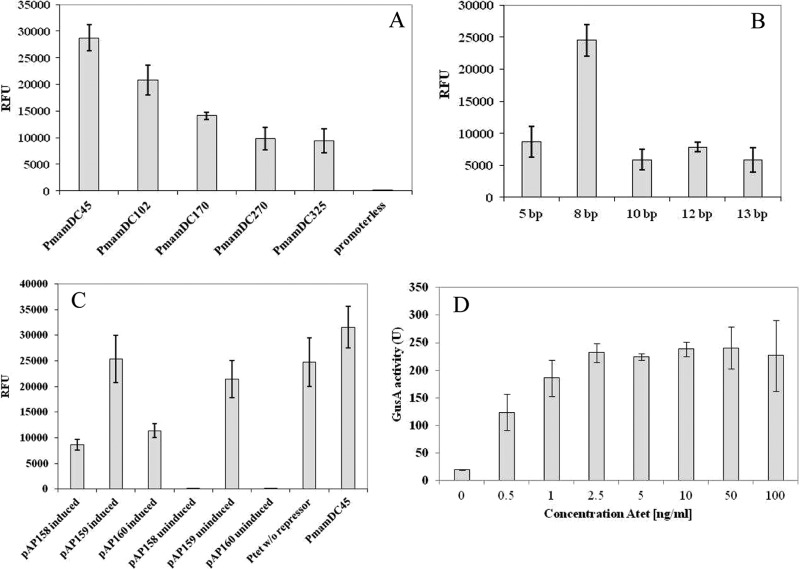
We used a stepwise approach (summarized in Fig. S1 in the supplemental material) to optimize the previously identified strong PmamDC, which is located within a 325-bp region upstream of the mamGFDC operon. We first attempted to identify the smallest yet transcriptionally active fragment (see Fig. S1A in the supplemental material). Several fragments that were gradually truncated from the 5′ end of the 325-bp upstream region of the mamG gene were cloned upstream of the egfp reporter, yielding the vectors pAP150 and pAP161-pAP164. Whereas a 270-bp fragment displayed the same fluorescence intensity as the untruncated 325-bp version, further truncation to 170, 102, and 45 bp increased the fluorescence 1.5-, 2.2-, and 3-fold, respectively (Fig. 1A). Truncation of the putative promoter region of the mamGFDC operon down to 45 bp (still comprising the −35 and −10 regions) increased expression of the reporter gene significantly, possibly because regulatory elements were excluded from the promoter region (22). Therefore, the 45-bp truncated version of PmamDC (designated PmamDC45) was chosen for all subsequent experiments.
FIG 1.
(A to C) Cellular EGFP fluorescence from various expression vectors in M. gryphiswaldense measured by fluorometry. Fluorescence was normalized to cell density and reported as relative fluorescence units (RFU). Error bars represent standard deviations, calculated from three independent experiments. (A) Effects of gradual truncation of the PmamDC promoter region from 325 bp down to 45 bp. A strain carrying a promoterless vector displays only weak background fluorescence, also observed for untransformed cells. (B) Effects of variation of the spacing from the SD sequence to the start codon from 13 to 5 bp in the RBS. (C) Influence of TetR expression from different promoters on the expression of the reporter EGFP from Ptet. (D) GusA activity of cell extract from M. gryphiswaldense expressing chromosomal GusA from Ptet promoter. GusA activity units are defined as nanomoles of product per minute per milligram of protein.
As optimal spacing between the Shine-Dalgarno (SD) sequence and the start codon has been shown to be crucial for bacterial gene expression (23, 24), we optimized the ribosomal binding site (RBS) for increased expression. To this end, we combined PmamDC45 with the original RBS of mamG, spaced by variable lengths from the SD sequence to the start codon of egfp (see Fig. S1B in the supplemental material). Whereas random single-base-pair substitutions within this region did not increase EGFP fluorescence (data not shown), an 8-bp spacing in combination with PmamDC45 caused 2.8-fold-higher fluorescence than the native RBS (13 bp), while spacings of 5, 10, or 12 bp did not increase fluorescence (Fig. 1B). Thus, a spacing-optimized RBS (AGGAGATCAGCATATG; RBS in italics and spacing in bold, followed by the start codon), referred to as oRBS, was used in all subsequent optimization steps.
Optimization of Tet-inducible expression.
In the next step, we wanted to generate an inducible expression system, which should exhibit tight repression in the absence of inducer while allowing high and tunable expression after induction. A hybrid promoter similar to that described by Yoshino et al. (14), consisting of the PmamDC45 promoter and two tetO sequences, yielded no expression in M. gryphiswaldense. Likewise, we failed to construct various hybrid promoters by combining operators from the Tet and the lactose systems with native M. gryphiswaldense promoters, including PmamDC45 and the nitrate-responsive PnirS (25). Also, we found none of the tested inducible expression systems reported for other alphaproteobacteria (26–29) to be sufficiently functional in M. gryphiswaldense. In a different approach, we failed to reconstruct a riboswitch that reportedly was functional in the closely related species M. magneticum (30) but exhibited high fluorescence in the absence of the inducer theophylline in M. gryphiswaldense (data not shown). The widely used Plac promoter yielded inducible yet very weak expression in M. gryphiswaldense (our unpublished data).
Therefore, we focused on optimization of the Tn10-derived Tet-inducible system (31), which in this study was found to be the only inducible expression system to be functional in M. gryphiswaldense. Cloning Ptet upstream of egfp (pAP160) yielded significant fluorescence in the presence of 70 ng ml−1 Atet while remaining tightly repressed (i.e., no fluorescence) in the absence of the inducer (Fig. 1C). Previous studies utilizing the tet system have shown that constitutive expression of TetR is more favorable for the tight repression of strong promoters than the original autoregulated expression approach derived from the Tn10 Tet resistance determinant (32); we tested pAP158, pAP159, and pAP160 for expression of the TetR repressor. However, only TetR expressed from the neomycin promoter Pneo resulted in tight repression (Fig. 1C). The construct carrying Pneo-TetR (pAP160) showed significant expression after induction (about 46% of the fluorescence observed for expression of egfp from Ptet without repressor). For further optimization, we combined Ptet containing two tetO sequences with the oRBS and cloned it upstream egfp as a reporter (see Fig. S1D in the supplemental material). Although Ptet reached only 30% of the fluorescence from constitutive PmamDC45, this expression level is sufficiently high for practical purposes, while still maintaining tight repression in the absence of inducer. Therefore, Pneo-TetR was chosen for further engineering of an inducible expression cassette.
Chromosomal insertion of an expression cassette with a codon-optimized egfp reporter gene results in brighter and uniform fluorescence.
Expression of foreign genes in M. gryphiswaldense (average G+C content = 62.2%) might be limited by different codon usage. We therefore explored the effect of codon optimization, by synthesis of an egfp variant based on the average codon usage of M. gryphiswaldense (see Fig. S1C in the supplemental material), designated Mag-egfp (for Magnetospirillum-optimized EGFP) (see Fig. S2 in the supplemental material). If expressed from constitutive PmamDC45 with oRBS, even minor adjustments increased fluorescence of the resulting Mag-EGFP by about 30% compared to EGFP (see Fig. S3A in the supplemental material). Western blots of M. gryphiswaldense cells expressing either Mag-EGFP (pSB1) or EGFP (pAP150) showed a more intense EGFP band for cells expressing Mag-EGFP than for cells expressing EGFP (see Fig. S3B in the supplemental material). Altogether, the combined optimizations amounted to an 8-fold-increased fluorescence and were cloned together into a single cassette, DC_Mag-EGFP, harbored on pSB1.
Cells expressing Mag-egfp from a medium-copy-number plasmid (pSB1) showed a highly heterogenous phenotype: while 50% of cells were not fluorescent at all, about 20% showed only weak and about 30% strong fluorescence (Fig. 2A). Therefore, we attempted single-copy chromosomal integration of the expression cassette Tet_Mag-EGFP_TetR by Tn5-mediated transposition from the pBAM1 (17)-derived insertion plasmids pSB6 (PmamDC45) and pSB7 (Ptet) (for insertion sites, see Table S4 in the supplemental material). Fluorescence microscopy of populations with insertions of pSB6 and pSB7 showed a uniform phenotype, with about 98% of the cells showing identical levels of intermediate fluorescence (Fig. 2B), while the untransformed wild-type (wt) negative control showed only background fluorescence (Fig. 2C). In Western blots with whole cells of identical cell numbers (Fig. 2D), Mag-EGFP bands of approximately the same intensity were obtained, indicating that overall expression yields were comparable for populations expressing Mag-EGFP either from the plasmid (pSB1) or chromosomally, although single cells displayed the greatest amplitudes and variation of fluorescence in the population expressing Mag-EGFP from pSB1. We failed to detect intermediate fluorescence levels by varying the inducer concentration between 50 ng ml−1 (= saturated fluorescence) and 2.5 ng ml−1 (= no fluorescence) but instead observed an all-or-none response. Although different induction kinetics were observed with different amounts of Atet (e.g., after induction for 6 h with 70 ng ml−1 Atet, the fluorescence was approximately half of that of the constitutive promoter PmamDC45), increasing induction time to 18 h resulted in nearly same expression levels of Mag-EGFP as constitutive conditions (data not shown).
FIG 2.
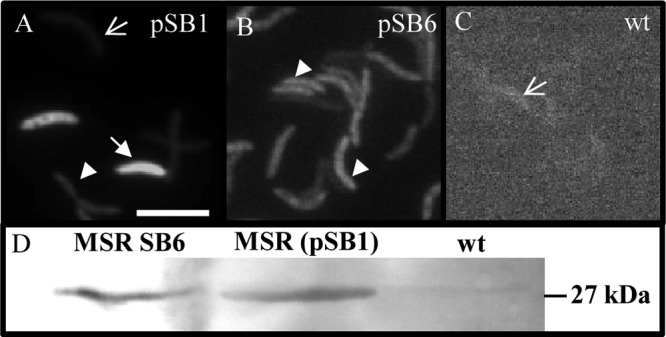
Fluorescence micrographs of M. gryphiswaldense expressing Mag-egfp under the control of PmamDC45 from pSB1 (A) or a chromosomal insertion via pSB6 (B) compared to nonfluorescent M. gryphiswaldense wt (C). Arrows indicate nonfluorescent cells, filled arrows indicate strongly fluorescent cells, and arrowheads indicate moderately fluorescent cells. Bar, 2 μm. (D) Western blot of whole M. gryphiswaldense cells expressing Mag-egfp from the chromosome (pSB6) or plasmid (pSB1). Wt cells were included as a negative control. Mag-EGFP was detected using rabbit anti-GFP IgG as the primary antibody and goat anti-rabbit IgG alkaline phosphatase antibodies as the secondary antibody.
Because of the known limitations of using GFP as the reporter (33), for estimation of induction kinetics we instead used the enzyme β-glucuronidase (GusA), which we recently demonstrated to be an efficient transcriptional reporter in M. gryphiswaldense (25). After replacement of Mag-egfp by gusA, we measured GusA activity in cells harboring a chromosomal copy of both the expression cassette and gusA (pSB8) in medium with and without Atet. As with Mag-EGFP, essentially no activity was detectable in the absence of inducer (Fig. 1D). While for Atet concentrations >2.5 ng ml−1 induction was maximal and could be not further enhanced by concentrations up to 100 ng ml−1, between 0.5 and 1 ng ml−1 a nearly linear response was observed (Fig. 1D). In summary, this demonstrated that transcriptional activity of Ptet is tunable within a narrow range of inducer concentrations. As observed with Mag-EGFP, maximum induction of Ptet yielded about one-third of the constitutively expressed GusA activity (data not shown).
Next, we attempted inducible expression of fusion proteins displayed on magnetosomes (7). Therefore, Mag-egfp was replaced by mamC–Mag-egfp, which was fused via a 10-glycine linker and cloned into pSB7. The resulting construct, pJH2, was chromosomally inserted into M. gryphiswaldense strain ΔC (harboring a single gene deletion of mamC [34]) cells to eliminate the background of nonfused MamC. Uninduced cells showed no fluorescence at all, while after addition of 70 ng ml−1 Atet, a linear fluorescence pattern characteristic of chain localization at midcell was observed after 6 to 18 h by fluorescence microscopy (see Fig. S4B and C in the supplemental material). Magnetosomes purified from this strain exhibited strong and even fluorescence under the microscope (see Fig. S5 in the supplemental material). This also demonstrates that the expression of magnetosome proteins and foreign proteins fused to them as well as their subsequent targeting to MM can be induced.
Optimized magnetosome expression of fusion proteins.
Because maximum expression levels obtained with both inducible and constitutive promoters proved to be limiting for even higher magnetosome expression, we attempted to further increase magnetosome expression of foreign genes by increasing their dosage. To this end, we fused the C terminus of our MamC anchor via 10 glycine residues to a sequence variant of egfp, connecting them to each other by an alpha helix linker. This was a precaution to reduce homologous recombination between the two copies, as egfp and Mag-egfp share only 89% nucleotide identity. The Mag-EGFP–EGFP construct was then cloned into pJH1, yielding pJH3, which carried the optimized constitutive expression cassette DC_MamC–Mag-EGFP–EGFP and was chromosomally inserted into M. gryphiswaldense ΔC (Fig. 3). Cells harboring Mag-EGFP–EGFP displayed much stronger fluorescence signals at midcell than cells with only a single Mag-EGFP fusion (see Fig. S4D in the supplemental material). Expression of the Mag-EGFP–EGFP fusion affected neither the biomineralization of magnetosomes nor the thickness and appearance of the MM (Fig. 4A and B). To estimate the amount of MamC–Mag-EGFP and MamC–Mag-EGFP–EGFP, we performed quantitative Western blots on extracted MM. As expected, immunostaining of Mag-EGFP–EGFP (pJH3) yielded a significantly stronger 74-kDa band than that of single Mag-EGFP (Mag-EGFP–EGFP was diluted 10× for quantitative Western blot analysis) (Fig. 4C). Using a GFP standard (Fig. 4D), we estimated that magnetosomes isolated from strain JH1 (displaying MamC–Mag-EGFP expressed from PmamDC45) contained approximately 33 ng Mag-EGFP per μg magnetite (as measured by iron content). If expressed from Ptet (strain JH2), 9 ng Mag-EGFP was detected per μg magnetite. Magnetosomes expressing a MamC–Mag-EGFP–EGFP fusion from PmamDC45 (strain JH3) displayed a much stronger band than the other samples, corresponding to 83 ng (Mag-)EGFP per μg magnetite (Fig. 4C; for details, see Fig. S7A and B in the supplemental material).
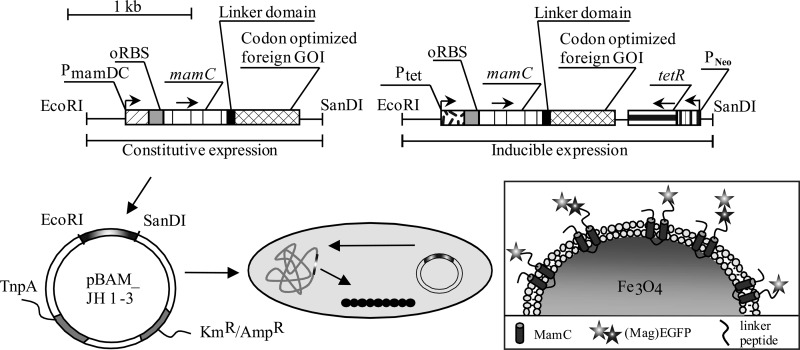
FIG 3.
Schematics of optimized expression vectors (DC_Mag-EGFP and Tet_Mag-EGFP_TetR or Mag-EGFP–EGFP fusion) and chromosomal insertion. Expression vectors contained either the strong optimized PmamDC45 or the inducible Ptet promoter, the optimized oRBS, and the magnetosome anchor mamC, which can be fused via a linker domain to any codon-optimized heterologous gene of interest (GOI). Insertion into the chromosome is via pBAM1-derived insertional plasmids and chromosome expression of fusion proteins.
FIG 4.
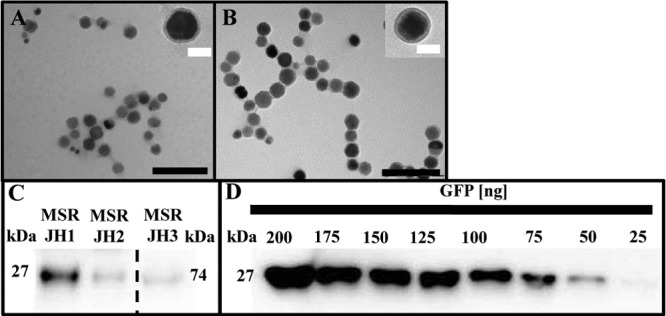
(A and B) Transmission electron micrographs of purified magnetosomes from M. gryphiswaldense ΔC JH3 (A) and wt (B), showing no effect on MM or magnetite crystals. Black scale bar, 200 nm; white scale bar, 40 nm. (C) Quantitative Western blot of (Mag-)EGFP in the MM, isolated from M. gryphiswaldense strains expressing different chromosomal mamC–Mag-egfp fusions from PmamDC45 (JH1) and Ptet (JH2) and mamC–Mag-egfp–egfp fusions (JH3) from PmamDC45. (Mag-)EGFP was detected by rabbit anti-GFP IgG as the primary antibody and goat anti-rabbit IgG horseradish peroxidase antibodies as secondary antibodies. The sample containing the Mag-EGFP–EGFP fusion was diluted 10× for quantitative Western blot analysis. Band sizes are as follows: GFP, 27 kDa; 2× GFP, 74 kDa. (D) Recombinant GFP was used as a standard.
DISCUSSION
We optimized and constructed versatile cassettes that allow either inducible or high-level constitutive expression and magnetosome display of foreign proteins in M. gryphiswaldense.
Increased constitutive expression was accomplished by the combined effect of various optimization steps, which altogether yielded an 8-fold-higher expression of the cytoplasmic Mag-EGFP reporter than previously available systems. The truncation also yielded a compact, easy-to-clone gene cassette, whose extension of 58 bp is within the typical range of other prokaryotic promoters (40 to 65 bp) (35).
None of the several tested inducible expression systems from other alphaproteobacteria were found to be functional in M. gryphiswaldense, because of lack of either expression or repression. We also failed to construct a functional tetracycline-responsive hybrid promoter by combining the optimized PmamDC45 with operators (tetO) and the repressor (TetR) from the well-characterized tet system (31), which is functional in a vast variety of bacteria (36, 37). Although a similar system was reported for the related M. magneticum (14), in M. gryphiswaldense different variants of hybrid promoters lacked functionality, possibly due to the absence of further regulatory elements in the genetic neighborhood of PmamDC45 (38).
However, we found that in M. gryphiswaldense the original Tn10 Ptet promoter is tightly repressed but can be induced to reasonably high expression levels in the presence of saturating Atet concentrations as low as 2.5 ng ml−1. This is 40-, 80-, 160-, and even 200-fold lower than in Helicobacter pylori (36), E. coli (39), Bacillus subtilis (37), and M. magneticum (14), respectively, while the regulatory range (up to 12-fold with the reporter GusA) is comparable to tet-responsive systems in other bacteria (S. aureus, 50- to 100-fold; S. pneumoniae, 5-fold [40, 41]). We also found that a chromosomal insertion of Tet_MamC–Mag-EGFP_TetR from vector pJH2 provides tight TetR-mediated silencing of MamC–Mag-EGFP, while induction in magnetosome-containing wt cells caused Mag-EGFP to be reasonably expressed on magnetosomes after only 6 h. This implies that insertion of newly synthesized MamC–Mag-EGFP fusion proteins into the MM of pre-existing magnetosome particles is possible. In addition to magnetosome display, the TetR-controlled expression system could also be used for the generation of conditional knockouts and gene depletion studies and thus extends the genetic toolbox available in M. gryphiswaldense. Despite these improvements, maximum expression of fully induced Ptet reaches only 30% of constitutive PmamDC45-driven expression. However, this level is sufficient and appropriate for many practical purposes.
Inhomogeneous expression (that is, uneven expression levels varying between individuals) of the reporter gene from plasmids in isogenic cultures is frequently observed in bacteria (42, 43). We achieved a much more homogenous Mag-EGFP expression by chromosomal insertion of single copies than did previous attempts with multicopy expression (7, 8). Tn5-mediated transposition allows straightforward, single-site integration into the host chromosome, despite the possible disadvantage of random insertion. One caveat of using Tn5-mediated transposition is that the expression cassette integrates randomly into genomic loci of unknown function, possibly causing unwanted mutations. However, all insertants lacked obvious phenotypes with respect to growth and magnetosome formation (compare Fig. S6A and B in the supplemental material), indicating the absence of effects on host metabolism. On the other hand, reporter expression in the absence or presence of inducer were similar in all insertants, suggesting that no interference such as read-through from external promoters occurred.
In addition to increased transcription, using GFP as a model we explored two strategies to enhance translation of foreign proteins. First, we demonstrated that even minor adjustments of the codon usage closer to that of M. gryphiswaldense (62.2% G+C) increased the fluorescence of the resulting synthetic Mag-EGFP (Magnetospirillum-optimized EGFP) by 30%, which thus represents a reporter with increased sensitivity for future tagging and localization studies. Codon optimization seems promising also for boosting expression of other foreign proteins, as demonstrated in other bacteria (44).
Second, we showed that magnetosome expression of foreign proteins can also be enhanced by increasing their copy number. In similar approaches, Choi and coworkers integrated double copies of the cym repressor into Methylobacterium extorquens, thereby achieving tight repression of an inducible promoter (45). In the same organism, expression of five chromosomal copies of gfp resulted in 20-fold-higher expression than single-copy expression (46). Multicopy insertion of recombinant pathways can increase gene expression by 60% in contrast to plasmid expression of the same pathway in E. coli (47). In our study, duplication of (Mag-)egfp fused in tandem to mamC resulted in strong fluorescence and 2.5-fold increased expression of the (Mag-)EGFP reporter on magnetosomes. Mag-EGFP–EGFP fusions displayed proteolytic stability, as no cleavage products could be detected via Western blotting. These engineered magnetosomes represent magnetic nanoparticles with greatly enhanced fluorescence, which could be of immediate relevance for a number of applications, such as, for instance, as bimodal contrast agents for both magnetic resonance imaging (MRI) and near-infrared fluorescence optical (NIRF) imaging (48). In addition, magnetosome-expressed single and tandem EGFP fusions with enhanced fluorescence intensity and uniformity can be used as fluorescent tags to follow intracellular protein localization or to study the intricate cell biology of this and other magnetic bacteria. MamC–Mag-EGFP expression driven by PmamDC45 resulted in 33 ng Mag-EGFP per μg magnetite, which was 3.6-fold higher than that from Ptet (9 ng μg−1 magnetite). The amount of (Mag-)EGFP obtained with Mag-EGFP–EGFP fusion per μg magnetite was 83 ng and thus 2.5-fold higher than single copy MamC–Mag-EGFP expressed constitutively. Based on these data, we attempted to estimate the copy number of GFP proteins expressed on single magnetosome particles. Assuming a diameter of 37.5 nm for a single magnetite crystal as determined by transmission electron microscopy, a density of 5.24 g/cm3 for magnetite, and for simplicity an approximately spherical shape, this would result in a volume of 2.76 × 10−17 cm3 and mass of 1.45 × 10−16 g for an average single magnetosome crystal (see Fig. S7C in the supplemental material). For MamC–Mag-EGFP expressed from PmamDC45, we thus can estimate about 100 Mag-EGFP copies per magnetosome, while only about 30 copies were present if the same construct was expressed from Ptet. 250 (Mag-)EGFP copies per particle were calculated for the Mag-EGFP–EGFP fusion. This more-than-double amount of (Mag-)EGFP might be due to increased stability of the Mag-EGFP–EGFP fusion or, alternatively, just to the variability of magnetosome sizes, which to some extent depend on the growth stage of the cells. Assuming a lower size of only 35 nm and an upper size of 48 nm (as found within the typical range of variation [49]), the same calculations would yield GFP copy numbers of 80 to 210 in strain JH1 and 200 to 520 (Mag-EGFP–EGFP fusion) in strain JH3. Assuming MamC-to-Mag-EGFP ratios of 1:1 for the single protein and 1:2 for the Mag-EGFP–EGFP fusion, the number of MamC copies per magnetosome particle is most likely within the range of 80 to 260. Assuming a surface area of 4,417 nm2 for a 37.5-nm magnetosome particle and a diameter of approximately 3.45 nm for the 12.5-kDa MamC protein (7), the theoretical number of MamC copies that would cover the entire particle surface would be 1,280. However, as previous estimations revealed MamC to be only a part (relative abundance, 16.3% [50]) of the MM, which contains about 20 different proteins (50), the estimated 80 to 250 copies occupying about 6 to 20% of the surface seem to be a realistic range. Thus, the number of MamC molecules that can serve as fusion anchors is unlikely to be further increased without disturbing MM function. Instead, increasing the number of protein units fused to a single MamC anchor, as shown by our Mag-EGFP–EGFP fusion, is a more appropriate route to increase yields of heterologous proteins expressed per particle.
Supplementary Material
ACKNOWLEDGMENTS
We thank J. Armitage, S. K. Farrand, P. S. Poole, and M. Thanbichler for providing several expression plasmids, V. de Lorenzo for providing the pBAM1 vector, and K. T. Silva for help with arbitrary PCR.
This project was funded by DFG grants Schu 12-1 and 15-1.
Footnotes
Published ahead of print 14 February 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.00192-14.
REFERENCES
- 1.Jogler C, Schüler D. 2009. Genomics, genetics, and cell biology of magnetosome formation. Annu. Rev. Microbiol. 63:501–521. 10.1146/annurev.micro.62.081307.162908 [DOI] [PubMed] [Google Scholar]
- 2.Komeili A. 2012. Molecular mechanisms of compartmentalization and biomineralization in magnetotactic bacteria. FEMS Microbiol. Rev. 36:232–255. 10.1111/j.1574-6976.2011.00315.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lang C, Schüler D, Faivre D. 2007. Synthesis of magnetite nanoparticles for bio- and nanotechnology: genetic engineering and biomimetics of bacterial magnetosomes. Macromol. Biosci. 7:144–151. 10.1002/mabi.200600235 [DOI] [PubMed] [Google Scholar]
- 4.Faivre D, Schüler D. 2008. Magnetotactic bacteria and magnetosomes. Chem. Rev. 108:4875–4898. 10.1021/cr078258w [DOI] [PubMed] [Google Scholar]
- 5.Ceyhan B, Alhorn P, Lang C, Schüler D, Niemeyer CM. 2006. Semisynthetic biogenic magnetosome nanoparticles for the detection of proteins and nucleic acids. Small 2:1251–1255. 10.1002/smll.200600282 [DOI] [PubMed] [Google Scholar]
- 6.Wacker R, Ceyhan B, Alhorn P, Schüler D, Lang C, Niemeyer CM. 2007. Magneto Immuno-MR: a novel immunoassay based on biogenic magnetosome nanoparticles. Biochem. Biophys. Res. Commun. 357:391–396. 10.1016/j.bbrc.2007.03.156 [DOI] [PubMed] [Google Scholar]
- 7.Lang C, Schüler D. 2008. Expression of green fluorescent protein fused to magnetosome proteins in microaerophilic magnetotactic bacteria. Appl. Environ. Microbiol. 74:4944–4953. 10.1128/AEM.00231-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ohuchi S, Schüler D. 2009. In vivo display of a multisubunit enzyme complex on biogenic magnetic nanoparticles. Appl. Environ. Microbiol. 75:7734–7738. 10.1128/AEM.01640-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pollithy A, Romer T, Lang C, Müller FD, Helma J, Leonhardt H, Rothbauer U, Schüler D. 2011. Magnetosome expression of functional camelid antibody fragments (nanobodies) in Magnetospirillum gryphiswaldense. Appl. Environ. Microbiol. 77:6165–6171. 10.1128/AEM.05282-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lang C, Pollithy A, Schüler D. 2009. Identification of promoters for efficient gene expression in Magnetospirillum gryphiswaldense. Appl. Environ. Microbiol. 75:4206–4210. 10.1128/AEM.02906-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schübbe S, Würdemann C, Peplies J, Heyen U, Wawer C, Glöckner FO, Schüler D. 2006. Transcriptional organization and regulation of magnetosome operons in Magnetospirillum gryphiswaldense. Appl. Environ. Microbiol. 72:5757–5765. 10.1128/AEM.00201-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yoshino T, Kato F, Takeyama H, Nakai M, Yakabe Y, Matsunaga T. 2005. Development of a novel method for screening of estrogenic compounds using nano-sized bacterial magnetic particles displaying estrogen receptor. Anal. Chim. Acta 532:105–111. 10.1016/j.aca.2004.10.074 [DOI] [Google Scholar]
- 13.Yoshino T, Takahashi M, Takeyama H, Okamura Y, Kato F, Matsunaga T. 2004. Assembly of G protein-coupled receptors onto nanosized bacterial magnetic particles using Mms16 as an anchor molecule. Appl. Environ. Microbiol. 70:2880–2885. 10.1128/AEM.70.5.2880-2885.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yoshino T, Shimojo A, Maeda Y, Matsunaga T. 2010. Inducible expression of transmembrane proteins on bacterial magnetic particles in Magnetospirillum magneticum AMB-1. Appl. Environ. Microbiol. 76:1152–1157. 10.1128/AEM.01755-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Heyen U, Schüler D. 2003. Growth and magnetosome formation by microaerophilic Magnetospirillum strains in an oxygen-controlled fermentor. Appl. Microbiol. Biotechnol. 61:536–544. 10.1007/s00253-002-1219-x [DOI] [PubMed] [Google Scholar]
- 16.Sambrook J, Russell D. 2001. Molecular cloning: a laboratory manual , 3rd ed, p 1–44 Cold Spring Harbor Laboratory Press, New York, NY [Google Scholar]
- 17.Martinez-Garcia E, Calles B, Arevalo-Rodriguez M, de Lorenzo V. 2011. pBAM1: an all-synthetic genetic tool for analysis and construction of complex bacterial phenotypes. BMC Microbiol. 11:38. 10.1186/1471-2180-11-38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bryksin AV, Matsumura I. 2010. Overlap extension PCR cloning: a simple and reliable way to create recombinant plasmids. Biotechniques 48:463–465. 10.2144/000113418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schüler D, Rainer U, Bäuerlein E. 1995. A simple light-scattering method to assay magnetism in Magnetospirillum gryphiswaldense. FEMS Microbiol. Lett. 132:139–145. 10.1111/j.1574-6968.1995.tb07823.x [DOI] [Google Scholar]
- 20.Wilson KJ, Hughes SG, Jefferson RA. 1992. The Escherichia coli gus operon: induction and expression of the gus operon in E. coli and the occurrence and use of GUS in other bacteria, p 7–22 In Gallagher SR. (ed), Using the GUS gene as reporter of gene expression. Academic Press Inc., San Diego, CA [Google Scholar]
- 21.Laemmli UK. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680–685. 10.1038/227680a0 [DOI] [PubMed] [Google Scholar]
- 22.Haugen SP, Ross W, Gourse RL. 2008. Advances in bacterial promoter recognition and its control by factors that do not bind DNA. Nat. Rev. Microbiol. 6:507–519. 10.1038/nrmicro1912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen HY, Bjerknes M, Kumar R, Jay E. 1994. Determination of the optimal aligned spacing between the Shine-Dalgarno sequence and the translation initiation codon of Escherichia coli messenger RNAs. Nucleic Acids Res. 22:4953–4957. 10.1093/nar/22.23.4953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ma J, Campbell A, Karlin S. 2002. Correlations between Shine-Dalgarno sequences and gene features such as predicted expression levels and operon structures. J. Bacteriol. 184:5733–5745. 10.1128/JB.184.20.5733-5745.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li YJ, Katzmann E, Borg S, Schüler D. 2012. The periplasmic nitrate reductase Nap is required for anaerobic growth and involved in redox control of magnetite biomineralization in Magnetospirillum gryphiswaldense. J. Bacteriol. 194:4847–4856. 10.1128/JB.00903-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ind AC, Porter SL, Brown MT, Byles ED, de Beyer JA, Godfrey SA, Armitage JP. 2009. Inducible-expression plasmid for Rhodobacter sphaeroides and Paracoccus denitrificans. Appl. Environ. Microbiol. 75:6613–6615. 10.1128/AEM.01587-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Khan SR, Gaines J, Roop RM, Farrand SK. 2008. Broad-host-range expression vectors with tightly regulated promoters and their use to examine the influence of TraR and TraM expression on Ti plasmid quorum sensing. Appl. Environ. Microbiol. 74:5053–5062. 10.1128/AEM.01098-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tett AJ, Rudder SJ, Bourdes A, Karunakaran R, Poole PS. 2012. Regulatable vectors for environmental gene expression in Alphaproteobacteria. Appl. Environ. Microbiol. 78:7137–7140. 10.1128/AEM.01188-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thanbichler M, Iniesta AA, Shapiro L. 2007. A comprehensive set of plasmids for vanillate- and xylose-inducible gene expression in Caulobacter crescentus. Nucleic Acids Res. 35:e137. 10.1093/nar/gkm818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Topp S, Reynoso CMK, Seeliger JC, Goldlust IS, Desai SK, Murat D, Shen A, Puri AW, Komeili A, Bertozzi CR, Scott JR, Gallivan JP. 2010. Synthetic riboswitches that induce gene expression in diverse bacterial species. Appl. Environ. Microbiol. 76:7881–7884. 10.1128/AEM.01537-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hillen W, Berens C. 1994. Mechanisms underlying expression of Tn10 encoded tetracycline resistance. Annu. Rev. Microbiol. 48:345–369. 10.1146/annurev.mi.48.100194.002021 [DOI] [PubMed] [Google Scholar]
- 32.Kamionka A, Bertram R, Hillen W. 2005. Tetracycline-dependent conditional gene knockout in Bacillus subtilis. Appl. Environ. Microbiol. 71:728–733. 10.1128/AEM.71.2.728-733.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cubitt AB, Heim R, Adams SR, Boyd AE, Gross LA, Tsien RY. 1995. Understanding, improving and using green fluorescent proteins. Trends Biochem. Sci. 20:448–455. 10.1016/S0968-0004(00)89099-4 [DOI] [PubMed] [Google Scholar]
- 34.Scheffel A, Gärdes A, Grünberg K, Wanner G, Schüler D. 2008. The major magnetosome proteins MamGFDC are not essential for magnetite biomineralization in Magnetospirillum gryphiswaldense but regulate the size of magnetosome crystals. J. Bacteriol. 190:377–386. 10.1128/JB.01371-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hawley DK, McClure WR. 1983. Compilation and analysis of Escherichia coli promoter DNA sequences. Nucleic Acids Res. 11:2237–2255. 10.1093/nar/11.8.2237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Debowski AW, Verbrugghe P, Sehnal M, Marshall BJ, Benghezal M. 2013. Development of a tetracycline-inducible gene expression system for the study of Helicobacter pylori pathogenesis. Appl. Environ. Microbiol. 79:7351–7359. 10.1128/AEM.02701-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Geissendörfer M, Hillen W. 1990. Regulated expression of heterologous genes in Bacillus subtilis using the Tn10 encoded Tet regulatory elements. Appl. Microbiol. Biotechnol. 33:657–663. 10.1007/BF00604933 [DOI] [PubMed] [Google Scholar]
- 38.Guss AM, Rother M, Zhang JK, Kulkkarni G, Metcalf WW. 2008. New methods for tightly regulated gene expression and highly efficient chromosomal integration of cloned genes for Methanosarcina species. Archaea 2:193–203. 10.1155/2008/534081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Skerra A. 1994. Use of the tetracycline promoter for the tightly regulated production of a murine antibody fragment in Escherichia coli. Gene 151:131–135. 10.1016/0378-1119(94)90643-2 [DOI] [PubMed] [Google Scholar]
- 40.Ji YD, Marra A, Rosenberg M, Woodnutt G. 1999. Regulated antisense RNA eliminates alpha-toxin virulence in Staphylococcus aureus infection. J. Bacteriol. 181:6585–6590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stieger M, Wohlgensinger B, Kamber M, Lutz R, Keck W. 1999. Integrational plasmids for the tetracycline-regulated expression of genes in Streptococcus pneumoniae. Gene 226:243–251. 10.1016/S0378-1119(98)00561-7 [DOI] [PubMed] [Google Scholar]
- 42.Davidson CJ, Surette MG. 2008. Individuality in bacteria. Annu. Rev. Genet. 42:253–268. 10.1146/annurev.genet.42.110807.091601 [DOI] [PubMed] [Google Scholar]
- 43.Siegele DA, Hu JC. 1997. Gene expression from plasmids containing the araBAD promoter at subsaturating inducer concentrations represents mixed populations. Proc. Natl. Acad. Sci. U. S. A. 94:8168–8172. 10.1073/pnas.94.15.8168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Grosjean H, Fiers W. 1982. Preferential codon usage in prokaryotic genes: the optimal codon-anticodon interaction energy and the selective codon usage in efficiently expressed genes. Gene 18:199–209. 10.1016/0378-1119(82)90157-3 [DOI] [PubMed] [Google Scholar]
- 45.Choi YJ, Morel L, Bourque D, Mullick A, Massie B, Miguez CB. 2006. Bestowing inducibility on the cloned methanol dehydrogenase promoter (PmxaF) of Methylobacterium extorquens by a applying regulatory elements of Pseudomonas putida F1. Appl. Environ. Microbiol. 72:7723–7729. 10.1128/AEM.02002-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Choi YJ, Bourque D, Morel L, Groleau D, Míguez CB. 2006. Multicopy integration and expression of heterologous genes in Methylobacterium extorquens ATCC 55366. Appl. Environ. Microbiol. 72:753–759. 10.1128/AEM.72.1.753-759.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tyo KEJ, Ajikumar PK, Stephanopoulos G. 2009. Stabilized gene duplication enables long-term selection-free heterologous pathway expression. Nat. Biotechnol. 27:760–765. 10.1038/nbt.1555 [DOI] [PubMed] [Google Scholar]
- 48.Lisy MR, Hartung A, Lang C, Schüler D, Richter W, Reichenbach JR, Kaiser WA, Hilger I. 2007. Fluorescent bacterial magnetic nanoparticles as bimodal contrast agents. Invest. Radiol. 42:235–241. 10.1097/01.rli.0000255832.44443.e7 [DOI] [PubMed] [Google Scholar]
- 49.Lohße A, Ullrich S, Katzmann E, Borg S, Wanner G, Richter M, Voigt B, Schweder T, Schüler D. 2011. Functional analysis of the magnetosome island in Magnetospirillum gryphiswaldense: the mamAB operon is sufficient for magnetite biomineralization. PLoS One 6:e25561. 10.1371/journal.pone.0025561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Grünberg K, Müller EC, Otto A, Reszka R, Linder D, Kube M, Reinhardt R, Schüler D. 2004. Biochemical and proteomic analysis of the magnetosome membrane in Magnetospirillum gryphiswaldense. Appl. Environ. Microbiol. 70:1040–1050. 10.1128/AEM.70.2.1040-1050.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.