Abstract
Coprolites are fossilized fecal material that can reveal information about ancient intestinal and environmental microbiota. Viral metagenomics has allowed systematic characterization of viral diversity in environmental and human-associated specimens, but little is known about the viral diversity in fossil remains. Here, we analyzed the viral community of a 14th-century coprolite from a closed barrel in a Middle Ages site in Belgium using electron microscopy and metagenomics. Viruses that infect eukaryotes, bacteria, and archaea were detected, and we confirmed the presence of some of them by ad hoc suicide PCR. The coprolite DNA viral metagenome was dominated by sequences showing homologies to phages commonly found in modern stools and soil. Although their phylogenetic compositions differed, the metabolic functions of the viral communities have remained conserved across centuries. Antibiotic resistance was one of the reconstructed metabolic functions detected.
INTRODUCTION
Viral metagenomics is a sequencing-based analysis of all of viral genomes isolated from a sample. It has promoted the characterization of viral community diversity. Viral metagenomics has already been successfully applied to the exploration of modern environmental specimens sampled from marine water, freshwater, stromatolites and thrombolites, and soil (1–4) and to modern human-associated specimens collected from the liver, blood, nasopharyngeal aspirates, and stool (5–9). The DNA viromes generated from modern stools have been demonstrated to be dominated by bacteriophages (10, 11) and to be less diverse than environmental samples (8, 12).
Viral metagenomics does not require culturing viruses or a priori knowledge of the sequences that will be targeted, which allows for the identification of new, unknown or unexpected viruses and for the global assessment of the virome. Viral metagenomics is thus particularly suitable for paleomicrobiological studies, as little is known about which viruses are characteristic of ancient specimens. Indeed, the majority of ancient DNA (aDNA) studies are based on the analysis of human and bacterial aDNA (13–15), and viral persistence and its detectability in ancient specimens remain unclear. Electron microscopy has previously revealed that viral particles can persist for over 400 years, but their viability was lost (16). Moreover, PCR amplifications yielded positive results for viral aDNA in ancient specimens, such as mummified soft tissues, bones, and teeth. The amplification products varied between 100 and 570 bp in size, which indicated that viral aDNA can be detected for at least 1,500 years (17–20).
Here, we used electron microscopy and, for what may be the first time, viral metagenomics to characterize the viral community of an ancient stool specimen. A viral DNA metagenome was generated from a 14th-century coprolite sample that was recovered from a Middle Ages site in Namur, Belgium.
MATERIALS AND METHODS
VLP isolation, TEM, and DNA extraction.
First, 5.8 g of the interior of the coprolite was aseptically removed and solubilized overnight at 4°C under continuous rotation in 40 ml of phosphate-buffered saline (PBS), pH 7.4 (bioMérieux, Marcy l'Etoile, France), which had previously been passed through a 0.02-μm-pore filter. The coprolite solution was centrifuged for 10 min at 500 × g, and then the upper layer was removed and filtered in stages using sterile Whatman filters (pore sizes of 0.8, 0.45, and 0.22 μm [Whatman division of GE Healthcare, Dassel, Germany]). Twenty-five milliliters of the coprolite filtrate was used to precipitate and purify viral particles onto a cesium chloride density gradient using ultracentrifugation, and DNase treatment was then performed (21). A 40-μl aliquot of the purified viral particles was stained with 1.5% ammonium molybdate (Euromedex) and observed by transmission electron microscopy (TEM) using a Philips Morgagni 268D electron microscope (FEI Co., Eindhoven, Netherlands). To isolate the nucleic acids from the purified viral particles, the formamide procedure previously described by Thurber et al. (21) was used. A standard 18S ribosomal DNA (rDNA) PCR was performed to verify the absence of human DNA contamination.
Viral metagenomic library preparation and sequencing.
Nucleic acids were amplified in duplicate reactions using the Illustra GenomiPhi V2 DNA amplification kit (GE Healthcare Life Sciences, Freiburg, Germany). Amplification products were pooled and ethanol purified.
A shotgun strategy was chosen for high-throughput pyrosequencing on a 454 Life Sciences Genome FLX sequencer using titanium chemistry (Genome Sequencer RLX; Roche). Sequencing was performed using 1/16 of a picotiter plate.
Preprocessing of sequencing data.
The reads were screened for quality using mothur (22). Only reads longer than 50 bp and with an average quality score greater than 21 were kept. Reads with more than two ambiguous base calls and/or reads with homopolymers longer than 10 bases were eliminated. Identical sequences artificially generated by the pyrosequencing technology were also excluded using the “unique.seqs” mothur command. The preprocessed viral metagenome is publicly available on the Metavir server (http://metavir-meb.univ-bpclermont.fr) with the identifier “NAMUR_viral” under the project “HumanCoprolite” and on the NCBI Sequence Read Archive (http://www.ncbi.nlm.nih.gov/sra) under accession no. SRP033437.
Annotation of reads.
A BLASTN search against the nonredundant NCBI database (E value, <1e−5) was performed. Reads with no significant similarity to sequences stored in the NCBI database were classified as “unknown reads.” The virome taxonomic composition was estimated using GAAS (23), which is based on a BLASTX search against the RefSeq Viral Genomes database (E value, <1e−5) and normalizes the number of reads matching each viral genotype by the length of the genome.
Functional annotation was performed on the MG-RAST server (24) using the nonredundant SEED database (E value, <1e−5). A stringent search of virulence factors was also performed using BLASTX on the Virulence Factor Database (25), with 60% as the minimum identity and a cutoff E value of <1e−5.
Assembly and contig annotation.
The reads were assembled into contigs using the Newbler de novo assembler (Roche) with at least 98% identity and 35 bp of overlap. Only contigs longer than 400 bp were used in subsequent analyses.
Known and unknown contigs were identified on the basis of the BLASTN search against the nonredundant NCBI database (E value, <1e−5). The taxonomic and functional contig classification was based on a BLASTX search against the nonredundant NCBI database (E value, <1−5). A specific search for contigs encoding antibiotic resistance genes was also performed using BLAST on the ARDB (Antibiotic Resistance Genes Database) with an E value of <1e−5 (26). Significant hits were manually verified.
Phylogenetic trees.
When possible, phylogenetic trees of the contigs encoding antibiotic resistance genes were built. The program Prodigal was used to search for open reading frames (ORFs) in these contigs (27). Homologs to the translated ORFs were searched against the nonredundant NCBI database using BLASTP. A multiple alignment was constructed using MUSCLE (28) and curated using Gblocks (29). The phylogenetic tree was then built using the PhyML algorithm (30) with a bootstrap value of 100. These tasks were all performed using the pipeline freely available at www.phylogeny.fr (31). The trees were visualized using MEGA v.4 (32).
Evidence of temperate bacteriophages.
Contigs generated from the assembly were analyzed to search for indicators of temperate bacteriophages, as previously described (33). We searched for three indicators: (i) nucleotide identity to bacterial genomes (BLASTN; E value, <1e−5; 90% minimum identity; 90% minimum query coverage), (ii) the presence of integrase-encoding genes using annotations from the COG and PFAM databases (E value, <1e−5), and (iii) significant similarity to prophage proteins available on the ACLAME database (BLASTX on the ACLAME prophages database; E value, <1e−5). Data were graphically represented using the R package “VennDiagram.”
Comparative metagenomics.
The coprolite-associated virome was taxonomically and functionally compared to 21 published viromes of modern stools from healthy adult humans (12, 33), which had been generated using multiple-displacement amplification (MDA), as was the case with the coprolite virome. All viromes were taxonomically (GenBank database; E value, <1e−5) and functionally annotated (SEED database; E value, <1e−5) using MG-RAST. Annotations were performed on reads using amino-acid-level comparisons. The taxonomic and functional virome profiles were compared using principal component analysis on the MG-RAST server (normalized data, Bray-Curtis measure of distance). Species richness estimations were obtained from the MG-RAST server. Functional diversity (measured by the Shannon-Wiener index) was calculated using the “estimateDiversity” function of the ShotGunFunctionalizeR package on the SEED-based functional metagenome annotations (E value, <1e−5) (34).
Specific PCR amplifications and sequencing.
Suicide PCR amplifications were performed to confirm high-throughput pyrosequencing results. To perform suicide PCR, the primer pairs were used only once in working areas, and no positive controls were used (35). For giant virus detection, primer pairs targeting the nonfunctional B-family DNA polymerase were used. Additional primer pairs were designed to specifically target ORFs identified in some viral contigs assembled de novo from the virome and matching cyanophages, Mycobacterium phages, Bacillus phages, Burkholderia phages, Celeribacter phages, and Clostridium phages (see Table S1 and Section 4 in the supplemental material).
RESULTS
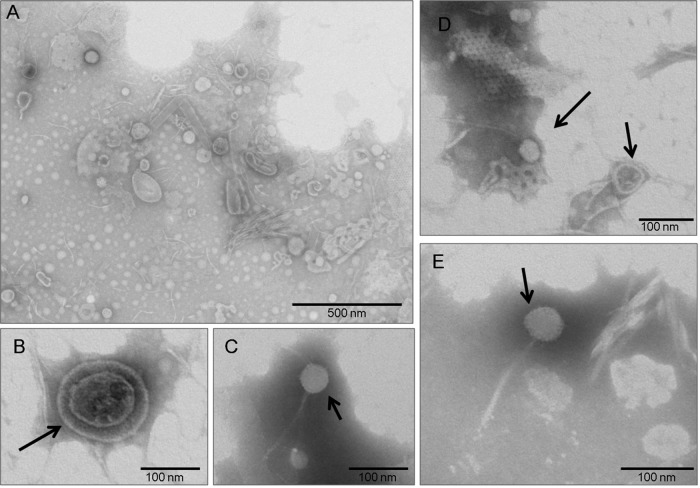
The specimen was excavated in 1996 and collected from the interior of a closed barrel, which was commonly used during this period as a pit or latrine (36). The barrel was buried at a depth of 3.80 m. The 121.4-g coprolite specimen was dark brown and well preserved under anaerobic taphonomic conditions. Extensive precautions were undertaken to avoid contaminating the coprolite specimen in our laboratory environment: no positive control was used (15), and suicide PCR protocols were applied (35). All negative controls, used in a 1:4 control/specimen ratio, were consistent with current recommendations for paleomicrobiological and paleoparasitological studies (13, 15, 37, 38) and remained negative. Virus-like particles (VLPs) purified from the internal region of the coprolite, after the external layer was removed, were morphologically diverse and varied in size and shape. Oval particles of different lengths (up to 200 nm) and diameters (up to 100 nm), as well as rod-shaped structures (up to 250 nm in length), were observed (Fig. 1A). We identified a VLP with a dense core and a diameter of approximately 150 nm, apparently surrounded by an envelope-like structure (Fig. 1B). Viral particles exhibiting characteristics typical of the Siphoviridae bacteriophage family (icosahedral head, long tail) were also observed (Fig. 1C to E).
FIG 1.
Transmission electron microscopy of negatively stained viral particles. (A) Overview of stained viral particles, which vary in size and shape, isolated from the Middle Ages coprolite. (B to E) A representative virion (B) and virus-like particles (C to E) with icosahedral nucleocapsids and a long filament tail characteristic of Siphoviridae bacteriophages.
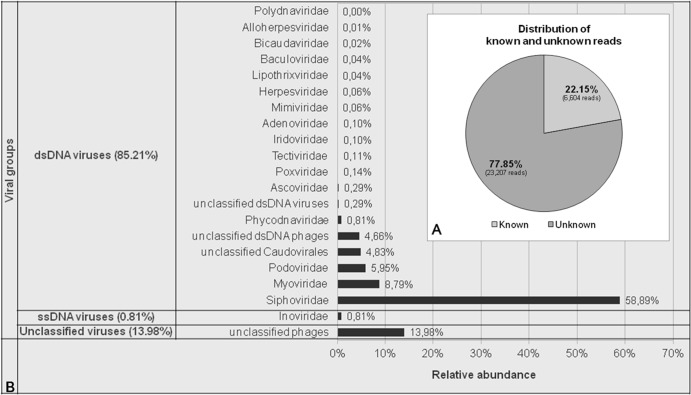
High-throughput sequencing generated 30,654 reads corresponding to approximately 10.8 million bp. After quality trimming and duplicate removal, 29,811 reads remained (see Table S2 in the supplemental material). The preprocessed read lengths ranged between 77 bp and 574 bp and had an average GC content of 47% (see Fig. S2 in the supplemental material). Finally, 41.93% of the reads were assembled into 1,464 contigs that ranged from 421 to 12,500 bp (see Table S2). In total, 22.15% of all reads and 17.28% of all contigs were significantly similar to known sequences from public databases (Fig. 2A; also see Table S2). Genome-length normalized counts of viral reads showed that about 85.21% and 0.81% of viral similarities were to double-stranded DNA (dsDNA) viruses and single-stranded DNA (ssDNA) viruses, respectively (Fig. 2B). Among the double-stranded DNA viral reads, we mostly observed Siphoviridae (58.89%), Myoviridae (8.79%), and Podoviridae (5.95%). Overall, we found reads to viral families that can infect eukaryotes (Ascoviridae, Poxviridae, Iridoviridae, Adenoviridae, Mimiviridae, Herpesviridae, Baculoviridae, Polydnaviridae, and Phycodnaviridae), archaea (Lipothrixiviridae, Tectiviridae, and Bicaudaviridae), and bacteria (Siphoviridae, Myoviridae, and Podoviridae) (Fig. 2B).
FIG 2.
(A) The proportion of known and unknown reads (in percent). Reads were defined as “unknown” if they lacked homology to the nonredundant NCBI database according to a BLASTN search (E value, <1e−5) and as “known” otherwise. (B) Relative abundance of viral families. The relative abundance of identified viral families was estimated using the GAAS software.
Few reads showed similarities to eukaryotic viruses, and among them, those belonging to Phycodnaviridae were the most abundant (0.81%) (Fig. 2B). We also identified a contig encoding a hypothetical protein of invertebrate iridescent virus 3 (IIV-3). IIV-3 is a member of the Iridoviridae family, genus Chloriridovirus, which has a large particle size (180 nm) and infects mosquitoes (see Table S3 in the supplemental material). Metagenomic results were confirmed by ad hoc suicide PCR (35). In the presence of negative controls, a 167-bp fragment of a Mimiviridae-like nonfunctional B-family DNA polymerase was amplified and sequenced, revealing 84% identity to that of the Moumouvirus of the Mimiviridae family (GenBank accession no. GU265560.1).
Only a small proportion of viral reads were related to viral families infecting archaea. These families corresponded to Lipothrixiviridae (0.04%), Tectiviridae (0.11%), and Bicaudaviridae (0.02%) (Fig. 2B). One contig was found to have similarity to an environmental halophage, eHP-6, an unclassified bacteriophage that infects Haloarchaea (see Table S3 in the supplemental material).
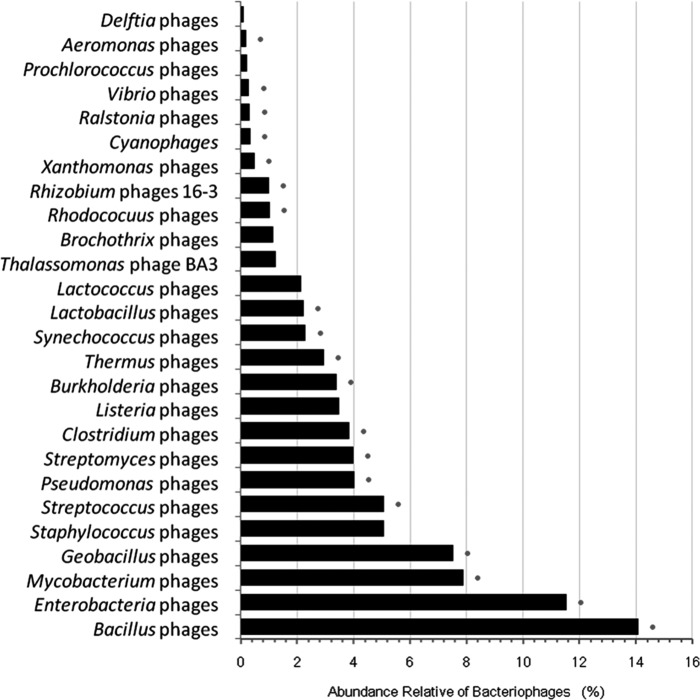
In contrast, the majority of the identifiable viral reads showed homology to genomes of viruses infecting bacteria (bacteriophages), especially those of the genus Bacillus (14.08%). We identified reads with homology to genomes of bacteriophages infecting as many as 37 different bacterial genera, including bacterial genera commonly associated with the human gut, such as Enterobacterium phages (11.54%), Lactobacillus phages (2.23%), and Lactococcus phages (2.14%) (Fig. 3). Other findings included reads with similarity to bacteriophages that infect typical soil-dwelling bacteria: Geobacillus phages (7.53%), Streptomyces phages (3.98%), and Delftia phages (0.11%). Several reads were found to show homology to bacteriophages whose bacterial hosts belong to genera that also include human pathogens, such as Mycobacterium phages (7.89%), Vibrio phages (0.29%), Pseudomonas phages (4.01%), Streptococcus phages (5.06%), Staphylococcus phages (5.07%), Listeria phages (3.48%), Burkholderia phages (3.38%), and Clostridium phages (3.83%) (Fig. 3). The presence of sequences homologous to genes of some of these bacteriophages (Bacillus, Clostridium, Mycobacterium, and Burkholderia phages) was further supported by contig reconstruction, ad hoc PCR amplification, and sequencing (see Tables S1 and S3 in the supplemental material). Moreover, contigs were found to harbor ORFs with similarity to genes from bacteriophages that are likely to infect hosts known to live in aquatic environments. In particular, we detected contigs matching the tail fiber protein coding gene of cyanophage S-TIM5, the tape measure protein coding gene of Planctomyces limnophilus DSM 3776, the gene coding for an unnamed protein product of Synechococcus phage S-CB53, and a hypothetical protein coding gene from an uncultured phage identified in a viral metagenomic study of water from the Mediterranean Sea. An ORF encoding a putative phage tail fiber protein of Celeribacter phage P12053L was also identified on one of these contigs, amplified by specific PCR, and the 280-bp amplicon was verified by Sanger sequencing. However, the weak similarities shown to some of these highly shared bacteriophage genes and the database bias toward genomes of marine viruses make it difficult to state if these specific aquatic bacteriophages or other populations of bacteriophages are present in the sample. At last, a 1,939-bp contig matched an unidentified phage previously described in a viral metagenomic study performed on modern human stools (33) (see Table S3). Only a scaffold is available for the unidentified phage, and the matched protein is annotated as a hypothetical protein; however, this hypothetical protein is predicted to contain a conserved domain corresponding to an N-acetylmuramoyl-l-alanine amidase. This domain is characteristic of autolysins that degrade peptidoglycans and is typically observed in bacteriophage, prophage, and bacterial genomes.
FIG 3.
Relative abundance of hits to known bacteriophages. The relative abundance of hits to known bacteriophages was estimated using the GAAS software. The hosts of the bacteriophages that were also identified in a previous study on the bacterial community associated with this specimen (Appelt et al., unpublished data) are noted to the right with dots.
Evidence of a temperate life cycle for the detected bacteriophage sequences was observed using three indicators (33): (i) nucleotide identity to bacterial genomes (an indicator of prophage formation), (ii) the presence of integrase-encoding genes (markers of temperate bacteriophages), and (iii) similarity to prophage sequences available in the ACLAME database (see Materials and Methods). We observed that 329 contigs (22.47%) significantly matched prophage proteins in the ACLAME database, 52 contigs (3.55%) had significant nucleotide identity to bacterial genomes (especially genomes of Escherichia coli strains), and 32 contigs (2.18%) harbored integrase-encoding genes (see Fig. S2 in the supplemental material). This strategy provides only a minimal estimate of the number of temperate bacteriophages, as stated by Minot et al. (33). Overall, 375 contigs (25.61%) presented at least one of the three indicators and could be tentatively classified as temperate bacteriophages (see Fig. S2 in the supplemental material).
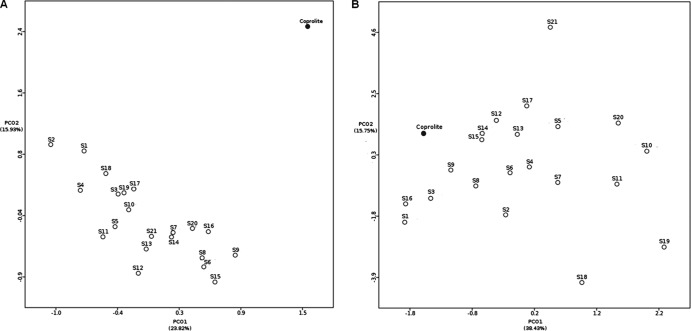
The coprolite-associated DNA virome was compared to the viromes of 21 modern human stool specimens (Fig. 4). At the taxonomic level, the coprolite virome did not group with modern stool viromes, whereas it was functionally more similar to some of the modern stool samples (Fig. 4). Overall, the coprolite virome displayed higher species richness (315.279) and seemed to be more functionally diverse (average Shannon-Wiener index of 4.8693) than modern stool viromes (average species richness, 77.824; average Shannon-Wiener index, 4.1264) (see Table S4 in the supplemental material). A more extensive functional analysis of the assembled contigs revealed that most of the identifiable ORFs harbored by these contigs carried genes involved in DNA metabolism (n = 80), as is typical of viromes, and virulence genes (n = 87). The most abundant virulence genes were those involved in resistance to antibiotics and toxic compounds. In particular, a contig encoding a chloramphenicol O-acetyltransferase gene that mediates chloramphenicol resistance was observed. This gene was found to belong to Chryseobacterium sp., and the BLAST-based annotation was further confirmed by a phylogenetic tree constructed from the ORF of this contig (see Fig. S3 in the supplemental material). To further investigate the presence of virulence genes, virome reads were examined using the Virulence Factor Database, which includes both conventional factors directly involved in pathogenesis and factors important to establishing infection. A stringent search allowed the identification of 166 reads encoding virulence factors. In particular, virulence factors of the bacterial genera Escherichia (n = 42), Salmonella (n = 39), and Shigella (n = 34) were observed (see Table S5 in the supplemental material). A pathway-centric analysis based on COG annotation revealed that virulence (defense mechanisms) was overrepresented in the coprolite compared to modern stools. Other differences included an overrepresentation of lipid transport and metabolism, fatty acid biosynthesis, and amino acid transport and metabolism (see Table S6 in the supplemental material). Indeed, 12 ORFs on annotated contigs were found to contain genes involved in lipid metabolism, in particular fatty acid biosynthesis (n = 3), glycerolipid and glycerophospholipid metabolism (n = 3), isoprenoid metabolism (n = 3), and polyhydroxybutyrate metabolism (n = 3). Annotated contigs also contained 36 ORFs encoding functions related to the metabolism of amino acids, especially lysine, threonine, methionine, and cysteine (n = 11) and arginine, urea, and polyamines (n = 10).
FIG 4.
Comparison between the modern human stool viromes and the coprolite virome. Principal component (PCO) analysis was used to compare the viral metagenomes associated with the coprolite (highlighted in red) to those associated with modern human stool samples (S1 to S21) at the taxonomic (A) and functional (B) levels.
DISCUSSION
We report what we believe to be the first metagenomic analysis of an ancient human DNA virome. The use of viral metagenomics allowed us to perform a systematic research of known and unknown viruses without a priori targeting of expected viruses.
Because minimizing contamination is vital in paleomicrobiology, extensive precautions established by previously published recommended protocols were implemented to avoid contamination of the coprolite specimen (13, 15, 37, 38). The coprolite studied here was recovered from a sealed barrel that was still intact at the time it was found, suggesting that the coprolite was protected from contamination by environmental material for centuries. Only the internal region of the coprolite was used in our experiments. We ascertained the presence of viruses by three independent approaches: i.e., electron microscopy, metagenomics, and suicide PCR. The PCR amplification product sequences were original (i.e., they had not been previously observed in our laboratory), and all negative controls remained negative.
The viral metagenome was generated using a multiple-displacement amplification of viral genomes via the ϕ29 polymerase. This method is known to preferentially amplify circular and single-stranded DNA (12). To potentially minimize this bias, a duplicate amplification reaction was performed as previously suggested (21).
The majority of the generated metagenomic sequences were of unknown origin. The taxonomic composition of the generated virome was estimated on the basis of the identifiable viral sequences (known sequences) of the DNA virome. The known sequences corresponded to DNA viruses that infect eukaryotes, bacteria, and archaea. Eukaryotic and archaeal viral sequences were detected only at low abundances, and their presence was supported by contig recovery or confirmed by ad hoc suicide PCR. The majority of the identifiable sequences recovered from the coprolite corresponded to bacteriophages, especially Siphoviridae. Contigs with significant similarity to characteristic bacteriophage genes were identified, such as genes coding for structural proteins (tail fiber proteins and capsid proteins) as well as proteins involved in replication (DNA polymerase) or DNA packaging (terminase). We could also identify reads with significant similarity to virulence factors associated with pathogenic bacteria and genes from bacteriophages that infect bacteria belonging to genera that include mammalian pathogens. These findings are consistent with those obtained for the previously generated bacterial metagenome associated with the coprolite (S. Appelt, F. Armougom, M. Le Bailly, C. Robert, and M. Drancourt, unpublished data). Comparative analyses to previously published viromes show that modern human stool viromes do not group with the coprolite virome at the taxonomic level. All previous works on viral communities associated with the stool from healthy individuals showed a high prevalence of bacteriophages, in particular double-stranded DNA bacteriophages of the Siphoviridae family (8, 11, 33) or single-stranded DNA bacteriophages of the Microviridae family (10, 12), with high interindividual variability. Accordingly, the coprolite virome shows a high prevalence of Siphoviridae (8, 11, 33). Moreover, as in modern stools, we found evidence for temperate bacteriophages (10, 33). However, we did not observe significant abundance of single-stranded DNA viruses (10, 12) or the same most abundant prophages identified in other modern stool viromes (33). At the functional level, no clear separation can be observed between the coprolite virome and modern stool viromes, and functions might be conserved between the coprolite and some modern stool samples. This finding is consistent with those of a recent study that demonstrated that despite interindividual taxonomic variability, the metabolic profile was significantly conserved within viromes from the same ecological niche (39). This persistence of metabolic functionalities across centuries may reinforce the crucial role of the viral community in the human gastrointestinal tract.
Finally, the coprolite virome is more functionally diverse and rich in virulence genes than modern stool sample viromes. One contig containing a gene for chloramphenicol resistance (the chloramphenicol O-acetyltransferase gene), a broad-spectrum antibiotic that inhibits bacterial protein synthesis, was identified. The presence of antibiotic resistance genes in viral metagenomes has been reported in modern human stools (33). Indeed, bacteriophages constitute a reservoir of resistance genes (40–42), and bacteriophage transduction represents one important mode of lateral transfer of resistance genes between bacterial species. Phylogenetic studies have demonstrated that the evolution and dissemination of resistance genes started well before the use of antibiotics (43–45). Accordingly, direct evidence for the presence of antibiotic resistance genes in pre-antibiotic-era specimens was provided by ad hoc PCR amplifications using DNA extracted from 30,000-year-old permafrost sediments in Canada (46). Here, we demonstrate that bacteriophages are an ancient reservoir of resistance genes associated with human samples that date back as far as the Middle Ages. Moreover, we provide evidence for the lysogenic lifestyle of these bacteriophages, which may support their role in the mobilization and lateral transfer of genes in bacterial communities.
Overall, this study furthers our understanding of past viral diversity and distribution and promotes the further exploration of ancient viral communities using coprolite specimens.
Supplementary Material
ACKNOWLEDGMENTS
We thank Sonia Monteil Bouchard and Catherina Robert for technical assistance.
C.D. and L.F. were funded by starting grant no. 242729 from the European Research Council to C.D.
The authors declare no competing interests.
Footnotes
Published ahead of print 7 February 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.03242-13.
REFERENCES
- 1.Angly FE, Felts B, Breitbart M, Salamon P, Edwards RA, Carlson C, Chan AM, Haynes M, Kelley S, Liu H, Mahaffy JM, Mueller JE, Nulton J, Olson R, Parsons R, Rayhawk S, Suttle CA, Rohwer F. 2006. The marine viromes of four oceanic regions. PLoS Biol. 4:e368. 10.1371/journal.pbio.0040368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lopez-Bueno A, Tamames J, Velazquez D, Moya A, Quesada A, Alcami A. 2009. High diversity of the viral community from an Antarctic lake. Science 326:858–861. 10.1126/science.1179287 [DOI] [PubMed] [Google Scholar]
- 3.Desnues C, Rodriguez-Brito B, Rayhawk S, Kelley S, Tran T, Haynes M, Liu H, Furlan M, Wegley L, Chau B, Ruan Y, Hall D, Angly FE, Edwards RA, Li L, Thurber RV, Reid RP, Siefert J, Souza V, Valentine DL, Swan BK, Breitbart M, Rohwer F. 2008. Biodiversity and biogeography of phages in modern stromatolites and thrombolites. Nature 452:340–343. 10.1038/nature06735 [DOI] [PubMed] [Google Scholar]
- 4.Kim KH, Chang HW, Nam YD, Roh SW, Kim MS, Sung Y, Jeon CO, Oh HM, Bae JW. 2008. Amplification of uncultured single-stranded DNA viruses from rice paddy soil. Appl. Environ. Microbiol. 74:5975–5985. 10.1128/AEM.01275-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Briese T, Paweska JT, McMullan LK, Hutchison SK, Street C, Palacios G, Khristova ML, Weyer J, Swanepoel R, Egholm M, Nichol ST, Lipkin WI. 2009. Genetic detection and characterization of Lujo virus, a new hemorrhagic fever-associated arenavirus from southern Africa. PLoS Pathog. 5:e1000455. 10.1371/journal.ppat.1000455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Anderson NG, Gerin JL, Anderson NL. 2003. Global screening for human viral pathogens. Emerg. Infect. Dis. 9:768–774. 10.3201/eid0907.030004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nakamura S, Yang CS, Sakon N, Ueda M, Tougan T, Yamashita A, Goto N, Takahashi K, Yasunaga T, Ikuta K, Mizutani T, Okamoto Y, Tagami M, Morita R, Maeda N, Kawai J, Hayashizaki Y, Nagai Y, Horii T, Iida T, Nakaya T. 2009. Direct metagenomic detection of viral pathogens in nasal and fecal specimens using an unbiased high-throughput sequencing approach. PLoS One 4:e4219. 10.1371/journal.pone.0004219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Breitbart M, Hewson I, Felts B, Mahaffy JM, Nulton J, Salamon P, Rohwer F. 2003. Metagenomic analyses of an uncultured viral community from human feces. J. Bacteriol. 185:6220–6223. 10.1128/JB.185.20.6220-6223.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Breitbart M, Rohwer F. 2005. Method for discovering novel DNA viruses in blood using viral particle selection and shotgun sequencing. Biotechniques 39:729–736. 10.2144/000112019 [DOI] [PubMed] [Google Scholar]
- 10.Reyes A, Haynes M, Hanson N, Angly FE, Heath AC, Rohwer F, Gordon JI. 2010. Viruses in the faecal microbiota of monozygotic twins and their mothers. Nature 466:334–338. 10.1038/nature09199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Breitbart M, Haynes M, Kelley S, Angly F, Edwards RA, Felts B, Mahaffy JM, Mueller J, Nulton J, Rayhawk S, Rodriguez-Brito B, Salamon P, Rohwer F. 2008. Viral diversity and dynamics in an infant gut. Res. Microbiol. 159:367–373. 10.1016/j.resmic.2008.04.006 [DOI] [PubMed] [Google Scholar]
- 12.Kim MS, Park EJ, Roh SW, Bae JW. 2011. Diversity and abundance of single-stranded DNA viruses in human feces. Appl. Environ. Microbiol. 77:8062–8070. 10.1128/AEM.06331-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pääbo S, Poinar H, Serre D, Jaenicke-Despres V, Hebler J, Rohland N, Kuch M, Krause J, Vigilant L, Hofreiter M. 2004. Genetic analyses from ancient DNA. Annu. Rev. Genet. 38:645–679. 10.1146/annurev.genet.37.110801.143214 [DOI] [PubMed] [Google Scholar]
- 14.Willerslev E, Cooper A. 2005. Ancient DNA. Proc. Biol. Sci. 272:3–16. 10.1098/rspb.2004.2813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Drancourt M, Raoult D. 2005. Palaeomicrobiology: current issues and perspectives. Nat. Rev. Microbiol. 3:23–35. 10.1038/nrmicro1063 [DOI] [PubMed] [Google Scholar]
- 16.Marennikova SS, Shelukhina EM, Zhukova OA, Yanova NN, Loparev VN. 1990. Smallpox diagnosed 400 years later: results of skin lesions examination of 16th century Italian mummy. J. Hyg. Epidemiol. Microbiol. Immunol. 34:227–231 [PubMed] [Google Scholar]
- 17.Bedarida S, Dutour O, Buzhilova AP, de Micco P, Biagini P. 2011. Identification of viral DNA (Anelloviridae) in a 200-year-old dental pulp sample (Napoleon's Great Army, Kaliningrad, 1812). Infect. Genet. Evol. 11:358–362. 10.1016/j.meegid.2010.11.007 [DOI] [PubMed] [Google Scholar]
- 18.Biagini P, Theves C, Balaresque P, Geraut A, Cannet C, Keyser C, Nikolaeva D, Gerard P, Duchesne S, Orlando L, Willerslev E, Alekseev AN, de Micco P, Ludes B, Crubezy E. 2012. Variola virus in a 300-year-old Siberian mummy. N. Engl. J. Med. 367:2057–2059. 10.1056/NEJMc1208124 [DOI] [PubMed] [Google Scholar]
- 19.Li HC, Fujiyoshi T, Lou H, Yashiki S, Sonoda S, Cartier L, Nunez L, Munoz I, Horai S, Tajima K. 1999. The presence of ancient human T-cell lymphotropic virus type I provirus DNA in an Andean mummy. Nat. Med. 5:1428–1432. 10.1038/71006 [DOI] [PubMed] [Google Scholar]
- 20.Sonoda S, Li HC, Cartier L, Nunez L, Tajima K. 2000. Ancient HTLV type 1 provirus DNA of Andean mummy. AIDS Res. Hum. Retroviruses 16:1753–1756. 10.1089/08892220050193263 [DOI] [PubMed] [Google Scholar]
- 21.Thurber RV, Haynes M, Breitbart M, Wegley L, Rohwer F. 2009. Laboratory procedures to generate viral metagenomes. Nat. Protoc. 4:470–483. 10.1038/nprot.2009.10 [DOI] [PubMed] [Google Scholar]
- 22.Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ, Sahl JW, Stres B, Thallinger GG, Van Horn DJ, Weber CF. 2009. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 75:7537–7541. 10.1128/AEM.01541-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Angly FE, Willner D, Prieto-Davo A, Edwards RA, Schmieder R, Vega-Thurber R, Antonopoulos DA, Barott K, Cottrell MT, Desnues C, Dinsdale EA, Furlan M, Haynes M, Henn MR, Hu Y, Kirchman DL, McDole T, McPherson JD, Meyer F, Miller RM, Mundt E, Naviaux RK, Rodriguez-Mueller B, Stevens R, Wegley L, Zhang L, Zhu B, Rohwer F. 2009. The GAAS metagenomic tool and its estimations of viral and microbial average genome size in four major biomes. PLoS Comput. Biol. 5:e1000593. 10.1371/journal.pcbi.1000593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Meyer F, Paarmann D, D'Souza M, Olson R, Glass EM, Kubal M, Paczian T, Rodriguez A, Stevens R, Wilke A, Wilkening J, Edwards RA. 2008. The metagenomics RAST server—a public resource for the automatic phylogenetic and functional analysis of metagenomes. BMC Bioinformatics 9:386. 10.1186/1471-2105-9-386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen L, Xiong Z, Sun L, Yang J, Jin Q. 2012. VFDB 2012 update: toward the genetic diversity and molecular evolution of bacterial virulence factors. Nucleic Acids Res. 40:D641–D645. 10.1093/nar/gkr989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu B, Pop M. 2009. ARDB—Antibiotic Resistance Genes Database. Nucleic Acids Res. 37:D443–D447. 10.1093/nar/gkn656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hyatt D, Chen GL, Locascio PF, Land ML, Larimer FW, Hauser LJ. 2010. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics 11:119. 10.1186/1471-2105-11-119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32:1792–1797. 10.1093/nar/gkh340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Talavera G, Castresana J. 2007. Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst. Biol. 56:564–577. 10.1080/10635150701472164 [DOI] [PubMed] [Google Scholar]
- 30.Guindon S, Delsuc F, Dufayard JF, Gascuel O. 2009. Estimating maximum likelihood phylogenies with PhyML. Methods Mol. Biol. 537:113–137. 10.1007/978-1-59745-251-9_6 [DOI] [PubMed] [Google Scholar]
- 31.Dereeper A, Guignon V, Blanc G, Audic S, Buffet S, Chevenet F, Dufayard JF, Guindon S, Lefort V, Lescot M, Claverie JM, Gascuel O. 2008. Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 36:W465–W469. 10.1093/nar/gkn180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tamura K, Dudley J, Nei M, Kumar S. 2007. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24:1596–1599. 10.1093/molbev/msm092 [DOI] [PubMed] [Google Scholar]
- 33.Minot S, Sinha R, Chen J, Li H, Keilbaugh SA, Wu GD, Lewis JD, Bushman FD. 2011. The human gut virome: inter-individual variation and dynamic response to diet. Genome Res. 21:1616–1625. 10.1101/gr.122705.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kristiansson E, Hugenholtz P, Dalevi D. 2009. ShotgunFunctionalizeR: an R-package for functional comparison of metagenomes. Bioinformatics 25:2737–2738. 10.1093/bioinformatics/btp508 [DOI] [PubMed] [Google Scholar]
- 35.Raoult D, Aboudharam G, Crubezy E, Larrouy G, Ludes B, Drancourt M. 2000. Molecular identification by “suicide PCR” of Yersinia pestis as the agent of medieval black death. Proc. Natl. Acad. Sci. U. S. A. 97:12800–12803. 10.1073/pnas.220225197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.RochaGCd 2003. Praça das Armas, Namur, Bélgica. Contribuição de um estudo paleoparasitológico. Escola Nacional de Saúde Pública-Fiocruz, Rio de Janeiro, Rio de Janeiro, Brazil [Google Scholar]
- 37.Cooper A, Poinar HN. 2000. Ancient DNA: do it right or not at all. Science 289:1139. 10.1126/science.289.5482.1139b [DOI] [PubMed] [Google Scholar]
- 38.Hofreiter M, Serre D, Poinar HN, Kuch M, Paabo S. 2001. Ancient DNA. Nat. Rev. Genet. 2:353–359. 10.1038/35072071 [DOI] [PubMed] [Google Scholar]
- 39.Willner D, Furlan M, Haynes M, Schmieder R, Angly FE, Silva J, Tammadoni S, Nosrat B, Conrad D, Rohwer F. 2009. Metagenomic analysis of respiratory tract DNA viral communities in cystic fibrosis and non-cystic fibrosis individuals. PLoS One 4:e7370. 10.1371/journal.pone.0007370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Muniesa M, Garcia A, Miro E, Mirelis B, Prats G, Jofre J, Navarro F. 2004. Bacteriophages and diffusion of β-lactamase genes. Emerg. Infect. Dis. 10:1134–1137. 10.3201/eid1006.030472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Colomer-Lluch M, Jofre J, Muniesa M. 2011. Antibiotic resistance genes in the bacteriophage DNA fraction of environmental samples. PLoS One 6:e17549. 10.1371/journal.pone.0017549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mazaheri Nezhad Fard R, Barton MD, Heuzenroeder MW. 2011. Bacteriophage-mediated transduction of antibiotic resistance in enterococci. Lett. Appl. Microbiol. 52:559–564. 10.1111/j.1472-765X.2011.03043.x [DOI] [PubMed] [Google Scholar]
- 43.Hall BG, Barlow M. 2004. Evolution of the serine β-lactamases: past, present and future. Drug Resist. Updates 7:111–123. 10.1016/j.drup.2004.02.003 [DOI] [PubMed] [Google Scholar]
- 44.Garau G, Di Guilmi AM, Hall BG. 2005. Structure-based phylogeny of the metallo-β-lactamases. Antimicrob. Agents Chemother. 49:2778–2784. 10.1128/AAC.49.7.2778-2784.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Aminov RI, Mackie RI. 2007. Evolution and ecology of antibiotic resistance genes. FEMS Microbiol. Lett. 271:147–161. 10.1111/j.1574-6968.2007.00757.x [DOI] [PubMed] [Google Scholar]
- 46.D'Costa VM, King CE, Kalan L, Morar M, Sung WW, Schwarz C, Froese D, Zazula G, Calmels F, Debruyne R, Golding GB, Poinar HN, Wright GD. 2011. Antibiotic resistance is ancient. Nature 477:457–461. 10.1038/nature10388 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.