Abstract
We sequenced the oldest blaKPC-2-bearing plasmid isolated in Brazil and another plasmid also carried by a Klebsiella pneumoniae strain of sequence type 442 (ST442), isolated 52 months later. Both plasmids present an IncN backbone and few acquired regions. Because the 2005 plasmid presented deletions and a truncated gene within Tn4401b compared to the 2009 plasmid, we can thus infer that IncN blaKPC-2-bearing plasmids pFCF1305 and pFCF3SP had a common ancestor circulating in Brazil prior to May 2005.
TEXT
Even though the first description of KPC-producing Klebsiella pneumoniae in Brazil was published in 2009, describing a strain isolated from a clinical sample in 2006 (1), a later publication reported the presence of a KPC-bearing strain isolated in May 2005 (2). The latter strain, FCF1305, was isolated in the state of São Paulo, Brazil, from a blood culture and belongs to sequence type 442 (ST442). We completely sequenced the blaKPC-2-bearing plasmid isolated from FCF1305 and also another blaKPC-2-bearing plasmid isolated from another K. pneumoniae strain, FCF3SP, also belonging to ST442 and also isolated from a blood culture from a patient in the state of São Paulo, but 52 months later, in September 2009. We chose to sequence the blaKPC-2-bearing plasmids of these strains because FCF1305 was the oldest strain-bearing KPC in Brazil and strain FCF3SP was the strain most geographically, clinically, and epidemiologically similar to FCF1305 that we could find, but isolated 52 months later, so that differences accumulated in the wild during this period could be evaluated.
Because strain FCF1305 bore three plasmids, whereas strain FCF3SP bore five, isolation of the blaKPC-2-bearing plasmids of each strain was carried out by transformation into Escherichia coli TOP10 (Invitrogen) as described previously (3). Plasmid DNA isolated from E. coli TOP10(pFCF1305) and E. coli TOP10(pFCF3SP) by alkaline lysis miniprep (3) was used to construct genomic fragment libraries (one for each plasmid) which were sequenced using the Roche GS-FLX sequencer (Roche Life Sciences, Branford, CT, USA) according to the manufacturer's instructions.
Sequencing adapters were clipped from the reads and used for de novo assembly using Mira 4.0 (4). Plasmid de novo contigs were then merged by assembling de novo using Geneious (5) with stringent parameters (maximum gap size = 1; minimum overlap identity = 95%). Circular (with large overlapping ends) contigs of ∼50 kbp were considered to be the plasmids. All other contigs generated which had no identity to the circular contigs were then subjected to a BLAST search (6) against GenBank's nonredundant (nr) database, and 100% of them had as top hits Enterobacteriaceae chromosomes or phages and so were discarded. The plasmid sequences were automatically annotated using the RAST server (7) and manually curated. Repeat regions were identified using UGENE (8) and insertion sequences were identified by online analysis using IS finder (9). The complete sequences of both plasmids were successfully obtained by de novo assembly yielding a 53,081-bp sequence for pFCF1305 (mean coverage of 261× ± 48×; minimum coverage of 39×; maximum coverage of 445×) and a 54,605-bp sequence for pFCF3SP (mean coverage of 1,013× ± 374×; minimum coverage of 67×; maximum coverage of 2,383×).
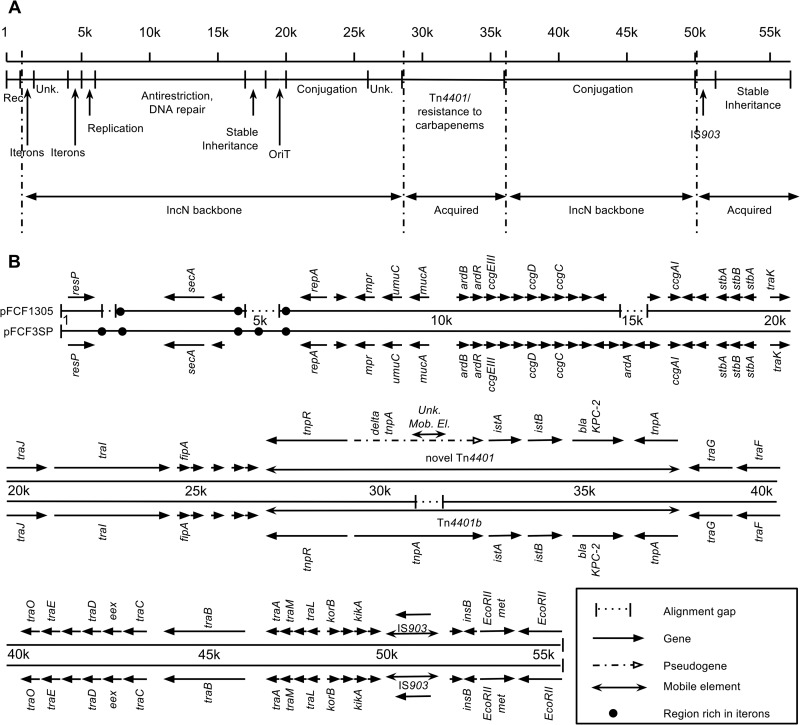
Both plasmids were found to have IncN plasmid backbones and carry a version of transposon Tn4401b (10). The general functional structures of both plasmids and a graphical comparison of the genetic architectures of the plasmids are presented in Fig. 1. The features present in both plasmid sequences are presented and compared to each other in Table S1 in the supplemental material.
FIG 1.
(A) Graphical representation of the general structures, origins, and functions of plasmids pFCF1305 (GenBank accession no. CP004366) and pFCF3SP (GenBank accession CP004367). Rec., recombination; Unk., unknown. (B) Representation of the alignment of the sequences of pFCF1305 and pFCF3SP showing features present and absent in each sequence.
The sequences of pFCF1305 and pFCF3SP have similar sizes and relatively minimal structures, inasmuch as both consist of a highly conserved IncN backbone with two acquired regions, one being a version of Tn4401b, which carries as the only resistance gene blaKPC-2, and a second region containing the small insertion sequence IS903B. Other IncN plasmids carrying blaKPC (GenBank accession numbers FJ223607, FJ223605, KC958437, and JX193301) are larger (∼65 kbp to ∼83 kbp) than pFCF1305 and pFCF3SP, with the exception of plasmid pKP1433 (JX397875), which is about the same size but contains several non-IncN backbone replication genes (11).
The sequences of pFCF1305 and pFCF3SP are highly conserved in spite of their having been isolated 52 months apart, with their difference in size being distributed between four regions of difference (Fig. 1). The region around ∼15 kbp missing in pFCF1305 contains ardA, an anti-restriction gene, and two small genes coding for hypothetical proteins. This deletion thus probably has a very small significance for the biological effects of inheriting either plasmid. Whereas pFCF3SP presented a Tn4401 sequence that is >99% identical to the prototype Tn4401b isoform (10) (EU176012), pFCF1305 presented a Tn4401b isoform with the tnpA gene truncated due to the insertion of a 256-bp sequence which seems to be a mobile genetic element. This unknown mobile element (UME), like insertion sequences, is limited by inverted repeats of 39 bp. Unlike common insertion sequences, it does not, however, possess a gene coding for a transposase between the inverted repeats. This is the first description of the truncation of the tnpA gene of Tn4401b, which warrants its designation as a novel Tn4401 isoform.
The novel Tn4401b isoform present in pFCF1305 with the truncated tnpA was unexpectedly present in the older molecule, pFCF1305, rather than the newer pFCF3SP. The perfect direct-repeat target site duplications generated in pFCF1305 and pFCF3SP due to transposition of Tn4401b are also identical in both plasmids (5′CTTCAG3′), which indicates that truncation of the tnpA found in pFCF1305 by the UME happened after Tn4401b was transposed into the common IncN ancestor of pFCF1305 and pFCF3SP and that this occurred earlier than 2005. Furthermore, because the two plasmids are different yet were both found in K. pneumoniae ST442 strains isolated in the same city from clinically similar cases, it can be inferred that the reason they are present in each strain is not due to vertical transmission. This can be argued because it is the plasmid from 2005 which presents small deletions of IncN backbone elements, which would have to have been “reacquired” by the later ST442 strain bearing pFCF3SP. Also, the UME truncating tnpA in pFCF1305 is not present in pFCF3SP, and although it is strictly possible that the element could be subsequently lost, it seems very unlikely that tnpA would have been restored to perfect functionality after such an event. We can thus infer that the IncN blaKPC-2-bearing plasmids pFCF1305 and pFCF3SP had a common ancestor circulating in Brazil prior to May 2005.
Nucleotide sequence accession numbers.
The nucleotide sequences for plasmids pFCF1305 and pFCF3SP have been deposited in GenBank under accession numbers CP004366 and CP004367, respectively.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the Centro de Inovações Tecnológicas do Instituto Evandro Chagas, SVS/MS, Ananindeua, Pará, Brazil. M.G.Q. was supported by the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES), Brazil. P.J.P.-C. was supported by Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP) grant 2012/20915-0. J.A.M. was supported by FAPESP grant 2013/12107-4.
We have no conflicts of interest to declare.
Footnotes
Published ahead of print 24 February 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AAC.02341-13.
REFERENCES
- 1.Monteiro J, Santos AF, Asensi MD, Peirano G, Gales AC. 2009. First report of KPC-2-producing Klebsiella pneumoniae strains in Brazil. Antimicrob. Agents Chemother. 53:333–334. 10.1128/AAC.00736-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pavez M, Mamizuka EM, Lincopan N. 2009. Early dissemination of KPC-2-producing Klebsiella pneumoniae strains in Brazil. Antimicrob. Agents Chemother. 53:2702. 10.1128/AAC.00089-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sambrook J, Russell DW. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 4.Chevreux B, Wetter T, Suhai S. 1999. Genome sequence assembly using trace signals and additional sequence information, p 45–56 In Computer science and biology: proceedings of the German Conference on Bioinformatics (GCB) 99 [Google Scholar]
- 5.Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, Thierer T, Ashton B, Meintjes P, Drummond A. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28:1647–1649. 10.1093/bioinformatics/bts199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403–410 [DOI] [PubMed] [Google Scholar]
- 7.Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, Edwards RA, Formsma K, Gerdes S, Glass EM, Kubal M, Meyer F, Olsen GJ, Olson R, Osterman AL, Overbeek RA, McNeil LK, Paarmann D, Paczian T, Parrello B, Pusch GD, Reich C, Stevens R, Vassieva O, Vonstein V, Wilke A, Zagnitko O. 2008. The RAST server: rapid annotations using subsystems technology. BMC Genomics 9:75. 10.1186/1471-2164-9-75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Okonechnikov K, Golosova O, Fursov M. 2012. Unipro UGENE: a unified bioinformatics toolkit. Bioinformatics 28:1166–1167. 10.1093/bioinformatics/bts091 [DOI] [PubMed] [Google Scholar]
- 9.Siguier P, Perochon J, Lestrade L, Mahillon J, Chandler M. 2006. ISfinder: the reference centre for bacterial insertion sequences. Nucleic Acids Res. 34:D32–D36. 10.1093/nar/gkj014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Naas T, Cuzon G, Villegas M-V, Lartigue M-F, Quinn JP, Nordmann P. 2008. Genetic structures at the origin of acquisition of the beta-lactamase bla KPC gene. Antimicrob. Agents Chemother. 52:1257–1263. 10.1128/AAC.01451-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Papagiannitsis CC, Miriagou V, Giakkoupi P, Tzouvelekis LS, Vatopoulos AC. 2013. Characterization of pKP1433, a novel KPC-2-encoding plasmid from Klebsiella pneumoniae sequence type 340. Antimicrob. Agents Chemother. 57:3427–3429. 10.1128/AAC.00054-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.