Abstract
The inherent drug susceptibility of microorganisms is determined by multiple factors, including growth state, the rate of drug diffusion into and out of the cell, and the intrinsic vulnerability of drug targets with regard to the corresponding antimicrobial agent. Mycobacterium tuberculosis, the causative agent of tuberculosis (TB), remains a significant source of global morbidity and mortality, further exacerbated by its ability to readily evolve drug resistance. It is well accepted that drug resistance in M. tuberculosis is driven by the acquisition of chromosomal mutations in genes encoding drug targets/promoter regions; however, a comprehensive description of the molecular mechanisms that fuel drug resistance in the clinical setting is currently lacking. In this context, there is a growing body of evidence suggesting that active extrusion of drugs from the cell is critical for drug tolerance. M. tuberculosis encodes representatives of a diverse range of multidrug transporters, many of which are dependent on the proton motive force (PMF) or the availability of ATP. This suggests that energy metabolism and ATP production through the PMF, which is established by the electron transport chain (ETC), are critical in determining the drug susceptibility of M. tuberculosis. In this review, we detail advances in the study of the mycobacterial ETC and highlight drugs that target various components of the ETC. We provide an overview of some of the efflux pumps present in M. tuberculosis and their association, if any, with drug transport and concomitant effects on drug resistance. The implications of inhibiting drug extrusion, through the use of efflux pump inhibitors, are also discussed.
INTRODUCTION
Tuberculosis (TB), caused by Mycobacterium tuberculosis, remains a global health problem, causing 8.8 million incident cases and 1.1 million deaths in 2012 (1, 2). In many countries, the TB epidemic continues unabated in the face of combination chemotherapy which involves the administration of at least four drugs. The most significant barrier to the eradication of TB is the rapid emergence of multidrug-resistant (MDR) and extensively drug-resistant (XDR) TB, which has rendered current treatments ineffective and placed an enormous patient management burden on TB control programs. Treatment of drug-resistant TB is costly and requires the use of highly toxic drugs, leading to numerous side effects. Moreover, the spread of MDR strains makes for an alarming situation which provides an ideal breeding ground for further, more advanced forms of drug resistance (3). Given the limited number of anti-TB drugs currently available and the duration of treatment required to achieve cure, there is a global need for the discovery of novel drugs with rapid sterilizing activity against active and dormant bacteria. These drugs should ideally shorten treatment duration and reduce the pill burden (2, 4). Moreover, maintaining the fidelity of the current antibiotics and further understanding the mechanism of emergence of drug resistance require immediate attention to address this growing problem.
Suboptimal intracellular concentrations of drugs often lead to transient drug tolerance, which may be a precursor to chromosomally encoded, stable drug resistance. There is a growing body of evidence that suggests that M. tuberculosis retains the capacity to extrude drugs from the cell, resulting in drug tolerance effects (reviewed in reference 5). In many cases, these processes are postulated to be dependent on the proton motive force (PMF) and the presence of sufficient ATP concentrations within the cell. This suggests that the activity of the mycobacterial electron transport chain (ETC) under different conditions plays a key role in determining the inherent susceptibility of M. tuberculosis to various drugs. In this review, we provide an overview of the mycobacterial ETC and discuss this component of M. tuberculosis metabolism as a target for novel antitubercular agents. We also highlight the importance of energy metabolism in mediating drug tolerance through efflux.
THE MYCOBACTERIAL ETC
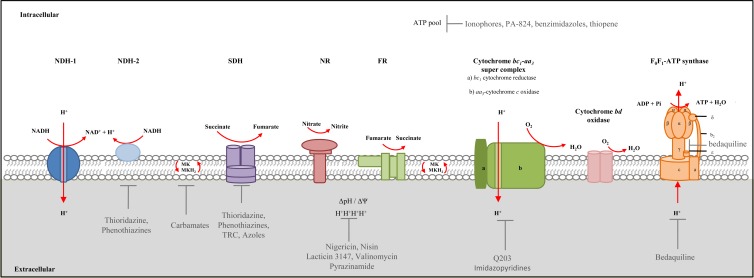
In bacteria, the ETC is integrally involved in the generation of energy via oxidative phosphorylation. The mycobacterial components involved in oxidative phosphorylation and energy production have been extensively reviewed, and the reader is referred to these for more-detailed information (6–12). Electrons enter and are shunted through the ETC in a variety of ways, depending on the source of growth substrates and the availability of terminal electron acceptors. Under aerobic conditions, oxygen is used in the final electron transfer steps, and under anaerobic conditions, nitrate or fumarate can be used (Fig. 1). Electron transport in mycobacteria is initiated through the activity of various NADH dehydrogenases (NDH) and succinate dehydrogenases (SDH), which transfer electrons to menaquinone, a lipophilic redox carrier (7, 13, 14). Electrons are then passed to various cytochrome oxidases, which are dependent on oxygen availability (Fig. 1) (7, 15, 16). Of particular note is the fact that the mycobacterial ETC, like many bacterial ETCs, is branched and displays an extensive capacity to utilize numerous electron donors and acceptors for adapting to decreasing levels of oxygen tension and the availability of different reducing equivalents. The roles of various components of the ETC are detailed below.
FIG 1.
The mycobacterial electron transport chain. Proposed aerobic and hypoxic/anaerobic pathways are shown (7, 9). NDH-1, NDH-2, and succinate dehydrogenase (SDH) are electron donors which reduce menaquinone (MK) to menaquinol (MKH2). Subsequently, MKH2 becomes oxidized, transferring electrons to the terminal electron acceptors through the activity of cytochrome oxidases, nitrate reductase (NR), and fumarate reductase (FR). ATP production via the F1F0-ATP synthase is fuelled by translocation of protons (H+). The F1F0-ATP synthase consists of two regions, namely, the hydrophobic integral membrane region (F0) composed of 3 subunits designated a, b, and c and the hydrophilic region (F1), which extends into the cytoplasm and is composed of 5 subunits designated α, β, δ, γ, and ε (7, 45–47). Energy production may be targeted by anti-TB compounds at multiple points involved in ATP synthesis as shown. These include the transmembrane proton gradient (ΔpH)—targeted by compounds such as nigericin, nisin, lacticin 3147, valinomycin, and pyrazinamide (PZA), resulting in disruption of ATP homeostasis. Compounds resulting in a depletion of the ATP pool include phenothiazines, ionophores (shown in the diagram), PA-824, benzimidazoles, thiophene, and imidazopyridines (Q203)/bedaquiline—through direct/indirect effects on the ETC. Other components of energy metabolism targeted by compounds include NDH-2 (thioridazine and phenothiazine), SDH (thioridazine, phenothiazine, triclosan, and azoles), MK biosynthesis (carbamates), Q203, and the F1F0-ATP synthase (bedaquiline). Vertical red arrows indicate proton pumping components of the ETC that directly translocate protons into the extracellular matrix.
ELECTRON DONORS
Analysis of the genome sequence of M. tuberculosis H37Rv reveals two types of NDHs in mycobacteria, namely, type I NDH (NDH-1), encoded by the nuoABCDEFGHIJKLMN operon, and type II NDH (NDH-2), encoded by either the ndh gene or the ndhA gene (17, 18). In mycobacteria, the reduction in expression of nuoB, encoding a subunit of NDH-1, during oxygen deprivation suggests that NDH-1 is required preferentially under aerobic conditions (19), while use of NDH-2 is favored under anaerobic/nonreplicating conditions (20). Treatment of mycobacteria with NDH-2-specific inhibitors, such as trifluoperazine, results in blockage of initiation of the ETC under anaerobic conditions, implying that NDH-2 is the dominant NDH involved in anaerobic respiration under the conditions tested (20). However, the dispensability of the entire nuoABCDEFGHIJKLMN operon under aerobic conditions (20) suggests that NDH-2 can operate as an electron donor irrespective of oxygen availability. The NuoG subunit of NDH-1 has been identified in a forward genetic screen to be essential for inhibiting macrophage apoptosis and in virulence in the murine model of TB infection (21). The precise mechanism of apoptosis inhibition is not yet understood, but more-recent work suggests that it involves the neutralization of NOX2-derived reactive oxygen species (22). In this regard, the suppression of apoptosis leads to reduced distribution of tubercle bacilli between cells found within the lung and this results in decreased proliferation of M. tuberculosis-specific naive T cells (23). Transposon mutagenesis has identified NDH-1 to be dispensable for growth in vitro, and in this context, the apparent essentiality of NuoG for macrophage apoptosis does not seem to be directly related to its role in energy metabolism (24). Saturating transposon mutagenesis predicts that NDH-1 and NDH-2 are dispensable for growth of M. tuberculosis in vitro (24). Furthermore, an ndhA mutant is able to colonize mouse lungs, confirming that this enzyme is not essential for pathogenesis (25).
SDH, or succinate:menaquinone oxidoreductase, is another enzyme responsible for the donation of electrons to the quinone pool (13). Transposon mutagenesis confirmed that SDH is essential for survival of M. tuberculosis in vitro (24). This enzyme predominantly acts in the citric acid cycle, where it oxidizes succinate to fumarate, thereby donating electrons to the quinone pool (26, 27). It has been recently demonstrated that, through remodelling of the tricarboxylic acid cycle, the activity of SDH is essential for the metabolic adaptation to hypoxia, maintenance of membrane potential, and ATP synthesis, indicating that this is a key enzyme for persistence and therefore represents a potential new drug target (27).
MENAQUINONE
Quinones are lipid soluble electron carriers that are responsible for the transfer of electrons between the components of the ETC (7, 28). In mycobacteria, the predominant quinones are menaquinones (MK), in contrast to the case in Escherichia coli, which utilizes both menaquinone and ubiquinone (14, 29–31). In M. tuberculosis, MK is synthesized from the precursor chorismate through a series of reactions catalyzed by enzymes encoded by the menABCDEFG operon. Recently, an array of MenA inhibitors which display bactericidal activity against nonreplicating M. tuberculosis was developed (32). These observations suggest that MK needs to be continuously resynthesized to maintain membrane potential. Of the compounds developed, carbamates have shown significant bacteriostatic activity against nonreplicating M. tuberculosis (Fig. 1) (32). Due to the importance of the MK pool in maintaining the ETC and subsequent ATP synthesis, this biosynthetic pathway represents an important target for anti-TB drug design.
ELECTRON ACCEPTORS
Electrons from reduced menaquinol can be transferred to either of two terminal oxidases: (i) the cytochrome bd-type menaquinol oxidase or (ii) the aa3-type cytochrome c oxidase (via the cytochrome bc1 reductase complex). It has been suggested that the mycobacterial aa3-type cytochrome c oxidase and cytochrome bc1 reductase form a supercomplex (7, 16). The M. tuberculosis genome encodes a putative cytochrome bd oxidase which, in Mycobacterium smegmatis, has been shown to play an important role in the adaption to a reduced oxygen environment (15, 17). The M. tuberculosis genome also encodes a membrane-bound respiratory/assimilatory narGHI-encoded nitrate reductase (NR) which is responsible for nitrate-associated respiration and assimilation (17, 33–36). Analysis of clinical M. tuberculosis isolates revealed that NarG plays an important role in fitness in macrophages (37), consistent with the detection of transcripts from this operon within granulomas from lungs of TB patients (38) and in the guinea pig model of TB infection (39). The NR is also required for survival of M. tuberculosis in vitro under anaerobic conditions of nonreplicating persistence (40) and for the protection of M. tuberculosis against acidic conditions during hypoxia (41). These data suggest that respiration using nitrate may be an important mechanism for adaption under various conditions of stress. In support of this hypothesis, the nirBD-encoded nitrite reductase has recently been shown to play an important role in nonreplicating persistence of M. tuberculosis (34, 42). Considering other alternate electron acceptors, transcriptional analysis revealed that frdA, a gene encoding the flavoprotein subunit of the fumarate reductase, is upregulated under hypoxic conditions in M. tuberculosis and plays a vital role in maintenance of an energized membrane (43).
ATP SYNTHESIS AND THE F1F0-ATP SYNTHASE
Substrate-level phosphorylation involves the production of ATP by the utilization of free energy produced during various steps in metabolic pathways and provides a fast source of ATP through a process that is not dependent on external electron acceptors (44). In contrast, during oxidative phosphorylation, ATP is produced through the activity of the F1F0-ATP synthase enzyme, which is coupled to the PMF. M. tuberculosis has been classified as an obligate aerobe; as such, it would be dependent on oxidative phosphorylation for growth and survival during pathogenesis. The structure of F1F0-ATP synthase in various prokaryotes is conserved and consists of two regions, namely, the hydrophobic integral membrane region (F0) and the hydrophilic region (F1), extending into the cytoplasm (Fig. 1) (45–47). The F0 region is composed of 3 subunits designated a, b, and c, whereas the F1 region is composed of 5 subunits designated α, β, δ, γ, and ε (45, 46). ATP is synthesized by the α3β3-hexamer through rotation of the γ-ε pair (48). Rotation of subunits ε and γ is coupled to rotation of the c-ring upon proton translocation (Fig. 1) (48, 49). The mycobacterial F1F0-ATP synthase is encoded by a single operon, Rv1303-atpBEFHAGDC-Rv1312 (17). Recently, BlaI (Rv1846c) was identified as a transcriptional regulator of this operon (50). Treatment with ATP synthase inhibitors results in increased expression of blaI, suggesting a role for this regulator in response to stress (51). The mode of regulation of blaI in Staphylococcus aureus has been elucidated. BlaI has a structure that is similar to that of penicillin binding proteins and is able to act as a transcriptional repressor in response to antibiotic treatment (52). In this regard, it has been shown that BlaI (Rv1846c) in M. tuberculosis responds to antibiotic treatment and is released from its cognate operator sequences to allow gene expression (50). BlaR (Rv1845c), a zinc-dependent metalloprotease, has been hypothesized to play a role in cleaving itself and BlaI during derepression of the operon (50, 52). More recently, transcriptional regulators Rv1773c and Rv3405c have been identified as regulators for Rv1303, the first gene of the atpBEFHAGDC operon (53).
DRUGS THAT TARGET THE ETC
Recent drug discovery efforts have led to numerous compounds which have shown great promise in the treatment of TB due to their ability to eliminate M. tuberculosis in various preclinical models and early clinical trials. Among these is the discovery of TMC207 (bedaquiline), which kills M. tuberculosis by inhibition of the membrane-bound F1F0-ATP synthase complex, resulting in depletion of cellular ATP levels and eventual death of the organism (49, 54–56). Use of this drug, commonly known as bedaquiline, resulted in decreased time to smear conversion during a phase IIb randomized trial (54, 57). Bedaquiline kills M. tuberculosis by interacting with the hydrophobic region of subunit c, as well as with subunit ε (48), of the F1F0-ATP synthase and does not cross-react with the human ATP synthase complex (56, 58). Inhibition of c-ring rotation due to disruption in the c-ring:ε subunit interaction results in inhibition of ATP production and subsequent cell death (48). The efficacy of bedaquiline in clinical trials confirms that targeting energy metabolism during TB infection may be promising, particularly for nonreplicating organisms, as there is documented evidence that ATP is essential for the viability of nonreplicating persistent mycobacteria (20, 55, 59). Moreover, the ability to eliminate subpopulations of persisting organisms provides an opportunity for tissue sterilization, thereby minimizing the risk for recrudescent disease through reactivation of persisting bacteria. The ability of these organisms to maintain an energized membrane potential in the face of prolonged quiescence is critical to their survival and highlights the importance of understanding the physiology of M. tuberculosis with regard to energy metabolism (9). In this context, ATP synthesis has become a focus area for the identification of new drug targets in mycobacteria (60).
The PMF is an important aspect in the final production of ATP via the F1F0-ATP synthase (20). The PMF is established through the development of the transmembrane proton gradient (ΔpH) which occurs when electrons move through the ETC and results in the establishment of membrane potential (Δψ) (reviewed in references 6 and 7). The proton gradient generated through oxidative phosphorylation drives ATP synthesis via the F1F0-ATP synthase which is responsible for the conversion of the electrochemical potential energy generated by the PMF into chemical energy in the form of ATP (45). Since oxidative phosphorylation is the main source of energy production in mycobacteria, ATP synthase represents the key enzyme involved in ATP production in mycobacteria.
Studies of valinomycin and nigericin treatment (inhibitors of Δψ and ΔpH, respectively) revealed that in both actively replicating and hypoxic nonreplicating bacilli, death occurs via decreased ATP levels in a dose-dependent manner (20). Nisin, a lantibiotic produced by Lactococcus lactis, has been shown to dissipate Δψ and ΔpH, thereby disrupting energy metabolism (20, 61). Lantibiotics such as nisin and lacticin 3147 form pores in the mycobacterial cell membrane which result in dissipation of Δψ (Fig. 1) (several studies have investigated the effect of these compounds in mycobacteria [61–65]). Although nisin is able to dissipate the membrane potential and decrease ATP levels in mycobacteria, the MIC values for various mycobacterial strains are very high and not comparable to those of current anti-TB drugs (62). This poor inhibitory activity of nisin has been attributed to its low solubility at pH 7, in contrast to lacticin 3147, which is soluble under such conditions and demonstrates greater activity against mycobacteria, thus warranting further investigation as a potential anti-TB drug (62). Another compound shown to target Δψ is pyrazinamide (PZA), where treatment results in a decrease in ATP levels, most likely the resulting effect of diminished membrane potential (66). Unlike current first- and second-line anti-TB drugs, PZA has been shown to be active against both replicating and nonreplicating mycobacteria (67). This was highlighted in a recent study, where nutrient-starved M. tuberculosis displayed increased susceptibility to PZA due to the decreased membrane potential (68). Furthermore, PZA treatment of mice infected with M. tuberculosis significantly reduces the release of proinflammatory cytokines and chemokines, suggesting that PZA has important host-directed effects (69).
Due to the importance of ATP for cellular viability, the components involved in the process of ATP production represent viable drug targets which, in combination with current anti-TB drugs, could be used for effective treatment. A number of existing compounds deplete cellular ATP levels and have subsequent bactericidal effects on replicating and nonreplicating mycobacteria (Fig. 1). These compounds include n-decanesulfonylacetamide (DSA) and nisin. DSA is the lead compound of the β-sulfonylacetamide class of antimicrobials and has been shown to be active in vitro against replicating M. tuberculosis as well as against anaerobic M. bovis BCG (70–73). It has been proposed that DSA interferes with components of the respiratory chain, thereby disrupting energy metabolism (73).
Another compound targeting energy metabolism is PA-824, which is currently in human clinical trials as an anti-TB drug (57, 74–76). PA-824, a bicyclic nitroimidazole, has the ability to kill both replicating and hypoxic, nonreplicating M. tuberculosis through a multifaceted mechanism that involves inhibition of mycolic acid biosynthesis and respiratory poisoning through intracellular release of nitric oxide, which is postulated to inhibit the final stages of electron transfer in cytochrome c oxidase (76).
More recent efforts have yielded a novel class of imidazo[1,2-a]pyridine amide (IPA) compounds (77, 78) that prevent proliferation of M. tuberculosis by inhibition of the cytochrome bc1 reductase complex in the mycobacterial respiratory chain (77, 79) (Fig. 1). These compounds bind the QcrB subunit and induce bacterial cell death by abrogating electron flow through the ETC, resulting in reduced ATP synthesis under aerobic and anaerobic conditions (79). Two independent studies identified QcrB as the target for IPAs through the generation of spontaneous resistant mutants carrying various substitutions at the Thr313 residue (77, 79). The lead compound from this series, Q203, displays potent killing of M. tuberculosis in axenic culture, in macrophages, and in the murine model of TB infection, with a spontaneous mutation rate in the order of 10−8 (79). Q203 is well tolerated in mice and now awaits further analysis in clinical trials.
Phenothiazines, such as chlorpromazine and thioridazine (THZ) (Fig. 1), are a group of clinically relevant compounds that are predicted to target the ETC through inhibition of NDH-2 (18). THZ is an old neuroleptic agent that has demonstrated activity in killing drug-susceptible and drug-resistant M. tuberculosis in various model systems in vitro and ex vivo and in the murine model of TB infection (80–84). In the promising development, THZ demonstrated therapeutic benefit in treatment of XDR-TB patients in Argentina and is currently being used in trials in India (reviewed in reference 85). In a recent study, THZ activity was shown to be independent of the bacterial growth phase; i.e., THZ is effective against actively replicating bacilli, semidormant bacilli, and nonreplicating persisters (86). That study also demonstrated a low mutation frequency in M. tuberculosis, suggesting a delay in the development of THZ resistance (86). While retaining the ability to inhibit the ETC in mycobacteria, THZ also has the potential to directly inhibit efflux of drugs (87). In addition to phenothiazines, which target the mycobacterial NDH-2, clofazimine (CFZ)—a rhiminophenazine—is subject to reduction by NDH-2 and, upon subsequent oxidation, leads to the formation of reactive oxygen species (88), which presumably contributes to its efficacy in mice (89).
Numerous studies have investigated the efficacy of novel combinations of drugs by incorporating new and existing compounds which have demonstrated activity against bacterial targets involved in energy metabolism. These combinations have demonstrated efficacy that is comparable or enhanced in comparison to the current anti-TB regimens, and the potential to shorten the current treatment period has been highlighted. A recent 14-day early bactericidal activity (EBA) study demonstrated the benefit of adding PA-824 to a regimen containing moxifloxacin (MXF) and PZA to shorten treatment duration (89). Bedaquiline–PZA–PA-824 and bedaquiline–PA-824–MFX combinations have demonstrated greater efficacy than rifampin (RIF)-isoniazid (INH)-PZA in reducing CFU (90). Tasneen et al. also demonstrated that drug combinations containing bedaquiline showed higher efficacy with respect to relapse prevention in a mouse model (89). In a separate study, numerous combinations of bedaquiline, PA-824, CFZ, and PNU-100480 were shown to be more effective than a RIF-INH-PZA combination (90).
ENERGY METABOLISM AND DRUG EFFLUX
Active transport of drugs and xenobiotics through the activity of efflux pumps (EPs) is well documented in bacteria and points to an important role for this process in phenotypic drug tolerance and the subsequent emergence of drug resistance due to reduced intracellular drug concentrations (5). A key feature of these systems is their dependence on the PMF or the availability of ATP, which inextricably links drug efflux to energy metabolism and the ETC (5, 91, 92). EPs adopt a diversity of structures in bacteria and can be classified into different families based on overall secondary structure and the molecules transported. These include, but are not limited to, the major facilitator superfamily (MFS), small multidrug resistance (SMR) family, resistance/nodulation/cell division (RND) family, the ATP-dependent ABC-type superfamily of transporters, and the multidrug and toxic compound extrusion (MATE) family (91, 93, 94).
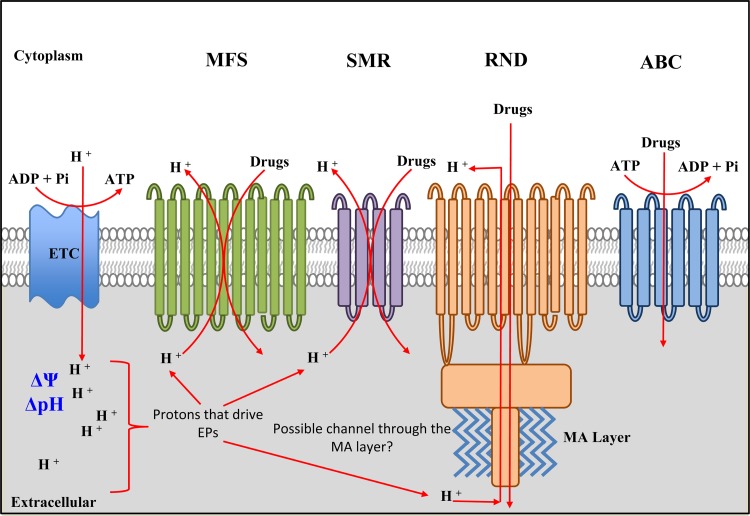
The widely distributed P-glycoprotein, encoded by the mdr gene, is one of the first ABC-type transporters implicated in drug efflux and has been implicated in the extrusion of numerous drugs in different organisms (91). The members of this family of transporters require the energy of ATP hydrolysis for active efflux of drugs (Fig. 2) (95, 96). In contrast, the MFS, SMR, RND, and MATE-type EPs require an energized membrane and the PMF (ΔpH and ΔΨ) for activity (reviewed in reference 91). The pumps often operate as drug-metabolite/proton symporters, antiporters, or uniporters. Schematic representations of these EPs are shown in Fig. 2. The MFS-type EPs can be characterized into 6 evolutionarily related subfamilies, including those containing 14 or 12 transmembrane segments, sugar importers, phosphate ester antiporters, and other transporters (97–99). It has been proposed that these large membrane-associated proteins have evolved via intragenic tandem gene duplication to give rise to a widely distributed, structurally diverse family of proteins (97). MFS EPs require the presence of protons in the periplasm and couple efflux with proton translocation to the cytoplasm (100) (Fig. 2). However, these systems differ in their dependence on the ΔpH and ΔΨ; for example, the bmr-encoded multidrug EP in Bacillus subtilis requires a strong ΔpH to drive the extrusion of ethidium bromide through a electroneutral drug/proton antiport mechanism (91).
FIG 2.
Efflux pumps in M. tuberculosis. The EPs dependent on the PMF and ATP are shown. The mycobacterial ETC generates a transmembrane proton gradient (ΔpH) resulting in membrane potential (Δψ); ΔpH and Δψ together constitute the PMF. Protons translocated into the pseudoperiplasmic space are used by EPs to extrude drugs. The major facilitator superfamily (MFS) EPs are made of integral membrane proteins with 12 to 14 transmembrane regions, while the small multidrug resistance (SMR) EPs contain 4 to 6 transmembrane domains. Both systems use protons from the periplasm, and the members differ in their requirements for ΔpH and Δψ (see the text for details). The resistance nodulation/cell division (RND) EPs are also integral membrane proteins, and members of this family from Gram-negative bacteria associate with other proteins to form a multisubunit complex that spans both the inner and outer membranes. Hence, there is a possibility that a similar structure occurs in mycobacteria, where the members of this group of EPs span the pseudoperiplasmic space and associate with the mycolic acid (MA) layer, possibly through OmpA (outer membrane protein A)-like homologues. RND proteins also require protons—although these may originate from outside the cell—and the PMF. ABC transporters require ATP for the active drug extrusion and, as such, are also dependent on energy production in the cell. Not shown in the figure are the members of the fifth class of EPS, termed the multidrug and toxic compound extrusion (MATE) proteins; while some of these are dependent on PMF and protons, the majority of characterized members operate via sodium influx (93).
The Smr staphylococcal multidrug efflux protein and E. coli EmrR represent well-characterized EPs that belong to the SMR family, which is constituted of four transmembrane-containing transporters (91, 101). Smr from Staphylococcus aureus requires the ΔpH and ΔΨ for efficient transport of a wide range of drugs and other small compounds (102). Reconstitution experiments with both Smr and Emr confirm that these EPs require the PMF and operate through a drug/proton antiport mechanism (103). The RND proteins constitute the third family of broad substrate EPs that require the PMF for activity and are structurally more complex than SMR EPs, containing 12 transmembrane domains and various loop regions (104, 105). Unlike the MFS family of EPs, these proteins are phylogenetically closely related, suggesting that they evolved from a single founding member (91). In addition to being autonomous transporters, the members of the RND family of proteins interact with other transport proteins in Gram-negative bacteria such as membrane fusion proteins (MFPs) and outer membrane factors (OMFs) to facilitate the transport of a variety of substrates (Fig. 1) (105–107).
The genome of M. tuberculosis retains multiple homologues of the major families of PMF-dependent EPs described above and, in addition, encodes numerous ABC-type transporters or hypothetical proteins with homology to transporters (17, 108, 109). This complex multiplicity of EPs, detailed in Table 1, illustrates the ability of the tubercle bacillus to transport a variety of toxic compounds or antibiotics and has important implications for drug resistance in TB infection. The key feature of note is that many of these transport systems require either the PMF or ATP, thus linking their activity with metabolic flux through the ETC and the maintenance of an energized membrane.
TABLE 1.
Putative efflux pumps in M. tuberculosisa
| Gene(s) | Description | Transporter familyb | Energy source | Drugs effluxed | Differentially regulated in clinical isolates | Differentially regulated by indicated drug(s) | Reference(s) |
|---|---|---|---|---|---|---|---|
| Rv0037c | Probable conserved integral membrane protein | MFS | ATP | Yes | 110 | ||
| Rv0194 | Drug transport transmembrane ATP-binding protein | ABC | ATP | 17, 111 | |||
| mmpS5 (Rv0677c) | Membrane protein MmpS5 | RND | PMF | TET | 17, 112 | ||
| Rv0849 | Probable conserved integral membrane protein | MFS | PMF | β-Lactams | 113 | ||
| Rv1218c | Probable tetronasin transport ATP-binding protein | ABC | ATP | β-Lactams | 113 | ||
| Rv1250 | Probable drug transport integral membrane protein | MFS | PMF | Yes | 110 | ||
| Rv1258c | Conserved membrane transport protein | MFS | PMF | INH, RIF, EMB, OFL, β-lactams | Yes | INH, RIF, OFL | 17, 114, 110, 115, 116, 113–117 |
| Rv1272c | Probable drug transport transmembrane ATP-binding protein | ABC | ATP | 17, 118 | |||
| Rv1273c | Probable drug transport transmembrane ATP-binding protein | ABC | ATP | Yes | 17, 110, 118 | ||
| itrA (Rv1348) | Probable drug transport transmembrane ATP-binding protein | ABC | ATP | 17, 118 | |||
| itrB (Rv1349) | Probable drug transport transmembrane ATP-binding protein | ABC | ATP | 17, 118 | |||
| Rv1410c | Aminoglycosides/tetracycline transport integral membrane protein | MFS | PMF | Yes | INH, RIF | 110, 115 | |
| Rv1456c-Rv1458c | Integral membrane proteins | ABC | ATP | Yes (in drug-resistant isolates) | 17, 119 | ||
| Rv1463 | Conserved transmembrane ATP-binding protein | ABC | ATP | 17 | |||
| Rv1634 | Drug efflux membrane protein | MFS | PMF | Yes | 17, 109, 110, 120, 121 | ||
| Rv1686c | Probable conserved ATP-binding protein | ABC | ATP | Yes (in drug-resistant isolates) | 122 | ||
| Rv1687c | Probable conserved ATP-binding protein | ABC | ATP | Yes | 110 | ||
| Rv1747 | Conserved transmembrane ATP-binding protein | ABC | ATP | INH | 17, 118 | ||
| Rv1877 | Conserved membrane protein | MFS | PMF | TET, KAN, erythromycin | 17, 120, 118, 121 | ||
| bacA (Rv1819c) | Drug transport transmembrane ATP-binding protein, vitamin B12 acquisition | ABC | ATP | INH, RIF | 17, 123, 124, 115 | ||
| Rv2209 | Probable conserved integral membrane protein | OFL | 124 | ||||
| Rv2333c | Conserved membrane transport protein | MFS | PMF | TET, spectinomycin | Yes | 17, 110, 120, 125 | |
| jefA (Rv2459) | Conserved integral membrane transport protein | MFS | PMF | INH, EMB | 17, 126, 124, 120, 127 | ||
| Rv2477c | Probable macrolide transport ATP-binding protein | ABC | ATP | OFL | 124 | ||
| Rv2686c | Antibiotic transport membrane leucine- and alanine- and valine-rich protein | ABC | ATP | CIP | 17, 118, 128 | ||
| Rv2687c | Antibiotic transport membrane leucine- and valine-rich protein | ABC | ATP | CIP | 17, 118, 128 | ||
| Rv2688c | Antibiotic transport ATP-binding protein | ABC | ATP | CIP | STR | 17, 124, 118, 128 | |
| Rv2994 | Conserved membrane protein | MFS | PMF | STR | 17, 124, 120, 121 | ||
| Rv3000 | Possible conserved transmembrane protein | ABC | ATP | Yes | 110 | ||
| Rv3239c | Conserved integral membrane transport protein | MFS | PMF | RIF | 17, 129, 120 | ||
| Rv3728 | Conserved two-domain membrane protein | MFS | PMF | INH, RIF, EMB | 17, 124, 120 | ||
| drrA (Rv2936) | Daunorubicin-dim (dimycocerosate) transport ATP-binding protein ABC transporter DrrA | ABC | ATP | TET, STR, EMB, RIF | Yes | 17, 110, 130, 118, 131 | |
| drrB (Rv2937) | Daunorubicin-dim transport membrane protein ABC transporter DrrB | ABC | ATP | TET, STR, EMB | Yes | 17, 110, 118, 131 | |
| drrC (Rv2938) | Daunorubicin-dim transport membrane protein ABC transporter DrrC | ABC | ATP | TET, STR, EMB | STR, EMB | 17, 124, 118, 131 | |
| efpA (Rv2846c) | Integral membrane efflux protein | MFS | PMF | possibly INH | INH | 17, 124, 121, 132–133 | |
| emrB (Rv0783) | Possible multidrug resistance integral membrane efflux protein EmrB | MFS | PMF | RIF | Yes | 17, 110, 132, 134 | |
| iniA (Rv0342) | Isoniazid inducible gene protein IniA | membrane protein | INH, EMB | Yes | 17, 110, 135, 136 | ||
| iniB (Rv0341) | Isoniazid-inducible gene protein IniB | membrane protein | INH | 17, 135, 136 | |||
| iniC (Rv0343) | Isoniazid-inducible gene protein IniC | Membrane protein | INH | 17, 135, 136 | |||
| mmpL3 (Rv0206c) | Probable conserved transmembrane transport protein MmpL3 | RND | PMF | 17, 137 | |||
| mmpL4 (Rv0450c) | Probable conserved transmembrane transport protein MmpL4 | RND | PMF | Yes | 110 | ||
| mmpL5 (Rv0676c) | Probable conserved transmembrane transport protein MmpL5 | RND | PMF | TET | 17, 112 | ||
| mmpL7 (Rv2942) | Probable conserved transmembrane transport protein MmpL7 | RND | PMF | INH | Yes | 17, 110, 128, 137 | |
| mmpL11 (Rv0202c) | Probable conserved transmembrane transport protein MmpL11 | RND | PMF | 17, 137 | |||
| mmr (Rv3065) | Integral membrane efflux protein | SMR | PMF | Erythromycin, β-lactams | INH, EMB | 17, 109, 124, 113, 121 | |
| pstB (Rv0933) | Phosphate transport ATP-binding protein | ABC | ATP | INH, RIF, EMB, CIP | 17, 130, 117, 118, 138, 139 |
CIP, ciprofloxacin; EMB, ethambutol; INH, isoniazid; KAN, kanamycin; OFL, ofloxacin; RIF, rifampin; STR, streptomycin; TET, tetracycline.
Transporter families were identified by literature searches (relevant cases are cited in the text) and from the genomic annotation/comparisons (http://genolist.pasteur.fr/TubercuList/ and http://tuberculist.epfl.ch/).
Further associations between the PMF and drug efflux in mycobacteria have been made through various studies which demonstrated that dissipation of the PMF by treatment with efflux pump inhibitors (108, 114, 126, 140) can reverse low levels of resistance to TB drugs. Early work with 14C-labeled RIF suggests that this drug may be extruded from mycobacteria by the activity of PMF-dependent efflux pumps—a process that can be marginally reversed by the addition of reserpine, an EP inhibitor (141). In drug-resistant strains, exposure to efflux pump inhibitors such as verapamil, reserpine, Phe-Arg-β-naphthylamide (PAβN), carbonyl cyanide m-chlorophenylhydrazone (CCCP), and 2,4-dinitrophenol (DNP) restores susceptibility to anti-TB drugs such as RIF and ofloxacin (OFL) (129, 142, 143). Inclusion of verapamil in drug susceptibility assays results in a marked decrease in the MIC for bedaquiline and clofazimine (144). Knockout studies have further implicated efflux pumps such as Rv1218c, Rv3065, Rv0849, and Rv1258c as important mechanisms of resistance to various chemical classes of drug compounds in M. tuberculosis (145, 146). A recent study demonstrated increased expression of 15 EP-encoding genes, from various classes, in drug-susceptible and drug-resistant strains compared to reference, laboratory grown strains (110). These data suggest that infection in the human host presumably drives expression of these genes to transport/detoxify noxious compounds during pathogenesis, with the concomitant benefit of drug extrusion. Similarly, the expression of 10 EP-encoding genes—Rv3065 (mmr), Rv2938 (drrC), Rv1819c (bacA—recently implicated in vitamin B12 acquisition [123]), Rv2209, Rv2459, Rv2477, Rv2688, Rv2846 (efpA), Rv2994, and Rv3728—is differentially induced during in vitro drug treatment of clinical isolates with standard TB drugs (124), as detailed further in Table 1, suggesting a complex induction pattern of these genes during short-course chemotherapy. While the regulatory mechanisms governing these gene expression changes have not been completely described, there is some evidence for transcriptional regulation of EPs. In this regard, it has been shown that WhiB7, a redox-sensitive transcriptional activator, plays an important role in mediating intrinsic drug resistance in mycobacteria, in some cases through direct regulation of EPs (147–149). Similarly, resistance to INH has been associated with the IniB-IniA-IniC efflux system (Table 1), which is regulated by the MtrAB two-component system (150), and the mmr-encoded EP is regulated by a TetR-type repressor (151). With respect to the evolution of drug resistance, transcriptional profiling of longitudinal isolates from drug-compliant patients, with MANU1, CAS, and Beijing spoligotypes, revealed upregulation of various EPs/multidrug resistance proteins, including Rv3065 (mmr), Rv2936 (drrA), Rv2397, Rv1686c, and Rv1687c, pointing to a role for these proteins in the evolution of drug resistance during treatment (122). Increased expression of genes encoding various efflux pumps upon exposure to RIF has been observed in M. tuberculosis which, in some cases, has been coupled with an increase in RIF and OFL tolerance. This phenomenon could be reversed by the addition of efflux pump inhibitors, highlighting the role of efflux in drug resistance (114–116, 129, 152). A similar result was observed during macrophage infection, where efflux pump activity was coupled to RIF and INH tolerance in M. tuberculosis (153). In RIF-monoresistant and -susceptible M. tuberculosis isolates, initial low-level resistance to INH (reversible by the addition of efflux pump inhibitors) preceded the development of drug resistance-conferring mutations (126). Recently, it has been demonstrated that two EP-encoding genes, those encoding Rv2936 and Rv0783 (Table 1), are associated with RIF resistance in RIF-monoresistant clinical isolates. Overexpression of these two genes, but not of that encoding Rv0933, resulted in increased RIF resistance in E. coli (130). Similarly, low-level efflux-induced azithromycin resistance in M. avium was followed by reversible high-level resistance, thought to be due to acquisition of mutations (140).
These data suggest that the initial efflux activity allows antibiotic tolerance in mycobacteria, enabling resistance-causing mutations to arise and/or be selected for. In this context, the production of ATP via the F1F0-ATP synthase would be essential for the function of these systems. Since efflux pumps seem to play an important role in contributing to drug tolerance, which may then lead to drug resistance, the driving energetic force behind these pumps represents an additional point of vulnerability with respect to targeted drug design in mycobacteria. Studies using PMF and ATP synthase inhibitors have demonstrated the role of the PMF and ATP in INH efflux in M. smegmatis (135). Although efflux has been studied in M. tuberculosis, the role of the driving force of these pumps in drug resistance remains poorly investigated. Direct coupling of PMF and ATP to drug efflux—and consequent drug resistance—has been investigated in E. coli, B. subtilis, L. lactis, Streptococcus pneumoniae, and a host of other organisms (reviewed in references 91, 97, 104, 105, 154, 155, 156, 157, and 158). Considering the success in targeting the ETC in TB drug development, together with the demonstrated importance of efflux as an active process contributing to drug resistance, the role of energy metabolism in drug resistance and inherent susceptibility in M. tuberculosis merits further investigation. The recent demonstration that iron-sulfur (Fe-S) cluster biogenesis is related to intrinsic susceptibility of bacteria to aminoglycosides due to the use of an alternate pathway for Fe-S biogenesis, which results in perturbations in the ETC, leading to reduced PMF and altered drug uptake (159), is consistent with this. Little is known about the energy requirements of mycobacteria during infection, although it has been demonstrated that ATP levels are important for survival in actively replicating as well as nonreplicating mycobacteria, demonstrating the importance of maintaining ATP levels under conditions of growth and survival (20, 55). These ATP levels and associated PMF are essential for M. tuberculosis to extrude drugs via PMF-dependent EPs.
The presence of numerous potential regulators involved in the control of cellular ATP levels suggests that there may be different mechanisms involved in the regulation of ATP synthesis. It has been demonstrated that, although ATP levels drop significantly during the shift down to a nonreplicating state, the decreased levels of cellular ATP are essential for viability since treatment with bedaquiline resulted in a loss of viability. M. smegmatis displays upregulation of F1F0-ATP synthase in response to antibiotic stress. For example, an increase in F1F0-ATP synthase abundance was observed upon the exposure of M. smegmatis to ethambutol (EMB). Furthermore, among the other proteins upregulated in response to EMB, 23% were related to energy metabolism (160). Similarly, treatment with β-lactam antibiotics resulted in increased expression of F1F0-ATP synthase in M. tuberculosis (51). These data suggest that antibiotic treatment may inflict physiological stresses on bacteria that impose a requirement for greater energy production. In the case of β-lactam antibiotics, it is unclear whether their effect on the cell wall results in a perturbation of the PMF which is compensated for by an upregulation of the genes encoding the ATP synthase. There have been no extensive studies showing the response of pathways involved in energy metabolism to anti-TB treatments such as RIF; this could provide novel insight into cellular responses or adaptations occurring within the cell leading to drug resistance or identification of targets for drug development.
CONCLUDING REMARKS
The ETC and associated PMF are essential components for energy production through the generation of ATP, which is required for metabolic processes within the cell. The ETC has gained recent prominence in TB drug development through the discovery of numerous compounds that target this pathway such as bedaquiline and Q203. However, in addition to the obvious effects of inhibiting the ETC, a secondary effect of targeting this pathway would be a reduction in the activity of the various PMF/ATP-dependent EPs present in M. tuberculosis. These effects may accelerate cell death through higher intracellular concentrations of drugs and reduced extrusion of toxic metabolites, with an added benefit of reducing transient drug tolerance and consequent drug resistance. Various studies now point to potentially positive therapeutic effects of using EP inhibitors such as verapamil to increase the potency of drugs and limit the acquisition of drug resistance. Energy metabolism, including the regulation thereof, represents an ideal component of metabolism to mine for new drug targets.
ACKNOWLEDGMENTS
This work was supported by grants from the Medical Research Council of South Africa, the National Research Foundation (NRF), and the University of the Witwatersrand and an International Early Career Scientist Award from Howard Hughes Medical Institute (to B.D.K.). Funding was also received from the Wellcome Trust (WT087383MA to T.C.V.) NRF (NRF81776 to T.C.V).
We thank Christopher Ealand, Bhavna Gordhan, Monique Williams, Edith Machowski, and Nicole Narrandes for helpful discussions and suggestions.
Footnotes
Published ahead of print 10 March 2014
REFERENCES
- 1.WHO. 2013. Global tuberculosis report 2013. http://www.who.int/tb/publications/global_report/en/
- 2.Zumla A, Raviglione M, Hafner R, von Reyn CF. 2013. Tuberculosis. N. Engl. J. Med. 368:745–755. 10.1056/NEJMra1200894 [DOI] [PubMed] [Google Scholar]
- 3.Marais BJ, Zumla A. 2013. History of tuberculosis and drug resistance. N. Engl. J. Med. 368:88–89. 10.1056/NEJMc1212308 [DOI] [PubMed] [Google Scholar]
- 4.Zumla A, Nahid P, Cole ST. 2013. Advances in the development of new tuberculosis drugs and treatment regimens. Nat. Rev. Drug Discov. 12:388–404. 10.1038/nrd4001 [DOI] [PubMed] [Google Scholar]
- 5.Sarathy JT, Dartois V, Lee EJD. 2012. The role of transport mechanisms in Mycobacterium tuberculosis drug resistance in tolerance. Pharmaceuticals (Basel) 5:1210–1235. 10.3390/ph5111210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cook GM, Berney M, Gebhard S, Heinemann M, Cox RA, Danilchanka O, Niederweis M. 2009. Physiology of mycobacteria. Adv. Microb. Physiol. 55:81–182, 318–319. 10.1016/S0065-2911(09)05502-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kana BD, Machowski E, Schechter N, Shin JT, Rubin H, Mizrahi V. 2009. Electron transport and respiration, p 35–64 In Parish T, Brown A. (ed), Mycobacterium: genomics and molecular biology. Horizon Press, London, United Kingdom [Google Scholar]
- 8.Bald D, Koul A. 2010. Respiratory ATP synthesis: the new generation of mycobacterial drug targets? FEMS Microbiol. Lett. 308:1–7. 10.1111/j.1574-6968.2010.01959.x [DOI] [PubMed] [Google Scholar]
- 9.Boshoff HIM, Barry CE., III 2005. Tuberculosis—metabolism and respiration in the absence of growth. Nat. Rev. Microbiol. 3:70–80. 10.1038/nrmicro1065 [DOI] [PubMed] [Google Scholar]
- 10.Neijssel OM, Teixeira de Mattos MJ. 1994. The energetics of bacterial growth: a reassessment. Mol. Microbiol. 13:172–182 [DOI] [PubMed] [Google Scholar]
- 11.Fillingame RH. 1997. Coupling H+ transport and ATP synthesis in F1F0-ATP synthases: glimpses of interacting parts in a dynamic molecular machine. J. Exp. Biol. 200:217–224 [DOI] [PubMed] [Google Scholar]
- 12.Unden G, Bongaerts J. 1997. Alternative respiratory pathways of Escherichia coli: energetics and transcriptional regulation in response to electron acceptors. Biochim. Biophys. Acta 1320:217–234. 10.1016/S0005-2728(97)00034-0 [DOI] [PubMed] [Google Scholar]
- 13.Lancaster CR, Kröger A. 2000. Succinate: quinone oxidoreductases: new insights from X-ray crystal structures. Biochim. Biophys. Acta 1459:422–431. 10.1016/S0005-2728(00)00180-8 [DOI] [PubMed] [Google Scholar]
- 14.Dhiman RK, Mahapatra S, Slayden RA, Boyne ME, Lenaerts A, Hinshaw JC, Angala SK, Chatterjee D, Biswas K, Narayanasamy P, Kurosu M, Crick DC. 2009. Menaquinone synthesis is critical for maintaining mycobacterial viability during exponential growth and recovery from non-replicating persistence. Mol. Microbiol. 72:85–97. 10.1111/j.1365-2958.2009.06625.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kana BD, Weinstein EA, Avarbock D, Dawes SS, Rubin H, Mizrahi V. 2001. Characterization of the cydAB-encoded cytochrome bd oxidase from Mycobacterium smegmatis. J. Bacteriol. 183:7076–7086. 10.1128/JB.183.24.7076-7086.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Matsoso LG, Kana BD, Crellin PK, Lea-Smith DJ, Pelosi A, Powell D, Dawes SS, Rubin H, Coppel RL, Mizrahi V. 2005. Function of the cytochrome bc1-aa3 branch of the respiratory network in mycobacteria and network adaptation occurring in response to its disruption. J. Bacteriol. 187:6300–6308. 10.1128/JB.187.18.6300-6308.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cole ST, Brosch R, Parkhill J, Garnier T, Churcher C, Harris D, Gordon SV, Eiglmeier K, Gas S, Barry CE, III, Tekaia F, Badcock K, Basham D, Brown D, Chillingworth T, Connor R, Davies R, Devlin K, Feltwell T, Gentles S, Hamlin N, Holroyd S, Hornsby T, Jagels K, Krogh A, McLean J, Moule S, Murphy L, Oliver K, Osborne J, Quail MA, Rajandream MA, Rogers J, Rutter S, Seeger K, Skelton J, Squares R, Squares S, Sulston JE, Taylor K, Whitehead S, Barrell BG. 1998. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393:537–544. 10.1038/31159 [DOI] [PubMed] [Google Scholar]
- 18.Weinstein EA, Yano T, Li L-S, Avarbock D, Avarbock A, Helm D, McColm AA, Duncan K, Lonsdale JT, Rubin H. 2005. Inhibitors of type II NADH:menaquinone oxidoreductase represent a class of antitubercular drugs. Proc. Natl. Acad. Sci. U. S. A. 102:4548–4553. 10.1073/pnas.0500469102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shi L, Sohaskey CD, Kana BD, Dawes S, North RJ, Mizrahi V, Gennaro ML. 2005. Changes in energy metabolism of Mycobacterium tuberculosis in mouse lung and under in vitro conditions affecting aerobic respiration. Proc. Natl. Acad. Sci. U. S. A. 102:15629–15634. 10.1073/pnas.0507850102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rao SPS, Alonso S, Rand L, Dick T, Pethe K. 2008. The protonmotive force is required for maintaining ATP homeostasis and viability of hypoxic, nonreplicating Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. U. S. A. 105:11945–11950. 10.1073/pnas.0711697105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Velmurugan K, Chen B, Miller JL, Azogue S, Gurses S, Hsu T, Glickman M, Jacobs WR, Jr, Porcelli SA, Briken V. 2007. Mycobacterium tuberculosis nuoG is a virulence gene that inhibits apoptosis of infected host cells. PLoS Pathog. 3:e110. 10.1371/journal.ppat.0030110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miller JL, Velmurugan K, Cowan MJ, Briken V. 2010. The type I NADH dehydrogenase of Mycobacterium tuberculosis counters phagosomal NOX2 activity to inhibit TNF-alpha-mediated host cell apoptosis. PLoS Pathog. 6:e1000864. 10.1371/journal.ppat.1000864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Blomgran R, Desvignes L, Briken V, Ernst JD. 2012. Mycobacterium tuberculosis inhibits neutrophil apoptosis, leading to delayed activation of naive CD4 T cells. Cell Host Microbe 11:81–90. 10.1016/j.chom.2011.11.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sassetti CM, Boyd DH, Rubin EJ. 2003. Genes required for mycobacterial growth defined by high density mutagenesis. Mol. Microbiol. 48:77–84. 10.1046/j.1365-2958.2003.03425.x [DOI] [PubMed] [Google Scholar]
- 25.McAdam RA, Quan S, Smith DA, Bardarov S, Betts JC, Cook FC, Hooker EU, Lewis AP, Woollard P, Everett MJ, Lukey PT, Bancroft GJ, Jacobs WR, Jr, Duncan K. 2002. Characterization of a Mycobacterium tuberculosis H37Rv transposon library reveals insertions in 351 ORFs and mutants with altered virulence. Microbiology 148:2975–2986 [DOI] [PubMed] [Google Scholar]
- 26.Kurokawa T, Sakamoto J. 2005. Purification and characterization of succinate:menaquinone oxidoreductase from Corynebacterium glutamicum. Arch. Microbiol. 183:317–324. 10.1007/s00203-005-0775-8 [DOI] [PubMed] [Google Scholar]
- 27.Eoh H, Rhee KY. 2013. Multifunctional essentiality of succinate metabolism in adaptation to hypoxia in Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. U. S. A. 110:6554–6559. 10.1073/pnas.1219375110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Truglio JJ, Theis K, Feng Y, Gajda R, Machutta C, Tonge PJ, Kisker C. 2003. Crystal structure of Mycobacterium tuberculosis MenB, a key enzyme in vitamin K2 biosynthesis. J. Biol. Chem. 278:42352–42360. 10.1074/jbc.M307399200 [DOI] [PubMed] [Google Scholar]
- 29.Collins MD, Goodfellow M, Minnikin DE, Alderson G. 1985. Menaquinone composition of mycolic acid-containing actinomycetes and some sporoactinomycetes. J. Appl. Microbiol. 58:77–86 [DOI] [PubMed] [Google Scholar]
- 30.Collins MD, Jones D. 1981. Distribution of isoprenoid quinone structural types in bacteria and their taxonomic implication. Microbiol. Rev. 45:316–354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pandya KP, King HK. 1966. Ubiquinone and menaquinone in bacteria: a comparative study of some bacterial respiratory systems. Arch. Biochem. Biophys. 114:154–157. 10.1016/0003-9861(66)90316-X [DOI] [PubMed] [Google Scholar]
- 32.Debnath J, Siricilla S, Wan B, Crick DC, Lenaerts AJ, Franzblau SG, Kurosu M. 2012. Discovery of selective menaquinone biosynthesis inhibitors against Mycobacterium tuberculosis. J. Med. Chem. 55:3739–3755. 10.1021/jm201608g [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sohaskey CD. 2005. Regulation of nitrate reductase activity in Mycobacterium tuberculosis by oxygen and nitric oxide. Microbiology 151:3803–3810. 10.1099/mic.0.28263-0 [DOI] [PubMed] [Google Scholar]
- 34.Malm S, Tiffert Y, Micklinghoff J, Schultze S, Joost I, Weber I, Horst S, Ackermann B, Schmidt M, Wohlleben W, Ehlers S, Geffers R, Reuther J, Bange FC. 2009. The roles of the nitrate reductase NarGHJI, the nitrite reductase NirBD and the response regulator GlnR in nitrate assimilation of Mycobacterium tuberculosis. Microbiology 155:1332–1339. 10.1099/mic.0.023275-0 [DOI] [PubMed] [Google Scholar]
- 35.Sohaskey CD. 2008. Nitrate enhances the survival of Mycobacterium tuberculosis during inhibition of respiration. J. Bacteriol. 190:2981–2986. 10.1128/JB.01857-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sohaskey CD, Wayne LG. 2003. Role of narK2X and narGHJI in hypoxic upregulation of nitrate reduction by Mycobacterium tuberculosis. J. Bacteriol. 185:7247–7256. 10.1128/JB.185.24.7247-7256.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Homolka S, Niemann S, Russell DG, Rohde KH. 2010. Functional genetic diversity among Mycobacterium tuberculosis complex clinical isolates: delineation of conserved core and lineage-specific transcriptomes during intracellular survival. PLoS Pathog. 6:e1000988. 10.1371/journal.ppat.1000988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fenhalls G, Stevens L, Moses L, Bezuidenhout J, Betts JC, van Helden P, Lukey PT, Duncan K. 2002. In situ detection of Mycobacterium tuberculosis transcripts in human lung granulomas reveals differential gene expression in necrotic lesions. Infect. Immun. 70:6330–6338. 10.1128/IAI.70.11.6330-6338.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kruh NA, Troudt J, Izzo A, Prenni J, Dobos KM. 2010. Portrait of a pathogen: the Mycobacterium tuberculosis proteome in vivo. PLoS One 5:e13938. 10.1371/journal.pone.0013938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Aly S, Wagner K, Keller C, Malm S, Malzan A, Brandau S, Bange FC, Ehlers S. 2006. Oxygen status of lung granulomas in Mycobacterium tuberculosis-infected mice. J. Pathol. 210:298–305. 10.1002/path.2055 [DOI] [PubMed] [Google Scholar]
- 41.Tan MP, Sequeira P, Lin WW, Phong WY, Cliff P, Ng SH, Lee BH, Camacho L, Schnappinger D, Ehrt S, Dick T, Pethe K, Alonso S. 2010. Nitrate respiration protects hypoxic Mycobacterium tuberculosis against acid- and reactive nitrogen species stresses. PLoS One 5:e13356. 10.1371/journal.pone.0013356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Akhtar S, Khan A, Sohaskey CD, Jagannath C, Sarkar D. 2013. Nitrite reductase NirBD is induced and plays an important role during in vitro dormancy of Mycobacterium tuberculosis. J. Bacteriol. 195:4592–4599. 10.1128/JB.00698-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Watanabe S, Zimmermann M, Goodwin MB, Sauer U, Barry CE, III, Boshoff HI. 2011. Fumarate reductase activity maintains an energized membrane in anaerobic Mycobacterium tuberculosis. PLoS Pathog. 7:e1002287. 10.1371/journal.ppat.1002287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Butlin JD, Cox GB, Gibson F. 1971. Oxidative phosphorylation in Escherichia coli K12. Mutations affecting magnesium ion- or calcium ion-stimulated adenosine triphosphatase. Biochem. J. 124:75–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Feniouk BA, Suzuki T, Yoshida M. 2007. Regulatory interplay between proton motive force, ADP, phosphate, and subunit epsilon in bacterial ATP synthase. J. Biol. Chem. 282:764–772. 10.1074/jbc.M606321200 [DOI] [PubMed] [Google Scholar]
- 46.Santana M, Ionescu MS, Vertes A, Longin R, Kunst F, Danchin A, Glaser P. 1994. Bacillus subtilis F0F1 ATPase: DNA sequence of the atp operon and characterization of atp mutants. J. Bacteriol. 176:6802–6811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Priya R, Biukovic G, Manimekalai MS, Lim J, Rao SP, Gruber G. 2013. Solution structure of subunit γ(γ1-204) of the Mycobacterium tuberculosis F-ATP synthase and the unique loop of γ165-178, representing a novel TB drug target. J. Bioenerg. Biomembr. 45:121–129. 10.1007/s10863-012-9486-4 [DOI] [PubMed] [Google Scholar]
- 48.Biukovic G, Basak S, Manimekalai MS, Rishikesan S, Roessle M, Dick T, Rao SP, Hunke C, Grüber G. 2013. Variations of subunit epsilon of the Mycobacterium tuberculosis F1F0 ATP synthase and a novel model for mechanism of action of the TB drug TMC207. Antimicrob. Agents Chemother. 57:168–176. 10.1128/AAC.01039-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Koul A, Dendouga N, Vergauwen K, Molenberghs B, Vranckx L, Willebrords R, Ristic Z, Lill H, Dorange I, Guillemont J, Bald D, Andries K. 2007. Diarylquinolines target subunit c of mycobacterial ATP synthase. Nat. Chem. Biol. 3:323–324. 10.1038/nchembio884 [DOI] [PubMed] [Google Scholar]
- 50.Sala C, Haouz A, Saul FA, Miras I, Rosenkrands I, Alzari PM, Cole ST. 2009. Genome-wide regulon and crystal structure of BlaI (Rv1846c) from Mycobacterium tuberculosis. Mol. Microbiol. 71:1102–1116. 10.1111/j.1365-2958.2008.06583.x [DOI] [PubMed] [Google Scholar]
- 51.Boshoff HIM, Myers TG, Copp BR, McNeil MR, Wilson MA, Barry CE., III 2004. The transcriptional responses of Mycobacterium tuberculosis to inhibitors of metabolism: novel insights into drug mechanisms of action. J. Biol. Chem. 279:40174–40184. 10.1074/jbc.M406796200 [DOI] [PubMed] [Google Scholar]
- 52.Wilke MS, Hills TL, Zhang H-Z, Chambers HF, Strynadka NCJ. 2004. Crystal structures of the Apo and penicillin-acylated forms of the BlaR1 beta-lactam sensor of Staphylococcus aureus. J. Biol. Chem. 279:47278–47287. 10.1074/jbc.M407054200 [DOI] [PubMed] [Google Scholar]
- 53.Lun DS, Sherrid A, Weiner B, Sherman DR, Galagan JE. 2009. A blind deconvolution approach to high-resolution mapping of transcription factor binding sites from ChIP-seq data. Genome Biol. 10:R142. 10.1186/gb-2009-10-12-r142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Diacon AH, Pym A, Grobusch M, Patientia R, Rustomjee R, Page-Shipp L, Pistorius C, Krause R, Bogoshi M, Churchyard G, Venter A, Allen J, Palomino JC, De Marez T, van Heeswijk RPG, Lounis N, Meyvisch P, Verbeeck J, Parys W, de Beule K, Andries K, Mc Neeley DF. 2009. The diarylquinoline TMC207 for multidrug-resistant tuberculosis. N. Engl. J. Med. 360:2397–2405. 10.1056/NEJMoa0808427 [DOI] [PubMed] [Google Scholar]
- 55.Koul A, Vranckx L, Dendouga N, Balemans W, Van den Wyngaert I, Vergauwen K, Göhlmann HWH, Willebrords R, Poncelet A, Guillemont J, Bald D, Andries K. 2008. Diarylquinolines are bactericidal for dormant mycobacteria as a result of disturbed ATP homeostasis. J. Biol. Chem. 283:25273–25280. 10.1074/jbc.M803899200 [DOI] [PubMed] [Google Scholar]
- 56.Andries K, Verhasselt P, Guillemont J, Göhlmann HWH, Neefs J-M, Winkler H, Van Gestel J, Timmerman P, Zhu M, Lee E, Williams P, de Chaffoy D, Huitric E, Hoffner S, Cambau E, Truffot-Pernot C, Lounis N, Jarlier V. 2005. A diarylquinoline drug active on the ATP synthase of Mycobacterium tuberculosis. Science 307:223–227. 10.1126/science.1106753 [DOI] [PubMed] [Google Scholar]
- 57.Diacon AH, Dawson R, von Groote-Bidlingmaier F, Symons G, Venter A, Donald PR, van Niekerk C, Everitt D, Winter H, Becker P, Mendel CM, Spigelman MK. 2012. 14-day bactericidal activity of PA-824, bedaquiline, pyrazinamide, and moxifloxacin combinations: a randomised trial. Lancet 380:986–993. 10.1016/S0140-6736(12)61080-0 [DOI] [PubMed] [Google Scholar]
- 58.Haagsma AC, Abdillahi-Ibrahim R, Wagner MJ, Krab K, Vergauwen K, Guillemont J, Andries K, Lill H, Koul A, Bald D. 2009. Selectivity of TMC207 towards mycobacterial ATP synthase compared with that towards the eukaryotic homologue. Antimicrob. Agents Chemother. 53:1290–1292. 10.1128/AAC.01393-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tran SL, Cook GM. 2005. The F1F0-ATP synthase of Mycobacterium smegmatis is essential for growth. J. Bacteriol. 187:5023–5028. 10.1128/JB.187.14.5023-5028.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Khan SR, Singh S, Roy KK, Akhtar MS, Saxena AK, Krishnan MY. 8 November 2012. Biological evaluation of novel substituted chloroquinolines targeting mycobacterial ATP synthase. Int. J. Antimicrob. Agents 10.1016/j.ijantimicag.2012.09.012 [DOI] [PubMed] [Google Scholar]
- 61.Chung HJ, Montville TJ, Chikindas ML. 2000. Nisin depletes ATP and proton motive force in mycobacteria. Lett. Appl. Microbiol. 31:416–420. 10.1046/j.1472-765x.2000.00840.x [DOI] [PubMed] [Google Scholar]
- 62.Carroll J, Draper LA, O'Connor PM, Coffey A, Hill C, Ross RP, Cotter PD, O'Mahony J. 2010. Comparison of the activities of the lantibiotics nisin and lacticin 3147 against clinically significant mycobacteria. Int. J. Antimicrob. Agents 36:132–136. 10.1016/j.ijantimicag.2010.03.029 [DOI] [PubMed] [Google Scholar]
- 63.Islam MR, Nagao J, Zendo T, Sonomoto K. 2012. Antimicrobial mechanism of lantibiotics. Biochem. Soc. Trans. 40:1528–1533. 10.1042/BST20120190 [DOI] [PubMed] [Google Scholar]
- 64.Montville TJ, Chung HJ, Chikindas ML, Chen Y. 1999. Nisin A depletes intracellular ATP and acts in bactericidal manner against Mycobacterium smegmatis. Lett. Appl. Microbiol. 28:189–193. 10.1046/j.1365-2672.1999.00511.x [DOI] [PubMed] [Google Scholar]
- 65.Ruhr E, Sahl HG. 1985. Mode of action of the peptide antibiotic nisin and influence on the membrane potential of whole cells and on cytoplasmic and artificial membrane vesicles. Antimicrob. Agents Chemother. 27:841–845. 10.1128/AAC.27.5.841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhang Y, Wade MM, Scorpio A, Zhang H, Sun Z. 2003. Mode of action of pyrazinamide: disruption of Mycobacterium tuberculosis membrane transport and energetics by pyrazinoic acid. J. Antimicrob. Chemother. 52:790–795. 10.1093/jac/dkg446 [DOI] [PubMed] [Google Scholar]
- 67.Zhang Y, Permar S, Sun Z. 2002. Conditions that may affect the results of susceptibility testing of Mycobacterium tuberculosis to pyrazinamide. J. Med. Microbiol. 51:42–49 [DOI] [PubMed] [Google Scholar]
- 68.Huang Q, Chen Z-F, Li Y-Y, Zhang Y, Ren Y, Fu Z, Xu S-Q. 2007. Nutrient-starved incubation conditions enhance pyrazinamide activity against Mycobacterium tuberculosis. Chemotherapy 53:338–343. 10.1159/000107723 [DOI] [PubMed] [Google Scholar]
- 69.Manca C, Koo MS, Peixoto B, Fallows D, Kaplan G, Subbian S. 2013. Host targeted activity of pyrazinamide in Mycobacterium tuberculosis infection. PLoS One 8:e74082. 10.1371/journal.pone.0074082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Jones PB, Parrish NM, Houston TA, Stapon A, Bansal NP, Dick JD, Townsend CA. 2000. A new class of antituberculosis agents. J. Med. Chem. 43:3304–3314. 10.1021/jm000149l [DOI] [PubMed] [Google Scholar]
- 71.Parrish NM, Houston T, Jones PB, Townsend C, Dick JD. 2001. In vitro activity of a novel antimycobacterial compound, N-octanesulfonylacetamide, and its effects on lipid and mycolic acid synthesis. Antimicrob. Agents Chemother. 45:1143–1150. 10.1128/AAC.45.4.1143-1150.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Parrish NM, Ko CG, Dick JD. 2009. Activity of DSA against anaerobically adapted Mycobacterium bovis BCG in vitro. Tuberculosis (Edinb) 89:325–327. 10.1016/j.tube.2009.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Parrish NM, Ko CG, Hughes MA, Townsend CA, Dick JD. 2004. Effect of n-octanesulphonylacetamide (OSA) on ATP and protein expression in Mycobacterium bovis BCG. J. Antimicrob. Chemother. 54:722–729. 10.1093/jac/dkh408 [DOI] [PubMed] [Google Scholar]
- 74.Barry CE, III, Boshoff HIM, Dowd CS. 2004. Prospects for clinical introduction of nitroimidazole antibiotics for the treatment of tuberculosis. Curr. Pharm. Des. 10:3239–3262. 10.2174/1381612043383214 [DOI] [PubMed] [Google Scholar]
- 75.Manjunatha U, Boshoff HI, Barry CE. 2009. The mechanism of action of PA-824: novel insights from transcriptional profiling. Commun. Integr. Biol. 2:215–218. 10.4161/cib.2.3.7926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Singh R, Manjunatha U, Boshoff HIM, Ha YH, Niyomrattanakit P, Ledwidge R, Dowd CS, Lee IY, Kim P, Zhang L, Kang S, Keller TH, Jiricek J, Barry CE., III 2008. PA-824 kills nonreplicating Mycobacterium tuberculosis by intracellular NO release. Science 322:1392–1395. 10.1126/science.1164571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Abrahams KA, Cox JA, Spivey VL, Loman NJ, Pallen MJ, Constantinidou C, Fernandez R, Alemparte C, Remuinan MJ, Barros D, Ballell L, Besra GS. 2012. Identification of novel imidazo[1,2-a]pyridine inhibitors targeting M. tuberculosis QcrB. PLoS One 7:e52951. 10.1371/journal.pone.0052951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Moraski GC, Markley LD, Cramer J, Hipskind PA, Boshoff H, Bailey M, Alling T, Ollinger J, Parish T, Miller MJ. 2013. Advancement of imidazo[1,2-a]pyridines with improved pharmacokinetics and nanomolar activity against Mycobacterium tuberculosis. ACS Med. Chem. Lett. 4:675–679. 10.1021/ml400088y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Pethe K, Bifani P, Jang J, Kang S, Park S, Ahn S, Jiricek J, Jung J, Jeon HK, Cechetto J, Christophe T, Lee H, Kempf M, Jackson M, Lenaerts AJ, Pham H, Jones V, Seo MJ, Kim YM, Seo M, Seo JJ, Park D, Ko Y, Choi I, Kim R, Kim SY, Lim S, Yim SA, Nam J, Kang H, Kwon H, Oh CT, Cho Y, Jang Y, Kim J, Chua A, Tan BH, Nanjundappa MB, Rao SP, Barnes WS, Wintjens R, Walker JR, Alonso S, Lee S, Oh S, Oh T, Nehrbass U, Han SJ, No Z, Lee J, Brodin P, Cho SN, Nam K. 2013. Discovery of Q203, a potent clinical candidate for the treatment of tuberculosis. Nat. Med. 19:1157–1160. 10.1038/nm.3262 [DOI] [PubMed] [Google Scholar]
- 80.Amaral L, Kristiansen JE, Viveiros M, Atouguia J. 2001. Activity of phenothiazines against antibiotic-resistant Mycobacterium tuberculosis: a review supporting further studies that may elucidate the potential use of thioridazine as anti-tuberculosis therapy. J. Antimicrob. Chemother. 47:505–511. 10.1093/jac/47.5.505 [DOI] [PubMed] [Google Scholar]
- 81.Martins M, Schelz Z, Martins A, Molnar J, Hajos G, Riedl Z, Viveiros M, Yalcin I, Aki-Sener E, Amaral L. 2007. In vitro and ex vivo activity of thioridazine derivatives against Mycobacterium tuberculosis. Int. J. Antimicrob. Agents 29:338–340. 10.1016/j.ijantimicag.2006.10.013 [DOI] [PubMed] [Google Scholar]
- 82.Martins M, Viveiros M, Kristiansen JE, Molnar J, Amaral L. 2007. The curative activity of thioridazine on mice infected with Mycobacterium tuberculosis. In Vivo 21:771–775 [PubMed] [Google Scholar]
- 83.Ordway D, Viveiros M, Leandro C, Bettencourt R, Almeida J, Martins M, Kristiansen JE, Molnar J, Amaral L. 2003. Clinical concentrations of thioridazine kill intracellular multidrug-resistant Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 47:917–922. 10.1128/AAC.47.3.917-922.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.van Ingen J, van der Laan T, Amaral L, Dekhuijzen R, Boeree MJ, van Soolingen D. 2009. In vitro activity of thioridazine against mycobacteria. Int. J. Antimicrob. Agents 34:190–191. 10.1016/j.ijantimicag.2009.02.015 [DOI] [PubMed] [Google Scholar]
- 85.Amaral L, Boeree MJ, Gillespie SH, Udwadia ZF, van Soolingen D. 2010. Thioridazine cures extensively drug-resistant tuberculosis (XDR-TB) and the need for global trials is now! Int. J. Antimicrob. Agents 35:524–526. 10.1016/j.ijantimicag.2009.12.019 [DOI] [PubMed] [Google Scholar]
- 86.Musuka S, Srivastava S, Dona CWS, Meek C, Leff R, Pasipanodya J, Gumbo T. 16 September 2013. Thioridazine pharmacokinetic-pharmacodynamic parameters “wobble” during treatment of tuberculosis: a theoretical basis for shorter-duration curative monotherapy with congeners. Antimicrob. Agents Chemother. 10.1128/AAC.00829-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Rodrigues L, Wagner D, Viveiros M, Sampaio D, Couto I, Vavra M, Kern WV, Amaral L. 2008. Thioridazine and chlorpromazine inhibition of ethidium bromide efflux in Mycobacterium avium and Mycobacterium smegmatis. J. Antimicrob. Chemother. 61:1076–1082. 10.1093/jac/dkn070 [DOI] [PubMed] [Google Scholar]
- 88.Yano T, Li L-S, Weinstein E, Teh J-S, Rubin H. 2006. Steady-state kinetics and inhibitory action of antitubercular phenothiazines on Mycobacterium tuberculosis type-II NADH-menaquinone oxidoreductase (NDH-2). J. Biol. Chem. 281:11456–11463. 10.1074/jbc.M508844200 [DOI] [PubMed] [Google Scholar]
- 89.Tasneen R, Li S-Y, Peloquin CA, Taylor D, Williams KN, Andries K, Mdluli KE, Nuermberger EL. 2011. Sterilizing activity of novel TMC207- and PA-824-containing regimens in a murine model of tuberculosis. Antimicrob. Agents Chemother. 55:5485–5492. 10.1128/AAC.05293-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Williams K, Minkowski A, Amoabeng O, Peloquin CA, Taylor D, Andries K, Wallis RS, Mdluli KE, Nuermberger EL. 2012. Sterilizing activities of novel combinations lacking first- and second-line drugs in a murine model of tuberculosis. Antimicrob. Agents Chemother. 56:3114–3120. 10.1128/AAC.00384-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Paulsen IT, Brown MH, Skurray RA. 1996. Proton-dependent multidrug efflux systems. Microbiol. Rev. 60:575–608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Chang G. 2003. Multidrug resistance ABC transporters. FEBS Lett. 555:102–105. 10.1016/S0014-5793(03)01085-8 [DOI] [PubMed] [Google Scholar]
- 93.Kuroda T, Tsuchiya T. 2009. Multidrug efflux transporters in the MATE family. Biochim. Biophys. Acta 1794:763–768. 10.1016/j.bbapap.2008.11.012 [DOI] [PubMed] [Google Scholar]
- 94.Moriyama Y, Hiasa M, Matsumoto T, Omote H. 2008. Multidrug and toxic compound extrusion (MATE)-type proteins as anchor transporters for the excretion of metabolic waste products and xenobiotics. Xenobiotica 38:1107–1118. 10.1080/00498250701883753 [DOI] [PubMed] [Google Scholar]
- 95.Hrycyna CA, Gottesman MM. 1998. Multidrug ABC transporters from bacteria to man: an emerging hypothesis for the universality of molecular mechanism and function. Drug Resist. Updat. 1:81–83. 10.1016/S1368-7646(98)80019-8 [DOI] [PubMed] [Google Scholar]
- 96.van Veen HW, Konings WN. 1997. Multidrug transporters from bacteria to man: similarities in structure and function. Semin. Cancer Biol. 8:183–191. 10.1006/scbi.1997.0064 [DOI] [PubMed] [Google Scholar]
- 97.Yan N. 2013. Structural advances for the major facilitator superfamily (MFS) transporters. Trends Biochem. Sci. 38:151–159. 10.1016/j.tibs.2013.01.003 [DOI] [PubMed] [Google Scholar]
- 98.Fluman N, Bibi E. 2009. Bacterial multidrug transport through the lens of the major facilitator superfamily. Biochim. Biophys. Acta 1794:738–747. 10.1016/j.bbapap.2008.11.020 [DOI] [PubMed] [Google Scholar]
- 99.Lewinson O, Adler J, Sigal N, Bibi E. 2006. Promiscuity in multidrug recognition and transport: the bacterial MFS Mdr transporters. Mol. Microbiol. 61:277–284. 10.1111/j.1365-2958.2006.05254.x [DOI] [PubMed] [Google Scholar]
- 100.Law CJ, Maloney PC, Wang DN. 2008. Ins and outs of major facilitator superfamily antiporters. Annu. Rev. Microbiol. 62:289–305. 10.1146/annurev.micro.61.080706.093329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Paulsen IT, Brown MH, Dunstan SJ, Skurray RA. 1995. Molecular characterization of the staphylococcal multidrug resistance export protein QacC. J. Bacteriol. 177:2827–2833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Grinius L, Dreguniene G, Goldberg EB, Liao CH, Projan SJ. 1992. A staphylococcal multidrug resistance gene product is a member of a new protein family. Plasmid 27:119–129. 10.1016/0147-619X(92)90012-Y [DOI] [PubMed] [Google Scholar]
- 103.Grinius LL, Goldberg EB. 1994. Bacterial multidrug resistance is due to a single membrane protein which functions as a drug pump. J. Biol. Chem. 269:29998–30004 [PubMed] [Google Scholar]
- 104.Alvarez-Ortega C, Olivares J, Martinez JL. 2013. RND multidrug efflux pumps: what are they good for? Front. Microbiol. 4:7. 10.3389/fmicb.2013.00007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Nikaido H. 2011. Structure and mechanism of RND-type multidrug efflux pumps. Adv. Enzymol. Relat. Areas Mol. Biol. 77:1–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Nikaido H, Takatsuka Y. 2009. Mechanisms of RND multidrug efflux pumps. Biochim. Biophys. Acta 1794:769–781. 10.1016/j.bbapap.2008.10.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Mima T, Sekiya H, Mizushima T, Kuroda T, Tsuchiya T. 2005. Gene cloning and properties of the RND-type multidrug efflux pumps MexPQ-OpmE and MexMN-OprM from Pseudomonas aeruginosa. Microbiol. Immunol. 49:999–1002. 10.1111/j.1348-0421.2005.tb03696.x [DOI] [PubMed] [Google Scholar]
- 108.Amaral L, Martins M, Viveiros M. 2007. Enhanced killing of intracellular multidrug-resistant Mycobacterium tuberculosis by compounds that affect the activity of efflux pumps. J. Antimicrob. Chemother. 59:1237–1246. 10.1093/jac/dkl500 [DOI] [PubMed] [Google Scholar]
- 109.De Rossi E, Ainsa JA, Riccardi G. 2006. Role of mycobacterial efflux transporters in drug resistance: an unresolved question. FEMS Microbiol. Rev. 30:36–52. 10.1111/j.1574-6976.2005.00002.x [DOI] [PubMed] [Google Scholar]
- 110.Calgin MK, Sahin F, Turegun B, Gerceker D, Atasever M, Koksal D, Karasartova D, Kiyan M. 2013. Expression analysis of efflux pump genes among drug-susceptible and multidrug-resistant Mycobacterium tuberculosis clinical isolates and reference strains. Diagn. Microbiol. Infect. Dis. 76:291–297. 10.1016/j.diagmicrobio.2013.02.033 [DOI] [PubMed] [Google Scholar]
- 111.Danilchanka O, Mailaender C, Niederweis M. 2008. Identification of a novel multidrug efflux pump of Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 52:2503–2511. 10.1128/AAC.00298-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Milano A, Pasca MR, Provvedi R, Lucarelli AP, Manina G, Ribeiro AL, Manganelli R, Riccardi G. 2009. Azole resistance in Mycobacterium tuberculosis is mediated by the MmpS5-MmpL5 efflux system. Tuberculosis (Edinb) 89:84–90. 10.1016/j.tube.2008.08.003 [DOI] [PubMed] [Google Scholar]
- 113.Dinesh N, Sharma S, Balganesh M. 2013. Involvement of efflux pumps in the resistance to peptidoglycan synthesis inhibitors in Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 57:1941–1943. 10.1128/AAC.01957-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Sharma S, Kumar M, Nargotra A, Koul S, Khan IA. 2010. Piperine as an inhibitor of Rv1258c, a putative multidrug efflux pump of Mycobacterium tuberculosis. J. Antimicrob. Chemother. 65:1694–1701. 10.1093/jac/dkq186 [DOI] [PubMed] [Google Scholar]
- 115.Jiang X, Zhang W, Zhang Y, Gao F, Lu C, Zhang X, Wang H. 2008. Assessment of efflux pump gene expression in a clinical isolate Mycobacterium tuberculosis by real-time reverse transcription PCR. Microb. Drug Resist. 14:7–11. 10.1089/mdr.2008.0772 [DOI] [PubMed] [Google Scholar]
- 116.Siddiqi N, Das R, Pathak N, Banerjee S, Ahmed N, Katoch VM, Hasnain SE. 2004. Mycobacterium tuberculosis isolate with a distinct genomic identity overexpresses a tap-like efflux pump. Infection 32:109–111. 10.1007/s15010-004-3097-x [DOI] [PubMed] [Google Scholar]
- 117.Gupta AK, Chauhan DS, Srivastava K, Das R, Batra S, Mittal M, Goswami P, Singhal N, Sharma VD, Venkatesan K, Hasnain SE, Katoch VM. 2006. Estimation of efflux mediated multi-drug resistance and its correlation with expression levels of two major efflux pumps in mycobacteria. J. Commun. Dis. 38:246–254 [PubMed] [Google Scholar]
- 118.Braibant M, Gilot P, Content J. 2000. The ATP binding cassette (ABC) transport systems of Mycobacterium tuberculosis. FEMS Microbiol. Rev. 24:449–467. 10.1111/j.1574-6976.2000.tb00550.x [DOI] [PubMed] [Google Scholar]
- 119.Hao P, Shi-Liang Z, Ju L, Ya-Xin D, Biao H, Xu W, Min-Tao H, Shou-Gang K, Ke W. 2011. The role of ABC efflux pump, Rv1456c-Rv1457c-Rv1458c, from Mycobacterium tuberculosis clinical isolates in China. Folia Microbiol. (Praha) 56:549–553. 10.1007/s12223-011-0080-7 [DOI] [PubMed] [Google Scholar]
- 120.De Rossi E, Arrigo P, Bellinzoni M, Silva PA, Martin C, Ainsa JA, Guglierame P, Riccardi G. 2002. The multidrug transporters belonging to major facilitator superfamily in Mycobacterium tuberculosis. Mol. Med. 8:714–724 [PMC free article] [PubMed] [Google Scholar]
- 121.Li XZ, Zhang L, Nikaido H. 2004. Efflux pump-mediated intrinsic drug resistance in Mycobacterium smegmatis. Antimicrob. Agents Chemother. 48:2415–2423. 10.1128/AAC.48.7.2415-2423.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Chatterjee A, Saranath D, Bhatter P, Mistry N. 2013. Global transcriptional profiling of longitudinal clinical isolates of Mycobacterium tuberculosis exhibiting rapid accumulation of drug resistance. PLoS One 8:e54717. 10.1371/journal.pone.0054717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Gopinath K, Venclovas C, Ioerger TR, Sacchettini JC, McKinney JD, Mizrahi V, Warner DF. 2013. A vitamin B12 transporter in Mycobacterium tuberculosis. Open Biol. 3:120175. 10.1098/rsob.120175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Gupta AK, Katoch VM, Chauhan DS, Sharma R, Singh M, Venkatesan K, Sharma VD. 2010. Microarray analysis of efflux pump genes in multidrug-resistant Mycobacterium tuberculosis during stress induced by common anti-tuberculous drugs. Microb. Drug Resist. 16:21–28. 10.1089/mdr.2009.0054 [DOI] [PubMed] [Google Scholar]
- 125.Ramón-García S, Martin C, De Rossi E, Ainsa JA. 2007. Contribution of the Rv2333c efflux pump (the Stp protein) from Mycobacterium tuberculosis to intrinsic antibiotic resistance in Mycobacterium bovis BCG. J. Antimicrob. Chemother. 59:544–547. 10.1093/jac/dkl510 [DOI] [PubMed] [Google Scholar]
- 126.Machado D, Couto I, Perdigao J, Rodrigues L, Portugal I, Baptista P, Veigas B, Amaral L, Viveiros M. 2012. Contribution of efflux to the emergence of isoniazid and multidrug resistance in Mycobacterium tuberculosis. PLoS One 7:e34538. 10.1371/journal.pone.0034538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Gupta AK, Reddy VP, Lavania M, Chauhan DS, Venkatesan K, Sharma VD, Tyagi AK, Katoch VM. 2010. jefA (Rv2459), a drug efflux gene in Mycobacterium tuberculosis confers resistance to isoniazid & ethambutol. Indian J. Med. Res. 132:176–188 [PubMed] [Google Scholar]
- 128.Pasca MR, Guglierame P, Arcesi F, Bellinzoni M, De Rossi E, Riccardi G. 2004. Rv2686c-Rv2687c-Rv2688c, an ABC fluoroquinolone efflux pump in Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 48:3175–3178. 10.1128/AAC.48.8.3175-3178.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Louw GE, Warren RM, Gey van Pittius NC, Leon R, Jimenez A, Hernandez-Pando R, McEvoy CRE, Grobbelaar M, Murray M, van Helden PD, Victor TC. 2011. Rifampicin reduces susceptibility to ofloxacin in rifampicin-resistant Mycobacterium tuberculosis through efflux. Am. J. Respir. Crit. Care Med. 184:269–276. 10.1164/rccm.201011-1924OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Pang Y, Lu J, Wang Y, Song Y, Wang S, Zhao Y. 2013. Study of rifampicin monoresistance mechanism in Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 57:893–900. 10.1128/AAC.01024-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Choudhuri BS, Bhakta S, Barik R, Basu J, Kundu M, Chakrabarti P. 2002. Overexpression and functional characterization of an ABC (ATP-binding cassette) transporter encoded by the genes drrA and drrB of Mycobacterium tuberculosis. Biochem. J. 367:279–285. 10.1042/BJ20020615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Doran JL, Pang Y, Mdluli KE, Moran AJ, Victor TC, Stokes RW, Mahenthiralingam E, Kreiswirth BN, Butt JL, Baron GS, Treit JD, Kerr VJ, Van Helden PD, Roberts MC, Nano FE. 1997. Mycobacterium tuberculosis efpA encodes an efflux protein of the QacA transporter family. Clin. Diagn. Lab. Immunol. 4:23–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Wilson M, DeRisi J, Kristensen HH, Imboden P, Rane S, Brown PO, Schoolnik GK. 1999. Exploring drug-induced alterations in gene expression in Mycobacterium tuberculosis by microarray hybridization. Proc. Natl. Acad. Sci. U. S. A. 96:12833–12838. 10.1073/pnas.96.22.12833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.De Rossi E, Branzoni M, Cantoni R, Milano A, Riccardi G, Ciferri O. 1998. mmr, a Mycobacterium tuberculosis gene conferring resistance to small cationic dyes and inhibitors. J. Bacteriol. 180:6068–6071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Choudhuri BS, Sen S, Chakrabarti P. 1999. Isoniazid accumulation in Mycobacterium smegmatis is modulated by proton motive force-driven and ATP-dependent extrusion systems. Biochem. Biophys. Res. Commun. 256:682–684. 10.1006/bbrc.1999.0357 [DOI] [PubMed] [Google Scholar]
- 136.Colangeli R, Helb D, Sridharan S, Sun J, Varma-Basil M, Hazbon MH, Harbacheuski R, Megjugorac NJ, Jacobs WR, Jr, Holzenburg A, Sacchettini JC, Alland D. 2005. The Mycobacterium tuberculosis iniA gene is essential for activity of an efflux pump that confers drug tolerance to both isoniazid and ethambutol. Mol. Microbiol. 55:1829–1840. 10.1111/j.1365-2958.2005.04510.x [DOI] [PubMed] [Google Scholar]
- 137.Domenech P, Reed MB, Barry CE., III 2005. Contribution of the Mycobacterium tuberculosis MmpL protein family to virulence and drug resistance. Infect. Immun. 73:3492–3501. 10.1128/IAI.73.6.3492-3501.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Banerjee SK, Bhatt K, Rana S, Misra P, Chakraborti PK. 1996. Involvement of an efflux system in mediating high level of fluoroquinolone resistance in Mycobacterium smegmatis. Biochem. Biophys. Res. Commun. 226:362–368. 10.1006/bbrc.1996.1362 [DOI] [PubMed] [Google Scholar]
- 139.Sarin J, Aggarwal S, Chaba R, Varshney GC, Chakraborti PK. 2001. B-subunit of phosphate-specific transporter from Mycobacterium tuberculosis is a thermostable ATPase. J. Biol. Chem. 276:44590–44597. 10.1074/jbc.M105401200 [DOI] [PubMed] [Google Scholar]
- 140.Schmalstieg AM, Srivastava S, Belkaya S, Deshpande D, Meek C, Leff R, van Oers NSC, Gumbo T. 2012. The antibiotic resistance arrow of time: efflux pump induction is a general first step in the evolution of mycobacterial drug resistance. Antimicrob. Agents Chemother. 56:4806–4815. 10.1128/AAC.05546-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Piddock LJ, Williams KJ, Ricci V. 2000. Accumulation of rifampicin by Mycobacterium aurum, Mycobacterium smegmatis and Mycobacterium tuberculosis. J. Antimicrob. Chemother. 45:159–165. 10.1093/jac/45.2.159 [DOI] [PubMed] [Google Scholar]
- 142.Louw GE, Warren RM, Gey van Pittius NC, McEvoy CRE, Van Helden PD, Victor TC. 2009. A balancing act: efflux/influx in mycobacterial drug resistance. Antimicrob. Agents Chemother. 53:3181–3189. 10.1128/AAC.01577-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Singh M, Jadaun GPS, Ramdas Srivastava K, Chauhan V, Mishra R, Gupta K, Nair S, Chauhan DS, Sharma VD, Venkatesan K, Katoch VM. 2011. Effect of efflux pump inhibitors on drug susceptibility of ofloxacin resistant Mycobacterium tuberculosis isolates. Indian J. Med. Res. 133:535–540 [PMC free article] [PubMed] [Google Scholar]
- 144.Gupta S, Cohen KA, Winglee K, Maiga M, Diarra B, Bishai WR. 2014. Efflux inhibition with verapamil potentiates bedaquiline in Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 58:574–576. 10.1128/AAC.01462-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Balganesh M, Dinesh N, Sharma S, Kuruppath S, Nair AV, Sharma U. 6 February 2012. Efflux pumps of Mycobacterium tuberculosis play a significant role in anti-tuberculosis activity of potential drug candidates. Antimicrob. Agents Chemother. 10.1128/AAC.06003-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Balganesh M, Kuruppath S, Marcel N, Sharma S, Nair A, Sharma U. 2010. Rv1218c, an ABC transporter of Mycobacterium tuberculosis with implications in drug discovery. Antimicrob. Agents Chemother. 54:5167–5172. 10.1128/AAC.00610-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Burian J, Ramon-Garcia S, Howes CG, Thompson CJ. 2012. WhiB7, a transcriptional activator that coordinates physiology with intrinsic drug resistance in Mycobacterium tuberculosis. Expert Rev. Anti Infect. Ther. 10:1037–1047. 10.1586/eri.12.90 [DOI] [PubMed] [Google Scholar]
- 148.Burian J, Ramón-García S, Sweet G, Gómez-Velasco A, Av-Gay Y, Thompson CJ. 2012. The mycobacterial transcriptional regulator whiB7 gene links redox homeostasis and intrinsic antibiotic resistance. J. Biol. Chem. 287:299–310. 10.1074/jbc.M111.302588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Morris RP, Nguyen L, Gatfield J, Visconti K, Nguyen K, Schnappinger D, Ehrt S, Liu Y, Heifets L, Pieters J, Schoolnik G, Thompson CJ. 2005. Ancestral antibiotic resistance in Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. U. S. A. 102:12200–12205. 10.1073/pnas.0505446102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Li Y, Zeng J, Zhang H, He ZG. 2010. The characterization of conserved binding motifs and potential target genes for M. tuberculosis MtrAB reveals a link between the two-component system and the drug resistance of M. smegmatis. BMC Microbiol. 10:242. 10.1186/1471-2180-10-242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Bolla JR, Do SV, Long F, Dai L, Su CC, Lei HT, Chen X, Gerkey JE, Murphy DC, Rajashankar KR, Zhang Q, Yu EW. 2012. Structural and functional analysis of the transcriptional regulator Rv3066 of Mycobacterium tuberculosis. Nucleic Acids Res. 40:9340–9355. 10.1093/nar/gks677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.de Knegt GJ, Bruning O, ten Kate MT, de Jong M, van Belkum A, Endtz HP, Breit TM, Bakker-Woudenberg IAJM, de Steenwinkel JEM. 2013. Rifampicin-induced transcriptome response in rifampicin-resistant Mycobacterium tuberculosis. Tuberculosis (Edinb) 93:96–101. 10.1016/j.tube.2012.10.013 [DOI] [PubMed] [Google Scholar]
- 153.Adams KN, Takaki K, Connolly LE, Wiedenhoft H, Winglee K, Humbert O, Edelstein PH, Cosma CL, Ramakrishnan L. 2011. Drug tolerance in replicating mycobacteria mediated by a macrophage-induced efflux mechanism. Cell 145:39–53. 10.1016/j.cell.2011.02.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.van Veen HW, Venema K, Bolhuis H, Oussenko I, Kok J, Poolman B, Driessen AJ, Konings WN. 1996. Multidrug resistance mediated by a bacterial homolog of the human multidrug transporter MDR1. Proc. Natl. Acad. Sci. U. S. A. 93:10668–10672. 10.1073/pnas.93.20.10668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Tanaka Y, Hipolito CJ, Maturana AD, Ito K, Kuroda T, Higuchi T, Katoh T, Kato HE, Hattori M, Kumazaki K, Tsukazaki T, Ishitani R, Suga H, Nureki O. 2013. Structural basis for the drug extrusion mechanism by a MATE multidrug transporter. Nature 496:247–251. 10.1038/nature12014 [DOI] [PubMed] [Google Scholar]
- 156.Bolhuis H, Molenaar D, Poelarends G, van Veen HW, Poolman B, Driessen AJ, Konings WN. 1994. Proton motive force-driven and ATP-dependent drug extrusion systems in multidrug-resistant Lactococcus lactis. J. Bacteriol. 176:6957–6964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Bolhuis H, van Veen HW, Brands JR, Putman M, Poolman B, Driessen AJ, Konings WN. 1996. Energetics and mechanism of drug transport mediated by the lactococcal multidrug transporter LmrP. J. Biol. Chem. 271:24123–24128. 10.1074/jbc.271.39.24123 [DOI] [PubMed] [Google Scholar]
- 158.Zeller V, Janoir C, Kitzis MD, Gutmann L, Moreau NJ. 1997. Active efflux as a mechanism of resistance to ciprofloxacin in Streptococcus pneumoniae. Antimicrob. Agents Chemother. 41:1973–1978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Ezraty B, Vergnes A, Banzhaf M, Duverger Y, Huguenot A, Brochado AR, Su SY, Espinosa L, Loiseau L, Py B, Typas A, Barras F. 2013. Fe-S cluster biosynthesis controls uptake of aminoglycosides in a ROS-less death pathway. Science 340:1583–1587. 10.1126/science.1238328 [DOI] [PubMed] [Google Scholar]
- 160.Jiang T, Zhan Y, Sun M, Liu S, Zang S, Ma Y, Xin Y. 2011. The novel responses of ethambutol against Mycobacterium smegmatis mc2155 revealed by proteomics analysis. Curr. Microbiol. 62:341–345. 10.1007/s00284-010-9711-5 [DOI] [PubMed] [Google Scholar]
- 161.Fu LM, Shinnick TM. 2007. Genome-wide exploration of the drug action of capreomycin on Mycobacterium tuberculosis using Affymetrix oligonucleotide GeneChips. J. Infect. 54:277–284. 10.1016/j.jinf.2006.05.012 [DOI] [PubMed] [Google Scholar]