Abstract
Linezolid is an antimicrobial agent for the treatment of multiresistant Gram-positive infections. We assessed the impact of linezolid on the microbiota and the emergence of resistance and investigated its relationship with plasma pharmacokinetics of the antibiotic. Twenty-eight patients were treated for the first time with linezolid administered orally (n = 17) or parenterally (n = 11) at 600 mg twice a day. Linezolid plasma pharmacokinetic analysis was performed on day 7. Colonization by fecal enterococci, pharyngeal streptococci, and nasal staphylococci were assessed using selective media with or without supplemental linezolid. The resistance to linezolid was characterized. The treatment led to a decrease of enterococci, staphylococci, and streptococci in the fecal (P = 0.03), nasal, and pharyngeal (P < 0.01) microbiotas. The appearance of resistant strains was observed only in enterococci from the fecal microbiota between the 7th and 21st days of treatment in four patients (14.3%). The resistance was mainly due for the first time to the mutation G2447T in the 23S rRNA gene. No pharmacokinetic parameters were significantly different between the patients, regardless of the appearance of resistance. The emergence of linezolid resistance during treatment was observed only in the intestinal microbiota and unrelated to pharmacokinetic parameters. However, colonization by Gram-positive bacteria was reduced as a result of treatment in all microbiotas.
INTRODUCTION
Linezolid, the only oxazolidinone antibiotic on the market, is an antibiotic for the treatment of multiresistant Gram-positive infections which inhibits the earliest initiation step of protein synthesis (1), while other antibiotics interfere later in the process. Linezolid resistance in Staphylococcus aureus, coagulase-negative staphylococci (CoNS), and enterococci is lower than 1% and appears stable (2, 3); however, the appearance of resistant strains could affect long-term efficacy. All resistance mechanisms described to date involve the 50S ribosomal subunit. Early reports described only nontransferable resistance (23S rRNA and ribosomal protein L3 and L4 mutations) appearing after prolonged exposure. However, the mobile cfr gene encoding a methyltransferase catalyzing the methylation of A2503 (Escherichia coli numbering) in the 23S rRNA can also confer resistance to linezolid and other antibiotics (4–8).
As a rule, antibiotic use alters the endogenous microbiota and facilitates colonization by exogenous, potentially pathogenic, and/or resistant strains or selects resistant microorganisms that are already present (9). Nonetheless, the extent to which resistance occurs with linezolid treatment remains unknown. After 7 days, linezolid suppressed enterococci in the volunteers' intestinal microbiotas. Simultaneously, linezolid decreased the concentration of bifidobacteria, lactobacilli, clostridia, and Bacteroides (10). In mice, linezolid promoted the colonization of Klebsiella pneumoniae with extended-spectrum β-lactamase (11). We demonstrated that in gnotobiotic mice, linezolid concentrations in the feces were critical in the appearance of resistant mutants; the dose administered to the mice that resulted in fecal concentrations similar to those observed during human treatments (10) was less selective than lower ones (12).
Here, we investigated the emergence of resistant strains in commensal microbiotas of patients treated with linezolid, and we studied the correlation to plasma pharmacokinetics parameters.
MATERIALS AND METHODS
Study.
We performed a four-center (centers A, B, C, and D) prospective study in inpatients older than 18 in whom linezolid was initiated for reasons independent of the study. Patients were excluded if they had previously received linezolid at any time. The dosages were 600 mg, administered twice a day, orally or intravenously (IV), for at least 7 days, as recommended (13). The exposures to antibiotics for 1 month before and during the linezolid therapy were recorded. The protocol was approved by an ad hoc ethics committee (protocol no. 04-022; committee name, CPP Henri Mondor). Written consent was obtained.
Microbiology.
Anterior naris, pharyngeal, and rectal swabs were obtained before treatment (D0), at day 7 (D7), and every 7 days until the end of the study or the discharge of the patient still undergoing treatment. The swabs were placed in 1 ml of brain heart infusion broth (bioMérieux, Marcy l'Etoile, France) with 10% glycerol and were stored in a blinded fashion at −80°C until processing. Samples were then defrosted in batches, and 100 μl was spread onto selective agar to count the number of target strains and resistant strains. We chose to follow three bacterial genera: Staphylococcus, Streptococcus, and Enterococcus, all of which are naturally susceptible to linezolid.
In nasal samples, total and linezolid-resistant staphylococci were counted on Chapman agar plates (bioMérieux) with or without 4 μg/ml linezolid.
In pharyngeal samples, total and resistant oral nongroupable streptococci (NGS) were counted on 5% sheep blood Columbia agar supplemented with 15 μg/ml nalidixic acid and 10 μg/ml colistin (bioMérieux) with or without 4 μg/ml linezolid.
In rectal samples, total and resistant enterococci were counted on bile-esculin agar plates (bioMérieux) with or without 4 μg/ml linezolid.
The plates were incubated at 37°C for 48 h under aerobic conditions to count the staphylococci and enterococci and under anaerobic conditions to count the streptococci. Five colonies from each positive plate were randomly picked and identified using conventional techniques. The antibiotic susceptibility was determined using the disk diffusion method, as recommended by EUCAST (http://www.eucast.org/antimicrobial_susceptibility_testing/disk_diffusion_methodology/), and the linezolid MIC was determined by the agar dilution method and interpreted using EUCAST criteria.
Screening for linezolid resistance mechanisms.
The linezolid-resistant Enterococcus faecalis or S. aureus isolates were screened to determine the presence of mutations in the V domain of the four and five individual copies of the 23S rRNA genes (rrl) (bp 2254 to 2683, E. coli numbering) and the presence of the cfr gene, as described previously (12, 14).
In E. faecalis, mutations in the rplC and rplD genes, encoding the ribosomal proteins L3 and L4, were also searched by amplification and sequencing using the primers L3/F (5′-CCAGTAACAGTAGTTGAAGCTACGCCA-3′), L3/R (5′-TCGCTCCAGGAATGTTTCCTTTGATT-3′), L4/F (5′-TGATGCAATCATCATGCAACGTGCT-3′), and L4/R (5′-TGAGTAAGAGCTGTTTGTGTTGCCA-3′). The linezolid-resistant enterococci were typed by pulsed-field gel electrophoresis (PFGE) as described previously (15).
Pharmacokinetic study.
Plasma samples were taken from each patient immediately before and 1.5, 4, and 8 h after the morning linezolid intake on D7. A fresh fecal sample was obtained from only 8 patients on D7 to determine linezolid concentration. All samples were stored in a blinded fashion at −20°C until analysis. The linezolid concentrations in the plasma and fecal samples at D7 were determined by liquid chromatography, as described previously (12).
Definitions.
The emergence of linezolid resistance was defined by the absence of resistant strains at D0 and the presence of linezolid-resistant strains at D7, D14, or D21. However, if patients were already colonized with resistant strains at D0, the emergence was defined by the presence of strains at D7, D14, and D21 with an MIC at least four times higher than the MICs of the resistant strains detected at D0.
A decrease of linezolid susceptibility was defined by the presence of strains at D7, D14, or D21 with an MIC at least four times higher than the MICs of the susceptible strains detected at D0.
Statistical analysis.
The pharmacokinetic (PK) population was analyzed from the linezolid concentrations of all patients at D7 using the SAEM algorithm in MONOLIX 3.1 (16). The switch from IV to oral administration before D7 was modeled. A one-compartment model was used with first-order absorption for oral administration. The PK parameters were absorption rate constant (ka), volume of distribution (V), clearance (CL), and bioavailability (F). Few measurements were performed during the absorption phase, and the ka was fixed at 2.7 h−1 (17). All parameters were assumed to have log normal distributions. Additive, proportional, and combined residual error models were tested. The final choice of the model was made on the Bayesian Information Criterion (BIC) and goodness-of-fit plots. Whether F was different from 1 was tested with a likelihood ratio test (LRT).
Factors such as height, weight, age, and sex were tested on each PK parameter. Screening was performed using univariate Wald tests (P < 0.1), followed by a forward selection performed using a LRT (P < 0.05).
Exact McNemar tests were performed to compare the proportion of patients at D0 and D7. A Fisher test was performed to compare the proportion of patients receiving coadministered antibiotics between those with and without emergence of linezolid resistance. Pharmacokinetic/pharmacodynamic (PK/PD) individual parameters at steady state were empirically estimated. The following Bayes individual parameters were determined: area under the concentration-time curve (AUC) (from 0 to 24 h), minimum concentration (Cmin), maximum concentration (Cmax) and half-life (t1/2). Several PK/PD parameters were derived using the MIC at D0, AUC/MIC, Cmax/MIC, AUC above MIC, and time above MIC in different microbiotas. These PK/PD parameters were compared between patients who were colonized at D7 and those who were not, using Wilcoxon tests. Similar tests were performed comparing patients with the emergence of linezolid resistance or a decrease of linezolid susceptibility to the others.
Descriptive statistics and tests were performed using the SAS software version 9.1 (SAS, Cary, NC, USA) with a type I error of 0.05.
RESULTS
Patients.
Twenty-eight patients were included (15, 6, 6, and 1 patients treated at centers A, B, C and D, respectively) (Table 1). All patients received other antibiotics in the month preceding the introduction of linezolid, and all patients received linezolid for at least 7 days, 16 patients received linezolid for at least 14 days, and 10 patients received linezolid for at least 21 days. Linezolid was administered orally to 18 patients and intravenously to 10 others. Three patients switched to oral administration at D5, D8, and D14. Seven patients were treated with linezolid only, 14 patients with one other antibiotic, five patients with two other antibiotics, and two patients with more than two other antibiotics. The coadministered antibiotics are presented in Table 1. The coadministered antibiotics were active against staphylococci in 19 patients, streptococci in 16 patients, and enterococci in 16 other patients. In 12 instances, the coadministered antibiotics were already taken during the month preceding inclusion.
TABLE 1.
Characteristics of the 28 enrolled patients treated for the first time with linezolid
| Characteristic | Value |
|---|---|
| Agea (yr) | 63 (27–85) |
| Wta (kg) | 70 (36–130) |
| Hta (cm) | 171 (150–195) |
| No. of males/no. of females (ratio)b | 13/15 (46.4) |
| No. treated via administration routeb | |
| Oral | 18 (64.3) |
| Intravenous | 7 (25.0) |
| Intravenous, then switched to oral | 3 (10.7) |
| Duration of linezolid treatmenta (days) | 13 (7–57) |
| No. with linezolid indicationb | |
| Pneumonia | 4 (14.3) |
| Bacteremia-endocarditis | 6 (21.7) |
| Skin and soft tissue infections | 3 (10.7) |
| Mediastinitis | 5 (17.8) |
| Osteoarticular infections | 6 (21.4) |
| Intra-abdominal infections | 4 (14.3) |
| No. receiving antibiotics in addition to linezolidb | 21 (75.0) |
| Glycopeptides | 5 (23.8) |
| Penicillins | 3 (14.3) |
| Cephalosporins | 1 (4.7) |
| Carbapenem | 2 (9.5) |
| Macrolides | 1 (4.7) |
| Fluoroquinolones | 4 (19.0) |
| Rifampin | 8 (38.1) |
| Aminoglycosides | 3 (14.3) |
| Other antibioticsc | 3 (14.3) |
Data are medians (ranges).
Data are absolute numbers (%).
Includes metronidazole and trimethoprim-sulfamethoxazole.
Safety and tolerability.
Five (17.9%) patients developed mild adverse events, including diarrhea (n = 2), thrombocytopenia (n = 1), paresthesia (n = 1), and moderate pleural effusion (n = 1). No severe adverse event was reported, and linezolid was never discontinued because of an adverse event. No Clostridium difficile infection was reported during the study.
Microbiology.
In the patients' nares, CoNS colonization rates decreased from 92.8% (26/28) at D0 to 53.6% (15/28) at D7 (P = 0.0009), to 43.8% (7/16) at D14, and to 0% (0/4) at D21 (Table 2). In the patients treated with linezolid alone or in combination with antibiotics not active against CoNS, the rates decreased from 100% (9/9) at D0 to 33.3% (3/9) at D7 and 0% (0/5) at D14.
TABLE 2.
Colonization by CoNS, S. aureus, nongroupable streptococci, or enterococci in the nasal, oropharyngeal, and intestinal microbiotas before (D0) and after 7 days (D7) of linezolid therapy
| Type of microbiota (no. of patients studied) | Organisma | No. (%) of patients colonized at: |
|
|---|---|---|---|
| D0 | D7 | ||
| Nose (28) | CoNS | 26 (92.8) | 15 (53.6) |
| Linezolid-resistant CoNS | 0 | 0 | |
| S. aureus | 7 (25.0) | 0 | |
| Linezolid-resistant S. aureus | 3 | 0 | |
| Oropharynx (27) | NGS | 17 (62.9) | 7 (25.9) |
| Linezolid-resistant NGS | 0 | 0 | |
| Intestinal (27) | Enterococci | 26 (96.3) | 21 (77.8) |
| Linezolid-resistant enterococci | 2 (7.4) | 1 (6.7) | |
Strains are defined as linezolid resistant when the linezolid MIC is >4 μg/ml. CoNS, coagulase-negative staphylococci; NGS, nongroupable streptococci.
All CoNS strains remained susceptible to linezolid according to breakpoints (linezolid MIC ≤ 4 μg/ml); however, the median MIC increased from 1 μg/ml (range, 0.5 to 4 μg/ml) in D0 strains to 3 μg/ml (range, 1 to 4 μg/ml) in D7 strains.
Nasal S. aureus colonization decreased from 25.0% (7/28) to 0% (0/28 at D7, 0/16 at D14, and 0/4 at D21) (Table 2). Three patients were colonized with methicillin- and linezolid-resistant S. aureus strains. One and two isolates had linezolid MICs of 32 μg/ml and 64 μg/ml, respectively. All three resistant strains harbored a G2576T mutation in three or four copies of the rrl genes; however, none harbored the cfr gene. At D7, D14, and D21, none remained colonized by S. aureus, including the three patients with linezolid-resistant strains at inclusion, although two of these patients were treated with linezolid only.
The oropharyngeal microbiota was studied in 27 patients. The NGS colonization rates decreased from 62.9% (17/27) at D0 to 25.9% (7/27) at D7 (P = 0.0075) but increased to 31.2% (5/16) at D14 and 100% (4/4) at D21 (Table 2). In patients treated with linezolid only or in combination with an antibiotic not active against streptococci, the NGS colonization rates decreased from 41.7% (5/12) at D0 to 33.3% (4/12) at D7 but later increased to 50% (3/6) at D14. The median linezolid MICs increased from 1 μg/ml (range 0.5 to 4 μg/ml) at D0 to 2 μg/ml after D7. No linezolid-resistant streptococci were isolated.
Intestinal enterococcus colonization rates decreased from 96.3% (26/27) at D0 to 77.8% (21/27) at D7 and 62.5% (10/16) at D14 (P = 0.02) but increased to 75.0% (3/4) at D21 (Table 2). The median linezolid MIC was 4 μg/ml (range, 2 to 32 μg/ml) at D0 and remained unchanged at D7. Six linezolid-resistant E. faecalis strains (range of linezolid MICs, 8 to 32 μg/ml) were isolated from five patients (4 and 1 patients at centers A and C, respectively). The resistance was stable in all linezolid-resistant E. faecalis isolates (data not shown). Two and four strains were isolated at D0 and during treatment (D7, D9, D10, and D21), respectively (Table 3). All strains harbored the wild-type sequence for the L3 and L4 ribosomal proteins, and no strains carried the cfr gene. Four strains harbored the mutation G2447T in 1 or 3 rrl genes. The resistance mechanism remained undetected for two strains.
TABLE 3.
PFGE profiles of linezolid-resistant E. faecalis strains isolated in intestinal microbiota of 5 patients before and during treatment in centers A and C
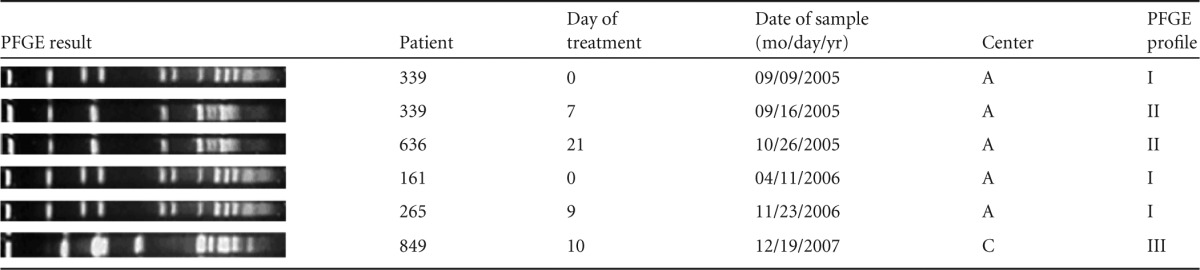
In center A, three linezolid-resistant E. faecalis isolates shared an identical SmaI digestion pattern (I), while two isolates shared a different pattern (II). The patterns were different from the unique strain found at the center C, suggesting that only local cross-transmission occurred (Table 3). In one patient, two isolates with different patterns were isolated at D0 and D7 (Table 3).
Only 25% (1/4) of patients with emergence of linezolid resistance had coadministered antibiotics, whereas in patients without emergence of linezolid resistance, 83.3% (20/24) had coadministered antibiotics (P = 0.038).
Pharmacokinetics.
A one-compartment model with a first-order elimination and absorption for oral administration adequately described the plasma concentrations of linezolid in all patients. The best error model was a combined one. Pharmacokinetic parameter estimates and their relative standard errors (RSEs) with the basic model are shown in Table 4. All RSEs were <30% for fixed effects and <50% for the other parameters. The goodness-of-fit plots provided good evidence of the model's adequacy.
TABLE 4.
Linezolid pharmacokinetic parameters for the population in the basic and final model estimates and relative standard errors (RSEs)
| Parameter | Basic model |
Final model |
||
|---|---|---|---|---|
| Estimate | RSE (%) | Estimate | RSE (%) | |
| ka (h−1) | 2.7 | 2.7 | ||
| V (liters) | 47.5 | 8 | 47.1 | 8 |
| CL (liters/h) | 4.37 | 12 | 4.31 | 9 |
| βCL_weight | 1.33 | 31 | ||
| βCL_age | −0.81 | 33 | ||
| ωka | 1.92 | 28 | 2.67 | 22 |
| ωV | 0.242 | 39 | 0.243 | 40 |
| ωCL | 0.611 | 14 | 0.445 | 14 |
| a (μg/ml) | 1.16 | 26 | 1.15 | 26 |
| b | 0.07 | 37 | 0.07 | 37 |
ka, absorption rate constant; CL, clearance; V, volume of distribution; βCL_weight, weight effect on CL centered to the mean; βCL_age, age effect on CL centered to the mean; ωka, ωV, and ωCL, standard deviation for interpatient variability; a and b, parameters of error model.
Bioavailability (F) was estimated at 1.17, with a standard error of 0.14 and a 95% confidence interval of 0.89 to 1.44. F was not significantly different from 1 (P = 0.31) and was consequently fixed at 1 in the chosen model. From the model, we derived the exposure pharmacokinetic parameters at steady state: AUC over 24 h (median, 339.1, and range, 68.6 to 953.7 mg/liter · h for the oral route; median, 273.9, and range, 105.4 to 727.0 mg · h/liter for the IV route) and Cmax (median, 19.3, and range, 4.3 to 44.6 μg/ml for the oral route; median, 17.6, and range, 13.0 to 36.2 μg/ml for the IV route).
Covariate univariate analysis found significant effects of weight (P = 0.002) and age (P = 0.0025) on CL. Following a forward selection based on LRT, the final model had a weight effect (P = 0.0013) and an age effect (P = 0.0026) on CL. The population parameter estimates of this final model and their RSEs are given in Table 4.
The median linezolid concentration in feces, determined in 8 patients on D7, was 7.0 μg/g (range, 4.5 to 10.3 μg/g).
Relationships between colonization and pharmacokinetics.
The distribution of AUC/MIC of linezolid was not significantly different between the patients who were and those who were not colonized by linezolid-resistant strains or between patients with a decrease and those without a decrease in linezolid susceptibility during the treatment in any of the microbiotas studied (Tables 5 and 6). Similar results were obtained for all PK/PD variables.
TABLE 5.
Comparison of pharmacokinetic/pharmacodynamic parameters between inpatients colonized and those not colonized at D7 by coagulase-negative staphylococci (CoNS) in the nose, nongroupable streptococci (NGS) in the oropharynx, or enterococci in the intestine
| Type of microbiota (no. of patients colonized at D0) and variable | Colonization on D7 (median [range]) |
P (Wilcoxon test) | |
|---|---|---|---|
| No | Yes | ||
| Nasal (26) | n = 11 | n = 15 | |
| AUC/MIC (h) | 121.4 (17.1–378.2) | 80.8 (34.8–953.7) | 0.65 |
| Cmax/MIC | 6.0 (1.1–20.5) | 6.6 (3.0–44.6) | 0.92 |
| AUC >MIC (mg/liter · h) | 300.0 (10.4–679.0) | 177.9 (61.7–929.7) | 0.15 |
| Time >MIC (h) | 24.0 (5.1–24.0) | 24.0 (14.8–24.0) | 0.97 |
| Oropharyngeal (17) | n = 12 | n = 5 | |
| AUC/MIC (h) | 138.2 (26.3–953.7) | 261.5 (83.8–775.3) | 0.23 |
| Cmax/MIC | 10.4 (3.1–44.6) | 15.2 (4.9–41.9) | 0.33 |
| AUC >MIC (mg/liter · h) | 118.3 (36.9–929.7) | 250.3 (237.5–582.0) | 0.02 |
| Time >MIC (h) | 24.0 (10.2–24.0) | 24.0 (24.0–24.0) | 0.51 |
| Intestinal (26) | n = 5 | n = 21 | |
| AUC/MIC (h) | 87.9 (22.7–476.9) | 68.8 (17.1–315.0) | 0.66 |
| Cmax/MIC | 7.3 (1.1–22.3) | 4.4 (1.1–15.2) | 0.66 |
| AUC >MIC (mg/liter · h) | 213.0 (34.9–905.7) | 179.4 (10.4–582.0) | 0.49 |
| Time >MIC (h) | 24.0 (8.1–24.0) | 24.0 (5.1–24.0) | 0.90 |
TABLE 6.
Comparison of the pharmacokinetic/pharmacodynamic parameters between patients who developed linezolid resistance or decreased susceptibility in nasal, oropharyngeal, and intestinal microbiotas under treatment and patients who did not
| Type of microbiota (no. of patients colonized at D0) and variable | Resistance or decrease of susceptibility to linezolid during treatment, median value (range) |
P (Wilcoxon test) | |
|---|---|---|---|
| No | Yes | ||
| Nasal (26) | n = 24 | n = 2 | |
| AUC/MIC (h) | 98.0 (17.1–488.8) | 560.5 (167.2–953.7) | 0.12 |
| Cmax/MIC | 5.6 (1.1–35.2) | 28.5 (12.4–44.6) | 0.10 |
| AUC >MIC (mg/liter · h) | 223.7 (10.4–679.0) | 536.5 (143.2–929.7) | 0.55 |
| Time >MIC (h) | 24.0 (5.1–24.0) | 24.0 (14.8–24.0) | 0.88 |
| Oropharyngeal (17) | n = 14 | n = 3 | |
| AUC/MIC (h) | 142.3 (26.3–661.2) | 775.3 (137.2–953.7) | 0.12 |
| Cmax/MIC | 10.4 (3.1–35.6) | 41.5 (8.6–44.6) | 0.16 |
| AUC >MIC (mg/liter · h) | 168.6 (36.9–582.0) | 375.7 (56.6–929.7) | 0.43 |
| Time >MIC (h) | 24.0 (10.2–24.0) | 24.0 (24.0–24.0) | 0.73 |
| Intestinal (26) | n = 22 | n = 4 | |
| AUC/MIC (h) | 69.8 (17.1–476.9) | 131.3 (32.7–198.0) | 0.56 |
| Cmax/MIC | 4.4 (1.1–22.3) | 7.3 (2.6–10.4) | 0.71 |
| AUC >MIC (mg/liter · h) | 178.7 (10.4–905.7) | 259.5 (106.4–348.0) | 0.51 |
| Time >MIC (h) | 24.0 (5.1–24.0) | 24.0 (14.8–24.0) | 0.77 |
DISCUSSION
In this prospective, observational study including linezolid-naive patients, our most striking result was that linezolid treatment reduced colonization by staphylococci, streptococci, and enterococci in the different microbiotas. The emergence of linezolid resistance in microbiotas was a less frequent phenomenon than emergence of other antibiotic resistances (18, 19).
We focused on the analysis of these bacteria because they are present in everyone's microbiota, can cause infections, and are a source of horizontal gene transfer between commensal bacteria (20). Similar findings were previously reported, however, for healthy volunteers and only for intestinal enterococci (10). This reduction in colonization could be due to the antibiotics coadministered with linezolid. However, this hypothesis is not plausible, because we also observed a decrease of colonization in patients treated with linezolid only or with antibiotics that are not active for the three bacteria genera studied. It was noticed that S. aureus nasal colonization disappeared at D7, regardless of the linezolid MIC of the strain. This result might be due to the high linezolid concentration in nasal tissues (21), which could have exceeded the MIC of linezolid-resistant S. aureus, thus allowing this eradication. Previous studies reported such an eradication of nasal S. aureus colonization in patients treated with linezolid (22), as well as skin colonization by methicillin-resistant S. aureus (23). This finding is interesting, because S. aureus nasal decontamination is indicated to prevent postoperative infections in high-risk patients (24). Currently, topical mupirocin is the antibiotic recommended for that purpose; however, its use is associated with a 1% risk of acquiring a drug-resistant strain during treatment (25), and this resistance is associated with drug failure (26). Whether linezolid could be an alternative in such patients remains hypothetical.
Strikingly, the selection of linezolid-resistant strains during treatment was observed in only the intestinal microbiota and in only four patients between D7 and D21 of treatment. This result agrees with our observation of gnotobiotic mice exposed to intestinal concentrations of linezolid, similar to the results measured in humans receiving 1,200 mg/day (12). Interestingly, the time between the initiation of treatment and the emergence of linezolid-resistant enterococci in the intestinal microbiota in our patients was similar to the time reported for the emergence of linezolid-resistant enterococci in clinical infections (27). The rarity of the emergence of linezolid resistance in the microbiota might be related to the relatively short duration of treatment used (median, 13 days), the selected patients without previous linezolid exposure and the high percentage of patients (21/28 [75%]) treated with other concomitant antibiotics. Our results suggest that coadministration of other antibiotics with linezolid decreased the selection of linezolid-resistant strains in the microbiota. This is important because although antibiotic combinations are well known to reduce the emergence of mutants at the site of infection (28, 29), this is, to the best of our knowledge, the first time that this has been demonstrated for the emergence of resistance in the intestinal microbiota. We did not report the emergence of linezolid resistance in the oropharynx, unlike the observation made with volunteers treated either with macrolides or ciprofloxacin using similar microbiological methods (18, 19). This result could be because patients were often treated with antibiotic combinations, which is not the case when volunteers are studied. However, we cannot exclude the possibility that a more frequent emergence of resistance would arise in the subsequent days or weeks following the end of the treatment, as we observed with ciprofloxacin in healthy volunteers (18). The specific genetics underlying the resistance to each type of antibiotic may also be involved.
Linezolid resistance was found in E. faecalis but not in E. faecium, according to results obtained in vitro, where it is more likely to generate linezolid-resistant E. faecalis than E. faecium (30). Linezolid resistance was conferred by a mutation in the 23S rRNA genes in two of the enterococcus isolates that emerged during treatment. In clinical enterococci, linezolid resistance was mainly linked to the G2576T mutation (4). In gnotobiotic mice, we described the emergence of two types of mutants with G2576T or G2505A mutations, depending on the linezolid regimen (12). Indeed, this is the first report of a G2447T mutation in E. faecalis. This mutation was detected in linezolid-resistant Staphylococcus clinical isolates, in mutants of E. coli, and in Mycobacterium smegmatis strains selected in vitro (31, 32). The two strains with unknown resistance mechanisms may be related to either a decrease of drug uptake into bacterial cells, an increase of active efflux of the drug (31), or a decrease of methylation of G2445 in 23S rRNA (33).
We observed the spread of two clones of linezolid-resistant E. faecalis in center A. Indeed, hospital dissemination of clonal linezolid-resistant enterococci and staphylococci has been previously reported, including by us, stressing the nosocomial burden that can be associated with resistance to linezolid (27, 34, 35).
Linezolid bioavailability was 100%, similar to what has been reported elsewhere (36); however, no PK/PD parameter was significantly different between patients who were and those who were not colonized by strains exhibiting a decreased susceptibility to linezolid. This result is similar to the finding we obtained in healthy volunteers receiving ciprofloxacin (18). This lack of correlation between the pharmacokinetic parameters and the emergence of resistance in the microbiota sharply contrasts with what was observed between these parameters and the emergence of resistance in bacterial infections of patients treated with fluoroquinolones (37) or linezolid (38). This result provides an additional argument to the theory that the microbiota is the epicenter of resistance (39) and signifies that this theory requires careful attention when control strategies are being designed.
In conclusion, we demonstrated that the emergence of resistance to linezolid in the microbiota was rare during treatment but unrelated to pharmacokinetic parameters. However, a cross-transmission of resistant strains was significant. The judicious use of this antibiotic and the application of appropriate infection control measures should be combined to preserve the activity of this valuable antibiotic.
ACKNOWLEDGMENTS
This work was supported by INSERM-RBM 04-03, by FP7 EvoTAR, by Paris-Descartes University and by Paris Sud University. N.B.-N. was supported in part by a grant from Assistance Publique des Hôpitaux de Paris. G.D. was supported in part by a grant from the French National Academy of Medicine.
We acknowledge Agnes Certain and Emmanuelle Papy for their contribution to this study.
No conflict of interest is related to this work.
Footnotes
Published ahead of print 24 February 2014
REFERENCES
- 1.Wilson DN, Schluenzen F, Harms JM, Starosta AL, Connell SR, Fucini P. 2008. The oxazolidinone antibiotics perturb the ribosomal peptidyl-transferase center and effect tRNA positioning. Proc. Natl. Acad. Sci. U. S. A. 105:13339–13344. 10.1073/pnas.0804276105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Flamm RK, Farrell DJ, Mendes RE, Ross JE, Sader HS, Jones RN. 2012. LEADER surveillance program results for 2010: an activity and spectrum analysis of linezolid using 6801 clinical isolates from the United States (61 medical centers). Diagn. Microbiol. Infect. Dis. 74:54–61. 10.1016/j.diagmicrobio.2012.05.012 [DOI] [PubMed] [Google Scholar]
- 3.Flamm RK, Mendes RE, Ross JE, Sader HS, Jones RN. 2013. An international activity and spectrum analysis of linezolid: ZAAPS program results for 2011. Diagn. Microbiol. Infect. Dis. 76:206–213. 10.1016/j.diagmicrobio.2013.01.025 [DOI] [PubMed] [Google Scholar]
- 4.Long KS, Vester B. 2012. Resistance to linezolid caused by modifications at its binding site on the ribosome. Antimicrob. Agents Chemother. 56:603–612. 10.1128/AAC.05702-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mendes RE, Deshpande L, Rodriguez-Noriega E, Ross JE, Jones RN, Morfin-Otero R. 2010. First report of Staphylococcal clinical isolates in Mexico with linezolid resistance caused by cfr: evidence of in vivo cfr mobilization. J. Clin. Microbiol. 48:3041–3043. 10.1128/JCM.00880-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mendes RE, Deshpande LM, Castanheira M, DiPersio J, Saubolle MA, Jones RN. 2008. First report of cfr-mediated resistance to linezolid in human staphylococcal clinical isolates recovered in the United States. Antimicrob. Agents Chemother. 52:2244–2246. 10.1128/AAC.00231-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Morales G, Picazo JJ, Baos E, Candel FJ, Arribi A, Peláez B, Andrade R, de la Torre MA, Fereres J, Sánchez-García M. 2010. Resistance to linezolid is mediated by the cfr gene in the first report of an outbreak of linezolid-resistant Staphylococcus aureus. Clin. Infect. Dis. 50:821–825. 10.1086/650574 [DOI] [PubMed] [Google Scholar]
- 8.Kehrenberg C, Schwarz S. 2006. Distribution of florfenicol resistance genes fexA and cfr among chloramphenicol-resistant Staphylococcus isolates. Antimicrob. Agents Chemother. 50:1156–1163. 10.1128/AAC.50.4.1156-1163.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sullivan A, Edlund C, Nord CE. 2001. Effect of antimicrobial agents on the ecological balance of human microflora. Lancet Infect. Dis. 1:101–114. 10.1016/S1473-3099(01)00066-4 [DOI] [PubMed] [Google Scholar]
- 10.Lode H, Von Der HN, Ziege S, Borner K, Nord CE. 2001. Ecological effects of linezolid versus amoxicillin/clavulanic acid on the normal intestinal microflora. Scand. J. Infect. Dis. 33:899–903. 10.1080/00365540110076714 [DOI] [PubMed] [Google Scholar]
- 11.Pultz NJ, Stiefel U, Donskey CJ. 2005. Effects of daptomycin, linezolid, and vancomycin on establishment of intestinal colonization with vancomycin-resistant enterococci and extended-spectrum-beta-lactamase-producing Klebsiella pneumoniae in mice. Antimicrob. Agents Chemother. 49:3513–3516. 10.1128/AAC.49.8.3513-3516.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bourgeois-Nicolaos N, Massias L, Couson B, Butel MJ, Andremont A, Doucet-Populaire F. 2007. Dose dependence of emergence of resistance to linezolid in Enterococcus faecalis in vivo. J. Infect. Dis. 195:1480–1488. 10.1086/513876 [DOI] [PubMed] [Google Scholar]
- 13.Dryden MS. 2011. Linezolid pharmacokinetics and pharmacodynamics in clinical treatment. J. Antimicrob. Chemother. 66:iv7–iv15 [DOI] [PubMed] [Google Scholar]
- 14.Pillai SK, Sakoulas G, Wennersten C, Eliopoulos GM, Moellering RC, Jr, Ferraro MJ, Gold HS. 2002. Linezolid resistance in Staphylococcus aureus: characterization and stability of resistant phenotype. J. Infect. Dis. 186:1603–1607. 10.1086/345368 [DOI] [PubMed] [Google Scholar]
- 15.Barbier N, Saulnier P, Chachaty E, Dumontier S, Andremont A. 1996. Random amplified polymorphic DNA typing versus pulsed-field gel electrophoresis for epidemiological typing of vancomycin-resistant enterococci. J. Clin. Microbiol. 34:1096–1099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kuhn E, Lavielle M. 2005. Maximum likelihood estimation in nonlinear mixed effects model. Comput. Stat. Data Anal. 49:1020–1038. 10.1016/j.csda.2004.07.002 [DOI] [Google Scholar]
- 17.McGee B, Dietze R, Hadad DJ, Molino LP, Maciel EL, Boom WH, Palaci M, Johnson JL, Peloquin CA. 2009. Population pharmacokinetics of linezolid in adults with pulmonary tuberculosis. Antimicrob. Agents Chemother. 53:3981–3984. 10.1128/AAC.01378-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fantin B, Duval X, Massias L, Alavoine L, Chau F, Retout S, Andremont A, Mentré F. 2009. Ciprofloxacin dosage and emergence of resistance in human commensal bacteria. J. Infect. Dis. 200:390–398. 10.1086/600122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Crémieux AC, Muller-Serieys C, Panhard X, Delatour F, Tchimichkian M, Mentre F, Andremont A. 2003. Emergence of resistance in normal human aerobic commensal flora during telithromycin and amoxicillin-clavulanic acid treatments. Antimicrob. Agents Chemother. 47:2030–2035. 10.1128/AAC.47.6.2030-2035.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bourgeois-Nicolaos N, Moubareck C, Mangeney N, Butel MJ, Doucet-Populaire F. 2006. Comparative study of vanA gene transfer from Enterococcus faecium to Enterococcus faecalis and to Enterococcus faecium in the intestine of mice. FEMS Microbiol. Lett. 254:27–33. 10.1111/j.1574-6968.2005.00004.x [DOI] [PubMed] [Google Scholar]
- 21.Stein GE, Wells EM. 2010. The importance of tissue penetration in achieving successful antimicrobial treatment of nosocomial pneumonia and complicated skin and soft-tissue infections caused by methicillin-resistant Staphylococcus aureus: vancomycin and linezolid. Curr. Med. Res. Opin. 26:571–588. 10.1185/03007990903512057 [DOI] [PubMed] [Google Scholar]
- 22.Hyatt JM, Ballow CH, Forrest A, Tumak MR, Stakker DJ, Schentag JJ. 1998. Safety and efficacy of linezolid in the eradication of nasal Staphylococcus aureus, abstr. A-4 Abstr. 38th Intersci. Conf. Antimicrob. Agents Chemother [Google Scholar]
- 23.Hayman S, Wilson AP, Singer M, Bellingan G. 2007. Effect of linezolid and teicoplanin on skin staphylococci. J. Antimicrob. Chemother. 59:1281–1282. 10.1093/jac/dkm093 [DOI] [PubMed] [Google Scholar]
- 24.Perl TM, Cullen JJ, Wenzel RP, Zimmerman MB, Pfaller MA, Sheppard D, Twombley J, French PP, Herwaldt LA, Mupirocin and the Risk of Staphylococcus aureus Study Team 2002. Intranasal mupirocin to prevent postoperative Staphylococcus aureus infections. N. Engl. J. Med. 346:1871–1877. 10.1056/NEJMoa003069 [DOI] [PubMed] [Google Scholar]
- 25.Jones JC, Rogers TJ, Brookmeyer P, Dunne WM, Jr, Storch GA, Coopersmith CM, Fraser VJ, Warren DK. 2007. Mupirocin resistance in patients colonized with methicillin-resistant Staphylococcus aureus in a surgical intensive care unit. Clin. Infect. Dis. 45:541–547. 10.1086/520663 [DOI] [PubMed] [Google Scholar]
- 26.Ammerlaan HS, Kluytmans JA, Wertheim HF, Nouwen JL, Bonten MJ. 2009. Eradication of methicillin-resistant Staphylococcus aureus carriage: a systematic review. Clin. Infect. Dis. 48:922–930. 10.1086/597291 [DOI] [PubMed] [Google Scholar]
- 27.Ntokou E, Stathopoulos C, Kristo I, Dimitroulia E, Labrou M, Vasdeki A, Makris D, Zakynthinos E, Tsakris A, Pournaras S. 2012. Intensive care unit dissemination of multiple clones of linezolid-resistant Enterococcus faecalis and Enterococcus faecium. J. Antimicrob. Chemother. 67:1819–1823. 10.1093/jac/dks146 [DOI] [PubMed] [Google Scholar]
- 28.Miller K, O'Neill AJ, Wilcox MH, Ingham E, Chopra I. 2008. Delayed development of linezolid resistance in Staphylococcus aureus following exposure to low levels of antimicrobial agents. Antimicrob. Agents Chemother. 52:1940–1944. 10.1128/AAC.01302-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vilchèze C, Jacobs WR. 2012. The combination of sulfamethoxazole, trimethoprim, and isoniazid or rifampin is bactericidal and prevents the emergence of drug resistance in Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 56:5142–5148. 10.1128/AAC.00832-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Prystowsky J, Siddiqui F, Chosay J, Shinabarger DL, Millichap J, Peterson LR, Noskin GA. 2001. Resistance to linezolid: characterization of mutations in rRNA and comparison of their occurrences in vancomycin-resistant enterococci. Antimicrob. Agents Chemother. 45:2154–2156. 10.1128/AAC.45.7.2154-2156.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sander P, Belova L, Kidan YG, Pfister P, Mankin AS, Bottger EC. 2002. Ribosomal and non-ribosomal resistance to oxazolidinones: species-specific idiosyncrasy of ribosomal alterations. Mol. Microbiol. 46:1295–1304. 10.1046/j.1365-2958.2002.03242.x [DOI] [PubMed] [Google Scholar]
- 32.Miller K, Dunsmore CJ, Fishwick CW, Chopra I. 2008. Linezolid and tiamulin cross-resistance in Staphylococcus aureus mediated by point mutations in the peptidyl transferase center. Antimicrob. Agents Chemother. 52:1737–1742. 10.1128/AAC.01015-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Feng J, Lupien A, Gingras H, Wasserscheid J, Dewar K, Légaré D, Ouellette M. 2009. Genome sequencing of linezolid-resistant Streptococcus pneumoniae mutants reveals novel mechanisms of resistance. Genome Res. 19:1214–1223. 10.1101/gr.089342.108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gómez-Gil R, Romero-Gómez MP, García-Arias A, Ubeda MG, Busselo MS, Cisterna R, Gutiérrez-Altés A, Mingorance J. 2009. Nosocomial outbreak of linezolid-resistant Enterococcus faecalis infection in a tertiary care hospital. Diagn. Microbiol. Infect. Dis. 65:175–179. 10.1016/j.diagmicrobio.2009.06.010 [DOI] [PubMed] [Google Scholar]
- 35.Mihaila L, Defrance G, Levesque E, Ichai P, Garnier F, Derouin V, Decousser JW, Doucet-Populaire F, Bourgeois-Nicolaos N. 2012. A dual outbreak of bloodstream infections with linezolid-resistant Staphylococcus epidermidis and Staphylococcus pettenkoferi in a liver intensive care unit. Int. J. Antimicrob. Agents 40:472–474. 10.1016/j.ijantimicag.2012.06.014 [DOI] [PubMed] [Google Scholar]
- 36.Welshman IR, Sisson TA, Jungbluth GL, Stalker DJ, Hopkins NK. 2001. Linezolid absolute bioavailability and the effect of food on oral bioavailability. Biopharm. Drug Dispos. 22:91–97. 10.1002/bdd.255 [DOI] [PubMed] [Google Scholar]
- 37.Louie A, Heine HS, VanScoy B, Eichas A, Files K, Fikes S, Brown DL, Liu W, Kinzig-Schippers M, Sörgel F, Drusano GL. 2011. Use of an in vitro pharmacodynamic model to derive a moxifloxacin regimen that optimizes kill of Yersinia pestis and prevents emergence of resistance. Antimicrob. Agents Chemother. 55:822–830. 10.1128/AAC.00818-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Boak LM, Li J, Rayner CR, Nation RL. 2007. Pharmacokinetic/pharmacodynamic factors influencing emergence of resistance to linezolid in an in vitro model. Antimicrob. Agents Chemother. 51:1287–1292. 10.1128/AAC.01194-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Carlet J. 2012. The gut is the epicentre of antibiotic resistance Antimicrob. Resist. Infect. Control. 1:39. 10.1186/2047-2994-1-39 [DOI] [PMC free article] [PubMed] [Google Scholar]


