Abstract
When microbes lack the nutrients necessary for growth, they enter stationary phase. In cases when energy sources are still present in the environment, they must decide whether to continue to use their metabolic program to harvest the available energy. Here we characterized the metabolic response to a variety of types of nutrient starvation in Escherichia coli and Bacillus subtilis. We found that E. coli exhibits a range of phenotypes, with the lowest metabolic rates under nitrogen starvation and highest rates under magnesium starvation. In contrast, the phenotype of B. subtilis was dominated by its decision to form metabolically inactive endospores. While its metabolic rates under most conditions were thus lower than those of E. coli, when sporulation was suppressed by a genetic perturbation or an unnatural starvation condition, the situation was reversed. To further probe stationary-phase metabolism, we used quantitative metabolomics to investigate possible small-molecule signals that may regulate the metabolic rate of E. coli and initiate sporulation in B. subtilis. We hypothesize a role for phosphoenolpyruvate (PEP) in regulating E. coli glucose uptake and for the redox cofactors NAD(H) and NADP(H) in initiation of sporulation. Our work is directly relevant to synthetic biology and metabolic engineering, where active metabolism during stationary phase, which uncouples production from growth, remains an elusive goal.
INTRODUCTION
Microbes have evolved to survive in extreme environments. A particularly ubiquitous stress condition is one where absence of nutrients prevents growth and proliferation. Whether in terrestrial, aquatic, or host environments, microbes are constantly faced with conditions where key elements, such as carbon, nitrogen, phosphorus, or sulfur, are scarce (1–3). While most organisms have evolved intricate methods for scavenging the few available resources, they must also survive the scenario when scavenging fails and they are forced to enter stationary phase or quiescence. This nongrowing state can lead to a fascinating diversity of behavior, from cannibalism (4) to differentiation programs like sporulation (5). A great deal of interest in quiescent bacteria is due to their enhanced resistance to antibiotics (6, 7) combined with the fact that many pathogens may stay in a quiescent state inside the host for extended periods (8). The fact that bacteria maintained in stationary phase quickly develop mutations that enhance survival (9) points to the importance of regulation of stationary-phase processes.
The majority of laboratory stationary-phase studies are done under conditions where carbon is limiting, which includes stationary phase in complex media, such as LB (see Fig. S1 in the supplemental material). For the many bacteria that use carbon catabolism for energy generation, carbon starvation implies a lack of energy, and consequently, the cell has a limited array of stationary-phase responses. On the other hand, if noncarbon nutrients are limiting for growth, a full catabolic program is possible (10), and the harvested energy could be invested in a variety of ways. Still, metabolic activity without growth could have potential downsides, such as the accumulation of toxic intermediates or decreased resistance to antibiotics, and it is not obvious how microbes resolve this trade-off (11).
Stationary phase under excess-carbon conditions is also of great interest for industrial microbiology (12). Production of heterologous metabolites during exponential growth requires a costly trade-off between biomass synthesis and product synthesis (13), while production during stationary phase would in theory maximize product synthesis and ideally could be part of a continuous production process (14). The potential of cells to continue active metabolism in the absence of growth has been explored but not fully realized. Starvation for nitrogen, for instance, is a standard technique for lipid production in algae (15), but attempts to increase the metabolic rate in model organisms through selection have yielded only minor improvements (16).
In this study, we characterized stationary-phase metabolism in the model Gram-positive and Gram-negative species Bacillus subtilis and Escherichia coli. Despite their ancient divergence and significant morphological differences, they have a great deal of similarity in the regulation of their metabolism during exponential growth (17). However, these organisms possess very different programs to respond to nongrowth conditions, which might be expected given their wildly different ecological niches: E. coli primarily inhabits the intestines of mammalian hosts, while B. subtilis is commonly found in the soil. The E. coli response to starvation is largely mediated by the transcription factor σS, which induces a program that remodels the cell into a more resistant state but also prepares for fast outgrowth should environmental conditions improve (18). While B. subtilis encodes an analogous program via the transcription factor SigB (19), it can also initiate a complex differentiation program, involving almost one-third of its genome, that results in the formation of an endospore (20). These spore cells are highly stress resistant and may endure long periods of unfavorable environmental conditions but are metabolically inert and resume growth less readily. The specifics of these stationary-phase responses may depend strongly on the precise environmental conditions (21, 22). Moreover, the extent to which cells adjust their metabolism during stationary phase and the mechanisms by which they accomplish this are largely unclear.
By examining carbon-excess stationary phase, where a large metabolic program is available to the cell, we found significant differences between the metabolic phenotypes of E. coli and B. subtilis. While E. coli exhibited a range of metabolic activity in stationary phase that depended on the limiting nutrient, the response of B. subtilis was defined almost completely by its decision to form metabolically inactive spores. When the sporulation response was inhibited, either by a genetic perturbation or by using an artificial starvation condition, B. subtilis exhibited extended metabolic activity that was significantly stronger than the activity of E. coli under most conditions.
MATERIALS AND METHODS
Strains and media.
E. coli BW25113 was used for all growth, physiology, and metabolomics experiments, except those involving tryptophan and leucine starvation, which used KEIO collection (23) ΔtrpC and ΔleuC strains, respectively (the ΔtrpE and ΔleuB strains were also tested and gave identical results). E. coli pptsG-GFP was obtained from a promoter-green fluorescent protein (GFP) library (24). B. subtilis TF8A (25) was used for all growth, physiology, and metabolomics experiments, except leucine starvation, which used a ΔleuCD mutant in the same background (26). Other genetic backgrounds used for comparative sporulation frequencies are identified and referenced in Fig. 3.
FIG 3.
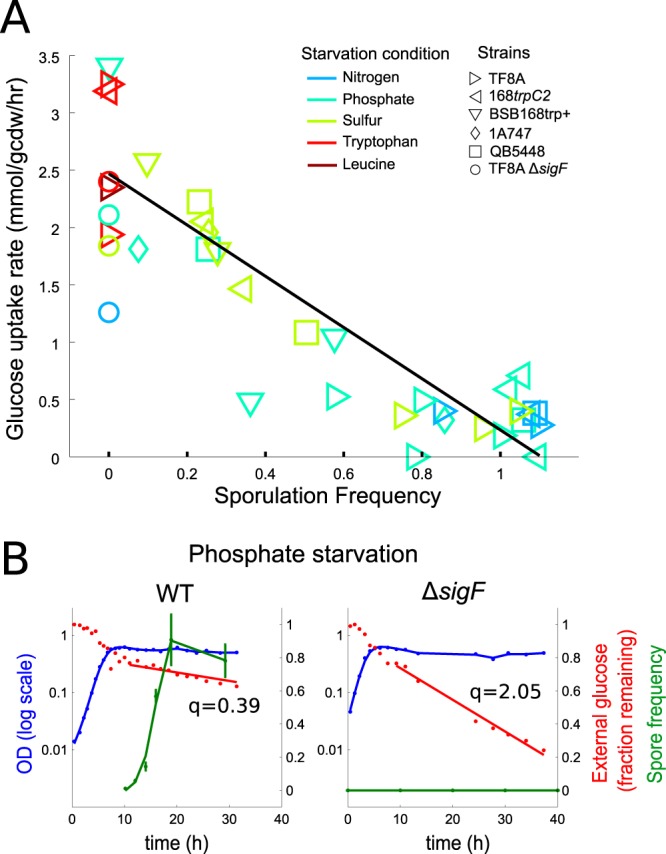
Sporulation frequency and glucose uptake in B. subtilis. (A) Sporulation frequencies and uptake rates for different strains of B. subtilis under various carbon-excess stationary-phase conditions. The TF8A genetic background strain (25) used for the majority of this work was compared to other standard lab strains, 168trpC2 and 1A747 (38), as well as the prototrophic strain BSB168trp+ (37) and strain QB5448 (36), which was used as a control for the ptsG overexpression strains. While all strains sporulated at near-100% rates under nitrogen starvation (blue) and carbon starvation (not shown), there was more variability in sporulation frequency under phosphate (aqua) and sulfate (yellow) starvation. In some cases, stationary-phase physiology could be separated into early and late stages, in which case the two stages are shown as separate points. Biological replicates are also shown as separate points. (B) Comparison of stationary-phase physiology for the wild type (TF8A) and the sporulation-deficient ΔsigF mutant under phosphate limitation.
M9 medium with glucose as a carbon source was used for all experiments except phosphate starvation, which used a modified version of defined low-phosphate medium (27). Complete medium composition is provided in Table S1 in the supplemental material. All experiments were performed at 37°C with 25 to 50 ml of culture in 500-ml flasks. For all experiments, exponential-phase LB precultures were diluted into standard M9 glucose medium to an optical density at 600 nm (OD600) of ∼0.01. After 4 to 6 doublings, cells were spun down, residual medium was washed off, and cultures in nutrient-limited medium were inoculated at an OD600 of ∼10−4 to ensure steady-state exponential growth before the start of sampling.
Physiology.
Glucose in culture supernatants was measured using the Gopod enzymatic assay (Megazyme, Bray, Ireland) using a microplate absorbance reader (Tecan, Männedorf, Switzerland), typically with three technical replicates per data point. Acetate, pyruvate, lactate, and other organic by-products were measured using high-performance liquid chromatography (HPLC) with a UV detector (Agilent, Santa Clara, CA,USA) (10). A conversion factor of 0.41 g cell dry weight (gcdw)/liter/OD600 was used for all rate calculations.
ATP synthesis rates were estimated using the simplified E. coli metabolic model (28), modified to eliminate the succinate/H+ antiporter and allow reversibility of the succinate/H+ symporter. Biomass production was fixed to zero, and the objective was set to maximization of ATP maintenance use. For each condition, 1,000 simulations were done, each one consisting of choosing substrate uptake and secretion rates from a normal distribution determined by the means and standard deviations given below (see Table 1) and providing them as hard constraints. The means and standard deviations of the resulting distribution of ATP yield are given below (see Table 1).
TABLE 1.
Physiology of E. coli in carbon-excess stationary phasea
| Parameter | Value with indicated limiting nutrient |
|||||
|---|---|---|---|---|---|---|
| Nitrogen | Phosphate | Sulfur | Magnesium | Tryptophan | Leucine | |
| qglucose | −0.46 ± 0.06 | −0.90 ± 0.07 | −1.30 ± 0.10 | −4.27 ± 0.34 | −0.75 ± 0.09 | −0.53 ± 0.07 |
| qpyruvate | 0.04 ± 0.08 | 0 | 0.43 ± 0.08 | 3.15 ± 0.05 | 0 | 0 |
| qlactate | 0.05 ± 0.03 | 0 | 0 | 0 | 0 | 0 |
| qsuccinate | 0.03 ± 0.02 | 0 | 0.14 ± 0.04 | 0.37 ± 0.04 | 0 | 0 |
| qacetate | 0.09 ± 0.03 | 0.52 ± 0.07 | 0.19 ± 0.03 | 0.25 ± 0.03 | 0.59 ± 0.12 | 0.39 ± 0.07 |
| qATPb | 5.14 ± 1.15 | 9.25 ± 1.12c | 12.77 ± 1.90 | 26.64 ± 5.23d | 8.33 ± 0.78 | 5.92 ± 0.67 |
Uptake and secretion rates (q) are given in units of (1/6) × mmol C · gcdw−1 · h−1. Errors are SEM. Unaccounted-for carbon is assumed to be combusted to CO2. The ATP synthesis rate, qATP, is expressed in units of mmol/gcdw/h.
The ATP synthesis rate is estimated from physiology with a stoichiometric model (see Materials and Methods).
This number represents the energy available from carbon catabolism. A lack of intracellular phosphate could lower ATP synthesis rates. However, there are relatively few other phosphate-consuming reactions in the cell, so it may be possible for ATP synthase to compete efficiently for turned-over phosphate.
This number represents the energy available from carbon catabolism. Mg2+ is required for full ATP synthase activity (48), and thus true ATP synthesis rates may be lower.
Metabolomics.
Samples for metabolite quantification were collected by vacuum filtering 1 ml of culture onto a 0.22-μm Durapore filter (Millipore) and immediately placing it in a cold (−20°C) 40/40/20 (vol/vol/vol) acetonitrile-methanol-water mixture for 30 min (29). A standard solution of previously extracted 13C-labeled biomass was added concurrently with the sample. Cell debris was removed by centrifugation at 14,000 × g at 0°C for 10 min, and samples were then dried in a speed vac setup (Christ, Osterode am Harz, Germany) and stored at −80°C until analysis. For mass spectrometry analysis, samples were resuspended in 100 μl water, and liquid chromatography-tandem mass spectrometry (LC-MS/MS) was performed as described previously (30). Metabolites were quantified by computing the ratio of the 12C and 13C peaks, and absolute concentrations were obtained by comparing to a standard curve produced by pure compounds.
Sporulation frequency.
B. subtilis cultures were diluted in 0.9% NaCl to approximately 1,000 to 5,000 cells/ml and divided into two tubes; one was placed in an 80°C water bath for 25 min, while the other was kept at room temperature. One hundred microliters from each tube was spread on each of three LB agar plates, and colonies were counted after 18 to 24 h. Sporulation frequency was determined as the ratio of the average colony counts of heated cells to the average colony counts of nonheated cells.
RESULTS
Diverse starvation environments lead to quantitative differences in metabolic activity.
To quantify the effect of starvation for essential noncarbon nutrients on metabolic activity, we grew cultures in minimal glucose medium with reduced levels of nitrogen, sulfur, phosphorus, or magnesium. Unlike a chemostat, in which medium is replenished to allow a defined growth rate, the limiting nutrient was completely exhausted in our experiments, leading to stationary phase with extended metabolic activity. The concentrations of the limiting nutrients (see Table S1 in the supplemental material) were chosen to allow growth until an OD600 of 0.5 to 1.0 was reached, when less than 25% of the original glucose (4 g/liter initial concentration) had been depleted. In both E. coli and B. subtilis, the growth rate during the exponential phase was largely unaffected by the lower concentration of the limiting nutrient. Upon growth arrest, the metabolic activity of the culture, defined by the glucose uptake rate, quickly declined and within a few hours usually reached a new steady state that persisted for more than 24 h.
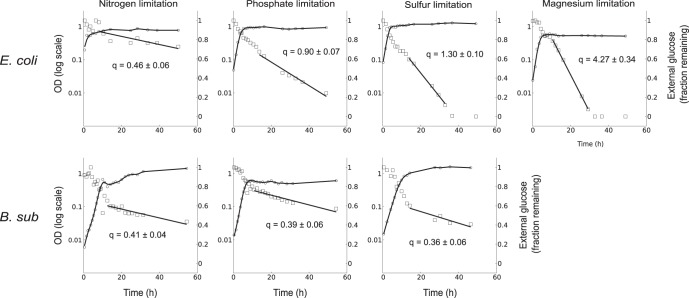
All starvation conditions led to measurable stationary-phase metabolic activity (Fig. 1). E. coli showed a continuum of uptake rates, with the lowest rate under nitrogen starvation (0.46 mmol/g cell dry weight [gcdw]/h) and the highest under magnesium starvation (4.27 mmol/gcdw/h). These represent approximately 5% and 50%, respectively, of the uptake rate during exponential growth (8.72 mmol/gcdw/h). In contrast, B. subtilis was less metabolically active, and its physiology was more consistent across conditions. While the glucose uptake rate in B. subtilis under nitrogen starvation was similar to that of E. coli, B. subtilis also exhibited similar rates under phosphate and sulfur limitation, which were lower than the corresponding rates for E. coli. This is despite the fact that its uptake rate during exponential growth is similar to that of E. coli (9.75 mmol/gcdw/h). We could not achieve a defined magnesium-limited stationary phase for B. subtilis, presumably due to its known low affinity for Mg2+ (31).
FIG 1.
Physiology of stationary phase caused by natural starvation. E. coli and B. subtilis (B. sub) were grown under limiting concentrations of ammonium, phosphate, sulfate, and magnesium (E. coli only). The OD600 (○) and residual glucose (□) for a typical time course are shown. Residual glucose is shown as a fraction of the starting 4 g/liter. The glucose uptake rate during stationary phase (q) is shown inside each plot, in units of mmol/gcdw/h, calculated from at least 3 biological replicates.
Distinction between natural and artificial starvation conditions.
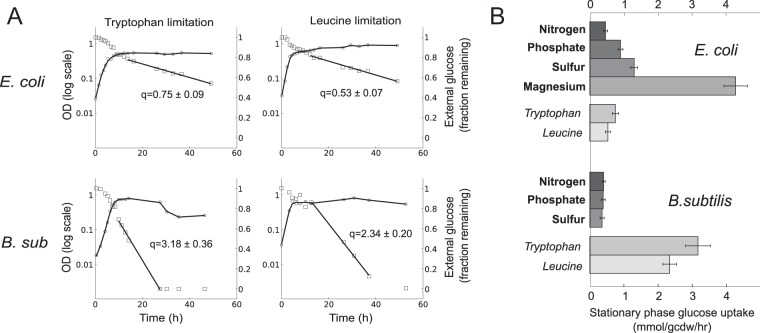
Previous studies in yeast have elucidated key differences between “natural” starvation, in which nutrients like carbon, nitrogen, phosphorus, or sulfur are absent from the environment, and “artificial” starvation imposed by the combination of a genetic mutation, for instance, in an amino acid biosynthesis pathway, and the absence of an appropriate supplement in the environment. In Saccharomyces cerevisiae, natural starvation led to increased survival and lower metabolic activity, as well as significant differences in cell size and morphology, compared to findings for artificial starvation conditions (32, 33). To determine the extent to which the distinction between natural and artificial starvation played a role in the stationary-phase metabolic phenotypes of E. coli and B. subtilis, we subjected mutants deficient in biosynthesis of tryptophan or leucine to starvation for the respective nutrient. Similar to the situation for natural starvation, growth during the exponential growth phase was largely unaffected but stopped upon exhaustion of the limiting nutrient.
In parallel to yeast, we found that B. subtilis starved for leucine or tryptophan and unable to synthesize the amino acid exhibited behavior entirely opposite to its phenotype during nitrogen, phosphate, or sulfate starvation (Fig. 2). The glucose uptake rate of 2.3 mmol/g/h (leucine starvation) and 3.2 mmol/g/h (tryptophan starvation) was 5-fold to 10-fold higher than what was observed for the natural-starvation cases. However, in E. coli, the metabolic activity under leucine and tryptophan starvation was in the same range as that observed for natural starvation, suggesting an entirely different strategy of metabolic control in stationary phase.
FIG 2.
Physiology of stationary phase caused by artificial amino acid starvation. (A) E. coli ΔtrpC and ΔleuC strains were grown in limiting tryptophan and leucine, respectively, along with the B. subtilis (B. sub) TF8A (tryptophan auxotroph) and ΔleuCD strains. OD600 (○) and residual glucose (□) for typical time courses are shown. Residual glucose is shown as a fraction of the starting 4 g/liter. The glucose uptake rate during stationary phase (q) is shown inside each plot, in units of mmol/gcdw/h, calculated from at least 3 biological replicates. (B) Comparison of glucose uptake rates during stationary phase between natural and unnatural starvation in B. subtilis and E. coli. Starvation conditions are identified according to their limiting nutrient; bold text labels signify natural starvation, while italics signify unnatural starvation.
Sporulation frequency dominates stationary-phase behavior in B. subtilis.
One of the defining features of B. subtilis and closely related species is the ability to form highly stress-resistant and metabolically inactive endospores. While carbon starvation is the most common laboratory condition used to induce sporulation, starvation for nitrogen and phosphate has also been linked to sporulation (5, 21). To determine the role that the sporulation program plays in regulating metabolic activity under our carbon-excess stationary-phase conditions, we quantified spore formation by counting heat-resistant cells at various time points after starvation. We found that for all three natural starvation conditions (nitrogen, phosphate, and sulfur), viable spores began to appear within about 4 to 6 h of the end of exponential growth, and a plateau of 80 to 100% of cells forming spores was reached after approximately 8 to 10 h (Fig. 3). This corresponds to the time period thought to be required to form a mature spore (5), suggesting that the onset of the sporulation program begins soon after growth arrest for most cells. Since mature spores are known to be essentially metabolically inactive (34), this large fraction of spores is very likely to be the key mechanistic explanation of the very low metabolic activity by B. subtilis under nitrogen, phosphate, or sulfur starvation. In contrast, auxotrophic strains under tryptophan or leucine starvation formed no spores, in agreement with the high metabolic activity under those conditions. Lack of sporulation is unlikely to be caused by a requirement for the missing amino acid for spore formation, since previous studies showed that leucine auxotrophs do sporulate in carbon starvation media also lacking leucine (35).
To further characterize the relationship between sporulation frequency and metabolic activity, we performed the same experiments with a number of genetic variants of B. subtilis. The experiments described above were performed with strain TF8A (25), a variant of B. subtilis 168 with several large and apparently nonfunctional genomic regions removed. No phenotypic differences between TF8A and the more common lab strain 168trpC2 were previously reported (25). We assayed the original 168trpC2 strain, along with several other derivatives (36–38) for sporulation frequency and glucose uptake rate during stationary phase. Two remarkable findings are evident in the results shown in Fig. 3A. First, even closely related strains whose genetic differences do not have any obvious relationship to the conditions tested differ greatly in their sporulation frequencies. While all strains sporulated with high frequency under nitrogen starvation, more variability was present under phosphate starvation, and only TF8A formed a high number of spores under sulfur starvation. Second, there is a clear inverse linear relationship between sporulation frequency and glucose uptake rate in stationary phase, further corroborating the hypothesis that the sporulation decision determines the metabolic phenotype of stationary-phase B. subtilis.
An alternative explanation for the relationship between sporulation frequency and metabolic activity is that the starvation environment leads to lower glucose uptake rates and that the lower metabolic activity in turn induces sporulation. To analyze this hypothesis, we assayed stationary-phase metabolism in a number of genetic mutants deficient in sporulation. Figure 3B shows the stationary-phase physiology of a ΔsigF mutant (in the TF8A background) compared to that of the wild type. Since sigF encodes a key transcription factor in the sporulation program, this strain arrests the sporulation program before mother cell lysis. The ΔsigF strain continues to take up glucose for more than 24 h after starvation, with a metabolic rate more than 5 times that of the wild type. Under other starvation conditions, the glucose uptake rates of this strain are close to the intercept of the linear fit in Fig. 3A, i.e., the predicted uptake rate of a wild-type strain that does not form any spores. The same experiment was performed with deletions of other genes crucial for sporulation (Δspo0A, Δspo0H, ΔsigE, ΔsigG, and ΔspoVS) and in other genetic backgrounds, with identical results. These results suggest that sporulation is induced directly by the environment and that the low metabolic rate is a consequence.
Factors involved in regulation of stationary-phase glucose uptake.
Sporulation represents a major developmental program with a clear connection to control of stationary-phase metabolism in B. subtilis. Without a comparable program in E. coli, the question of what factors influence metabolic rates during stationary phase in E. coli remains. One candidate mechanism by which E. coli could control its uptake of glucose would be the expression of the glucose transport genes. To determine if transcriptional control of transporter expression is responsible for the range of observed metabolic activity phenotypes, we assayed expression of the major glucose transport protein PtsG (Fig. 4A) using a promoter-GFP construct (24). We found that PtsG expression under all glucose-excess stationary-phase conditions was essentially identical to its expression during exponential growth on glucose (Fig. 4B), showing that transporter expression cannot be limiting for glucose uptake during stationary phase and uptake must instead be controlled at the metabolite level. An analogous construct was used to assay B. subtilis PtsG expression (39), with similar results. Furthermore, strains constitutively expressing PtsG in B. subtilis (36) were not deficient in sporulation under any condition.
FIG 4.
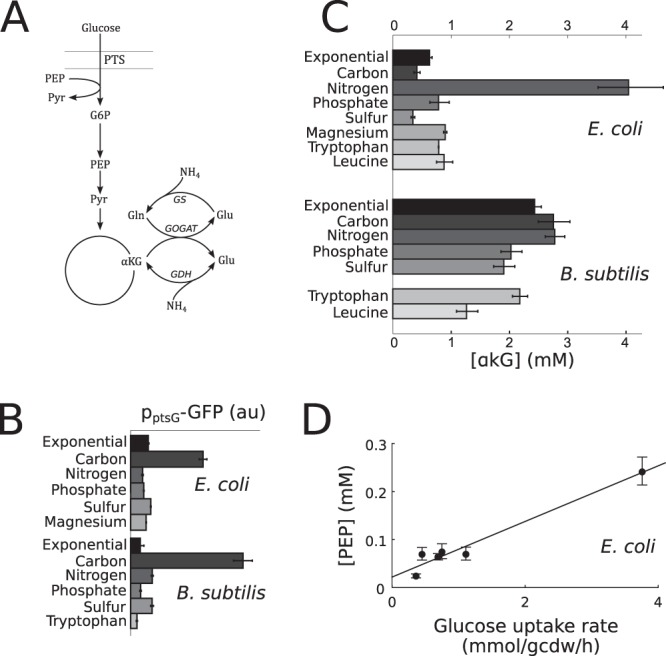
Factors involved in regulation of glucose uptake and central metabolism during stationary phase. (A) Schematic diagram of central metabolism common to both E. coli and B. subtilis. (B) Expression of the major glucose transport protein PtsG assayed by a promoter-GFP reporter under exponential-growth and stationary-phase conditions. Stationary-phase conditions are identified by the limiting nutrient. (C) Intracellular concentration of α-ketoglutarate under exponential-growth and stationary-phase conditions. Stationary-phase conditions are identified by the limiting nutrient. (D) Correlation of intracellular PEP concentration and glucose uptake rate in E. coli (r2 = 0.96; r2 = 0.36 with the outlier of magnesium starvation removed). In panels B to D, reported values are averages of at least 3 measurements from each of two biological replicates, taken between 2 h and 8 h after the end of exponential growth.
To find out what metabolic factors could be responsible for the control of glucose uptake in stationary phase, we quantified metabolite concentrations under the previously analyzed stationary-phase conditions and under carbon starvation and exponential-growth conditions. Many effects were both intuitive and largely conserved across both E. coli and B. subtilis. In both species, carbon limitation led to an increase in phosphoenolpyruvate (PEP) and decreases in other glycolytic metabolites, most notably 1,6-fructose-bis-phosphate (FBP). Phosphate limitation led to significant decreases in phosphorylated glycolytic intermediates as well as nucleotides, while nitrogen limitation caused severe decreases in amino acids (see Tables S2 and S3 in the supplemental material). These effects correspond well to previous characterization of these starvation states in E. coli and S. cerevisiae (40).
One of the strongest metabolic effects of nitrogen starvation previously characterized in E. coli is the buildup of α-ketoglutarate (αKG), the substrate of the major nitrogen assimilation pathway. Indeed, we see a 7-fold increase in the αKG concentration uniquely under nitrogen starvation (Fig. 4C). Consistent with this increase mediating allosteric inhibition of the phosphotransferase system (PTS) glucose transporter (41), glucose uptake rates during nitrogen starvation were lower than those under any other noncarbon starvation condition for E. coli. However, for B. subtilis, we did not observe a significant increase in the αKG concentration at any point during nitrogen starvation. This unexpected finding may be due to a combination of two factors: first, intracellular glutamate and glutamine concentrations in B. subtilis are about 5-fold and 10-fold higher, respectively, than those in E. coli, and second, the glutamate dehydrogenase (GDH) enzyme in B. subtilis has a very low affinity for ammonium, and thus B. subtilis relies exclusively on glutamine synthase (the GS-GOGAT cycle) for ammonium assimilation (42) (Fig. 4A). Thus, one possibility is that B. subtilis uses the large pool of glutamine to continue running the GOGAT reaction for some time in the absence of ammonium, preventing αKG accumulation. It is unclear what happens after the glutamine pool is depleted (as occurs after 1 to 2 h; see Table S3 in the supplemental material), but it is possible that nitrogen is released from protein degradation, incoming carbon is diverted before reaching αKG, or αKG consumption by other pathways, like the tricarboxylic acid (TCA) cycle, increases.
The major outlying condition for E. coli metabolic activity was magnesium starvation, when glucose uptake rates were significantly higher than those under any other stationary-phase condition. Further investigation into the physiology of magnesium-starved E. coli revealed a massive accumulation and secretion of pyruvate, with pyruvate secretion accounting for about 75% of consumed carbon (Table 1). This unusual phenotype (acetate and lactate are more common E. coli fermentation products [43]) is likely to be due to inactive pyruvate dehydrogenase (PDH), an enzyme that absolutely requires Mg2+ as a cofactor (44). However, given that several upstream enzymes in glycolysis also bind Mg2+ and require it for full activity, it is unclear why the metabolic block should manifest itself specifically at PDH. An attractive hypothesis was that flux during magnesium starvation is shuttled to the Entner-Doudoroff pathway, whose enzymes may potentially function without Mg2+. However, we did not observe any accumulation of the Entner-Doudoroff intermediate 2-keto-3-deoxy-gluconate-phosphate under magnesium starvation, and dynamic 13C labeling experiments were consistent with the majority of glucose entering the Embden-Meyerhoff (classical glycolysis) pathway (data not shown). As such, a more likely explanation is that PDH has a lower affinity for Mg2+ than glycolytic enzymes or has less potential for substitution by other metal ions.
The block at PDH did propagate somewhat to the upstream metabolites PEP, bis-phosphoglycerate, and phosphoglycerate, leading to higher concentrations of those metabolites than observed under any other condition (except for PEP under carbon starvation). Curiously, conditions with intermediate glucose uptake rates also showed intermediate PEP levels, leading to a correlation of 0.98 between the uptake rate and the PEP concentration across conditions (Fig. 4D). Six other metabolites also showed correlation above 0.9, but only PEP also showed correlation above 0.5 if the outlying magnesium starvation condition was removed. A role for PEP in activating glucose uptake is a promising hypothesis for two reasons. First, PEP is a substrate of the glucose phosphorylation reaction that is coupled to uptake through the PTS system, and thus high concentrations could increase activity directly. Second, increases in intracellular PEP have consistently been associated with carbon starvation (40, 45), a condition under which the cell may be trying to maximize uptake of the little available carbon.
An alternative hypothesis for the observation of high glucose uptake rates under magnesium limitation relates to energy demand. Glucose uptake rates in E. coli during exponential growth have been linked to ATP demand (46), and production of pyruvate would provide significantly less ATP than complete combustion of glucose to CO2 through the TCA cycle. Thus, cells may compensate by increasing the total amount of glucose catabolism. To determine if the total amounts of ATP produced during stationary phase were comparable under the diverse stationary-phase conditions, we estimated ATP production using flux balance analysis with a simplified stoichiometric model of E. coli metabolism (28). Biomass production rates were set to zero, substrate uptake and product secretion rates were constrained by observed data (Table 1), and net ATP synthesis was maximized with a P-to-O ratio of approximately 1.2. The observed rates for most conditions were consistent with current estimates of 8 to 10 mmol ATP/gcdw/h for non-growth-associated maintenance costs (47) (Table 1). The ATP production estimated under magnesium limitation exceeded this threshold, suggesting that energy demand cannot fully explain the high uptake rates observed under this condition. However, two caveats could lead to an overestimation of energy yields: first, the lack of Mg2+ may inhibit ATP synthase itself (48), and second, increased membrane fluidity in magnesium-starved cells (49) could increase proton permeability and thus lower the P-to-O ratio. Other caveats fundamental to stoichiometric analysis also remain; for instance, ATP-dissipating futile cycles could lead to lower than estimated ATP yields.
Sporulation signals in B. subtilis.
As noted previously, the dominating factor in B. subtilis stationary-phase behavior was the decision to sporulate. While the network of protein phosphorylation and transcriptional regulation events that drives the sporulation program is extremely well characterized (5), less is known about how B. subtilis senses the starvation signal that initiates sporulation. Much evidence points to a decrease in the GTP concentration as a key trigger (50, 51), but a complete model of how the GTP concentration affects sporulation is still lacking (52, 53). To investigate the extent to which changes in GTP and other associated metabolites predicted sporulation, we analyzed the time course of metabolite concentrations upon entry into starvation and compared conditions that led to sporulation with those that did not. As seen in Fig. 5A, GTP levels dropped sharply during carbon starvation, consistent with findings in previous work (21). However, nitrogen starvation, which led to equally high sporulation frequencies, actually induced a slight increase in the GTP concentration, while tryptophan starvation, which prevented sporulation, led to an even larger drop in GTP. Similarly, considering other proposed signals of sporulation, such as the guanylate energy charge (a measure of the fraction of phosphorylation state of guanine nucleotides), ATP (54), or the ATP-to-GTP ratio, could not clearly distinguish conditions that led to a high sporulation frequency from those that did not (Fig. 5B to E).
FIG 5.
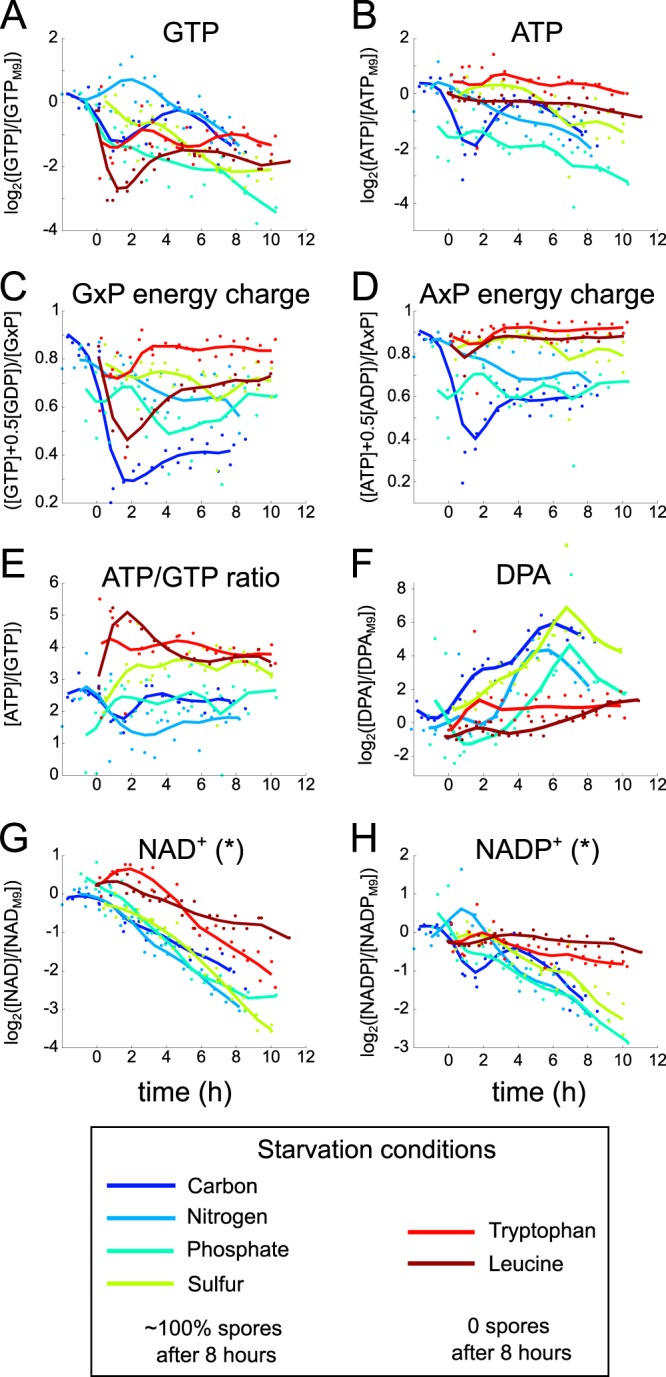
Time courses of intracellular metabolite concentrations in B. subtilis. T = 0 corresponds approximately to the end of the exponential growth phase. Metabolite concentrations (A, B, and F to H) are given in log2 scale relative to the concentrations during exponential growth in M9 medium, while ratios (C to E) are given in absolute quantities. Dipicolinic acid (DPA) (F) is a major component of the spore coat, shown to demonstrate induction of sporulation. Lines are cubic spline fits. “*” indicates that the methods used in this work cannot accurately distinguish reduced and oxidized cofactor forms. While the best estimate of [NAD+] is shown, the observed decrease can be due to either a decrease in the total NAD(H) pool or a decrease in the NAD+/NADH ratio, and the same caveat applies to NADP+.
In fact, no single signal was absolutely consistent with driving the sporulation decision. The closest examples of metabolic changes that mirrored the division between sporulating and nonsporulating conditions were NAD(H) and NADP(H) concentrations, which dropped more slowly and by a smaller magnitude under tryptophan and leucine starvation than under natural starvation conditions (Fig. 5G to H). These redox cofactors, particularly NAD+, have recently been implicated in regulation of the KinA and KinB kinases upstream of the master sporulation regulator Spo0A (55). While the extraction and quantification methods used in this work do not allow distinction of total pool fluctuations from redox balance fluctuations, our results are consistent with the hypothesis that a major sporulation trigger is inhibition of respiratory activity, which leads to a drop in NAD+, activating KinA and the downstream program (56, 57).
DISCUSSION
Control of metabolic activity in stationary phase.
In this work, we explored stationary-phase behavior in two distantly related bacteria, E. coli and B. subtilis. In contrast to most studies of starvation, which focus on conditions where carbon is limiting and little active metabolism is possible, we have turned our focus to carbon-excess stationary phase. We find that the rate of metabolic activity, measured as the substrate uptake rate, depends strongly on the limiting nutrient for both E. coli and B. subtilis. E. coli exhibited a continuum of stationary-phase behavior, ranging from very low metabolic activity under nitrogen starvation (∼5% of the uptake rate during exponential growth) to extremely high activity under magnesium starvation (∼50% of the uptake rate during exponential growth). On the other hand, the phenotype of B. subtilis was almost entirely determined by the sporulation decision. This led the metabolic activity under natural starvation conditions to be generally lower than that of E. coli. However, when the sporulation phenotype was suppressed, either by genetic perturbation or unnatural starvation conditions, B. subtilis actually exhibited significantly higher metabolic activity than E. coli (with the exception of the magnesium starvation condition). This higher rate was also previously observed in wild-type cells under nitrogen limitation when a somewhat different experimental protocol led to no spore formation (10).
Currently, understanding of how glucose uptake is controlled during stationary phase in E. coli is sparse. This is in large part because, as in many other aspects of metabolic regulation, the key regulatory interactions likely do not modify gene expression but rather enzyme activity through allosteric regulation or protein modification. One instance of such regulation is the inhibition of glucose uptake by αKG, which accumulates during nitrogen starvation (41). Our data, which show nitrogen starvation with the lowest glucose uptake rate and highest αKG concentrations among all conditions tested, are consistent with this key role for αKG in E. coli (though not in B. subtilis). However, αKG concentrations do not explain the variability in E. coli metabolic rates observed under conditions other than nitrogen starvation. A plausible candidate for another regulatory metabolite is PEP. PEP levels correlated well with the glucose uptake rate in stationary phase, due in large part to the high PEP concentrations observed under magnesium starvation, which led to the highest glucose uptake rate among all conditions. PEP acts as a phosphate donor for the PTS glucose uptake system, and as such, it is easy to see that PEP concentrations could positively affect glucose uptake. Such a mechanism would also be consistent with the PEP concentration acting as a signal of carbon starvation, as has been proposed in several studies (40, 45).
Implications for synthetic biology and metabolic engineering.
The finding of high glucose uptake during magnesium starvation opens the door to a critical direction in synthetic biology and metabolic engineering: the decoupling of growth and metabolic activity. Production of heterologous molecules in the absence of growth (i.e., during stationary phase) would eliminate the costly trade-off between biomass production and product synthesis. However, as shown in this work, overall metabolic rates during stationary phase tend to be an order of magnitude lower than metabolic rates during exponential growth, and thus increasing and extending the metabolic rates in stationary phase are of great interest. Previous efforts using directed evolution to increase the metabolic rate in E. coli under nitrogen starvation led to only minor improvements (16). The fact that magnesium starvation in E. coli led to significantly higher rates than previously observed suggests that there is no inherent obstacle to high metabolic rates during stationary phase. The overflow of pyruvate under magnesium starvation means that engineered pathways that start at pyruvate or higher points in glycolysis could benefit immediately from this condition (subject to the caveat that many heterologous enzymes may also require magnesium). However, the major implication is that high metabolic activity during carbon-excess stationary phase is clearly possible, and decoding the regulatory networks that normally shut down metabolism under these conditions could allow similar rates under other conditions, which would be a major breakthrough in metabolic engineering.
Supplementary Material
ACKNOWLEDGMENTS
We thank K. Bych, E. Denham, F. Escartin, I. Gruner, and M. Jules for providing strains, M. Hörl for assistance with dynamic labeling experiments, and H. Link and M. Zimmerman for other technical assistance.
Funding for this work was provided by the European Commission project BaSynthec (FP7-244093).
Footnotes
Published ahead of print 28 February 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.00061-14.
REFERENCES
- 1.Hobbie JE, Hobbie EA. 2013. Microbes in nature are limited by carbon and energy: the starving-survival lifestyle in soil and consequences for estimating microbial rates. Front. Microbiol. 4:324. 10.3389/fmicb.2013.00324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.McArthur JV. 2006. Microbial ecology: an evolutionary approach. Academic Press, New York, NY [Google Scholar]
- 3.Menge DNL, Hedin LO, Pacala SW. 2012. Nitrogen and phosphorus limitation over long-term ecosystem development in terrestrial ecosystems. PLoS One 7:e42045. 10.1371/journal.pone.0042045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.González-Pastor JE. 2011. Cannibalism: a social behavior in sporulating Bacillus subtilis. FEMS Microbiol. Rev. 35:415–424. 10.1111/j.1574-6976.2010.00253.x [DOI] [PubMed] [Google Scholar]
- 5.Stragier P, Losick R. 1996. Molecular genetics of sporulation in Bacillus subtilis. Annu. Rev. Genet. 30:297–341. 10.1146/annurev.genet.30.1.297 [DOI] [PubMed] [Google Scholar]
- 6.Eng RH, Padberg FT, Smith SM, Tan EN, Cherubin CE. 1991. Bactericidal effects of antibiotics on slowly growing and nongrowing bacteria. Antimicrob. Agents Chemother. 35:1824–1828. 10.1128/AAC.35.9.1824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lewis K. 2007. Persister cells, dormancy and infectious disease. Nat. Rev. Microbiol. 5:48–56. 10.1038/nrmicro1557 [DOI] [PubMed] [Google Scholar]
- 8.Gengenbacher M, Kaufmann SHE. 2012. Mycobacterium tuberculosis: success through dormancy. FEMS Microbiol. Rev. 36:514–532. 10.1111/j.1574-6976.2012.00331.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Finkel SE. 2006. Long-term survival during stationary phase: evolution and the GASP phenotype. Nat. Rev. Microbiol. 4:113–120. 10.1038/nrmicro1340 [DOI] [PubMed] [Google Scholar]
- 10.Rühl M, Coq DL, Aymerich S, Sauer U. 2012. 13C-flux analysis reveals NADPH-balancing transhydrogenation cycles in stationary phase of nitrogen-starving Bacillus subtilis. J. Biol. Chem. 287:27959–27970. 10.1074/jbc.M112.366492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Benaroudj N, Lee DH, Goldberg AL. 2001. Trehalose accumulation during cellular stress protects cells and cellular proteins from damage by oxygen radicals. J. Biol. Chem. 276:24261–24267. 10.1074/jbc.M101487200 [DOI] [PubMed] [Google Scholar]
- 12.Shaikh AS, Tang YJ, Mukhopadhyay A, Martín HG, Gin J, Benke PI, Keasling JD. 2010. Study of stationary phase metabolism via isotopomer analysis of amino acids from an isolated protein. Biotechnol. Prog. 26:52–56. 10.1002/btpr.325 [DOI] [PubMed] [Google Scholar]
- 13.Peralta-Yahya PP, Zhang F, del Cardayre SB, Keasling JD. 2012. Microbial engineering for the production of advanced biofuels. Nature 488:320–328. 10.1038/nature11478 [DOI] [PubMed] [Google Scholar]
- 14.Boender LGM, de Hulster EAF, van Maris AJA, Daran-Lapujade PAS, Pronk JT. 2009. Quantitative physiology of Saccharomyces cerevisiae at near-zero specific growth rates. Appl. Environ. Microbiol. 75:5607–5614. 10.1128/AEM.00429-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Siaut M, Cuiné S, Cagnon C, Fessler B, Nguyen M, Carrier P, Beyly A, Beisson F, Triantaphylidès C, Li-Beisson Y, Peltier G. 2011. Oil accumulation in the model green alga Chlamydomonas reinhardtii: characterization, variability between common laboratory strains and relationship with starch reserves. BMC Biotechnol. 11:7. 10.1186/1472-6750-11-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sonderegger M, Schümperli M, Sauer U. 2005. Selection of quiescent Escherichia coli with high metabolic activity. Metab. Eng. 7:4–9. 10.1016/j.ymben.2004.05.005 [DOI] [PubMed] [Google Scholar]
- 17.Chubukov V, Gerosa L, Kochanowski K, Sauer U. Coordination of microbial metabolism. Nat. Rev. Microbiol., in press [DOI] [PubMed] [Google Scholar]
- 18.Kolter R, Siegele DA, Tormo A. 1993. The stationary phase of the bacterial life cycle. Annu. Rev. Microbiol. 47:855–874. 10.1146/annurev.mi.47.100193.004231 [DOI] [PubMed] [Google Scholar]
- 19.Hecker M, Pané-Farré J, Völker U. 2007. SigB-dependent general stress response in Bacillus subtilis and related Gram-positive bacteria. Annu. Rev. Microbiol. 61:215–236. 10.1146/annurev.micro.61.080706.093445 [DOI] [PubMed] [Google Scholar]
- 20.Piggot PJ, Hilbert DW. 2004. Sporulation of Bacillus subtilis. Curr. Opin. Microbiol. 7:579–586. 10.1016/j.mib.2004.10.001 [DOI] [PubMed] [Google Scholar]
- 21.Lopez JM, Marks CL, Freese E. 1979. The decrease of guanine nucleotides initiates sporulation of Bacillus subtilis. Biochim. Biophys. Acta 587:238–252. 10.1016/0304-4165(79)90357-X [DOI] [PubMed] [Google Scholar]
- 22.Peterson CN, Mandel MJ, Silhavy TJ. 2005. Escherichia coli starvation diets: essential nutrients weigh in distinctly. J. Bacteriol. 187:7549–7553. 10.1128/JB.187.22.7549-7553.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, Mori H. 2006. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 2:2006.0008. 10.1038/msb4100050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zaslaver A, Bren A, Ronen M, Itzkovitz S, Kikoin I, Shavit S, Liebermeister W, Surette MG, Alon U. 2006. A comprehensive library of fluorescent transcriptional reporters for Escherichia coli. Nat. Methods 3:623–628. 10.1038/nmeth895 [DOI] [PubMed] [Google Scholar]
- 25.Westers H, Dorenbos R, van Dijl JM, Kabel J, Flanagan T, Devine KM, Jude F, Séror SJ, Beekman AC, Darmon E, Eschevins C, de Jong A, Bron S, Kuipers OP, Albertini AM, Antelmann H, Hecker M, Zamboni N, Sauer U, Bruand C, Ehrlich DS, Alonso JC, Salas M, Quax WJ. 2003. Genome engineering reveals large dispensable regions in Bacillus subtilis. Mol. Biol. Evol. 20:2076–2090. 10.1093/molbev/msg219 [DOI] [PubMed] [Google Scholar]
- 26.Tanaka K, Henry CS, Zinner JF, Jolivet E, Cohoon MP, Xia F, Bidnenko V, Ehrlich SD, Stevens RL, Noirot P. 2013. Building the repertoire of dispensable chromosome regions in Bacillus subtilis entails major refinement of cognate large-scale metabolic model. Nucleic Acids Res. 41:687–699. 10.1093/nar/gks963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Müller JP, An Z, Merad T, Hancock IC, Harwood CR. 1997. Influence of Bacillus subtilis phoR on cell wall anionic polymers. Microbiology 143:947–956. 10.1099/00221287-143-3-947 [DOI] [PubMed] [Google Scholar]
- 28.Orth JD, Fleming RM, Palsson BØ. 1 February 2010. Chapter 10.2.1. Reconstruction and use of microbial metabolic networks: the core Escherichia coli metabolic model as an educational guide. In Karp PD. (ed), EcoSal— Escherichia coli and Salmonella: cellular and molecular biology. ASM Press, Washington, DC. 10.1128/ecosalplus.10.2.1 [DOI] [PubMed] [Google Scholar]
- 29.Rabinowitz JD, Kimball E. 2007. Acidic acetonitrile for cellular metabolome extraction from Escherichia coli. Anal. Chem. 79:6167–6173. 10.1021/ac070470c [DOI] [PubMed] [Google Scholar]
- 30.Büscher JM, Moco S, Sauer U, Zamboni N. 2010. Ultrahigh performance liquid chromatography-tandem mass spectrometry method for fast and robust quantification of anionic and aromatic metabolites. Anal. Chem. 82:4403–4412. 10.1021/ac100101d [DOI] [PubMed] [Google Scholar]
- 31.Tempest DW, Dicks JW, Meers JL. 1967. Magnesium-limited growth of Bacillus subtilis, in pure and mixed cultures, in a chemostat. J. Gen. Microbiol. 49:139–147. 10.1099/00221287-49-1-139 [DOI] [PubMed] [Google Scholar]
- 32.Brauer MJ, Huttenhower C, Airoldi EM, Rosenstein R, Matese JC, Gresham D, Boer VM, Troyanskaya OG, Botstein D. 2008. Coordination of growth rate, cell cycle, stress response, and metabolic activity in yeast. Mol. Biol. Cell 19:352–367. 10.1091/mbc.E07-08-0779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Slavov N, Botstein D. 2013. Decoupling nutrient signaling from growth rate causes aerobic glycolysis and deregulation of cell size and gene expression. Mol. Biol. Cell 24:157–168. 10.1091/mbc.E12-09-0670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nicholson WL, Munakata N, Horneck G, Melosh HJ, Setlow P. 2000. Resistance of Bacillus endospores to extreme terrestrial and extraterrestrial environments. Microbiol. Mol. Biol. Rev. 64:548–572. 10.1128/MMBR.64.3.548-572.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Doering JL, Bott KF. 1972. Differential amino acid requirements for sporulation in Bacillus subtilis. J. Bacteriol. 112:345–355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bachem S, Stülke J. 1998. Regulation of the Bacillus subtilis GlcT antiterminator protein by components of the phosphotransferase system. J. Bacteriol. 180:5319–5326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Büscher JM, Liebermeister W, Jules M, Uhr M, Muntel J, Botella E, Hessling B, Kleijn RJ, Le Chat L, Lecointe F, Mäder U, Nicolas P, Piersma S, Rügheimer F, Becher D, Bessieres P, Bidnenko E, Denham EL, Dervyn E, Devine KM, Doherty G, Drulhe S, Felicori L, Fogg MJ, Goelzer A, Hansen A, Harwood CR, Hecker M, Hubner S, Hultschig C, Jarmer H, Klipp E, Leduc A, Lewis P, Molina F, Noirot P, Peres S, Pigeonneau N, Pohl S, Rasmussen S, Rinn B, Schaffer M, Schnidder J, Schwikowski B, Van Dijl JM, Veiga P, Walsh S, Wilkinson AJ, Stelling J, Aymerich S, Sauer U. 2012. Global network reorganization during dynamic adaptations of Bacillus subtilis metabolism. Science 335:1099–1103. 10.1126/science.1206871 [DOI] [PubMed] [Google Scholar]
- 38.Zeigler DR, Pragai Z, Rodriguez S, Chevreux B, Muffler A, Albert T, Bai R, Wyss M, Perkins JB. 2008. The origins of 168, W23, and other Bacillus subtilis legacy strains. J. Bacteriol. 190:6983–6995. 10.1128/JB.00722-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Botella E, Fogg M, Jules M, Piersma S, Doherty G, Hansen A, Denham EL, Chat LL, Veiga P, Bailey K, Lewis PJ, Dijl JM, van Aymerich S, Wilkinson AJ, Devine KM. 2010. pBaSysBioII: an integrative plasmid generating gfp transcriptional fusions for high-throughput analysis of gene expression in Bacillus subtilis. Microbiology 156:1600–1608. 10.1099/mic.0.035758-0 [DOI] [PubMed] [Google Scholar]
- 40.Brauer MJ, Yuan J, Bennett BD, Lu W, Kimball E, Botstein D, Rabinowitz JD. 2006. Conservation of the metabolomic response to starvation across two divergent microbes. Proc. Natl. Acad. Sci. U. S. A. 103:19302–19307. 10.1073/pnas.0609508103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Doucette CD, Schwab DJ, Wingreen NS, Rabinowitz JD. 2011. α-Ketoglutarate coordinates carbon and nitrogen utilization via enzyme I inhibition. Nat. Chem. Biol. 7:894–901. 10.1038/nchembio.685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Commichau FM, Gunka K, Landmann JJ, Stülke J. 2008. Glutamate metabolism in Bacillus subtilis: gene expression and enzyme activities evolved to avoid futile cycles and to allow rapid responses to perturbations of the system. J. Bacteriol. 190:3557–3564. 10.1128/JB.00099-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Clark DP. 1989. The fermentation pathways of Escherichia coli. FEMS Microbiol. Lett. 63:223–234. 10.1111/j.1574-6968.1989.tb03398.x [DOI] [PubMed] [Google Scholar]
- 44.Kale S, Arjunan P, Furey W, Jordan F. 2007. A dynamic loop at the active center of the Escherichia coli pyruvate dehydrogenase complex E1 component modulates substrate utilization and chemical communication with the E2 component. J. Biol. Chem. 282:28106–28116. 10.1074/jbc.M704326200 [DOI] [PubMed] [Google Scholar]
- 45.Xu Y-F, Amador-Noguez D, Reaves ML, Feng X-J, Rabinowitz JD. 2012. Ultrasensitive regulation of anapleurosis via allosteric activation of PEP carboxylase. Nat. Chem. Biol. 8:562–568. 10.1038/nchembio.941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Koebmann BJ, Westerhoff HV, Snoep JL, Nilsson D, Jensen PR. 2002. The glycolytic flux in Escherichia coli is controlled by the demand for ATP. J. Bacteriol. 184:3909–3916. 10.1128/JB.184.14.3909-3916.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Orth JD, Conrad TM, Na J, Lerman JA, Nam H, Feist AM, Palsson BØ 2011. A comprehensive genome-scale reconstruction of Escherichia coli metabolism—2011. Mol. Syst. Biol. 7:535. 10.1038/msb.2011.65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ko YH, Hong S, Pedersen PL. 1999. Chemical mechanism of ATP synthase: magnesium plays a pivotal role in formation of the transition state where ATP is synthesized from ADP and inorganic phosphate. J. Biol. Chem. 274:28853–28856. 10.1074/jbc.274.41.28853 [DOI] [PubMed] [Google Scholar]
- 49.Ebel H, Günther T. 1980. Magnesium metabolism: a review. J. Clin. Chem. Clin. Biochem. 18:257–270 [DOI] [PubMed] [Google Scholar]
- 50.Ochi K, Kandala J, Freese E. 1982. Evidence that Bacillus subtilis sporulation induced by the stringent response is caused by the decrease in GTP or GDP. J. Bacteriol. 151:1062–1065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ratnayake-Lecamwasam M, Serror P, Wong K-W, Sonenshein AL. 2001. Bacillus subtilis CodY represses early-stationary-phase genes by sensing GTP levels. Genes Dev. 15:1093–1103. 10.1101/gad.874201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sonenshein AL. 2000. Control of sporulation initiation in Bacillus subtilis. Curr. Opin. Microbiol. 3:561–566. 10.1016/S1369-5274(00)00141-7 [DOI] [PubMed] [Google Scholar]
- 53.Stephens C. 1998. Bacterial sporulation: a question of commitment? Curr. Biol. 8:R45–R48. 10.1016/S0960-9822(98)70031-4 [DOI] [PubMed] [Google Scholar]
- 54.Stephens C, Singer M, Shapiro L. 1994. Bacterial sporulation: an ATP/ADP switch. Curr. Biol. 4:630–632. 10.1016/S0960-9822(00)00139-1 [DOI] [PubMed] [Google Scholar]
- 55.Kolodkin-Gal I, Elsholz AKW, Muth C, Girguis PR, Kolter R, Losick R. 2013. Respiration control of multicellularity in Bacillus subtilis by a complex of the cytochrome chain with a membrane-embedded histidine kinase. Genes Dev. 27:887–899. 10.1101/gad.215244.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Eswaramoorthy P, Duan D, Dinh J, Dravis A, Devi SN, Fujita M. 2010. The threshold level of the sensor histidine kinase KinA governs entry into sporulation in Bacillus subtilis. J. Bacteriol. 192:3870–3882. 10.1128/JB.00466-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tojo S, Hirooka K, Fujita Y. 2013. Expression of kinA and kinB of Bacillus subtilis, necessary for sporulation initiation, is under positive stringent transcription control. J. Bacteriol. 195:1656–1665. 10.1128/JB.02131-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.