Abstract
We investigated a novel Japanese isolate of sequence type 11 (ST11), the Klebsiella pneumoniae carbapenemase-2 (KPC-2)-producing K. pneumoniae strain Kp3018, which was previously obtained from a patient treated at a Brazilian hospital. This strain was resistant to various antibiotic classes, including carbapenems, and harbored the gene blaKPC-2, which was present on the transferable plasmid of ca. 190 kb, in addition to the blaCTX-M-15 gene. Furthermore, the ca. 2.3-kb sequences (ISKpn8-blaKPC-2–ISKpn6-like), encompassing blaKPC-2, were found to be similar to those of K. pneumoniae strains from China.
TEXT
The increase in carbapenemase-producing Enterobacteriaceae is of serious concern globally, including in Japan, because these organisms are resistant to the carbapenems that are used to treat severe infections caused by multidrug-resistant bacteria (1–3). Ambler class A (Klebsiella pneumoniae carbapenemase [KPC]), class B (IMP, VIM, and NDM-1), and class D (OXA-48) β-lactamases are known as carbapenemases (1, 3). In particular, since KPC-producing K. pneumoniae isolates were first reported in the United States in 2001 (4), KPC-producing Enterobacteriaceae have been isolated as a cause of nosocomial and community-acquired infections in various countries (1, 3). However, the isolation of KPC-producing bacteria has not yet been described in Japan. In the present study, we investigated a novel Japanese isolate of KPC-2-producing K. pneumoniae strain Kp3018, isolated from a patient previously treated at a Brazilian hospital.
A 73-year-old Japanese man was admitted to a Brazilian hospital for a sudden onset of cerebral hemorrhage on 23 May 2012. On 4 July 2012, he was transferred to Tokyo and admitted to the Medical Hospital of Tokyo Medical and Dental University. Kp3018, a K. pneumoniae strain, which is resistant to carbapenems, was isolated from blood, stool, and decubitus ulcer samples from the patient. The MICs of antibiotics were determined using the MicroScan WalkAway system (Siemens Healthcare Diagnostics, Inc., Tokyo, Japan) and interpreted according to the Clinical and Laboratory Standards Institute (CLSI) guidelines (5). The MICs of cefotaxime and ceftazidime with clavulanic acid (4 μg/ml) and of imipenem with dipicolinic acid (400 μg/ml) were also determined by the broth microdilution method. Quality control for the MICs was performed using the reference strains Escherichia coli ATCC 25922 and K. pneumoniae ATCC 700603. Carbapenemase production was screened for by the modified Hodge test (5). The presence of blaTEM, blaSHV, blaCTX-M, and blaNDM-1 genes was determined using previously published methods (6, 7). Moreover, the presence of blaIMP, blaVIM, and blaKPC genes was screened for using previously published primer sets (8) as follows: 2 min of initial denaturation at 94°C, 30 cycles of PCR, each consisting of 30 s at 94°C and 45 s at 68°C, and 3 min of final extension at 72°C. Furthermore, the KPC type was identified using the primers KPC-F1 (5′-ATCGCCGTCTAGTTCTGCTG-3′) and KPC-R1 (5′-CCCTCGAGCGCGAGTCTA-3′), constructed using the KPC-2-producing K. pneumoniae sequence (GenBank accession no. AY034847). Additionally, the presence of plasmid-mediated ampC genes was investigated by multiplex PCR, as described previously (9). For molecular typing, we used multilocus sequence typing (MLST) (see http://www.pasteur.fr/recherche/genopole/PF8/mlst/Kpneumoniae.html).
Conjugation experiments were performed in LB broth with strain Kp3018 as a donor and the rifampin-resistant strain E. coli C600 as the recipient. Transconjugants were selected on Drigalski agar (bromothymol blue [BTB] agar) plates containing 80 μg/ml rifampin and 1 μg/ml imipenem (10). The MICs of the transconjugants were determined as described above. Plasmid DNA was extracted from donors and transconjugants using a NucleoBond Xtra midi (TaKaRa Bio, Shiga, Japan), according to the manufacturer's instructions, and was then analyzed by electrophoresis on a 0.7% (wt/vol) agarose gel. The size of transferred plasmids was estimated using a OneSTEP ladder supercoiled plasmid (Nippon Gene, Tokyo, Japan). Southern hybridization experiments were performed using a DIG-High Prime DNA labeling and detection starter kit I (Roche Diagnostics, Tokyo, Japan). Probes (850 bp) were generated by PCR using the primers KPC-F1 and KPC-R1. The genetic structures encompassing blaKPC in K. pneumoniae Kp3018 and its transconjugant were investigated by PCR mapping and subsequent sequencing, as described previously (11). A partial sequence (2,323 bp) encompassing blaKPC has been deposited in the DDBJ/EMBL/GenBank database under the accession no. AB854326.
In the present study, K. pneumoniae strain Kp3018 was found to be resistant to 11 antibiotics, including imipenem and meropenem, but not to amikacin and trimethoprim-sulfamethoxazole, as shown in Table 1. Furthermore, it phenotypically produced carbapenemases and harbored blaKPC-2, blaCTX-M-15, and blaSHV-11. In contrast, no other β-lactamase genes of classes A (blaTEM) and B (blaIMP, blaVIM, and blaNDM) were detected. These results are similar to those of previous studies, which demonstrated that KPC-producing isolates are resistant to not only all β-lactam antibiotics, including carbapenems, but also to some non-β-lactam antibiotics, such as aminoglycosides and fluoroquinolones (1, 3). Moreover, Kp3018 was grouped into sequence type 11 (ST11). These KPC-2-producing K. pneumoniae isolates of ST11 have been reported from various geographic regions, such as China (12), Greece (13), Poland (14), the United Kingdom (15), and Brazil (16, 17). Isolates producing blaCTX-M-15 and blaSHV-11 have also been reported in Singapore (18). Furthermore, the coproduction of KPC-2 and CTX-M-15 has been described in K. pneumoniae ST437 (clonal complex 11) isolated from patients with bloodstream infections in Brazil (19).
TABLE 1.
MICs of K. pneumoniae Kp3018, its transconjugant TcKp3018, and strain E. coli C600
| Antibiotic(s)a | MIC (μg/ml) against: |
||
|---|---|---|---|
| K. pneumoniae Kp3018 | E. coli TcKp3018 | E. coli C600 | |
| Imipenem | >8 | 4 | ≤1 |
| Imipenem + DPA | >8 | 4 | NDb |
| Meropenem | >8 | 4 | ≤1 |
| Ceftazidime | >16 | 16 | ≤1 |
| Ceftazidime + CLA | >4 | >4 | ND |
| Cefotaxime | >32 | >32 | ≤8 |
| Cefotaxime + CLA | 4 | 4 | ND |
| Aztreonam | >16 | >16 | ≤8 |
| Cefpirome | >16 | >16 | ≤8 |
| Cefmetazole | >32 | ≤4 | ≤4 |
| Piperacillin-tazobactam | >64 | >64 | ≤8 |
| Gentamicin | >8 | >8 | ≤1 |
| Amikacin | ≤4 | ≤4 | ≤4 |
| Levofloxacin | >4 | ≤0.5 | ≤0.5 |
| TMP-STX | ≤2 | ≤2 | ≤2 |
| Fosfomycin | >16 | ≤4 | ≤4 |
DPA, dipicolinic acid; CLA, clavulanic acid; TMP-STX, trimethoprim-sulfamethoxazole.
ND, not determined.
Plasmid analysis detected two plasmids of ca. 50 kb and ca. 190 kb in Kp3018 (Fig. 1A). Among these, the plasmid of ca. 190 kb was transferred successfully to E. coli C600 by conjugation. A transconjugant, E. coli C600 TcKp3018, showed a carbapenem-resistant phenotype and possessed blaKPC-2 in addition to blaCTX-M-15. Furthermore, Southern hybridization revealed that blaKPC-2 was present on the plasmid of ca. 190 kb in both Kp3018 and TcKp3018 (Fig. 1B). To investigate the sequence of the blaKPC-2 genetic environment, PCR mapping was performed using the previously published primers 816U and 4714 (11). The ca. 2.3-kb sequence (ISKpn8-blaKPC-2–ISKpn6-like) encompassing blaKPC-2 was analyzed and was found to be identical to that of plasmid pKP048 (GenBank accession no. FJ628167) in a K. pneumoniae isolate from China (20), except for a deletion of 11 bp among the 38-bp complete right invert repeat of Tn3 located between downstream of ISKpn8 and upstream of the blaKPC-2 gene. A previous study demonstrated that Tn4401 without ISKpn7 is present in KPC-2-producing K. pneumoniae isolates of ST11 in Brazil (16). Although we did not investigate the overall structure of the transmissible plasmid in Kp3018, we speculate that ISKpn8 was independently inserted downstream of the Tn3-based element by transposition. However, in this study, we cannot exclude the possibility that this KPC-2-positive strain of ST11 was acquired outside Brazil, since KPC-producing K. pneumoniae isolates with ISKpn8 have not yet been reported in Brazil.
FIG 1.
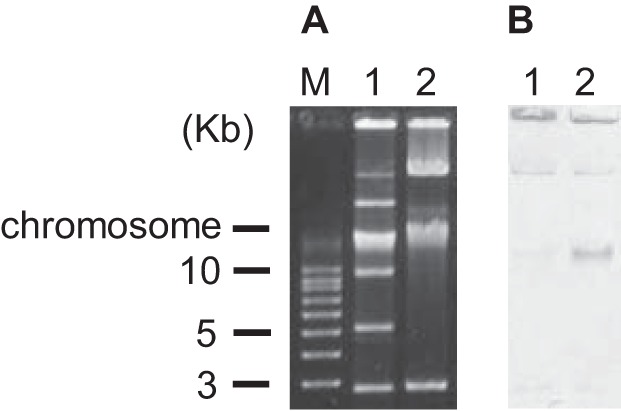
Plasmid profiles (A) and the results of Southern hybridization analysis carried out with a blaKPC-2 probe (850 bp) (B). Lane M, molecular weight; lane 1, K. pneumoniae strain Kp3018; lane 2, E. coli C600 transconjugant strain TcKp3018.
In conclusion, we identified a KPC-2-producing K. pneumoniae strain, Kp3018, for the first time in Japan and demonstrated that diversity of the blaKPC-2 genetic environment within the ca. 190-kb transferable plasmid was conferred by Tn3-based transposition. Since the dissemination of Enterobacteriaceae possessing carbapenemase activities, including KPC, pose a significant threat to the management of those infections worldwide, it will be important to continuously monitor the prevalence of carbapenemase-producing Enterobacteriaceae in Japan.
ACKNOWLEDGMENTS
We thank Makiko Kiyosuke and Dongchon Kang, who provided KPC-3-producing K. pneumoniae.
The study did not receive financial support from any third party. We declare no conflicts of interest.
Footnotes
Published ahead of print 24 February 2014
REFERENCES
- 1.Nordmann P, Poirel L. 2013. Strategies for identification of carbapenemase-producing Enterobacteriaceae. J. Antimicrob. Chemother. 68:487–489. 10.1093/jac/dks426 [DOI] [PubMed] [Google Scholar]
- 2.Koyano S, Saito R, Nagai R, Tatsuno K, Okugawa S, Okamura N, Moriya K. 2013. Molecular characterization of carbapenemase-producing clinical isolates of Enterobacteriaceae in a teaching hospital, Japan. J. Med. Microbiol. 62:446–450. 10.1099/jmm.0.050708-0 [DOI] [PubMed] [Google Scholar]
- 3.Tzouvelekis LS, Markogiannakis A, Psichogiou M, Tassios PT, Daikos GL. 2012. Carbapenemases in Klebsiella pneumoniae and other Enterobacteriaceae: an evolving crisis of global dimensions. Clin. Microbiol. Rev. 25:682–707. 10.1128/CMR.05035-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yigit H, Queenan AM, Anderson GJ, Domenech-Sanchez A, Biddle JW, Steward CD, Alberti S, Bush K, Tenover FC. 2001. Novel carbapenem-hydrolyzing beta-lactamase, KPC-1, from a carbapenem-resistant strain of Klebsiella pneumoniae. Antimicrob. Agents Chemother. 45:1151–1161. 10.1128/AAC.45.4.1151-1161.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.CLSI. 2011. Performance standards for antimicrobial susceptibility testing; 21st informational supplement. CLSI document M100-S21. Clinical and Laboratory Standards Institute, Wayne, PA [Google Scholar]
- 6.Ode T, Saito R, Kumita W, Sato K, Okugawa S, Moriya K, Koike K, Okamura N. 2009. Analysis of plasmid-mediated multidrug resistance in Escherichia coli and Klebsiella oxytoca isolates from clinical specimens in Japan. Int. J. Antimicrob. Agents 34:347–350. 10.1016/j.ijantimicag.2009.05.007 [DOI] [PubMed] [Google Scholar]
- 7.Wachino J, Yoshida H, Yamane K, Suzuki S, Matsui M, Yamagishi T, Tsutsui A, Konda T, Shibayama K, Arakawa Y. 2011. SMB-1, a novel subclass B3 metallo-beta-lactamase, associated with ISCR1 and a class 1 integron, from a carbapenem-resistant Serratia marcescens clinical isolate. Antimicrob. Agents Chemother. 55:5143–5149. 10.1128/AAC.05045-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dallenne C, Da Costa A, Decré D, Favier C, Arlet G. 2010. Development of a set of multiplex PCR assays for the detection of genes encoding important beta-lactamases in Enterobacteriaceae. J. Antimicrob. Chemother. 65:490–495. 10.1093/jac/dkp498 [DOI] [PubMed] [Google Scholar]
- 9.Perez-Perez FJ, Hanson ND. 2002. Detection of plasmid-mediated AmpC β-lactamase genes in clinical isolates by using multiplex PCR. J. Clin. Microbiol. 40:2153–2162. 10.1128/JCM.40.6.2153-2162.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Saito R, Kumita W, Sato K, Chida T, Okamura N, Moriya K, Koike K. 2007. Detection of plasmid-mediated quinolone resistance associated with qnrA in an Escherichia coli clinical isolate producing CTX-M-9 β-lactamase in Japan. Int. J. Antimicrob. Agents 29:600–602. 10.1016/j.ijantimicag.2006.11.016 [DOI] [PubMed] [Google Scholar]
- 11.Naas T, Cuzon G, Villegas MV, Lartigue MF, Quinn JP, Nordmann P. 2008. Genetic structures at the origin of acquisition of the β-lactamase blaKPC gene. Antimicrob. Agents Chemother. 52:1257–1263. 10.1128/AAC.01451-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Qi Y, Wei Z, Ji SJ, Du X, Shen P, Yu Y. 2011. ST11, the dominant clone of KPC-producing Klebsiella pneumoniae in China. J. Antimicrob. Chemother. 66:307–312. 10.1093/jac/dkq431 [DOI] [PubMed] [Google Scholar]
- 13.Giakkoupi P, Papagiannitsis CC, Miriagou V, Pappa O, Polemis M, Tryfinopoulou K, Tzouvelekis LS, Vatopoulos AC. 2011. An update of the evolving epidemic of blaKPC-2-carrying Klebsiella pneumoniae in Greece (2009–10). J. Antimicrob. Chemother. 66:1510–1513. 10.1093/jac/dkr166 [DOI] [PubMed] [Google Scholar]
- 14.Baraniak A, Grabowska A, Izdebski R, Fiett J, Herda M, Bojarska K, Żabicka D, Kania-Pudło M, Młynarczyk G, Żak-Puławska Z, Hryniewicz W, Gniadkowski M, KPC-PL Study Group 2011. Molecular characteristics of KPC-producing Enterobacteriaceae at the early stage of their dissemination in Poland, 2008–2009. Antimicrob. Agents Chemother. 55:5493–5499. 10.1128/AAC.05118-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Virgincar N, Iyer S, Stacey A, Maharjan S, Pike R, Perry C, Wyeth J, Woodford N. 2011. Klebsiella pneumoniae producing KPC carbapenemase in a district general hospital in the UK. J. Hosp. Infect. 78:293–296. 10.1016/j.jhin.2011.03.016 [DOI] [PubMed] [Google Scholar]
- 16.Pereira PS, de Araujo CF, Seki LM, Zahner V, Carvalho-Assef AP, Asensi MD. 2013. Update of the molecular epidemiology of KPC-2-producing Klebsiella pneumoniae in Brazil: spread of clonal complex 11 (ST11, ST437 and ST340). J. Antimicrob. Chemother. 68:312–316. 10.1093/jac/dks396 [DOI] [PubMed] [Google Scholar]
- 17.Andrade LN, Curiao T, Ferreira JC, Longo JM, Clímaco EC, Martinez R, Bellissimo-Rodrigues F, Basile-Filho A, Evaristo MA, Del Peloso PF, Ribeiro VB, Barth AL, Paula MC, Baquero F, Cantón R, Darini AL, Coque TM. 2011. Dissemination of blaKPC-2 by the spread of Klebsiella pneumoniae clonal complex 258 clones (ST258, ST11, ST437) and plasmids (IncFII, IncN, IncL/M) among Enterobacteriaceae species in Brazil. Antimicrob. Agents Chemother. 55:3579–3583. 10.1128/AAC.01783-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Balm MND, Ngan G, Jureen R, Lin RTP, Teo J. 2012. Molecular characterization of newly emerged blaKPC-2-producing Klebsiella pneumoniae in Singapore. J. Clin. Microbiol. 50:475–476. 10.1128/JCM.05914-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Seki LM, Pereira PS, de Souza Conceição M, Souza MJ, Marques EA, Carballido JM, de Carvalho MES, Assef APDC, Asensi MD. 2013. Molecular epidemiology of CTX-M producing Enterobacteriaceae isolated from bloodstream infections in Rio de Janeiro, Brazil: emergence of CTX-M-15. Braz. J. Infect. Dis. 17:640–646. 10.1016/j.bjid.2013.03.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shen P, Wei Z, Jiang Y, Du X, Ji S, Yu Y, Li L. 2009. Novel genetic environment of the carbapenem-hydrolyzing beta-lactamase KPC-2 among Enterobacteriaceae in China. Antimicrob. Agents Chemother. 53:4333–4338. 10.1128/AAC.00260-09 [DOI] [PMC free article] [PubMed] [Google Scholar]


