Abstract
The carbapenem-hydrolyzing class D β-lactamase OXA-253 was identified in an Acinetobacter baumannii clinical isolate belonging to sequence type 113 (ST113) in Brazil. OXA-253 shares 93.8% amino acid identity with OXA-143. The blaOXA-253 gene is located on a ca. 20-kb plasmid. The genetic environment of the blaOXA-253 gene shares the highest identity with ubiquitous GR2 group plasmids usually carrying blaOXA-24/-40 genes.
TEXT
Acinetobacter baumannii strains are frequently associated with nosocomial infections, and their resistance to carbapenems has risen in recent decades (1, 2). The main mechanisms of resistance to carbapenems of A. baumannii are efflux pumps, porin mutations, overexpression of the chromosomally encoded OXA-51-like β-lactamase, and the biosynthesis of acquired carbapenem-hydrolyzing class D β-lactamases (CHDLs) (oxacillinases) (3). To date, five main groups of CHDLs have been identified in A. baumannii, the intrinsic chromosomally encoded OXA-51-like enzyme and the acquired chromosomally encoded and plasmid-encoded OXA-23-like, OXA-40-like, OXA-58-like, and OXA-143-like enzymes (3). OXA-143 was first described in 2009 from an A. baumannii strain isolated in Brazil in 2004 (2). In this study, we report an A. baumannii clinical strain producing OXA-253, a variant of OXA-143. The genetic environment of the plasmid-borne blaOXA-253 gene is detailed.
The clinical isolate A. baumannii 25 was recovered from a perineal swab in the intensive care unit in Minas Gerais Hospital, Brazil. MICs were determined by Etest (bioMérieux, La Balme-les-Grottes, France). Isolate 25 was resistant to all β-lactams, including carbapenems, and had reduced susceptibility to cefepime according to the CLSI guidelines (4) (Table 1). It was also resistant to fluoroquinolones (ciprofloxacin), and susceptible to aminoglycosides (gentamicin, tobramycin, amikacin, and netilmicin), and tetracycline. A modified version of the CarbaNP test (5) and the CarbAcinetoNP test (P. Nordmann, personal communication) was performed and showed a carbapenemase activity. PCR experiments, performed as previously described for screening of blaOXA-23, blaOXA-40, blaOXA-58 and blaOXA-143 genes (6), identified a blaOXA-143-like gene. Further sequencing of the PCR product identified the blaOXA-253 gene (GenBank number KC479324) and showed 95% nucleotide identity with the blaOXA-143 gene. The deduced protein sequence showed 17 amino acid differences between OXA-143 and OXA-253 (93.8% identity) (Fig. 1). Those amino acid changes do not occur in the conserved residues STFK (position 70 to 73) and KSG (position 216 to 218) of class D β-lactamases or in the FGN structural element (position 144 to 146) of carbapenem-hydrolyzing class D β-lactamases (CHDLs) (Fig. 1).
TABLE 1.
MICs of β-lactams for strains tested
| β-Lactam(s) | MIC (μg/ml)a |
|||||
|---|---|---|---|---|---|---|
| A. baumannii 25 | A. baumannii(pOXA-253) | A. baumannii CIP7010 | E. coli TOP10(pTOPO-OXA-253) | E. coli TOP10(pTOPO-OXA-143) | E. coli TOP10 | |
| Amoxicillin | >256 | >256 | 32 | >256 | >256 | 4 |
| Amoxicillin + CLAb | >256 | >256 | 32 | 128 | 64 | 4 |
| Ticarcillin | >256 | >256 | 8 | >256 | 128 | 4 |
| Ticarcillin + CLA | >256 | >256 | 8 | 128 | 64 | 4 |
| Piperacillin | >256 | >256 | 16 | 8 | 4 | 1 |
| Ceftazidime | >256 | 4 | 4 | 0.38 | 0.38 | 0.06 |
| Cefotaxime | >256 | 12 | 2 | 0.12 | 0.12 | 0.12 |
| Cefepime | 4 | 3 | 2 | 0.12 | 0.12 | 0.12 |
| Aztreonam | >256 | 24 | 8 | 0.06 | 0.09 | 0.06 |
| Imipenem | >32 | 32 | 0.25 | 1 | 0.38 | 0.06 |
| Ertapenem | NDc | ND | ND | 1 | 0.75 | 0.06 |
| Meropenem | >32 | >32 | 0.25 | 0.5 | 0.19 | 0.01 |
The MICs were determined by Etest.
CLA, clavulanic acid at a fixed concentration of 4 μg/ml.
ND, not determinable due to the natural resistance of these Gram-negative rods to ertapenem.
FIG 1.

Alignment of the OXA-253 amino acid sequence with that of OXA-143. Conserved residues among class D β-lactamases (DBLs) are shaded. Dashes represent conserved amino acids. β-Lactamases are numbered according to the DBL numbering system (12).
Plasmid extraction from A. baumannii 25 performed by the Kieser technique yielded two plasmids of ca. 70 and 20 kb (7). The gene encoding the OXA-253 determinant from A. baumannii 25 was transferred to Escherichia coli TOP10 by electroporation. The resulting E. coli TOP10 strain harboring the plasmid of ca. 20 kb (pAb25) was selected on ticarcillin (50 μg/ml) containing Trypticase soy agar. Plasmid DNA from E. coli TOP10 (pAb25) was used as the template for sequencing the 3.59-kb sequence encompassing the blaOXA-253 gene by primer walking, i.e., by using primers sequentially designed on the obtained sequence from the inside of the blaOXA-253 gene to the outside. Surprisingly, the genetic environment of the blaOXA-253 gene, with the open reading frames (ORFs) tonB, ORF2, ORF1, repAci2, and repB, differed greatly from that of the blaOXA-143 gene described previously (2) but showed 95% identity with that of the blaOXA-24/-40 gene of pABVA01 plasmid (GR2 group) from an A. baumannii strain from Italy (8), although the blaOXA-253 gene shares only 88% nucleotide identity with the blaOXA-24/-40 gene (Fig. 2). The blaOXA-253 gene was most probably integrated via the XerC/XerD recognition site, located between the blaOXA-253 gene and the replicase-coding gene repAci2, as recently described for other GR2 group plasmids carrying blaOXA-24/-40-like genes in A. baumannii isolates (9) (Fig. 2). The integration of the blaOXA-253 gene on this ubiquitous plasmid may facilitate the dissemination of this carbapenemase. In order to determine the role of OXA-253 in the carbapenem resistance of A. baumannii isolate 25, the natural plasmid harboring the blaOXA-253 gene was electroporated into the A. baumannii reference strain CIP 7010, and MICs of β-lactams for the resulting strain A. baumannii CIP 7010(pOXA-253) were determined by Etest (Table 1). MICs of carbapenems were the same for A. baumannii CIP 7010(pOXA-253) and for A. baumannii 25, confirming that OXA-253 on its own was able to confer carbapenem resistance.
FIG 2.
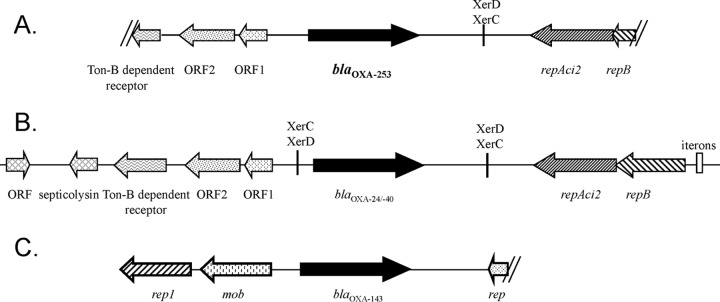
Schematic representation of the β-lactamase genes' genetic environment. (A) Genetic environment of the blaOXA-253 gene in A. baumannii isolate 25; (B) previously identified structure surrounding the blaOXA-24/-40 gene in pABVA01 (8); (C) previously identified structure surrounding the blaOXA-143 gene in A. baumannii 135040 (2) (C).
In order to compare the β-lactam resistance profile conferred by each oxacillinase, OXA-143 and OXA-253 were produced in the same genetic background. The PCR amplicon encompassing the entire sequence of the blaOXA-143 gene was obtained with primers OXA-143A (5′-TACCTTCGGACGTTTGAAAGTTC-3′) and OXA-143B (5′-TAGCTCCCAATTTCCGTTTGG-3′), and that of the blaOXA-253 gene with primers OXA-253A (5′-AAGCCGACTTGTTTCAAAGTCGGC-3′) and OXA-253B (5′-ACTATTCGCATGTTTAAGTGGCAC-3′). The corresponding genes were cloned in the same pTOPO vector (Qiagen, Courtaboeuf, France) under the same promoter and expressed in E. coli TOP10. MICs of the resulting strains E. coli TOP10(pTOPO-OXA-143) and E. coli TOP10(pTOPO-OXA-253) were determined by Etest. Comparison of MICs showed that OXA-253 conferred a higher level of resistance to carbapenems to E. coli TOP10 only than that conferred by OXA-143, with MICs being only 2-fold higher (Table 1).
Multilocus sequence typing was performed using specific primers and conditions described in the A. baumannii multilocus sequence type (MLST) database of the Pasteur Institute (www.pasteur.fr/recherche/genopole/PF8/mlst/Abaumannii.html). MLST analysis showed that A. baumannii 25 belonged to ST113. Until now, this ST was not commonly recovered. Indeed, some A. baumannii isolates belonging to ST113 have been identified in Kuwait in 2013, containing plasmids harboring the blaGES-11 carbapenemase gene alone or coharboring the blaOXA-23 gene (10). More recently, several A. baumannii isolates belonging to ST113 or clonal complex 113 (CC113) (ST99 and ST227) were recovered from southeast Brazil, almost the same area as the hospital of Minas Gerais where this A. baumannii 25 isolate came from (11). Moreover, Clímaco et al. recovered the blaOXA-143 gene from multiple STs of carbapenem-resistant A. baumannii strains (CRABs) (ST109, ST405, and ST406) (11), emphasizing that the dissemination of this carbapenemase gene is not related to a single clone.
CRABs have become increasingly isolated in Brazil, with those carrying blaOXA-23-like and blaOXA-143-like genes being most prevalent (11). As opposed to what occurs in the rest of the world, with the predominance of ST92/OXA-23 A. baumannii isolates, it seems that CRABs carrying blaOXA-23-like and blaOXA-143-like genes mainly belong to multiple STs and clonal complexes CC104, CC109, and CC113 in Latin American countries.
Nucleotide sequence accession number.
The nucleotide sequence of the blaOXA-253 gene is available under GenBank accession number KF824909.
ACKNOWLEDGMENTS
This work was funded by a grant from the Ministère de l'Education Nationale et de la Recherche, Université Paris XI, and from the INSERM.
Footnotes
Published ahead of print 3 March 2014
REFERENCES
- 1.Roca I, Espinal P, Vila-Farrés X, Vila J. 2012. The Acinetobacter baumannii oxymoron: commensal hospital dweller turned pan-drug-resistant menace. Front. Microbiol. 3:148. 10.3389/fmicb.2012.00148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Higgins PG, Poirel L, Lehmann M, Nordmann P, Seifert H. 2009. OXA-143, a novel carbapenem-hydrolyzing class D beta-lactamase in Acinetobacter baumannii. Antimicrob. Agents Chemother. 53:5035–5038. 10.1128/AAC.00856-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Poirel L, Nordmann P. 2006. Carbapenem resistance in Acinetobacter baumannii: mechanisms and epidemiology. Clin. Microbiol. Infect. 12:826–836. 10.1111/j.1469-0691.2006.01456.x [DOI] [PubMed] [Google Scholar]
- 4.Clinical and Laboratory Standards Institute. 2014. Performance standards for antimicrobial susceptibility testing: 24th informational supplement M100-S24. Clinical and Laboratory Standards Institute, Wayne, PA [Google Scholar]
- 5.Nordmann P, Poirel L, Dortet L. 2012. Rapid detection of carbapenemase producing Enterobacteriaceae. Emerg. Infect. Dis. 18:1503–1507. 10.3201/eid1809.120355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Higgins PG, Lehmann M, Seifert H. 2010. Inclusion of OXA-143 primers in a multiplex polymerase chain reaction (PCR) for genes encoding prevalent OXA carbapenemases in Acinetobacter spp. Int. J. Antimicrob. Agents 35:305. 10.1016/j.ijantimicag.2009.10.014 [DOI] [PubMed] [Google Scholar]
- 7.Kieser T. 1984. Factors affecting the isolation or CCC DNA from Streptomyces lividans and Escherichia coli. Plasmid 12:19–36. 10.1016/0147-619X(84)90063-5 [DOI] [PubMed] [Google Scholar]
- 8.D'Andrea MM, Giani T, D'Arezzo S, Capone A, Petrosillo N, Visca P, Luzzaro F, Rossolini GM. 2009. Characterization of pABVA01, a plasmid encoding the OXA-24 carbapenemase from Italian isolates of Acinetobacter baumannii. Antimicrob. Agents Chemother. 53:3528–3533. 10.1128/AAC.00178-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Povilonis J, Seputiene V, Krasauskas R, Juskaite R, Miskinyte M, Suziedelis K, Suziedeliene E. 2013. Spread of carbapenem-resistant Acinetobacter baumannii carrying a plasmid with two genes encoding OXA-72 carbapenemase in Lithuanian hospitals. J. Antimicrob. Chemother. 68:1000–1006. 10.1093/jac/dks499 [DOI] [PubMed] [Google Scholar]
- 10.Bonnin RA, Rotimi VO, Al Hubail M, Gasiorowski E, Al Sweih N, Nordmann P, Poirel L. 2013. Wide dissemination of GES-type carbapenemases in Acinetobacter baumannii isolates in Kuwait. Antimicrob. Agents Chemother. 57:183–188. 10.1128/AAC.01384-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Clímaco EC, de Oliveira ML, Pitondo-Silva A, Oliveira MG, Medeiros M, Lincopan N, da Costa Darini AL. 2013. Clonal complexes 104, 109 and 113 playing a major role in the dissemination of OXA-carbapenemase-producing Acinetobacter baumannii in southeast Brazil. Infect. Genet. Evol. 19:127–133. 10.1016/j.meegid.2013.06.024 [DOI] [PubMed] [Google Scholar]
- 12.Couture F, Lachapelle J, Levesque RC. 1992. Phylogeny of LCR-1 and OXA-5 with class A and class D beta-lactamases. Mol. Microbiol. 6:1693–1705. 10.1111/j.1365-2958.1992.tb00894.x [DOI] [PubMed] [Google Scholar]


