Abstract
This study exploited the possibility to detect Citrobacter freundii-derived CMY-2-like cephalosporinases in Enterobacteriaceae clinical isolates using matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS). Periplasmic proteins were prepared using a modified sucrose method and analyzed by MALDI-TOF MS. A ca. 39,850-m/z peak, confirmed to represent a C. freundii-like β-lactamase by in-gel tryptic digestion followed by MALDI-TOF/TOF MS, was observed only in CMY-producing isolates. We have also shown the potential of the assay to detect ACC- and DHA-like AmpC-type β-lactamases.
TEXT
Matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS) is increasingly used as a procedure for identification of pathogenic bacteria and fungi due to its time- and cost-effectiveness (1, 2). Recently, further applications of MALDI-TOF MS focusing on antimicrobial resistance mechanisms, including detection of carbapenemase activity in Enterobacteriaceae, Pseudomonas spp., and Acinetobacter spp., have been described (3–7).
In 2007, Camara and Hays described for the first time the use of MALDI-TOF MS for differentiating wild-type Escherichia coli from ampicillin-resistant (Ampr) plasmid-transformed E. coli strains by the direct visualization of a β-lactamase (8). In a recent MALDI-TOF MS study, Schaumann et al. were not able to distinguish Enterobacteriaceae and P. aeruginosa isolates producing extended-spectrum β-lactamases (ESBLs) or metallo-β-lactamases (MBLs) from nonproducers (9). Consequently, so far the attempts to visualize native β-lactamases by MALDI-TOF MS in wild-type bacteria have been mostly unsuccessful.
We describe here a new assay for the identification of CMY-2-like β-lactamases in clinical enterobacterial isolates by MALDI-TOF MS. These enzymes are the most prevalent acquired AmpC-type cephalosporinases in Enterobacteriaceae (10). The method is based on the extraction of periplasmic proteins and the detection of CMY-2-like β-lactamases by MALDI-TOF MS according to their molecular weight.
Thirty-eight characterized Enterobacteriaceae strains from collections of the Faculty of Medicine and University Hospital in Plzen, Czech Republic, the Hellenic Pasteur Institute in Athens, Greece, and the National Medicines Institute in Warsaw, Poland, were used (Table 1) (11, 12). The group included 29 CMY-2-positive clinical isolates, two E. coli transconjugants/transformants with CMY-2-like enzymes (E. coli A15 or DH5α), and seven non-CMY-producing isolates (13–21). E. coli ATCC 25922 and Klebsiella pneumoniae ATCC 13883 were used as negative controls. Purified CMY-2 enzyme (13) was used as a positive control for MALDI-TOF MS measurements.
TABLE 1.
Summary of the MALDI-TOF MS analysis of the periplasmic extractsa
| Strain | Country | β-Lactamase(s) produced | Peak at m/z 39,850 | Reference or source |
|---|---|---|---|---|
| E. coli pB-cmy2 | Greece | Cloned CMY-2 | + | 13 |
| E. coli S95 | Greece | CMY-6 | + | 14 |
| E. coli S208 | Greece | LAT-1, SHV-5, TEM-1 | + | 14 |
| E. coli T27 | Greece | CMY-2, CTX-M-3, TEM-1 | − | 15 |
| E. coli AK-3281 | Greece | CMY-2, TEM-1 | + | This study |
| E. coli AK-5231 | Greece | CMY-2, TEM-1 | + | This study |
| E. coli AK-5495 | Greece | CMY-2, TEM-1 | + | This study |
| E. coli PL 5143/09 | Poland | CMY-2 | + | 16 |
| E. coli PL 5138/09 | Poland | CMY-4 | + | 16 |
| E. coli PL 6691/10 | Poland | CMY-42 | + | 16 |
| E. coli Cz 9162 | Czech Republic | CMY-2, CTX-M-15 | + | This study |
| Trc E. coli Cz 9162 | Czech Republic | CMY-2 | + | This study |
| E. coli Cz 9178 | Czech Republic | CMY-2 | + | This study |
| E. coli Cz 9261 | Czech Republic | CTX-M-14 | − | This study |
| E. coli Cz 9309 | Czech Republic | CTX-M-27 | − | This study |
| E. coli Cz 9355 | Czech Republic | CTX-M-15 | − | This study |
| E. coli A15 | NT | − | ||
| E. coli ATCC 25922 | NT | − | ||
| E. aerogenes Y15 | Greece | CMY-2 | + | 14 |
| E. aerogenes Y25 | Greece | CMY-2, SHV-5 | + | 14 |
| K. pneumoniae P20 | Greece | LAT-1, SHV-5 | + | 17 |
| K. pneumoniae L67 | Greece | CMY-2 | − | 14 |
| K. pneumoniae N1 | Greece | CMY-2, SHV-5, TEM-1 | + | 14 |
| K. pneumoniae N2 | Greece | CMY-2, SHV-5, TEM-1 | + | 14 |
| K. pneumoniae T80 | Greece | CMY-2 | + | 14 |
| K. pneumoniae HP205 | Greece | CMY-36, SHV-5, TEM-1 | + | 18 |
| K. pneumoniae PL 7246/10 | Poland | CMY-2 | + | 19 |
| K. pneumoniae PL 6185/11 | Poland | CMY-4, VIM-19 | − | This study |
| K. pneumoniae Cz 1006 | Czech Republic | CMY-2 | + | This study |
| K. pneumoniae Cz 3602 | Czech Republic | CMY-2, NDM-1 | + | This study |
| K. pneumoniae Cz 431 | Czech Republic | VIM-1, SHV-5 | − | This study |
| K. pneumoniae Cz 597 | Czech Republic | KPC-2, OXA-9, SHV-12, TEM-1 | − | This study |
| K. pneumoniae Cz 163243 | Czech Republic | SHV-5 | − | This study |
| K. pneumoniae ATCC 13883 | NT | − | ||
| P. mirabilis PL 6735/99 | Poland | CMY-14, TEM-1 | − | 20 |
| P. mirabilis PL 27/00 | Poland | CMY-12, TEM-2 | + | 20 |
| P. mirabilis PL 1662/00 | Poland | CMY-15, TEM-2 | + | 20 |
| P. mirabilis PL 864/01 | Poland | CMY-4, TEM-1 | − | 20 |
| P. mirabilis PL 1376/01 | Poland | CMY-45, TEM-1 | − | 20 |
| P. mirabilis PL 1455/04 | Poland | CMY-38, TEM-2 | + | 20 |
| P. mirabilis PM91 | Greece | VEB-1, VIM-1 | − | 21 |
+, peak at m/z 39,850 observed; −, peak at m/z 39,850 not observed. Trc, transconjugant; NT, not tested.
Isolates were inoculated into 50 ml of Mueller-Hinton broth (Oxoid Ltd.) and incubated at 35°C for 16 h. Cultures were centrifuged at 5,000 × g for 20 min, and the cell pellet was used for the extraction of the periplasmic proteins, performed essentially as described by Naglak and Wang (22). Briefly, the pellet was resuspended in 360 μl of 40% sucrose and incubated for 2 h at 4°C. After centrifugation (5,000 × g, 5 min), the supernatant was discarded and the cell pellet was resuspended in 360 μl of ice-cold double-distilled water (ddH2O). After a 30-min incubation at 4°C, 40 μl of 1 M Tris-HCl buffer (pH 7.8) and 12 μl of lysozyme (10 mg/liter) were added to the suspension, which was then incubated for 90 min at 35°C. Spheroplasts were removed by centrifugation (14,000 × g, 5 min), leaving the periplasmic fraction in the supernatant.
A 200-μl volume of the periplasmic proteins was added to 1 ml of ice-cold ethanol (95%) supplemented with trifluoroacetic acid (TFA; 0.1%). After 20 min of incubation at −20°C, the solution was centrifuged at 14,000 × g for 20 min. The supernatant was removed, and the pellet was allowed to dry. The pellet was resuspended in 50 μl of TFA-acetonitrile-water (0.1:50:49.9 [volume fraction]), using a vortex device for 1 min, and centrifuged (14,000 × g, 2 min) to obtain the supernatant extract. Subsequently, 1 μl of each supernatant was applied on a stainless steel MALDI target plate (MSP 96 Target; Bruker Daltonics). After air drying, each sample was overlaid with 1 μl of matrix (sinapinic acid as a saturated solution in 50% ethanol). The matrix/sample spots were allowed to crystallize at room temperature. Each sample was spotted in triplicate. The MALDI-TOF mass spectra were obtained using a Microflex LT mass spectrometer with flexControl 3.3 software (Bruker Daltonics), operating in the positive linear ion mode within the m/z range 20,000 to 45,000. The parameters were set up as follows: ion source 1, 20 kV; ion source 2, 16.7 kV; lens, 7 kV; pulsed ion extraction, 170 ns; detection gain, 50×; electronic gain, enhanced (100 mV); sample rate, 2.0 GS/s; mass range selector, medium range; laser frequency, 30 Hz; digitizer trigger level, 2,500 mV; and laser range, 100%. Spectra were measured manually in at least 10 positions with 500 laser shots. Spectra were analyzed using flexAnalysis 3.0 software (Bruker Daltonics).
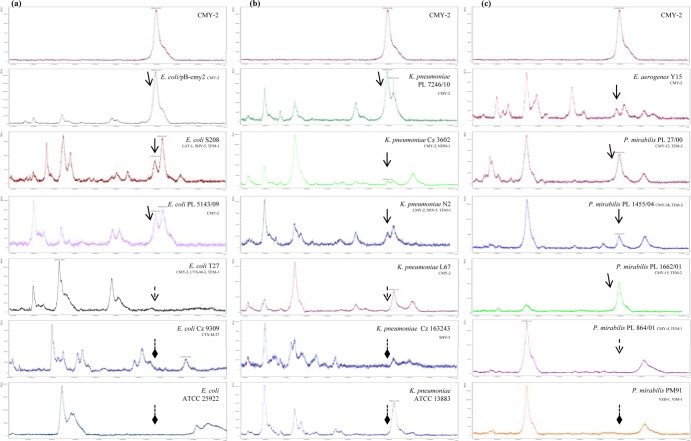
The MALDI-TOF MS measurement of the molecular mass of the purified CMY-2 detected one major peak with m/z of 39,852 (Fig. 1), slightly differing from the expected value for the mature CMY-2 protein (39,854 [23]). The presence of a peak with a m/z of ca. 39,850 was also observed in the mass spectrum of the CMY-2-producing E. coli DH5α pB-cmy2 transformant. In the mass spectra of the tested isolates, the ∼39,850-m/z peak was found in most of the E. coli isolates (11/12) and K. pneumoniae isolates (8/10) and in all Enterobacter aerogenes isolates (2/2) producing CMY-2-like enzymes (Table 1). Of six Proteus mirabilis isolates, the peak was identified in three. The latter isolates carried two copies of the blaCMY-2-like gene in their chromosomes, while the false-negative ones carried a single copy of the gene (20). The lack of the ∼39,850-m/z peak was observed for E. coli and K. pneumoniae ATCC strains and for all of the non-CMY-producing isolates. Mass spectra of representative isolates are shown in Fig. 1.
FIG 1.
Mass spectra of the purified CMY-2 enzyme and the periplasmic extracts of representative CMY- and non-CMY-producing E. coli (a), K. pneumoniae (b), and E. aerogenes and P. mirabilis (c) isolates. Peaks corresponding to CMY β-lactamases are indicated with arrows with solid lines. The absence of the ca. 39,850-m/z peaks, representing CMY β-lactamases, is indicated with arrows with dotted lines for CMY producers and diamond-shaped arrows with dotted lines for non-CMY-producing isolates.
The protein content of periplasmic extracts was characterized by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) (8). Protein bands of around 40,000 g/mol were detected in extracts of all the CMY-producing strains that were positive in the MALDI-TOF MS assay. These bands comigrated with the purified CMY-2 β-lactamase and were not found in the non-CMY-producing strains and the CMY producers that were negative in the MALDI-TOF MS assay. Identification of proteins observed in SDS-PAGE at approximately 40,000 g/mol was performed by in-gel tryptic digestion followed by MALDI-TOF/TOF MS (24). The identification of the ∼40,000-g/mol bands revealed multiple tryptic peptides, being fragments of CMY-2-like polypeptides, for all of the isolates that were positive in the MALDI-TOF MS assay (Table 2). Consistently, such peptides were not detected in extracts from the corresponding gel fragments for all the isolates that were negative by the MALDI-TOF MS assay. The absence of CMY-2-like tryptic peptides in the extracts of CMY producers that were negative in the MALDI-TOF MS assay might be explained by a low concentration of CMY-2-like enzymes in the periplasmic extracts of the respective isolates. However, these results suggested that the presence of the ∼39,850-m/z peak can be used as an indicator of the presence of the C. freundii-derived CMY-2-like group of acquired AmpC β-lactamases (10).
TABLE 2.
β-Lactamase peptides detected by in-gel tryptic and MALDI-TOF/TOF MS analysis
| Observed m/z value | Expected m/z value | Calculated m/z value | ppm | Amino acid sequencea | Amino acid positionb |
|---|---|---|---|---|---|
| 930.4312 | 929.4239 | 929.4032 | 22.4 | k-DYAWGYR-e | 217–225 |
| 1,285.7499 | 1,284.7426 | 1,284.7150 | 21.5 | k-TLQQGIALAQSR-y | 266–279 |
| 1,544.8279 | 1,543.8206 | 1,543.7895 | 20.1 | k-SYPNPVRVEAAWR-i | 362–376 |
| 1,556.8695 | 1,555.8622 | 1,555.8318 | 19.6 | -AAKTEQQIADIVNR-t | 21–35 |
| 1,658.8035 | 1,657.7962 | 1,657.7518 | 26.8 | r-WVQANMDASHVQEK-t | 252–267 |
| 1,664.9054 | 1,663.8981 | 1,663.8682 | 18.0 | k-LAHTWITVPQNEQK-d | 203–218 |
| 1,827.1019 | 1,826.0946 | 1,826.0665 | 15.4 | k-VALAALPAVEVNPPAPAVK-a | 310–330 |
| 2,081.1052 | 2,080.0979 | 2,080.0589 | 18.8 | r-EGKPVHVSPGQLDAEAYGVK-s | 224–245 |
Hyphens indicate tryptic restriction sites. Small capital letters correspond to amino acids found outside the restriction sites. Underlined residues were modified by oxidation.
CMY-2 peptide from K. pneumoniae HEL-1 (GenBank accession no. CAA62957) was used as a reference for the alignment of β-lactamase tryptic peptides.
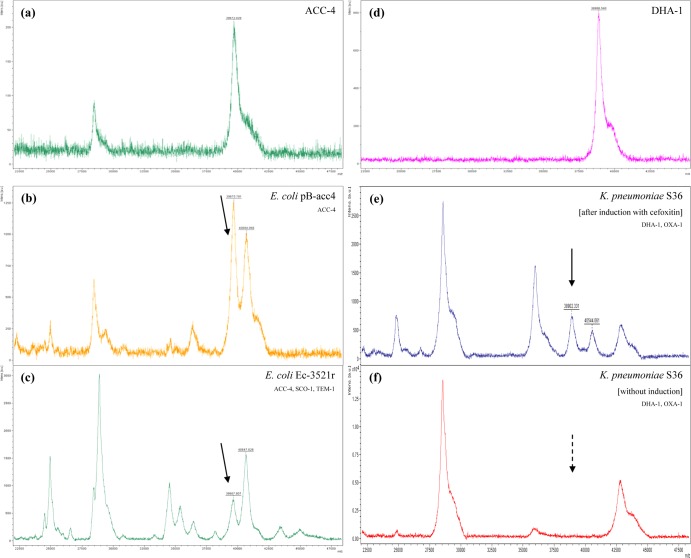
In the preliminary analysis of other AmpC-type β-lactamases, a ca. 39,670-m/z peak was observed in the mass spectra of the previously purified ACC-4 enzyme (theoretical relative molecular mass, 39,673 Da) and of the periplasmic extracts of the ACC-4-producing E. coli EC-3521r and pB-acc4/DH5α strains (Fig. 2) (25). The MALDI-TOF MS measurement of the molecular mass of the purified DHA-1 β-lactamase detected a 38,887-m/z peak (theoretical relative molecular mass, 38,881 Da). In the DHA-1-producing isolate K. pneumoniae S36 strain (26) with the functional ampC-ampR system (10), a corresponding peak of ca. 38,900-m/z was observed but only when the AmpC production was induced by adding cefoxitin at 50 μg/ml in broth cultures 3 h before harvesting the cells (Fig. 2). These data suggested that the assay can be used for the detection of other AmpC-type β-lactamases. Additionally, the observation of the 39,850-m/z, ∼39,670-m/z, and ∼38,900-m/z peaks for CMY-2-like, ACC-4, and DHA-1 enzymes, respectively, indicated that MALDI-TOF MS may discriminate the diverse groups of acquired AmpC-type cephalosporinases.
FIG 2.
(a to c) Mass spectra of the purified ACC-4 enzyme (a) and the periplasmic extracts of ACC-producing E. coli pB-acc4 (b) and EC-3521r (c). (d to f) Mass spectra of the purified DHA-1 enzyme (d) and the periplasmic extracts of DHA-producing K. pneumoniae S36 after induction with cefoxitin (e) and without induction (f). Peaks corresponding to β-lactamases are indicated with arrows with solid lines. In the mass spectra of K. pneumoniae S36, the dotted arrow indicates the absence of the ca. 38,900-m/z peak, representing DHA-1 β-lactamase, in periplasmic extract prepared without adding cefoxitin in broth culture.
In this study, we showed for the first time that MALDI-TOF MS has the potential to detect the most clinically important acquired AmpC β-lactamases, such as the CMY-2-like, ACC, and DHA types, in clinical isolates of Enterobacteriaceae. The described MALDI-TOF MS assay worked well with most of the CMY-producing isolates. However, the method performed poorly for P. mirabilis. It might be hypothesized that, in that case, increased production of a CMY-2-like enzyme upon gene duplication is important for the visualization of the ∼39,850-m/z peak.
In agreement with previous studies illustrating that MALDI-TOF MS applications are quick and cheap procedures (3), the described protocol exhibits an 22-h turnaround time, which is comparable to that of molecular techniques only if considering PCR plus sequencing of the amplicon in order to identify the specific allelic variant of the β-lactamase gene. The use of classic PCR and real-time PCR (RT-PCR) assays in clinical settings is more expensive than the use of the described MALDI-TOF (not considering the initial cost of investment for the equipment) but is less labor intensive and with a shorter turnaround time. Detection of β-lactamases by MALDI-TOF MS is a proteomic approach allowing the study of the behavior of the tested strains and should complement techniques already used for characterization of β-lactamases such as PCR and isoelectric focusing (IEF). The fact that MALDI-TOF MS can directly detect class A (9) and class C β-lactamases, as well as other mechanisms such as methylation of rRNA and cell wall components (3, 27), indicates the feasibility of establishing a MALDI-TOF supplementary database of resistance mechanisms that would promote research in this field. Notwithstanding the aforementioned problems, we strongly believe that proper modifications and validation of the described MALDI-TOF assay will easily lead to acceptance of its future application in diagnostic laboratories and reference centers.
ACKNOWLEDGMENTS
This work was supported by research project grants NT11032-6/2010 from the Ministry of Health of the Czech Republic and by the Charles University Research Fund (project number P36). C.C.P. was supported by the project. The establishment, development, and mobility of quality research teams at the Charles University, registration number CZ.1.07/2.3.00/30.0022, was financed by The Education for Competitiveness Operational Programme (ECOP) funded by the ESF and the government budget of the Czech Republic.
A patent application corresponding to this test has been sent on behalf of Charles University.
Footnotes
Published ahead of print 24 February 2014
REFERENCES
- 1.Seng P, Rolain JM, Fournier PE, La Scola B, Drancourt M, Raoult D. 2010. MALDI-TOF-mass spectrometry applications in clinical microbiology. Future Microbiol. 5:1733–1754. 10.2217/fmb.10.127 [DOI] [PubMed] [Google Scholar]
- 2.Wieser A, Schneider L, Jung J, Schubert S. 2012. MALDI-TOF MS in microbiological diagnostics-identification of microorganisms and beyond (mini review). Appl. Microbiol. Biotechnol. 93:965–974. 10.1007/s00253-011-3783-4 [DOI] [PubMed] [Google Scholar]
- 3.Hrabák J, Chudácková E, Walková R. 2013. Matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) mass spectrometry for detection of antibiotic resistance mechanisms: from research to routine diagnosis. Clin. Microbiol. Rev. 26:103–114. 10.1128/CMR.00058-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hrabák J, Walková R, Studentová V, Chudácková E, Bergerová T. 2011. Carbapenemase activity detection by matrix-assisted laser desorption ionization-time of flight mass spectrometry. J. Clin. Microbiol. 49:3222–3227. 10.1128/JCM.00984-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Burckhardt I, Zimmermann S. 2011. Using matrix-assisted laser desorption ionization-time of flight mass spectrometry to detect carbapenem resistance within 1 to 2.5 hours. J. Clin. Microbiol. 49:3321–3324. 10.1128/JCM.00287-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kempf M, Bakour S, Flaudrops C, Berrazeg M, Brunnel JM, Drissi M, Msli E, Touati A, Rolain JM. 2012. Rapid detection of carbapenem resistance in Acinetobacter baumannii using matrix-assisted laser desorption ionization-time of flight mass spectrometry. PLoS One 7:e31676. 10.1371/journal.pone.0031676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hrabák J, Studentová V, Walková R, Zemlicková H, Jakubu V, Chudácková E, Gniadkowski M, Pfeifer Y, Perry JD, Wilkinson K, Bergerová T. 2012. Detection of NDM-1, VIM-1, KPC, OXA-48, and OXA-162 carbapnemases by matrix-assisted laser desorption ionization-time of flight mass spectrometry. J. Clin. Microbiol. 50:2441–2443. 10.1128/JCM.01002-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Camara JE, Hays FA. 2007. Discrimination between wild-type and ampicillin-resistant Escherichia coli by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Anal. Bioanal. Chem. 389:1633–1638. 10.1007/s00216-007-1558-7 [DOI] [PubMed] [Google Scholar]
- 9.Schaumann R, Knoop N, Gnzel GH, Losensky K, Rosenkranz C, Stîngu CS, Schellenberger W, Rodloff AC, Eschrich K. 2012. A step towards the discrimination of beta-lactamase-producing clinical isolates of Enterobacteriaceae and Pseudomonas aeruginosa by MALDI-TOF mass spectrometry. Med. Sci. Monit. 18:MT1–MT77. 10.12659/MSM.883339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jacoby GA. 2009. AmpC beta-lactamases. Clin. Microbiol. Rev. 22:161–182. 10.1128/CMR.00036-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Empel J, Baraniak A, Literacka E, Mrówka A, Fiett J, Sadowy E, Hryniewicz W, Gniadkowski M; Beta-PL Study Group. 2008. Molecular survey of beta-lactamases conferring resistance to newer beta-lactams in Enterobacteriaceae isolates from Polish hospitals. Antimicrob. Agents Chemother. 52:2449–2454. 10.1128/AAC.00043-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pérez-Pérez FJ, Hanson ND. 2002. Detection of plasmid-mediated AmpC beta-lactamase genes in clinical isolates by using multiplex PCR. J. Clin. Microbiol. 40:2153–2162. 10.1128/JCM.40.6.2153-2162.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kotsakis SD, Papagiannitsis CC, Tzelepi EE, Tzouvelekis LS, Miriagou V. 2009. Extended-spectrum properties of CMY-30, a Val211Gly mutant of CMY-2 cephalosporinase. Antimicrob. Agents Chemother. 53:3520–3523. 10.1128/AAC.00219-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gazouli M, Tzouvelekis LS, Prinarakis E, Miriagou V, Tzelepi E. 1996. Transferable cefoxitin resistance in enterobacteria from Greek hospitals and characterization of a plasmid-mediated group 1 beta-lactamase (LAT-2). Antimicrob. Agents Chemother. 40:1736–1740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mavroidi A, Tzelepi E, Miriagou V, Gianneli D, Legakis NJ, Tzouvelekis LS. 2002. CTX-M-3 beta-lactamase-producing Escherichia coli from Greece. Microb. Drug Resist. 8:35–37. 10.1089/10766290252913737 [DOI] [PubMed] [Google Scholar]
- 16.Izdebksi R, Baraniak A, Fiett J, Adler A, Kazma M, Salomon J, Lawrence C, Rossini A, Salvia A, Hryniewicz W, Brun-Buisson C, Carmeli Y, Gniadkowski M; MOSAR WP2 and WP5 Study Groups. 2013. Clonal structure, extended-spectrum β-lactamases, and acquired AmpC-type cephalosporinase of Escherichia coli populations colonizing patients in rehabilitation centers in four countries. Antimicrob. Agents Chemother. 57:309–316. 10.1128/AAC.01656-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tzouvelekis LS, Tzelepi E, Mentis AF. 1994. Nucleotide sequence of a plasmid-mediated cephalosporinase gen (blaLAT-1) found in Klebsiella pneumoniae. Antimicrob. Agents Chemother. 38:2207–2209. 10.1128/AAC.38.9.2207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zioga A, Whichard JM, Kotsakis SD, Tzouvelekis Tzelepi LS E, Miriagou V. 2009. CMY-31 and CMY-36 cephalosporinases encoded by ColE1-like plasmids. Antimicrob. Agents Chemother. 53:1256–1259. 10.1128/AAC.01284-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Baraniak A, Izdebski R, Fiett J, Sadowy E, Adler A, Kazma M, Salomon J, Lawrence C, Rossini A, Salvia A, Vidal Samso J, Fierro J, Paul M, Lerman Y, Malhotra-Kumar S, Lammens C, Goossens H, Hryniewicz W, Brun-Buisson C, Carmeli Y, Gniadkowski M; MOSAR WP2 and WP5 Study Groups. 2013. Comparative population analysis of Klebsiella pneumoniae strains with extended-spectrum β-lactamases colonizing patients in rehabilitation centers in four countries. Antimicrob. Agents Chemother. 57:1992–1997. 10.1128/AAC.02571-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.D'Andrea MM, Leteracka E, Zioga A, Giani T, Baraniak A, Fiett Sadowy JE, Tassios PT, Rossolini GM, Gniadkowski M, Miriagou V. 2011. Evolution and spread of a multidrug-resistant Proteus mirabilis clone with chromosomal AmpC-type cephalosporinase in Europe. Antimicrob. Agents Chemother. 55:2735–2742. 10.1128/AAC.01736-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Papagiannitsis CC, Miriagou V, Kotsakis SD, Tzelepi E, Vatopoulos AC, Petinaki E, Tzouvelekis LS. 2012. Characterization of a transmissible plasmid encoding VEB-1 and VIM-1 in Proteus mirabilis. Antimicrob. Agents Chemother. 56:4024–4025. 10.1128/AAC.00470-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Naglak TJ, Wang HY. 1990. Recovery of a foreign protein from the periplasm of Escerichia coli by chemical permeabilization. Enzyme Microb. Technol. 12:603–611. 10.1016/0141-0229(90)90134-C [DOI] [PubMed] [Google Scholar]
- 23.Kotsakis SD, Caselli E, Tzouvelekis LS, Petinaki E, Prati F, Miriagou V. 2013. Interactions of oximino-substituted boronic acids and β-lactams with the CMY-2-derived extended-spectrum cephalosporinases CMY-30 and CMY-42. Antimicrob. Agents Chemother. 57:968–976. 10.1128/AAC.01620-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mares J, Richtrova P, Hricinova A, Tuma Z, Moravec J, Jysak D, Matejovic M. 2010. Proteomic profiling of blood-dialyzer interactome reveals involvement of lectin complement pathway in hemodialysis-induced inflammatory response. Proteomics Clin. Appl. 4:829–838. 10.1002/prca.201000031 [DOI] [PubMed] [Google Scholar]
- 25.Papagiannitsis CC, Tzouvelekis LS, Tzelepi E, Miriagou V. 2007. Plasmid encoded ACC-4, an extended-spectrum cephalosporinase variant from Escherichia coli. Antimicrob. Agents Chemother. 51:3763–3767. 10.1128/AAC.00389-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Empel J, Hrabak J, Kozinska A, Bergerova T, Ubraskova P, Kern-Zdanowicz I, Gniadkowski M. 2010. DHA-1-producing Klebsiella pneumoniae in a teaching hospital in the Czech Republic. Microb. Drug Resist. 16:291–295. 10.1089/mdr.2010.0030 [DOI] [PubMed] [Google Scholar]
- 27.Cai JC, Hu YY, Zhang R, Zhou HW, Chen GX. 2012. Detection of OMPK36 proin loss in Klebsiella spp. by matrix-assisted laser desorption ionization-time of flight mass spectrometry. J. Clin. Microbiol. 50:2179–2182. 10.1128/JCM.00503-12 [DOI] [PMC free article] [PubMed] [Google Scholar]