Abstract
Moxifloxacin-resistant Mycobacterium tuberculosis mutants were selected in vitro using different concentrations of moxifloxacin. gyrA mutations at codons 88 and 94 were associated with resistance (defined as an MIC of ≥2 μg/ml) (P < 0.0001 and P = 0.0053, respectively). Despite the presence of gyrA mutations, moxifloxacin significantly impedes bacterial growth, supporting its use for the treatment of ofloxacin-resistant M. tuberculosis.
TEXT
Moxifloxacin, a broad-spectrum fluoroquinolone (1), is significantly more potent than earlier narrow-spectrum fluoroquinolones, such as ofloxacin, which has been widely used in the treatment of tuberculosis (TB) (2). Moxifloxacin shows improved in vitro activity against ofloxacin-susceptible Mycobacterium tuberculosis (3). Cross-resistance exists between fluoroquinolones, and mutations that emerge under ofloxacin treatment are usually associated with moxifloxacin MICs above the WHO-defined critical concentration of 0.25 μg/ml (4). However, 84 to 98% of isolates with mutations conferring fluoroquinolone resistance have MICs of ≤2 μg/ml for moxifloxacin, which does not necessarily reflect clinical resistance (3, 5–9). Based on this, an alternative critical concentration of 2 μg/ml has been suggested (5, 6, 10).
The WHO has recommended substituting moxifloxacin for ofloxacin in the treatment of multidrug-resistant TB (MDR-TB), defined as being resistant to at least isoniazid and rifampin, and extensively drug-resistant TB (XDR-TB), defined as MDR with additional resistance to a fluoroquinolone and an injectable antibiotic (6, 11). Moxifloxacin as a first-line drug to treat susceptible TB is also currently undergoing trials (12, 13) and has shown promise to shorten treatment duration (13). Since the use of moxifloxacin for the treatment of TB is not yet widespread, it is not clear which mutations could emerge. Mutations that facilitate increased moxifloxacin resistance (MICs of ≥2 μg/ml) have clinical relevance, given that the current standard dose leads to serum concentrations of 1.3 to 6.9 μg/ml (5, 6, 10, 14, 15). Knowledge of such mutations is critical for optimizing rapid molecular methods for detecting moxifloxacin resistance. This study therefore aimed to identify spontaneous moxifloxacin-resistant mutants selected in the presence of ≥2 μg/ml moxifloxacin.
A pansusceptible M. tuberculosis Beijing genotype strain was used as a progenitor for the selection of a wide range of independent mutants according to Morlock et al. (16). Briefly, four sets of 30 to 35 low-inoculum parallel cultures were set up in 5 ml of enriched 7H9 medium (supplemented with albumin-dextrose-catalase, 0.2% glycerol, and 0.05% Tween 80) and cultured at 37°C for 28 days. Subsequently, the entire culture was plated on 7H10 medium (supplemented with albumin-dextrose-catalase and 0.5% glycerol) containing 3 or 4 μg/ml moxifloxacin, while approximately 10% of the culture was plated onto 7H10 medium containing 0.5 or 2 μg/ml moxifloxacin. Where possible, two colonies were picked per plate and subcultured in 5 ml enriched 7H9 medium. The gyrA gene was subsequently amplified and sequenced from boiled aliquots of the respective cultures (6). For isolates with a wild-type gyrA gene, the gyrB gene was amplified and sequenced using the primers 5′-GTATCGCGGCACGTAAGG-3′ and 5′-CCACTTGAGTTTGTACAGC-3′ using a protocol similar to that for gyrA (6), except that the annealing temperature was adjusted to 53°C.
For 0.5, 2, 3, and 4 μg/ml moxifloxacin, 56, 55, 70, and 69 colonies, respectively, were selected and analyzed. An analysis of the sequencing results identified 9 gyrA and 4 gyrB mutations (Table 1). Two novel mutations, gyrA Ala90Lys (GCG/AAG) and Asp94Lys (GAC/AAA), were identified. Isolates harboring mutations in codons 89 and 90 of gyrA or any codon of gyrB were selected only on medium that contained 0.5 μg/ml of moxifloxacin. gyrA Gly88Cys and codon 94 mutations were associated with exposure to medium containing ≥2 μg/ml moxifloxacin (P < 0.0001 and P = 0.0053, respectively). The mutations were similar to those observed in a previous study (17). The proportion of clones with the Gly88Cys mutation increased as the concentration of moxifloxacin increased. The association of this mutation with resistance to newer fluoroquinolones was also shown in work on DC-159a (18).
TABLE 1.
Comparison between mutants selected on 0.5 μg/ml versus ≥2 μg/ml moxifloxacin in vitro
| Mutation |
Data for moxifloxacin at concn (μg/ml) of: |
|||||||
|---|---|---|---|---|---|---|---|---|
| Gene | Amino acid change | 0.5 |
2 |
3 |
4 |
|||
| No. (%)a | No. (%) | Pb | No. (%) | P | No. (%) | P | ||
| gyrA | Gly88Cys | 0 (0.0) | 32 (58.2) | <0.0001 | 48 (68.6) | <0.0001 | 61 (88.4) | <0.0001 |
| gyrA | Asp89Asn | 1 (1.8) | 0 (0.0) | 1 | 0 (0.0) | 0.4444 | 0 (0.0) | 0.4480 |
| gyrA | Ala90Lys | 1 (1.8) | 0 (0.0) | 1 | 0 (0.0) | 0.4444 | 0 (0.0) | 0.4480 |
| gyrA | Ala90Val | 1 (1.8) | 0 (0.0) | 1 | 0 (0.0) | 0.4444 | 0 (0.0) | 0.4480 |
| gyrA | Asp94Asn | 5 (8.9) | 10 (18.2) | 0.1757 | 14 (20.0) | 0.1315 | 7 (10.1) | 1 |
| gyrA | Asp94Gly | 5 (8.9) | 3 (5.5) | 0.7163 | 4 (5.7) | 0.5093 | 0 (0.0) | 0.0163 |
| gyrA | Asp94His | 3 (5.4) | 1 (1.8) | 0.6182 | 0 (0.0) | 0.0852 | 0 (0.0) | 0.0872 |
| gyrA | Asp94Lys | 0 (0.0) | 0 (0.0) | NAc | 0 (0.0) | NA | 1 (1.4) | 1 |
| gyrA | Asp94Tyr | 14 (25) | 9 (16.4) | 0.3497 | 4 (5.7) | 0.0038 | 0 (0.0) | <0.0001 |
| gyrB | Asn499Asp | 2 (3.6) | 0 (0.0) | 0.4955 | 0 (0.0) | 0.1956 | 0 (0.0) | 0.1987 |
| gyrBd | Asn499Lys | 17 (30.4) | 0 (0.0) | <0.0001 | 0 (0.0) | <0.0001 | 0 (0.0) | <0.0001 |
| gyrB | Thr500Asn | 6 (10.7) | 0 (0.0) | 0.0271 | 0 (0.0) | 0.0066 | 0 (0.0) | 0.0069 |
| gyrB | Glu501Asp | 1 (1.8) | 0 (0.0) | 1 | 0 (0.0) | 0.4444 | 0 (0.0) | 0.4480 |
Percentage of all mutants selected at the indicated moxifloxacin concentration.
Significance of proportion compared to that at 0.5 μg/ml by Fisher's exact test; values in bold type are significant (P < 0.05).
NA, not applicable.
Numbering for gyrB according to Maruri et al. (22).
In our study, the clones with the Gly88Cys mutation were not observed at 0.5 μg/ml moxifloxacin, suggesting a high fitness cost associated with this mutation. Conversely, the clones with codon 94 mutations were observed with all moxifloxacin concentrations tested, suggesting a low fitness cost with these mutations. This may explain why the codon 94 mutations but not Gly88Cys mutations were selected during moxifloxacin treatment of mice (14, 17) or emerged after treatment of patients with newer fluoroquinolones (which included moxifloxacin) prior to diagnosis of TB (19). Interestingly, the gyrA Asp94Asn mutation emerged in mice treated with moxifloxacin at a dose that produced similar pharmacodynamics/pharmacokinetics in patients (14), while in our study, this mutation was observed at equal proportions across all concentrations.
As a next step, we compared the mutation rate of moxifloxacin to that of ofloxacin. A fluctuation assay was set up according to Morlock et al. (16). Briefly, two sets of 15 parallel 7H9 cultures of the Beijing genotype strain were grown for 28 days at 37°C, and for 10 of these, the entire culture was plated on 7H10 medium containing 2 μg/ml of either ofloxacin or moxifloxacin. The average final population size was determined using the other 5 cultures after plating a 10-fold dilution series in 0.5% Tween 80 on 7H10 medium without antibiotics. The mutation rate was calculated using the Ma-Sandri-Sarkar maximum likelihood method (20). The spontaneous mutation rates for moxifloxacin and ofloxacin resistance were within the same logarithmic range (2.3 and 1.1 per 108 bacteria, respectively). This suggests that resistance to new fluoroquinolones is attained as easily as resistance to older fluoroquinolones.
However, in the first experiment (see Table 1), colonies with a reduced diameter were observed on solid medium containing 2, 3, or 4 μg/ml moxifloxacin compared with 0.5 μg/ml, suggesting that gyrA mutants are partially susceptible to ≥2 μg/ml moxifloxacin. To further investigate this, we monitored the growth rates of isolates harboring gyrA Gly88Cys, Asp94Asn, Asp94Tyr, Asp94His, and Asp94Gly mutations, cultured in stationary cultures containing enriched 7H9 and in the absence or in the presence of 2 μg/ml ofloxacin or moxifloxacin. We also monitored the growth of the progenitor strain without antibiotics. The mutants were selected randomly. For each isolate, a frozen 1-ml stock was inoculated into 9 ml enriched 7H9 medium. When growth reached an optical density (OD) of 0.8 to 1.0, the starter cultures were diluted to an OD of 0.05 in a total of 18 ml enriched 7H9 medium with or without the respective antibiotics. The OD of each culture was measured daily at 600 nm using a visible-light spectrophotometer. Each experiment was done in triplicate. Figure 1 shows that the growth of all mutants was significantly impeded by moxifloxacin but not by ofloxacin (P < 10−5), according to the double repeated-measures analysis of variance statistical test (Fig. 1A to E). The isolate with a Gly88Cys mutation was the least susceptible to moxifloxacin (Fig. 1A), followed by the isolate with an Asp94Asn mutation in gyrA (Fig. 1C). The remaining three mutants were almost completely inhibited.
FIG 1.
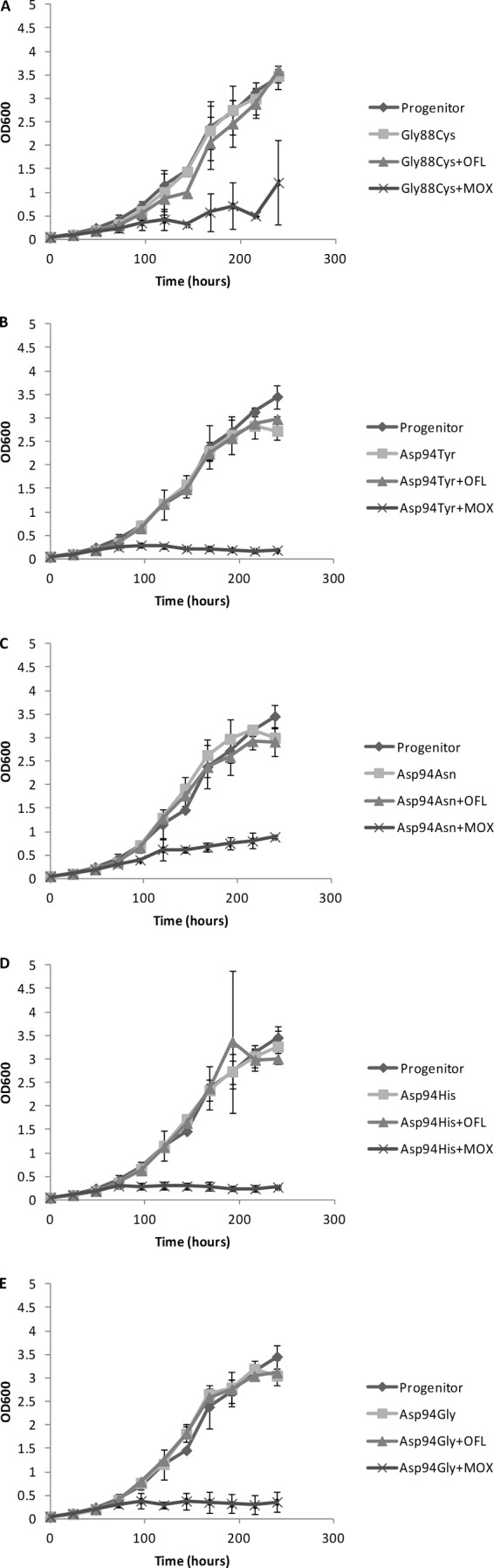
Comparison of growth of fluoroquinolone-resistant M. tuberculosis gyrA mutants and their sensitive progenitor in the presence and in the absence of antibiotics. (A) Gly88Cys; (B) Asp94Tyr; (C) Asp94Asn; (D) Asp94His; (E) Asp94Gly. For each culture, the optical density at 600 nm was measured daily in three independent experiments, and the average was plotted. MOX, moxifloxacin; OFL, ofloxacin. Error bars indicate standard deviations.
In conclusion, gyrA Gly88Cys and Asp94Gly/His/Tyr/Asn/Lys mutations are associated with high-level moxifloxacin resistance in vitro and may confer reduced susceptibility to moxifloxacin in patients treated with this drug. Rapid molecular drug resistance testing should therefore include these mutations. Encouragingly, however, the reduced in vitro growth rates of mutants in 2 μg/ml moxifloxacin suggest that the growth of these mutants in patients is likely to be suppressed at attainable moxifloxacin serum concentrations (5, 6, 10, 14, 15). This supports a previous meta-analysis, which showed that moxifloxacin was still effective for the treatment of XDR-TB despite the presence of fluoroquinolone resistance (21).
ACKNOWLEDGMENTS
Marieta McGrath was funded by the National Research Foundation and the Ernst and Ethel Eriksen Trust.
We thank Samantha Sampson for valuable input into the manuscript and Justin Harvey for statistical analysis.
Footnotes
Published ahead of print 10 February 2014
REFERENCES
- 1.World Health Organization. 2011. Guidelines for the programmatic management of drug-resistant tuberculosis—2011 update. WHO Press, Geneva, Switzerland: [DOI] [PubMed] [Google Scholar]
- 2.World Health Organization. 2010. Treatment of tuberculosis: guidelines, 4th ed. WHO Press, Geneva, Switzerland [Google Scholar]
- 3.Angeby KA, Jureen P, Giske CG, Chryssanthou E, Sturegård E, Nordvall M, Johansson AG, Werngren J, Kahlmeter G, Hoffner SE, Schön T. 2010. Wild-type MIC distributions of four fluoroquinolones active against Mycobacterium tuberculosis in relation to current critical concentrations and available pharmacokinetic and pharmacodynamic data. J. Antimicrob. Chemother. 65:946–952. 10.1093/jac/dkq091 [DOI] [PubMed] [Google Scholar]
- 4.World Health Organization. 2008. Policy guidance on drug-susceptibility testing (DST) of second-line antituberculosis drugs. WHO Press, Geneva, Switzerland: [PubMed] [Google Scholar]
- 5.Kam KM, Yip CW, Cheung TL, Tang HS, Leung OC, Chan MY. 2006. Stepwise decrease in moxifloxacin susceptibility amongst clinical isolates of multidrug-resistant Mycobacterium tuberculosis: correlation with ofloxacin susceptibility. Microb. Drug Resist. 12:7–11. 10.1089/mdr.2006.12.7 [DOI] [PubMed] [Google Scholar]
- 6.Sirgel FA, Warren RM, Streicher EM, Victor TC, van Helden PD, Böttger EC. 2012. gyrA mutations and phenotypic susceptibility levels to ofloxacin and moxifloxacin in clinical isolates of Mycobacterium tuberculosis. J. Antimicrob. Chemother. 67:1088–1093. 10.1093/jac/dks033 [DOI] [PubMed] [Google Scholar]
- 7.Cheng AFB, Yew WW, Chan EWC, Chin ML, Hui MMM, Chan RCY. 2004. Multiplex PCR amplimer conformation analysis for rapid detection of gyrA mutations in fluoroquinolone-resistant Mycobacterium tuberculosis clinical isolates. Antimicrob. Agents Chemother. 48:596–601. 10.1128/AAC.48.2.596-601.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Von Groll A, Martin A, Jureen P, Hoffner S, Vandamme P, Portaels F, Palomino JC, Da Silva PA. 2009. Fluoroquinolone resistance in Mycobacterium tuberculosis and mutations in gyrA and gyrB. Antimicrob. Agents Chemother. 53:4498–4500. 10.1128/AAC.00287-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nosova EY, Bukatina AA, Isaeva YD, Makarova MV, Galkina KY, Moroz AM. 2013. Analysis of mutations in the gyrA and gyrB genes and their association with the resistance of Mycobacterium tuberculosis to levofloxacin, moxifloxacin and gatifloxacin. J. Med. Microbiol. 62:108–113. 10.1099/jmm.0.046821-0 [DOI] [PubMed] [Google Scholar]
- 10.Poissy J, Aubry A, Fernandez C, Lott M-C, Chauffour A, Jarlier V, Farinotti R, Veziris N. 2010. Should moxifloxacin be used for the treatment of extensively drug-resistant tuberculosis? An answer from a murine model. Antimicrob. Agents Chemother. 54:4765–4771. 10.1128/AAC.00968-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.World Health Organization. 2008. Guidelines for the programmatic management of drug-resistant tuberculosis—emergency update 2008. WHO Press, Geneva, Switzerland [Google Scholar]
- 12.Clayden P, Collins S, Daniels C, Geffen N, Harrington M, Jefferys R, Jervis C, Kaplan K, Lessem E, Swan T. 2012. 2012 pipeline report: HIV, hepatitis C virus (HCV), and tuberculosis (TB) drugs, diagnostics, vaccines, and preventive technologies in development. i-BASE/Treatment Action Group, New York, NY. [Google Scholar]
- 13.Conde MB, Efron A, Loredo C, De Souza GRM, Graça NP, Cezar MC, Ram M, Chaudhary MA, Bishai WR, Kritski AL, Chaisson RE. 2009. Moxifloxacin versus ethambutol in the initial treatment of tuberculosis: a double-blind, randomised, controlled phase II trial. Lancet 373:1183–1189. 10.1016/S0140-6736(09)60333-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Almeida D, Nuermberger E, Tyagi S, Bishai WR, Grosset J. 2007. In vivo validation of the mutant selection window hypothesis with moxifloxacin in a murine model of tuberculosis. Antimicrob. Agents Chemother. 51:4261–4266. 10.1128/AAC.01123-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Manika K, Chatzika K, Zarogoulidis K, Kioumis I. 2012. Moxifloxacin in multidrug-resistant tuberculosis: is there any indication for therapeutic drug monitoring? Eur. Respir. J. 40:1051–1053. 10.1183/09031936.00202411 [DOI] [PubMed] [Google Scholar]
- 16.Morlock GP, Plikaytis BB, Crawford JT. 2000. Characterization of spontaneous, in vitro-selected, rifampin-resistant mutants of Mycobacterium tuberculosis strain H37Rv. Antimicrob. Agents Chemother. 44:3298–3301. 10.1128/AAC.44.12.3298-3301.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ginsburg AS, Sun R, Calamita H, Scott CP, Bishai WR, Grosset JH. 2005. Emergence of fluoroquinolone resistance in Mycobacterium tuberculosis during continuously dosed moxifloxacin monotherapy in a mouse model. Antimicrob. Agents Chemother. 49:3977–3979. 10.1128/AAC.49.9.3977-3979.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sekiguchi J, Disratthakit A, Maeda S, Doi N. 2011. Characteristic resistance mechanism of Mycobacterium tuberculosis to DC-159a, a new respiratory quinolone. Antimicrob. Agents Chemother. 55:3958-3960. 10.1128/AAC.00417-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Van der Heijden YF, Maruri F, Blackman A, Mitchel E, Bian A, Shintani AK, Eden S, Warkentin JV, Sterling TR. 2013. Fluoroquinolone susceptibility in Mycobacterium tuberculosis after prediagnosis exposure to older-versus newer-generation fluoroquinolones. Int. J. Antimicrob. Agents 42:232–237. 10.1016/j.ijantimicag.2013.04.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hall BM, Ma C-X, Liang P, Singh KK. 2009. Fluctuation analysis CalculatOR: a web tool for the determination of mutation rate using Luria-Delbruck fluctuation analysis. Bioinformatics 25:1564–1565. 10.1093/bioinformatics/btp253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jacobson KR, Tierney DB, Jeon CY, Mitnick CD, Murray MB. 2010. Treatment outcomes among patients with extensively drug-resistant tuberculosis: systematic review and meta-analysis. Clin. Infect. Dis. 51:6–14. 10.1086/653115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maruri F, Sterling TR, Kaiga AW, Blackman A, van der Heijden YF, Mayer C, Cambau E, Aubry A. 2012. A systematic review of gyrase mutations associated with fluoroquinolone-resistant Mycobacterium tuberculosis and a proposed gyrase numbering system. J. Antimicrob. Chemother. 67:819–831. 10.1093/jac/dkr566 [DOI] [PMC free article] [PubMed] [Google Scholar]


