Abstract
Carbazole 1,9a-dioxygenase (CARDO), a Rieske nonheme iron oxygenase (RO), is a three-component system composed of a terminal oxygenase (Oxy), ferredoxin, and a ferredoxin reductase. Oxy has angular dioxygenation activity against carbazole. Previously, site-directed mutagenesis of the Oxy-encoding gene from Janthinobacterium sp. strain J3 generated the I262V, F275W, Q282N, and Q282Y Oxy derivatives, which showed oxygenation capabilities different from those of the wild-type enzyme. To understand the structural features resulting in the different oxidation reactions, we determined the crystal structures of the derivatives, both free and complexed with substrates. The I262V, F275W, and Q282Y derivatives catalyze the lateral dioxygenation of carbazole with higher yields than the wild type. A previous study determined the crystal structure of Oxy complexed with carbazole and revealed that the carbonyl oxygen of Gly178 hydrogen bonds with the imino nitrogen of carbazole. In these derivatives, the carbazole was rotated approximately 15, 25, and 25°, respectively, compared to the wild type, creating space for a water molecule, which hydrogen bonds with the carbonyl oxygen of Gly178 and the imino nitrogen of carbazole. In the crystal structure of the F275W derivative complexed with fluorene, C-9 of fluorene, which corresponds to the imino nitrogen of carbazole, was oriented close to the mutated residue Trp275, which is on the opposite side of the binding pocket from the carbonyl oxygen of Gly178. Our structural analyses demonstrate that the fine-tuning of hydrophobic residues on the surface of the substrate-binding pocket in ROs causes a slight shift in the substrate-binding position that, in turn, favors specific oxygenation reactions toward various substrates.
INTRODUCTION
Rieske nonheme iron oxygenases (ROs) catalyze the initial oxygenation reaction of aromatic compounds (1). ROs are of interest for the biodegradation of aromatic pollutants and for synthetic applications requiring enantio- and regiospecific reactions. Some ROs break down toxic heterocyclic compounds by a single oxidization reaction. ROs are generally composed of two or three components, a terminal oxygenase (Oxy) and one or two electron transfer components. The Oxy is activated by the electron transfer component(s) and then catalyzes the oxygenation of the substrate. Recent biochemical and structural studies have revealed that Oxys typically exhibit an α3 or (αβ)3 configuration (2). The α subunit contains a Rieske [2Fe-2S] cluster and a mononuclear iron. The Rieske [2Fe-2S] cluster accepts electrons from the electron transfer component(s) and transfers electrons to the mononuclear iron. The mononuclear iron at the active site activates molecular oxygen, which attacks the substrate. The two oxygen atoms of the molecular oxygen bind to tandemly linked carbon atoms in an aromatic ring, creating two hydroxyl groups in the cis configuration (3). This process is called lateral dioxygenation. The amino acid residues that affect the substrate specificity of ROs have been identified (4–14). In these studies, the molecular mechanism by which substrate specificity is altered has been discussed on the basis of proposed structural similarities or molecular simulation. Homology modeling of protein structure also provides roughly satisfactory structural information, at least for the C-α backbone, but more detailed configurations, such as the orientations of the side chains and subtle differences in substrate binding, are difficult to simulate precisely. Recent reports of structural analyses of amino acid-substituted derivatives of the Oxy component of ROs provided insight into the structural basis of the substrate specificities (15–17). Detailed comparisons between wild-type (WT) naphthalene 1,2-dioxygenase from Pseudomonas sp. strain NCIB 9816-4 (NDO-O9816-4) and derivatives thereof in which Phe352 was replaced with Val revealed that regio- and stereoselectivity depend primarily on the orientation of the substrate binding at the active site (15). Structural comparisons of a series of amino acid-substituted derivatives of biphenyl dioxygenase from Burkholderia xenovorans strain LB400 demonstrated that mutations close to the catalytic cavity (T335A, T335A/F336M, and N338Q/L409F) constrain the substrate specificity to a greater or lesser degree (16, 17).
Carbazole 1,9a-dioxygenase (CARDO) is an RO that catalyzes the initial step of the carbazole (CAR) degradation pathway (18). A variety of CAR-degrading bacteria have been isolated, and their genes for CAR degradation have been identified (19–21). Among them, the CARDOs from Pseudomonas resinovorans CA10, Janthinobacterium sp. strain J3, Novosphingobium sp. strain KA1, and Nocardioides aromaticivorans IC177 have been structurally characterized in detail (T. Umeda et al., unpublished data) (22–26). CARDO catalyzes dioxygenation at angular (C-9a) and adjacent (C-1) carbon atoms to produce an unstable hemiaminal (1-hydro-1,9a-dihydroxycarbazole) that is spontaneously cleaved to form 2′-aminobiphenyl-2,3-diol (Fig. 1). This reaction has been termed angular dioxygenation and is involved in the degradation of heterocyclic aromatic compounds such as CAR, dibenzofuran, and dibenzo-p-dioxin (Fig. 1) (27). CARDO also catalyzes the lateral dioxygenation of biphenyl, naphthalene, and anthracene, as well as the monooxygenation of the methylene carbon of fluorene (FN) and the sulfur atom of dibenzothiophene (27, 28). CARDO consists of a ferredoxin reductase (Red), ferredoxin (Fd), and Oxy (19, 29). The substrate specificity of CARDO is determined mainly by the structure of the substrate-binding pocket in Oxy, which includes the mononuclear iron active site. The crystal structures of Oxy from strain J3 and Fd from strain CA10 have been determined (22, 23), and subsequently, the structures of the electron transfer complex of Oxy and Fd (Oxy-Fd binary complex) and its CAR-bound form (Oxy-Fd-CAR ternary complex) were determined (Fig. 2A) (24). The entrance of the substrate-binding pocket of Oxy is composed mainly of hydrophobic residues, while some hydrophilic residues are located deep inside the pocket (Fig. 2B) (22). The hydrophobic residues are Ile184, Ala259, Ile262, Leu270, Val272, Phe275, and Phe329, which are thought to play a role in determining substrate orientation by their hydrophobic interactions with the substrate (24). No significant direct interaction between the substrate and the hydrophilic residues Gln282, Glu284, and Asn330 was found, although the side chains of these hydrophilic residues are located close to the substrate (24). The carbonyl oxygen of Gly178 hydrogen bonds to the imino nitrogen of CAR, indicating that Gly178 is also involved in determining the substrate orientation (24).
FIG 1.
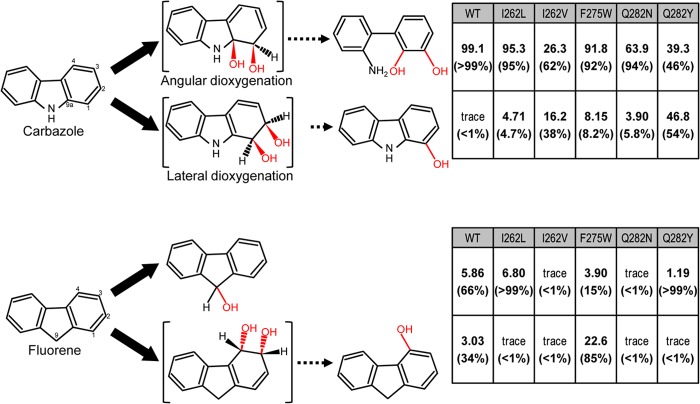
Reactions catalyzed by CARDO and its derivatives with the substrates CAR, dibenzo-p-dioxin, FN, and anthracene. The product formation rates of the biotransformation study previously performed with WT CARDO and its amino acid-substituted derivatives generated by site-directed mutagenesis (30) are indicated on the right. The product formation rates were calculated from the peak areas for the total ion current of the respective compounds extracted from the reaction mixtures containing E. coli cells expressing the WT or derivative CARDOs by gas chromatography-mass spectrometry analyses (30). Percentages of angular and lateral dioxygenation products are in parentheses. Unstable intermediate structures are bracketed.
FIG 2.
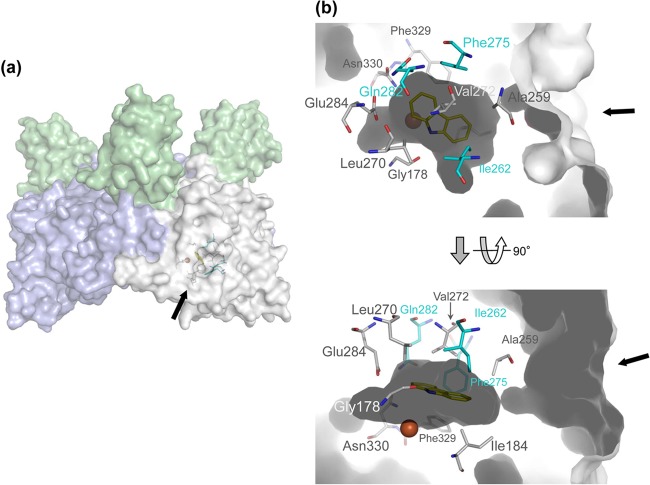
Crystal structure of WT CARDO (Oxy), Fd, and CAR ternary complex (PDB accession no. 2DE7) (24). Arrows indicate the entrance of the substrate-binding pocket. (a) Overall structure of the complex. Fd and chains B and C of Oxy are in green, white, and blue, respectively. (b) The substrate-binding pocket with CAR bound. Amino acid residues in cyan were substituted in this study. The mononuclear iron and CAR are shown as a brown sphere and yellow sticks, respectively.
To investigate the structural properties that determine the substrate specificity of CARDO, amino acid-substituted derivatives of Oxy have been generated by site-directed mutagenesis and their specificities for hetero- and polyaromatic substrates were determined (30). Four amino acid residues, Ile262, Phe275, Gln282, and Phe329, were selected for mutation on the basis of simulations of Oxy docking with substrates. The I262V and Q282Y derivatives converted CAR to 1-hydroxycarbazole with higher yields than the WT (Fig. 1). 1-Hydroxycarbazole probably results from the dehydration of cis-1,2-dihydroxy-1,2-dihydrocarbazole, formed by the lateral dioxygenation of CAR. The F275W derivative converted FN to 4-hydroxyfluorene (likely the dehydration product of cis-3,4-dihydroxy-3,4-dihydrofluorene) with significant activity, whereas the FN monooxygenation activity of WT Oxy was extremely low. The F275W derivative also oxidized fluoranthene to yield an unidentified cis-dihydrodiol. The Phe329-substituted F329A, F329I, F329L, F329L, F329V, and F329W derivatives showed only negligible oxygenation activity.
Detailed studies of the relationship between structure and substrate specificity may provide critical information regarding how to modify the substrate-binding pocket to enhance catalysis. This would be useful for bioremediation (degradation) of toxic pollutants, as well as for the enzymatic synthesis of chiral precursors in the production of specialty compounds. To apply ROs to bioremediation and to the regio- and stereoselective hydroxylation of aromatic compounds, a better understanding of the molecular mechanism of the catalysis process is needed. However, there are few reports of detailed structural studies with amino acid-substituted derivatives of ROs. In this study, we determined the crystal structures of derivatives of Oxy and its complexes with the substrates CAR and FN. Comparisons with the crystal structure of WT Oxy indicate that the substrate specificities of the derivatives are determined by altered hydrophobic interactions and hydrogen bonding between the substrates and the amino acid residues composing the surface of the substrate-binding pocket.
MATERIALS AND METHODS
Expression of a His-tagged form of each enzyme.
To express each amino acid-substituted derivative of Oxy as a C-terminally His-tagged protein, we used the pET system with the pET-26b(+) vector and Escherichia coli strain BL21(DE3) (Novagen, Madison, WI). Artificial NdeI and XhoI sites were created at the 5′ and 3′ ends of each carAa mutated gene, respectively, by PCR with template DNA containing the target mutant carAa gene (pUCARAJ3-I262L, -I262V, -F275W, -Q282N, or -Q282Y) (30) and primers 5′-CATATGGCGAACGTTGATGAGGCAAT-3′ and 5′-CTCGAGGCCCGAAACGTGCGCTTGGGTCTGAATACCCTG-3′ (the NdeI and XhoI restriction sites are italicized). The nucleotide sequences of the resultant PCR amplicons were confirmed by sequencing analysis. The mutated Oxy genes were ligated into the NdeI and XhoI sites of pET-26b(+) to produce pEI262L, pEI262V, pEF275W, pEQ282N, and pEQ282Y, respectively. E. coli strain BL21(DE3) (Novagen) harboring each expression plasmid was grown in lysogeny (L) medium (tryptone peptone at 10 g/liter, yeast extract at 5 g/liter, NaCl at 10 g/liter) (31) or SB medium (29). Both media were supplemented with kanamycin (50 μg/ml). E. coli cells were grown at 37°C with reciprocal shaking at 120 strokes/min to reach an optical density at 600 nm of 0.4 to 0.5 and subsequently cultivated for 15 h in the presence of isopropyl-β-d-thiogalactopyranoside (IPTG) at 25°C for the induction of protein expression. The final concentration of IPTG was 0.1 mM for the I262L, I262V, F275W, and Q282N derivatives or 0.25 mM for the Q282Y derivative.
Protein purification.
All purification procedures were performed at 4°C. E. coli cells were suspended in TI buffer (29), and the cell suspension was sonicated. After centrifugation, the supernatant was collected as a crude cell extract. The crude extract was applied to a HiTrap Chelation HP column (GE Healthcare, Tokyo, Japan) according to the manufacturer's recommendation. The fractions containing each protein were pooled and concentrated by ultrafiltration with Centriprep YM-10 (Millipore, Bedford, MA). The protein was further purified by gel filtration chromatography (GFC) with a Superdex 200 column (26 by 600 mm; GE Healthcare) and GFC buffer (20 mM Tris-HCl [pH 7.5], 0.2 M NaCl, 10% [vol/vol] glycerol). Fractions containing each protein were collected and concentrated by ultrafiltration. The buffer was replaced with 50 mM Tris-HCl (pH 7.5), and the resultant protein solutions were flash-frozen in liquid nitrogen and stored at −80°C until used for crystallization. WT Fd and Red were expressed and purified as described previously (23, 32, 33). Protein concentrations were measured with a protein assay kit (Bio-Rad) (34) with bovine serum albumin as the standard.
Crystallization and data collection.
The proteins were crystallized by the hanging-drop vapor diffusion method as described previously (32). In brief, each purified derivative Oxy protein was mixed with Fd at a molar ratio of 1:6. The crystallization conditions for the Oxy-Fd binary complex were 0.1 M ammonium acetate, 12.5% (vol/vol) polyethylene glycol 3350 in 0.05 M morpholineethanesulfonic acid (MES) at pH 5.5 (F275W, I262V, I262L, and Q282N derivatives) or pH 5.7 (Q282Y derivative), and 15 to 20 mg/ml protein at 20°C. The resultant crystals of the Oxy-Fd binary complex were cryocooled in liquid nitrogen until subjected to X-ray analysis.
The Oxy-Fd binary complex crystals containing the I262V, F275W, or Q282Y derivative were soaked in the reservoir solution containing 20% (vol/vol) glycerol and 0.5% (wt/wt) CAR or FN for 20 to 40 min at 20°C to obtain the Oxy-Fd-substrate ternary complex. The same procedure was used to obtain substrate- and oxygen-bound Oxy-Fd binary complex crystals, except that 20 mM sodium dithionite was included in the reservoir solution, and the crystals were subsequently incubated in an aerobic atmosphere for 10 min at 20°C. The crystals were cryocooled in liquid nitrogen.
The X-ray diffraction data for each crystal were collected at 100 K on the BL-5A, BL-17A, or AR-NW12 station at the Photon Factory (PF; High Energy Accelerator Research Organization, Tsukuba, Japan) and BL38B1 station at SPring-8 (Japan Synchrotron Radiation Institute, Hyogo Prefecture, Japan). All diffraction data were gathered at a wavelength of 1.0 Å and processed with the HKL2000 software (35). The data collection and processing statistics are presented in Table 1.
TABLE 1.
Summary of crystallographic data collection and refinement statistics
| Parameter | I262L-Fd | I262V-Fd | I262V-Fd-CAR | I262V-Fd-CAR-O2 | F275W-Fd (form1) | F275W-Fd-CAR (form2) | F275W-Fd-FN (form2) | Q282N-Fd | Q282Y-Fd | Q282Y-Fd-CAR |
|---|---|---|---|---|---|---|---|---|---|---|
| Beam line | AR-NW12 | AR-NW12 | BL38B1 | AR-NW12 | BL-17A | BL-5A | AR-NW12 | AR-NW12 | AR-NW12 | AR-NW12 |
| Wavelength (Å) | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 |
| Space group | P21 | P21 | P21 | P21 | P21 | P21 | P21 | P21 | P21 | P21 |
| Unit cell parameters (Å, °) | ||||||||||
| a | 98.3 | 98.1 | 98.3 | 98.4 | 98.5 | 98.3 | 98.5 | 98.3 | 98.3 | 98.2 |
| b | 89.8 | 89.2 | 90.0 | 90.1 | 90.1 | 88.2 | 88.3 | 89.5 | 89.9 | 90.0 |
| c | 105.2 | 104.9 | 105.3 | 105.3 | 105.4 | 108.1 | 108.6 | 105.1 | 105.3 | 105.2 |
| α = γ | 90.0 | 90.0 | 90.0 | 90.0 | 90.0 | 90.0 | 90.0 | 90.0 | 90.0 | 90.0 |
| β | 104.0 | 104.2 | 104.1 | 104.3 | 104.2 | 107.2 | 106.7 | 104.1 | 104.2 | 104.3 |
| Resolution range (Å)a | 50.0–2.00 (2.07–2.00) | 50.0–2.05 (2.12–2.05) | 50.0–2.10 (2.18–2.10) | 50.0–2.05 (2.12–2.05) | 50.0–1.95 (2.02–1.95) | 50.0–1.95 (2.02–1.95) | 50.0–2.10 (2.18–2.10) | 50.0–2.00 (2.07–2.00) | 50.0–1.85 (1.92–1.85) | 50.0–2.15 (2.23–2.15) |
| Total no. of reflections | 807,121 | 829,690 | 596,615 | 416,261 | 483,360 | 700,844 | 382,054 | 432,175 | 746,426 | 365,779 |
| No. of unique reflectionsa | 118,046 (11,386) | 108,437 (10,703) | 103,638 (10,301) | 110,854 (11,014) | 129,871 (12,825) | 128,061 (12,760) | 102,997 (10,306) | 119,101 (11,858) | 151,228 (15,073) | 96,341 (9586) |
| Completeness (%)a | 99.6 (96.5) | 98.4 (97.5) | 100.0 (100.0) | 99.7 (99.9) | 99.4 (98.5) | 99.5 (99.8) | 99.2 (99.9) | 99.9 (100.0) | 100.0 (100.0) | 100.0 (100.0) |
| Avg <I>/<σ(I)>a | 29.0 (3.2) | 43.5 (7.2) | 25.1 (4.5) | 33.3 (3.3) | 31.3 (5.8) | 49.1 (6.5) | 34.6 (4.1) | 33.9 (6.4) | 31.7 (4.6) | 22.5 (4.6) |
| Rmerge (%)a | 9.2 (28.6) | 7.0 (30.0) | 6.3 (28.5) | 5.5 (38.7) | 6.0 (28.2) | 8.9 (27.8) | 5.7 (36.9) | 14.5 (35.2) | 5.7 (29.8) | 7.4 (28.2) |
| Multiplicitya | 6.8 (4.4) | 7.7 (7.8) | 5.8 (5.7) | 3.8 (3.7) | 3.7 (3.7) | 5.5 (5.2) | 3.7 (3.6) | 3.6 (3.5) | 4.9 (4.9) | 3.8 (3.8) |
| Refinement | ||||||||||
| Resolution range (Å) | 47.9–2.01 | 40.4–2.05 | 32.8–2.10 | 33.6–2.05 | 39.3–1.95 | 32.6–1.95 | 33.8–2.10 | 31.2–2.00 | 39.2–1.85 | 40.9–2.15 |
| R factor (%)b | 19.0 | 18.2 | 19.0 | 19.5 | 18.8 | 20.5 | 21.7 | 18.6 | 18.9 | 18.3 |
| Rfreec (%) | 21.6 | 21.3 | 21.7 | 22.6 | 21.5 | 23.2 | 25.2 | 21.3 | 21.3 | 21.1 |
| RMSD (bond lengths) (Å) | 0.005 | 0.005 | 0.006 | 0.006 | 0.005 | 0.005 | 0.006 | 0.005 | 0.005 | 0.005 |
| RMSD (bond angles) (°) | 1.3 | 1.3 | 1.3 | 1.3 | 1.3 | 1.3 | 1.3 | 1.3 | 1.3 | 1.3 |
| B factors (Å2) | ||||||||||
| Avg | 31.6 | 31.3 | 26.8 | 39.8 | 26.6 | 42.4 | 42.8 | 30.5 | 23.8 | 27.9 |
| Proteins | 31.2 | 30.9 | 26.5 | 39.5 | 25.8 | 41.8 | 42.4 | 29.9 | 23.1 | 27.3 |
| Waters | 36.7 | 36.9 | 30.7 | 44.6 | 33.9 | 48.8 | 49.3 | 36.5 | 30.2 | 34.2 |
| Substrates | 37.8 | 49.6 | 46.4 | 48.3 | - | - | 40.0 | |||
| PDB accession no. | 4NB8 | 4NB9 | 4NBA | 4NBB | 4NBC | 4NBD | 4NBE | 4NBF | 4NBG | 4NBH |
Values in parentheses are for the outermost shell.
The R factor is defined as follows: R = Σ‖Fobs| − |Fcalc‖/Σ|Fobs|.
Rfree was calculated by using 5% of the unique reflections.
Three-dimensional structure determination and refinement.
Each complex structure was solved by the molecular replacement method with the program MOLREP (36) and the 1.9-Å resolution native WT Oxy-Fd binary complex structure (Protein Data Bank [PDB] accession no. 2DE5) (24) with the water molecules removed. Manual model building was performed where necessary with the Xtalview (37) or Coot (38) software package. Refinement was carried out with the Refmac5 software in CCP4 (39) and CNS 1.1 (40) by adding water molecules gradually. The refinement statistics are shown in Table 1. Superimposition of the protein structures was carried out with the QUANTA (Accelrys, San Diego, CA) or CCP4 software. Figures were generated with Xfit in the XtalView program suite, Coot, and PyMOL (41).
Simulation of docking between substrates and the substrate-binding pocket of amino acid-substituted CARDO-O enzymes.
The structure of the Oxy-Fd-dioxygen-substrate quaternary complex was modeled as described previously (22) with slight modification. The position of the dioxygen was calculated on the basis of the assumption that the dioxygen binds to the mononuclear iron at the active site of each Oxy derivative in a manner similar to that observed in the Oxy-Fd-CAR-O2 complex (PDB accession no. 3VMI) (26). The coordinates of the Oxy-Fd-CAR-O2 complex were translated and rotated to minimize the root mean square deviation (RMSD) value between the residues coordinating the active-site iron. The oxygen atom positions obtained were used as those in the active sites of the derivatives. The docking simulations of FN binding to the active site of the WT enzyme were performed with GOLD, version 3.1 (42). The search area was a sphere with a radius of 10 Å centered at the active-site position near the mononuclear iron, as determined by PASS, version 1.1 (43). All predicted docking structures were classified by using the RMSD between the nonhydrogen atoms of the substrate as a distance measure.
Steady-state kinetic measurements and data analysis.
Enzymatic activity was measured by monitoring the consumption rate of each substrate. The standard activity assay was performed with 200 μl of air-saturated 50 mM Tris-HCl (pH 7.5) at 30°C. The reaction mixture contained 5 to 625 μM CAR, 500 μM NADH, 500 μM Fe(NH4)2(SO4)2·6H2O, 1 mM flavin adenine dinucleotide (FAD), 5.7 μM Fd, 5.4 μM Red, and 0.57 to 2.25 μM Oxy. The assay was initiated by adding the protein mixture after equilibrating the reaction mixture with all of the other reagents. The enzymatic reaction was stopped by adding 10 μl of 1 N HCl, and then the reaction mixture was immediately cooled with liquid nitrogen. The samples were injected directly into a high-performance liquid chromatography (HPLC) system (Waters 600E Separations Module equipped with a Waters 996 Photodiode Array Detector) equipped with a C8 reverse-phase column (PEGASIL C8; Senshu Scientific) to quantify the remaining substrate. The HPLC analyses were operated at a flow rate of 1.0 ml/min. To elute CAR and FN, a linear gradient of acetonitrile (40 to 60%) was applied for 8 min, beginning 3 min after injection, followed by 100% acetonitrile for 4 min. An aliquot of 40 μl (for kinetic analyses) or 100 μl (for O2 uptake analyses) was injected, and the amount of CAR was determined from the area of the absorbance peak at 291 nm by using a standard curve. FN was determined from the area of the absorbance peak at 260 nm by using a standard curve. Negative controls without Fd in the system did not exhibited any enzymatic activity.
Measurement of O2 uptake.
The O2 uptake assay was performed with 1.7 ml of air-saturated 50 mM Tris-HCl (pH 7.5) at 30°C. The reaction mixture contained 100 μM substrate, 200 μM NADH, 58.8 μM Fe(NH4)2(SO4)2·6H2O, 1.18 μM FAD, 1.25 μM Fd, 0.75 μM Red, and 0.25 μM Oxy. Reactions were initiated by adding protein mixture and were quenched 0, 15, 30, 45, and 60 s later by adding 10 μl of 1 N HCl, and then 100 μl of reaction mixture was flash-frozen in liquid nitrogen. Oxygen consumption was monitored with a Clarke-type O2 electrode (Iijima Electronics, Aichi, Japan). The consumption of substrate was quantified directly by HPLC as described above.
Protein structure accession numbers.
The atomic coordinates and structure factors determined in this study have been deposited in the PDB (http://www.pdb.org) under accession no. 4NB8, 4NB9, 4NBA, 4NBB, 4NBC, 4NBD, 4NBE, 4NBF, 4NBG, and 4NBH for I262L-Fd, I262V-Fd, I262V-Fd-CAR, I262V-Fd-CAR-O2, F275W-Fd form 1, F275W-Fd-CAR form 2, F275W-Fd-FN form 2, Q282N-Fd, and Q282Y-Fd-CAR, respectively (Table 1).
RESULTS AND DISCUSSION
Determination of the crystal structures of amino acid-substituted derivatives of Oxy.
Of the previously generated Oxy derivatives derived from Oxy of strain J3 (30), the I262L, I262V, F275A, F275W, Q282N, Q282Y, and F329L derivatives were purified, mixed with the Fd component of strain CA10, and crystallized as the Oxy-Fd complex for three-dimensional structure determination. The I262L, I262V, F275W, Q282N, and Q282Y derivatives and the substrate-bound forms of the I262V, F275W, and Q282Y derivatives were crystallized as Oxy-Fd-substrate ternary complexes, and their structures were determined (Table 1). The F275W-Fd binary complex crystallized as two forms (forms 1 and 2 in Table 1). As crystal form 1, the complexes consisted of one Oxy molecule (three CarAa [Oxy component of CARDO] subunits; chains A, B, and C) and three Fd molecules (chains D, E, and F), similar to the WT Oxy-Fd binary complex (24). Crystal form 2 (Table 1), however, contained only two Fd molecules (chains D and E) per Oxy molecule. Among the structures, those of the I262V, F275W, and Q282Y derivatives complexed with CAR were determined. The crystal structure of the F275W derivative complexed with FN was also determined. The relationships between the substrate specificities and conformational changes were investigated by comparisons of the derivatives with the WT structures.
In the following sections, each substrate-bound Oxy complex designation has a dash separating the relevant components of the complex (e.g., I262V-CAR indicates the CAR-bound I262V Oxy-Fd complex).
Structures of Oxy molecules with amino acid replacements at Ile262.
Ile262 is located at the entrance of the substrate-binding pocket. This residue interacts hydrophobically with the substrates (Fig. 2B). The crystal structures of the Leu- and Val-substituted I262L and I262V derivatives, respectively, were determined.
In the previous study using E. coli cells expressing the I262L derivative, this derivative increased anthracene dioxygenation activity by approximately 3-fold over that of E. coli cells expressing WT Oxy, but the I262L derivative catalyzed CAR with a slightly higher yield of the lateral dioxygenation product (Fig. 1) (30). The I262L derivative crystal structure was compared with that of substrate-free WT Oxy (PDB accession no. 2DE5). The shape of the substrate-binding pocket was unchanged in the mutant protein, except for the side chain of the substituted amino acid. This subtle change may be responsible for the slight shift in the yield of the lateral dioxygenation product.
In the previous biotransformation assay, E. coli cells expressing the I262V derivative exhibited less (<50%) CAR dioxygenation activity than the WT. However, a substantial portion of the dioxygenation was lateral, unlike the WT, which yielded only trace amounts of the lateral dioxygenation product (Fig. 1) (30). The I262V derivative crystal structure shows that the substrate-binding pocket was enlarged by the amino acid substitution (see Fig. S1 in the supplemental material). The hydrophobic interactions between the substituted amino acid residue and CAR were probably weaker than in the WT because of the enlarged binding pocket, leading to less rigid substrate binding and more lateral dioxygenation of the substrate.
The crystal structure of I262V-CAR was obtained after soaking the I262V derivative crystals in CAR-containing mother liquor. In the I262V-CAR crystal structure, the electron density corresponding to CAR was observed in chains B and C, as in the WT (24). In the WT-CAR crystal structure, residues Leu202 to Thr214 and Asp229 to Val238, which surround the entrance to the substrate-binding pocket, are shifted toward the inside. This shift is not observed in the I262V-CAR crystal structure. Interestingly, superposition of the I262V-CAR and WT-CAR structures showed that CAR in chains B and C of I262V-CAR is rotated approximately 15° compared to that in WT-CAR (PDB accession no. 2DE7; Fig. 3A).
FIG 3.
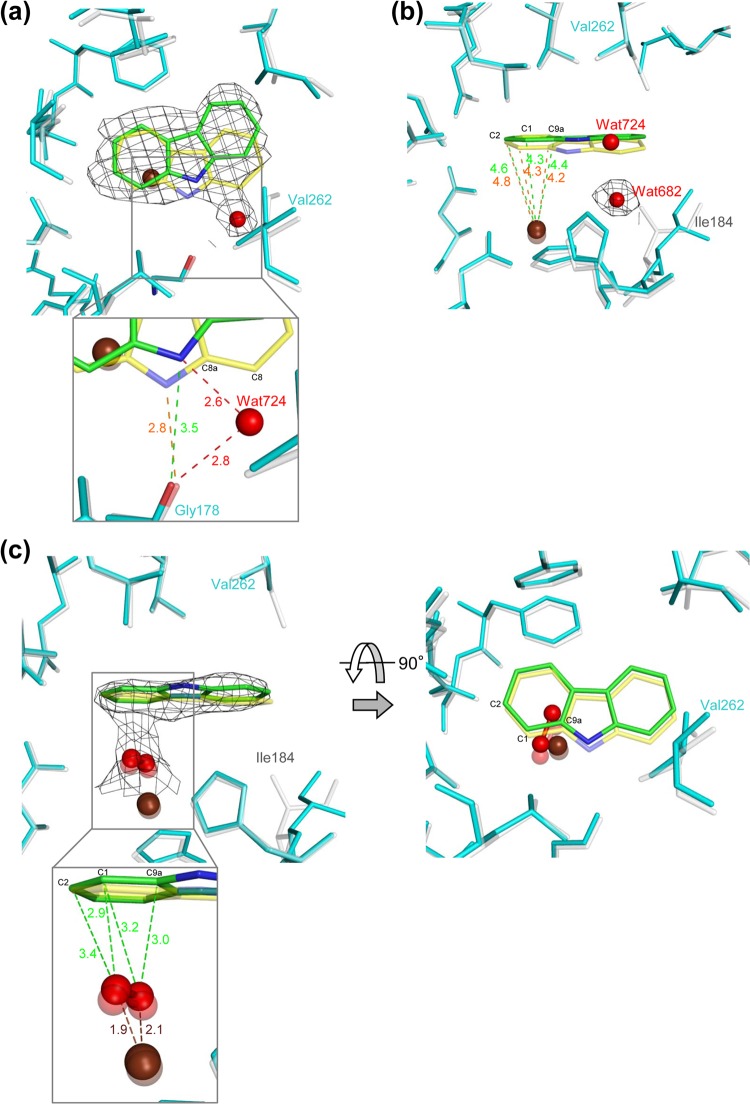
Substrate-binding pocket in chain B of the I262V CARDO (Oxy) derivative with bound CAR (PDB accession no. 4NBA) (a and b) and with bound CAR and peroxide (PDB accession no. 4NBB) (c). CAR is shown in the orientations observed in WT Oxy (yellow; PDB accession no. 2DE7 in panels a and b, 3VMI in panel c) and the I262V derivative (green). The mononuclear iron (brown sphere) and water (small red sphere) are also shown. (a) Overlay of WT Oxy (white) and the I262V derivative (cyan) complexed with CAR (I262V-CAR). The electron density of CAR (0.5 σ 2Fo-Fc) and a water molecule (Wat724) in the I262V derivative is shown as a black wire mesh. The distances between the carbonyl oxygen of Gly178 and the imino nitrogen of CAR in the WT and the I262V derivative are in orange and green (inset), respectively. The distances from Wat724 to the carbonyl oxygen of Gly178 and the imino nitrogen of CAR are also shown (dotted lines and red values). (b) Overlay of WT Oxy and I262V-CAR. The electron density of a water molecule (Wat682) in I262V-CAR is shown as a black wire mesh. The distances between the mononuclear iron and carbon atoms of CAR in the WT (orange) and the I262V derivative (green) are shown. (c) The distances from an oxygen species to the mononuclear iron (brown) and to the carbon atoms of CAR (green) in I262V-CAR (inset). The electron density of CAR (0.5 σ 2Fo-Fc) and the oxygen species in I262V-CAR is shown as a black wire mesh.
In the WT-CAR crystal structure, Ile184 is shifted toward the inside of the pocket and a water molecule is displaced by CAR binding (Fig. 3B). This shift of Ile184 likely minimizes interference with the substrate and enables the substrate to bind tightly (24). This shift of Ile184 in chains B and C is not present in the crystal structure of I262V-CAR, and the water molecule (Wat682 in chain B) is still present (Fig. 3B).
The 15° rotation of CAR in the I262V-CAR crystal structure alters the distances between the nonheme iron and the carbon atoms that could be attacked by oxygen. The distances between C-9a, C-1, and C-2 of CAR and the nonheme iron in chain B are 4.4, 4.3, and 4.6 Å, respectively, in the I262V-CAR crystal structure, as opposed to 4.2, 4.3 and 4.8 Å, respectively, in the WT-CAR crystal structure (Fig. 3B). Thus, in I262V-CAR, the sum of the distances of C-9a and C-1 from the nonheme iron (8.7 Å) is similar to that of C-1 and C-2 (8.9 Å), whereas in WT-CAR, the sum of the distances of C-9a and C-1 from the nonheme iron (8.5 Å) is significantly less than that of C-1 and C-2 (9.1 Å) (Fig. 3B). In addition, the CAR rotation creates open space in the substrate-binding pockets of chains B and C for water molecules (Wat724 and Wat734, respectively) close to C-8 of CAR (Fig. 3A). The water molecules hydrogen bond with the imino nitrogen of CAR and the carbonyl oxygen of Gly178, with bond distances of 2.6 and 2.8 Å, respectively. In the I262V-CAR crystal structure, the distance between the carbonyl oxygen of Gly178 and the imino nitrogen of CAR is 3.5 Å, as opposed to 2.8 Å in the WT-CAR structure (Fig. 3A). Thus, CAR orientation is likely stabilized by hydrogen bonding networks via water molecules in I262V, which is consistent with higher lateral dioxygenation activity (and consequently a higher yield of 1,2-dihydroxycarbazole) of the I262V derivative than of the WT.
The crystal structure of I262V-CAR with oxygen species close to the mononuclear iron was obtained after soaking the crystals in sodium dithionite and CAR and then exposing them to oxygen for 10 min as described by Ashikawa et al (26). (Fig. 3C). The distance between the oxygen atoms was 1.5 to 1.6 Å thus, the dioxygen species was likely hydrogen peroxide. The distances between the oxygen atoms and iron were 1.9 and 2.1 Å (Fig. 3C). This indicates that the oxygen species binds side on, as it does in other ROs, including WT Oxy (26, 44, 45). As with I262V-CAR without oxygen, the shift of Ile184 by substrate binding is not observed in the crystal structure of I262V-CAR with oxygen. However, with oxygen present, the CAR in I262V-CAR is not rotated and is oriented in the substrate-binding pocket very similarly to that in WT-CAR, with a slight shift toward the open space created by the amino acid substitution. Furthermore, the water molecule corresponding to Wat724, which was hydrogen bonded to the imino nitrogen of CAR in chain B of I262V-CAR, is not present in the crystal structure of I262V-CAR with oxygen (Fig. 3C). Thus, on the basis of the distance between CAR and the oxygen, angular dioxygenation is the most probable reaction in I262V-CAR with oxygen.
The shift of residues Leu202 to Thr214 and Asp229 to Val238 toward the substrate-binding pocket, seen in the WT-CAR crystal structure, is not seen in the I262V-CAR structure, irrespective of the presence of oxygen (24). This suggests that the entrance to the binding pocket in the I262V derivative is almost always open, which would leave the substrate bound more loosely than in the WT. Thus, oxygen may attack CAR at C-1 and C-9a, as well as at C-1 and C-2.
Structures of Oxy with Phe275 replaced with Trp.
Phe275 is located at the substrate-binding pocket close to the entrance and is likely to interact hydrophobically with aromatic substrates (Fig. 2). In the previous biotransformation assay, the F275W derivative also catalyzes the lateral dioxygenation of CAR (Fig. 1) (30). Moreover, the F275W derivative has unique catalytic properties with FN and anthracene. WT Oxy catalyzes the conversion of FN to 9-hydroxyfluorene and 4-hydroxyfluorene with 5.9 and 3.0% relative yields, respectively, whereas the F275W derivative yields significantly more of the latter product (3.9 and 22.6%, respectively; Fig. 1) (30). The products of the oxygenation of anthracene by WT Oxy are anthracene-1,2-dihydrodiol and putative anthracene-2,3-dihydrodiol with 20.8 and 6.2% relative yields, respectively. These yields are 18.5 and 22.7%, respectively, with the F275W derivative (Fig. 1). The F275W derivative products with an increased yield are likely the result of lateral dioxygenation of the aromatic substrates. The substituted Trp residue, being bulkier than the original Phe, slightly evened out a recessed region of the substrate-binding pocket (see Fig. S2 in the supplemental material); no other significant difference is apparent between the crystal structure of the F275W derivative and that of the WT.
Crystals of the F275W derivative were obtained in forms 1 and 2 (Table 1). Crystals of F275W-CAR were obtained from form 2 F275W crystals. Electron density maps showed that CAR was bound to all three Oxy chains, but the average temperature factor of CAR in chain C (27.7 Å2) was significantly lower than that in chain A (54.9 Å2) or B (58.0 Å2). In the crystal structure, Fd components are bound to chains A and B but not chain C, implying that the substrate is stabilized by disassembly of Fd. In F275W-CAR, residues Leu202 to Thr214 and Asp229 to Val238 around the substrate-binding pocket entrance are shifted in chains B and C but not in chain A. These results indicate that Fd binding may have a greater influence on substrate stability than the open or closed state of the substrate-binding pocket. Ile184 was shifted toward the inside of the pocket by substrate binding in all of the chains, similar to WT-CAR. In chains B and C, CAR was oriented in the substrate-binding site similarly to WT-CAR, while in chain A, CAR was rotated approximately 25° compared to that in WT-CAR. This is reminiscent of the manner in which CAR binds to chains B and C in I262V-CAR (Fig. 4A). Although the rotated CAR binding was observed only in chain A of the F275W derivative, this rotation may be sufficient to explain the increased yield of lateral dioxygenation products of the F275W derivative (Fig. 1).
FIG 4.
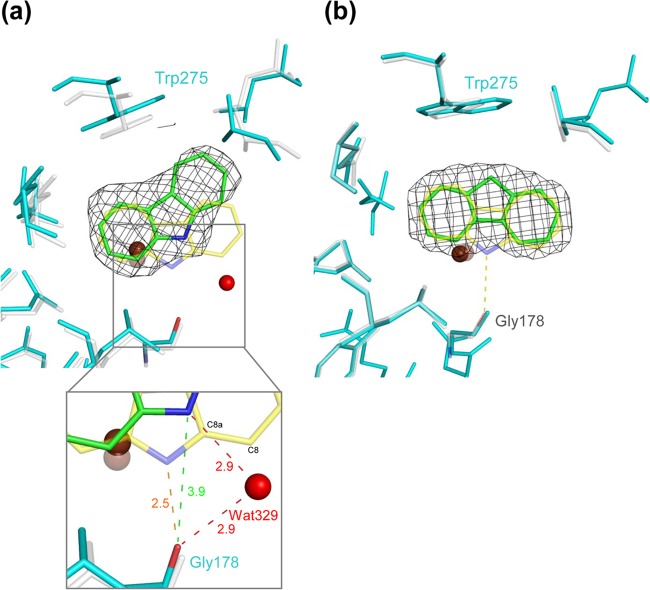
Substrate-binding pocket in chain C of the F275W derivative of CARDO (Oxy) complexed with FN overlaid on terminal Oxy structures. The mononuclear iron is shown as a brown sphere. (a) Overlaid structures of the F275W derivative (cyan) with CAR (green) (PDB accession no. 4NBD) and WT Oxy (white) with CAR (yellow) (PDB accession no. 2DE7). The electron density (1.0 σ 2Fo-Fc) of CAR in the F275W derivative is shown as a black wire mesh. The distances between the carbonyl oxygen of Gly178 and the imino nitrogen of CAR in the WT and the F275W derivative are orange and green (inset), respectively. (b) Overlaid structures of the F275W derivative (cyan) with FN (green) and WT Oxy (white) with CAR (yellow). The electron density (1.0 σ 2Fo-Fc) of FN in the F275W derivative is shown as a black wire mesh. The direction of C-9 of FN was the opposite of that of the imino nitrogen of CAR, which forms a hydrogen bond with the carbonyl oxygen of Gly178 in WT Oxy complexed with bound CAR.
The F275W-FN crystal structure was also determined. Bulky electron densities at an active site were present in only chains A and C in the Fourier map, and FN molecules were modeled to fit the configuration (Fig. 4B). However, the movement of residues L202-T214 and D229-V238 around the substrate-binding pocket entrance, as seen in the F275W-CAR crystal structure, is observed only in chain C of the F275W-FN crystal structure. This suggests that FN binds at the pocket differently than CAR. C-9 of FN is oriented toward mutated amino acid residue Trp275 and is likely stabilized by hydrophobic interaction with the residue (Fig. 4B; see Fig. S3 in the supplemental material). In contrast, the imino nitrogen of CAR hydrogen bonds with Gly178, located at the opposite side of the substrate-binding pocket from Phe275/Trp275.
Molecular modeling of oxygen on the active-site mononuclear iron in the F275W derivative indicates that the oxygen likely attacks C-3 and C-4 of FN rather than C-9; the distances from the oxygen atoms to C-3, C-4, and C-9 of FN were 3.0 (O-2 to C-4), 3.0 (O-1 to C-3), and 4.3 (O-2 to C-9) Å, respectively. Since dihydrodiol frequently undergoes a loss of water to yield a monohydroxylated compound (46), these structural observations are consistent with the increased yield of 4-hydroxyfluorene and reduced yield of 9-hydroxyfluorene of the F275W derivative.
Because the structure of WT Oxy-FN was not obtained, we modeled its structure by using a docking simulation. The docking poses varied, depending on the conditions of the simulation. When FN was docked into WT Oxy-CAR, the docking pose (pose 1) was similar to that in the crystal structure of F275W-FN. When FN was docked into the substrate-free form of WT Oxy, the docking pose (pose 2) was rotated approximately 90° compared to that in pose 1 while the atoms of the two poses lay in almost the same plane. Since the hydrogen atoms bound to C-9 of FN are weakly acidic, we performed the docking simulation enabling the donation of a hydrogen bond by C-9 of FN. In this case, the docking pose (pose 3) was flipped with respect to pose 2, and C-9 of FN formed a hydrogen bond with one of the oxygen atoms bound to the mononuclear iron. Because WT Oxy catalyzes FN to 4-hydroxyfluorene and 9-hydroxyfluorene and because poses 1 and 3 are suitable to produce them, respectively, we surmise that FN probably binds to WT Oxy in multiple poses, as predicted by the docking simulations.
In nitrobenzene dioxygenase (NBDO), the residue corresponding to Phe275 of Oxy is Asn258, which forms a hydrogen bond with one of the oxygen atoms in the nitro group of 3-nitrotoluene (47). Mutation of Asn258 to a Val in NBDO changed the regioselectivity of product formation (12). In Oxy, although the original and substituted residues are both hydrophobic, nevertheless, the mutation affected substrate specificity. These findings indicate that the residue at position 275 in Oxy and in the corresponding position of other ROs plays an important role in substrate specificity and product formation, not only by affecting hydrogen bonding to the substrate but also by hydrophobic interactions.
Structures of Oxy with amino acid replacement at Gln282.
Gln282 is one of the residues composing the substrate-interacting wall on the inner surface of the substrate-binding pocket of Oxy (Fig. 2). In this study, the crystal structures of the Asn- and Tyr-substituted Q282N and Q282Y derivatives, respectively, were determined. In the previous biotransformation assay, the oxygenation of CAR by the Q282N derivative yields less of the angular dioxygenation product and more of the lateral dioxygenation product than WT Oxy (Fig. 1) (30). The crystal structure of the Q282N derivative shows a small depression on the surface of the substrate-binding pocket, compared with WT Oxy (see Fig. S4 in the supplemental material). This depression widens part of the pocket, which may decrease the stability of the bound substrate and lead to more lateral dioxygenation of CAR. However, we cannot confirm this hypothesis at this time because crystals of the substrate-bound Q282N derivative were not obtained.
According to the previous biotransformation assay, the Q282Y derivative gave the highest yield of lateral dioxygenation product from CAR in a previous study of Oxy derivatives (Fig. 1) (30). The crystal structure of the Q282Y derivative shows that the hydroxyl group of the substituted Tyr hydrogen bonds with the O-ε atom of Glu284 and the N-ε atom of Gln298, although no significant difference from WT Oxy is apparent on the surface of the substrate-binding pocket (see Fig. S5 in the supplemental material).
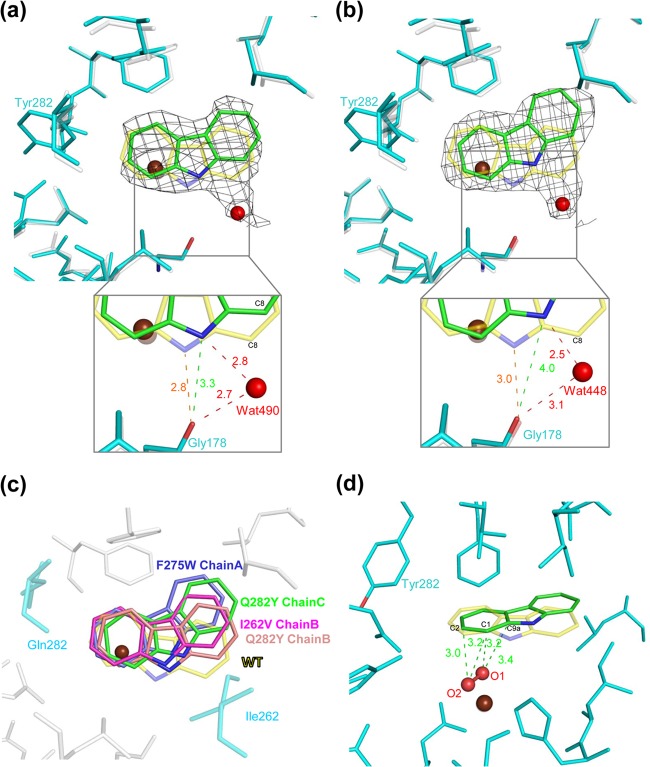
The crystal structure of Q282Y-CAR was determined. Electron density maps show that CAR is bound only in chains B and C of Q282Y-CAR, similar to WT-CAR (Fig. 5A and B) (24). However, like I262V-CAR, no movement of residues Leu202 to Thr214 and Asp229 to Val238 around the substrate-binding pocket entrance upon CAR binding is seen in Q282Y-CAR. The orientation of CAR in Q282Y-CAR differs from that in WT-CAR but is similar to that in chains B and C of I262V-CAR and chain A of F275W-CAR. CAR molecules in Q282Y-CAR and WT-CAR lie in almost the same plane, but the CAR in Q282Y-CAR is rotated approximately 15 and 25° in chains B and C, respectively, compared to that in WT-CAR (Fig. 5A and B). In Q282Y-CAR, water molecules (Wat490 and Wat448 in chains B and C, respectively) near C-8 (angular carbon) of CAR hydrogen bond with the imino nitrogen of CAR and the carbonyl oxygen of Gly178, stabilizing its orientation. As mentioned above, this CAR rotation is commonly found in the I262V derivative (Fig. 5C), which also gives a relatively high yield of the lateral dioxygenation product. To gain insight into the oxygenation mechanism, we modeled Q282Y-CAR chain C with oxygen on the basis of the crystal structure of WT-CAR with oxygen species (26). C-1 and C-2 were the closest carbon atoms in CAR to modeled oxygen atoms O-1 and O-2, respectively, in chain C of Q282Y-CAR, whereas the closest carbon atoms were C-9a and C-1, respectively, in WT-CAR (Fig. 5D). In chain B, however, C-1 was the closest carbon atom to both oxygen atoms (data not shown). These modeled structures suggest that the Q282Y derivative preferentially catalyzes the lateral dioxygenation of CAR by hydroxylating the C-1 and C-2 atoms. Taken together with the substrate-binding data for the I262V derivative, these results suggest that the slightly different orientation of CAR in the substrate-binding pocket, compared to that in the WT, is the reason for the increased lateral dioxygenation of CAR.
FIG 5.
Comparison of CAR binding in the substrate-binding pocket of WT CARDO (Oxy) and the Q282Y derivative of Oxy (PDB accession no. 2DE7 and 4NBH, respectively). Overlays of chains B and C of the Q282Y derivative (cyan) and WT Oxy (white) are shown in panels a and b, respectively. The Q282Y derivative and WT Oxy with bound CAR (PDB accession no. 2DE7) were superimposed with C-α. The CAR molecules are in the orientations observed in the Q282Y derivative and WT Oxy (green and yellow, respectively). The electron density (0.5 σ 2Fo-Fc) of CAR in the Q282Y derivative is shown as a black wire mesh. The water molecules that hydrogen bond with Gly178 and CAR are shown as red spheres (Wat490 and Wat448 for chains B and C, respectively). The distances between the carbonyl oxygen of Gly178 and the imino nitrogen and water molecules are also shown (insets). (c) The rotation of CAR in the substrate-binding pocket in WT Oxy and the I262V and Q282Y derivatives is shown. All of the structures, the I262V, F275W, and Q282Y derivatives and WT Oxy, complexed with CAR (PDB accession no. 4NBA, 4NBD, 4NBH, and 2DE7, respectively) are superimposed with C-α. CAR is shown in the substrate-binding pockets of the I262V (pink), F275W (blue), and Q282Y (chains B and C are in salmon and green, respectively) derivatives and WT Oxy (yellow). (d) Modeled oxygen species in the CAR-bound Q282Y derivative and the distances between carbon atoms C-1, C-2, and C-9 of CAR and the oxygen species. The oxygen species were modeled on the basis of WT Oxy complexed with CAR and oxygen species (3VMI) (26).
Steady-state utilization of substrates.
The structural analysis of the CARDO derivatives demonstrated the effects of mutations on substrate-binding orientation. To investigate the binding affinity between the substrate and the derivatives, steady-state kinetic analyses and oxygen uptake assays were performed with the purified proteins. In the previous biotransformation assay with whole E. coli cells, the substrate preferences were calculated on the basis of the relative abundance of the substrates and products, but in this assay, purified enzymes were used and the remaining substrate was measured quantitatively. The kinetic parameters of WT CARDO oxygenation of CAR were not obtained with sufficient precision because of the high catalytic activity of the WT enzyme, the low solubility of CAR in the reaction buffer, and the product detection limits. However, Km and kcat were determined for the oxygenation of CAR by the I262V, Q282N, and Q282Y derivatives by virtue of their lower reaction rates. FN, however, was not soluble enough for determination of the parameters (Table 2). The rates of oxygen uptake and substrate consumption for the oxygenation of CAR by the WT and the I262V, Q282N, and Q282Y derivatives and for the oxygenation of FN by the WT and the F275W derivative are presented in Table 2.
TABLE 2.
Steady-state substrate utilization by mutant CARDO enzymesa
| Oxy of CARDO | Substrate | Km (μM) | kcat (10−2 s−1) | kcat/Km (10−4 M−1 s−1) | O2 uptake (nmol s−1)c | Substrate consumption (nmol s−1) | Substrate/O2b |
|---|---|---|---|---|---|---|---|
| WT | CAR | NDd | ND | ND | 0.840 ± 0.002 | 0.651 ± 0.089 | 0.78 |
| WT | FN | ND | ND | ND | 0.361 ± 0.026 | 0.195 ± 0.071 | 0.54 |
| I262V derivative | CAR | 50.7 ± 12.3 | 4.62 ± 0.53 | 9.10 ± 1.28 | 0.250 ± 0.015 | 0.146 ± 0.074 | 0.58 |
| F275W derivative | FN | ND | ND | ND | 0.507 ± 0.030 | 0.264 ± 0.035 | 0.52 |
| Q282N derivative | CAR | 54.6 ± 4.4 | 4.00 ± 0.12 | 7.33 ± 0.41 | 0.310 ± 0.018 | 0.128 ± 0.022 | 0.41 |
| Q282Y derivative | CAR | 54.8 ± 21.1 | 5.49 ± 0.02 | 10.0 ± 2.2 | 0.880 ± 0.024 | 0.564 ± 0.064 | 0.64 |
The indicated values represent means and standard deviations obtained from three determinations. O2 consumption was corrected for the background activity observed in the absence of substrate.
Each value is the ratio of the rate of substrate consumption to the average rate of O2 consumption over the duration of the assay.
O2 concentrations without a substrate were measured by using catalase during the reaction as a negative control, and the oxygen concentration did not increase under any conditions.
ND, not determined.
There was no significant difference in the kcat/Km and Km values of the I262V, Q282N, and Q282Y derivatives, indicating that the affinity for CAR and turnover rates of these derivatives are similar. The ratio of the rate of substrate consumption to the rate of O2 uptake for WT CARDO was higher when CAR, rather than FN, was the substrate. This ratio was also higher for WT CARDO than for any of the derivatives, indicating that oxygen is effectively utilized by WT CARDO to degrade the substrate. The lower value of the ratio in the derivatives is likely due to an uncoupling of oxygen activation from substrate binding. This suggests that the activation of oxygen occurs even when the substrate is not properly positioned, explaining the unusual oxygenation products generated by the derivatives, such as lateral dioxygenation products from the I262V and Q282Y derivatives. Although the relative proportions of the two oxygenation products from FN, 9-hydroxyfluorene and 4-hydroxyfluorene, differed between the WT and the F275W derivative (the F275W derivative yielded more 4-hydroxyfluorene than 9-hydroxyfluorene, whereas the WT yields were similar), the ratios of the rate of FN consumption to the rate of O2 uptake were comparable for the WT and the F275W derivative. This suggests that FN is catalyzed by the F275W derivative at a rate similar to that of the WT but that a larger proportion of FN binds to the F275W derivative with an orientation in which C-9 of FN is positioned toward the substituted Trp residue, as shown in Fig. 4B.
Structural basis for substrate specificity.
Previous studies of CARDO demonstrated that site-directed mutagenesis of the hydrophobic amino acids located at the surface of the substrate-binding pocket alters the substrate specificity of the enzyme (30). In the crystal structure of WT-CAR, amino acid residues Leu202 to Thr214 and Asp229 to Val238 near the entrance of the substrate-binding pocket are shifted inward (26). This conformational change effectively closes the substrate-binding pocket once the substrate is bound and the substrate is located at the proper position in the pocket. The crystal structure of F275W-CAR, but not I262V-CAR, Q282Y-CAR, or F275W-FN, shows the same conformational change. The average temperature factors of the Leu202-to-Thr214 and Asp229-to-Val238 regions were significantly higher (32.0 and 30.8 Å2 in I262V-CAR, 35.2 and 33.3 Å2 in Q282Y-CAR, and 49.1 and 46.8 Å2 in F275W-FN, respectively) than those of the whole enzyme-substrate complex (26.8 Å2 in I262V-CAR, 27.9 Å2 in Q282Y-CAR, and 42.8 Å2 in F275W-FN; Table 1), indicating a relatively high degree of flexibility in these regions. This flexibility around the entrance to the substrate-binding pocket may facilitate the accommodation of various substrates (2). To further investigate the mechanisms governing the opening and closing of the entrance, we performed a hierarchical clustering according to the pairwise RMSD distance matrix for each chain in the structures determined (see Fig. S6 in the supplemental material). In the WT, the structures with and without a substrate are divided into different clusters. However, the substrate-binding pockets in the I262V and Q282Y derivatives fall in the same cluster as the open-state configurations of the WT, regardless of substrate binding. In F275W-CAR, the entrance was open in chain A, in which CAR was rotated, and closed in chains B and C, in which CAR was bound in an orientation similar to that in WT-CAR. In F275W-FN, the entrance was open in chain A and closed in chain C. Comparing the WT and I262V and Q282Y derivative structures with or without substrates, it appears that amino acid substitution at a position inside the substrate-binding pocket makes the entrance more likely to remain open after substrate binding. However, it is still unclear what controls the transition from the open to the closed configuration, and thus, further biochemical and structural analyses are needed to resolve this issue.
In this study, the crystal structures of amino acid-substituted derivatives of CARDO demonstrated novel substrate-binding modes in CARDO. The hydrophobic residues composing the substrate-binding pocket are altered in the I262V and F275W derivatives. The substituted residues in these derivatives are smaller or larger than the original residues but do not have significantly different polar and charge properties (Fig. 1 to 5). These substitutions altered substrate binding by modifying the patterns of hydrophobic interaction between the residues and the aromatic rings of the substrate. Generally, in ROs, a large part of the substrate-binding pocket surface consists of hydrophobic residues so that aromatic substrates can bind stably (2). The results of this study indicate that these hydrophobic residues that make up the surface of the substrate-binding pocket of ROs are potential targets for enzyme engineering to alter the specific site of oxygenation of aromatic rings. In the structure of the Q282Y derivative, there was an only slight difference in the shape of the surface of the substrate-binding pocket from that of WT Oxy, yet a water molecule was introduced that participates in a novel hydrogen bond network in the Q282Y derivative, causing CAR to bind in the pocket with a rotated orientation. This indicates that amino acid substitution, even without a drastic change in the shape of the substrate-binding pocket, can alter the substrate-binding orientation and enzymatic properties. The difference in opening and closing of the substrate-binding pocket was caused by the amino acid replacements. This implies that the slight change can also loosen substrate binding by keeping the entrance of the pocket open where proper substrate recognition and subsequent induced fit conformational changes are needed for the usual oxygenation reaction.
Supplementary Material
ACKNOWLEDGMENTS
Synchrotron radiation was used for this work with the approval of the PF Advisory Committee, the High Energy Accelerator Research Organization (proposals 2004G137, 2005G060, 2006G171, 2007G135, 2008G681, 2008G702, 2009G675, and 2010G663), and the Japan Synchrotron Radiation Research Institute (proposal 2005B0985).
Footnotes
Published ahead of print 28 February 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.04000-13.
REFERENCES
- 1.Parales RE, Resnick SM. 2006. Aromatic ring hydroxylating dioxygenases, p 287–340 In Ramos JL, Levesque RC. (ed), Pseudomonas, vol 4 Springer, New York, NY [Google Scholar]
- 2.Ferraro DJ, Gakhar L, Ramaswamy S. 2005. Rieske business: structure-function of Rieske non-heme oxygenases. Biochem. Biophys. Res. Commun. 338:175–190. 10.1016/j.bbrc.2005.08.222 [DOI] [PubMed] [Google Scholar]
- 3.Bugg TDH, Ramaswany S. 2008. Non-heme iron-dependent dioxygenases: unravelling catalytic mechanisms for complex enzymatic oxidations. Curr. Opin. Chem. Biol. 12:134–140. 10.1016/j.cbpa.2007.12.007 [DOI] [PubMed] [Google Scholar]
- 4.Parales RE, Lee K, Resnick SM, Jiang H, Lessner DJ, Gibson DT. 2000. Substrate specificity of naphthalene dioxygenase: effect of specific amino acids at the active site of the enzyme. J. Bacteriol. 182:1641–1649. 10.1128/JB.182.6.1641-1649.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Parales RE, Resnick SM, Yu CL, Boyd DR, Sharma ND, Gibson DT. 2000. Regioselectivity and enantioselectivity of naphthalene dioxygenase during arene cis-dihydroxylation: control by phenylalanine 352 in the alpha subunit. J. Bacteriol. 182:5495–5504. 10.1128/JB.182.19.5495-5504.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yu CL, Parales RE, Gibson DT. 2001. Multiple mutations at the active site of naphthalene dioxygenase affect regioselectivity and enantioselectivity. J. Ind. Microbiol. Biotechnol. 27:94–103. 10.1038/sj.jim.7000168 [DOI] [PubMed] [Google Scholar]
- 7.Suenaga H, Goto M, Furukawa K. 2001. Emergence of multifunctional oxygenase activities by random priming recombination. J. Biol. Chem. 276:22500–22506. 10.1074/jbc.M101323200 [DOI] [PubMed] [Google Scholar]
- 8.Suenaga H, Watanabe T, Sato M, Ngadiman, Furukawa K. 2002. Alteration of geospecificity in biphenyl dioxygenase by active-site engineering. J. Bacteriol. 184:3682–3688. 10.1128/JB.184.13.3682-3688.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Suenaga H, Goto M, Furukawa K. 2006. Active-site engineering of biphenyl dioxygenase: effect of substituted amino acids on substrate specificity and regiospecificity. Appl. Microbiol. Biotechnol. 71:168–176. 10.1007/s00253-005-0135-2 [DOI] [PubMed] [Google Scholar]
- 10.Zielinski M, Kahl S, Hecht HJ, Hofer B. 2003. Pinpointing biphenyl dioxygenase residues that are crucial for substrate interaction. J. Bacteriol. 185:6976–6980. 10.1128/JB.185.23.6976-6980.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zielinski M, Kahl S, Standfuss-Gabisch C, Camara B, Seeger M, Hofer B. 2006. Generation of novel-substrate-accepting biphenyl dioxygenases through segmental random mutagenesis and identification of residues involved in enzyme specificity. Apple. Environ. Microbiol. 72:2191–2199. 10.1128/AEM.72.3.2191-2199.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee KS, Parales JV, Friemann R, Parales RE. 2005. Active site residues controlling substrate specificity in 2-nitrotoluene dioxygenase from Acidovorax sp. strain JS42. J. Ind. Microbiol. Biotechnol. 32:465–473. 10.1007/s10295-005-0021-z [DOI] [PubMed] [Google Scholar]
- 13.Ju KS, Parales RE. 2006. Control of substrate specificity by active-site residues in nitrobenzene dioxygenase. Appl. Environ. Microbiol. 72:1817–1824. 10.1128/AEM.72.3.1817-1824.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Barriault D, Sylvestre M. 2004. Evolution of the biphenyl dioxygenase BphA from Burkholderia xenovorans LB400 by random mutagenesis of multiple sites in region III. J. Biol. Chem. 279:47480–47488. 10.1074/jbc.M406805200 [DOI] [PubMed] [Google Scholar]
- 15.Ferraro DJ, Okerlund AL, Mowers JC, Ramaswamy S. 2006. Structural basis for regioselectivity and stereoselectivity of product formation by naphthalene 1,2-dioxygenase. J. Bacteriol. 188:6986–6994. 10.1128/JB.00707-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kumar P, Mohammadi M, Viger JF, Barriault D, Gomez-Gil L, Eltis LD, Bolin JT, Sylvestre M. 2011. Structural insight into the expanded PCB-degrading abilities of a biphenyl dioxygenase obtained by directed evolution. J. Mol. Biol. 405:531–547. 10.1016/j.jmb.2010.11.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mohammadi M, Viger JF, Kumar P, Barriault D, Bolin JT, Sylvestre M. 2011. Retuning Rieske-type oxygenases to expand substrate range. J. Biol. Chem. 286:27612–27621. 10.1074/jbc.M111.255174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nojiri H. 2012. Structural and molecular genetic analyses of the bacterial carbazole degradation system. Biosci. Biotechnol. Biochem. 76:1–18. 10.1271/bbb.110620 [DOI] [PubMed] [Google Scholar]
- 19.Sato SI, Nam JW, Kasuga K, Nojiri H, Yamane H, Omori T. 1997. Identification and characterization of genes encoding carbazole 1,9a-dioxygenase in Pseudomonas sp. strain CA10. J. Bacteriol. 179:4850–4858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Inoue K, Widada J, Nakai S, Endoh T, Urata M, Ashikawa Y, Shintani M, Saiki Y, Yoshida T, Habe H, Omori T, Nojiri H. 2004. Divergent structures of carbazole degradative car operons isolated from Gram-negative bacteria. Biosci. Biotechnol. Biochem. 68:1467–1480. 10.1271/bbb.68.1467 [DOI] [PubMed] [Google Scholar]
- 21.Inoue K, Habe H, Yamane H, Nojiri H. 2006. Characterization of novel carbazole catabolism genes from Gram-positive carbazole degrader Nocardioides aromaticivorans IC177. Appl. Environ. Microbiol. 72:3321–3329. 10.1128/AEM.72.5.3321-3329.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nojiri H, Ashikawa Y, Noguchi H, Nam JW, Urata M, Fujimoto Z, Uchimura H, Terada T, Nakamura S, Shimizu K, Yoshida T, Habe H, Omori T. 2005. Structure of the terminal oxygenase component of angular dioxygenase, carbazole 1,9a-dioxygenase. J. Mol. Biol. 351:355–370. 10.1016/j.jmb.2005.05.059 [DOI] [PubMed] [Google Scholar]
- 23.Nam JW, Noguchi H, Fujimoto Z, Mizuno H, Ashikawa Y, Abo M, Fushinobu S, Kobashi N, Wakagi T, Iwata K, Yoshida T, Habe H, Yamane H, Omori T, Nojiri H. 2005. Crystal structure of the ferredoxin component of carbazole 1,9a-dioxygenase of Pseudomonas resinovorans strain CA10, a novel Rieske non-heme iron oxygenase system. Proteins 58:779–789. 10.1002/prot.20374 [DOI] [PubMed] [Google Scholar]
- 24.Ashikawa Y, Fujimoto Z, Noguchi H, Habe H, Omori T, Yamane H, Nojiri H. 2006. Electron transfer complex formation between oxygenase and ferredoxin components in Rieske nonheme iron oxygenase system. Structure 14:1779–1789. 10.1016/j.str.2006.10.004 [DOI] [PubMed] [Google Scholar]
- 25.Inoue K, Ashikawa Y, Umeda T, Abo M, Katsuki J, Usami Y, Noguchi H, Fujimoto Z, Terada T, Yamane H, Nojiri H. 2009. Specific interactions between the ferredoxin and terminal oxygenase components of a class IIB Rieske nonheme iron oxygenase, carbazole 1,9a-dioxygenase. J. Mol. Biol. 392:436–451. 10.1016/j.jmb.2009.07.029 [DOI] [PubMed] [Google Scholar]
- 26.Ashikawa Y, Fujimoto Z, Usami Y, Inoue K, Noguchi H, Yamane H, Nojiri H. 2012. Structural insight into the substrate- and dioxygen-binding manner in the catalytic cycle of Rieske nonheme iron oxygenase system, carbazole 1,9a-dioxygenase. BMC Struct. Biol. 12:15. 10.1186/1472-6807-12-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nojiri H, Nam JW, Kosaka M, Morii K, Takemura T, Furihata K, Yamane H, Omori T. 1999. Diverse oxygenations catalyzed by carbazole 1,9a-dioxygenase from Pseudomonas sp. strain CA10. J. Bacteriol. 181:3105–3113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Takagi T, Nojiri H, Yoshida T, Habe H, Omori T. 2002. Detailed comparison between the substrate specificities of two angular dioxygenases, dibenzofuran 4,4a-dioxygenase from Terrabacter sp. and carbazole 1,9a-dioxygenase from Pseudomonas resinovorans. Biotechnol. Lett. 24:2099–2106. 10.1023/A:1021325816766 [DOI] [Google Scholar]
- 29.Nam JW, Nojiri H, Noguchi H, Uchimura H, Yoshida T, Habe H, Yamane H, Omori T. 2002. Purification and characterization of carbazole 1,9a-dioxygenase, a three-component dioxygenase system of Pseudomonas resinovorans strain CA10. Appl. Environ. Microbiol. 68:5882–5890. 10.1128/AEM.68.12.5882-5890.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Uchimura H, Horisaki T, Umeda T, Noguchi H, Usami Y, Li L, Terada T, Nakamura S, Shimizu K, Takemura T, Habe H, Furihata K, Omori T, Yamane H, Nojiri H. 2008. Alteration of the substrate specificity of the angular dioxygenase carbazole 1,9a-dioxygenase. Biosci. Biotechnol. Biochem. 72:3237–3248. 10.1271/bbb.80512 [DOI] [PubMed] [Google Scholar]
- 31.Sambrook J, Russell DW. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 32.Ashikawa Y, Fujimoto Z, Noguchi H, Habe H, Omori T, Yamane H, Nojiri H. 2005. Crystallization and preliminary X-ray diffraction analysis of the electron-transfer complex between the terminal oxygenase component and ferredoxin in the Rieske non-haem iron oxygenase system carbazole 1,9a-dioxygenase. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 61:577–580. 10.1107/S1744309105014557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ashikawa Y, Uchimura H, Fujimoto Z, Inoue K, Noguchi H, Yamane H, Nojiri H. 2007. Crystallization and preliminary X-ray diffraction studies of the ferredoxin reductase component in the Rieske nonhaem iron oxygenase system carbazole 1,9a-dioxygenase. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 63:499–502. 10.1107/S174430910702163X [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bradford MM. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248–254. 10.1016/0003-2697(76)90527-3 [DOI] [PubMed] [Google Scholar]
- 35.Otwinowski Z, Minor W. 1997. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276:307–326. 10.1016/S0076-6879(97)76066-X [DOI] [PubMed] [Google Scholar]
- 36.Vagin A, Teplyakov A. 1997. MOLREP: an automated program for molecular replacement. J. Appl. Crystallogr. 30:1022–1025. 10.1107/S0021889897006766 [DOI] [Google Scholar]
- 37.McRee DE. 1999. XtalView/Xfit: a versatile program for manipulating atomic coordinates and electron density. J. Struct. Biol. 125:156–165. 10.1006/jsbi.1999.4094 [DOI] [PubMed] [Google Scholar]
- 38.Emsley P, Cowtan K. 2004. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60:2126–2132. 10.1107/S0907444904019158 [DOI] [PubMed] [Google Scholar]
- 39.Collaborative Computational Project, Number 4. 1994. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 50(Pt 5):760–763. 10.1107/S0907444994003112 [DOI] [PubMed] [Google Scholar]
- 40.Brünger AD, Adms PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW. 1998. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 54:905–921. 10.1107/S0907444998003254 [DOI] [PubMed] [Google Scholar]
- 41.DeLano WL. 2001. The PyMol user's manual. DeLano Scientific, San Carlos, CA [Google Scholar]
- 42.Jones G, Willett P, Glen RC, Leach AR, Tylor R. 1997. Development and validation of a genetic algorithm for flexible docking. J. Mol. Biol. 267:727–748. 10.1006/jmbi.1996.0897 [DOI] [PubMed] [Google Scholar]
- 43.Brady GP, Stouten PF. 2000. Fast prediction and visualization of protein binding pockets with PASS. J. Comput. Aided Mol. Des. 14:383–401. 10.1023/A:1008124202956 [DOI] [PubMed] [Google Scholar]
- 44.Karlsson A, Parales JV, Parales RE, Gibson DT, Eklund H, Ramaswammy S. 2003. Crystal structure of naphthalene dioxygenase: side-on binding of dioxygen to iron. Science 299:1039–1042. 10.1126/science.1078020 [DOI] [PubMed] [Google Scholar]
- 45.Dong X, Fushinobu S, Fukuda E, Terada T, Nakamura S, Shimizu K, Nojiri H, Omori T, Shoun H, Wakagi T. 2005. Crystal structure of the terminal oxygenase component of cumene dioxygenase from Pseudomonas fluorescens IP01. J. Bacteriol. 187:2483–2490. 10.1128/JB.187.7.2483-2490.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Boyd DR, Sharma ND, Allen CCR. 2001. Aromatic dioxygenases: molecular biocatalysis and applications. Curr. Opin. Biotechnol. 12:564–573. 10.1016/S0958-1669(01)00264-6 [DOI] [PubMed] [Google Scholar]
- 47.Friemann R, Ivkovic-Jansen MM, Lessner DJ, Yu CL, Gibson DT, Parales RE, Eklund H, Ramaswamy S. 2005. Structural insight into the dioxygenation of nitroarene compounds: the crystal structure of nitrobenzene dioxygenase. J. Mol. Biol. 248:1139–1151. 10.1016/j.jmb.2005.03.052 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.