Abstract
Fecal samples were obtained from a cohort of 330 healthy Danish infants at 9, 18, and 36 months after birth, enabling characterization of interbacterial relationships by use of quantitative PCR targeting 31 selected bacterial 16S rRNA gene targets representing different phylogenetic levels. Nutritional parameters and measures of growth and body composition were determined and investigated in relation to the observed development in microbiota composition. We found that significant changes in the gut microbiota occurred, particularly from age 9 to 18 months, when cessation of breastfeeding and introduction of a complementary feeding induce replacement of a microbiota characterized by lactobacilli, bifidobacteria, and Enterobacteriaceae with a microbiota dominated by Clostridium spp. and Bacteroides spp. Classification of samples by a proxy enterotype based on the relative levels of Bacteroides spp. and Prevotella spp. showed that enterotype establishment occurs between 9 and 36 months. Thirty percent of the individuals shifted enterotype between 18 and 36 months. The composition of the microbiota was most pronouncedly influenced by the time of cessation of breastfeeding. From 9 to 18 months, a positive correlation was observed between the increase in body mass index and the increase of the short-chain-fatty-acid-producing clostridia, the Clostridum leptum group, and Eubacterium hallii. Considering previously established positive associations between rapid infant weight gain, early breastfeeding discontinuation, and later-life obesity, the corresponding microbial findings seen here warrant attention.
INTRODUCTION
Establishment of the human intestinal microbiota during infancy is influenced by multiple factors, including delivery mode, sanitary conditions, administration of antibiotics to the infant or mother (1, 2), and level of breastfeeding (3). Breastfeeding has been shown to significantly increase the relative abundance of bifidobacteria and lactic acid bacteria, including lactobacilli and Enterococcus spp. (4, 5). The microbial composition within the first year of life is typically characterized by low species diversity and high instability (6–8). Nevertheless, a number of recent studies suggest that some of the bacteria that become part of the adult microbiota colonize the gut already during the first months of life (9, 10). A more complex, stable, and adult-like microbiota is established between 1 and 2 years after birth (11–13), the composition of which is believed to affect the risk of several lifestyle-related disorders, including obesity and type 2 diabetes (14, 15). Increased prevalence of childhood obesity is currently a major societal concern due to the high association with adult obesity (16, 17). It has also been proposed that obesity in adults may be related to the capacity of the gut microbiota to harvest energy through breakdown of indigestible polysaccharides (18, 19). Moreover, obesity and insulin resistance are often accompanied by a state of chronic low-grade inflammation (20). Since species-specific bacterial surface markers are involved in regulation of inflammation, differences in commensal bacterial composition between individuals may differentially predispose them to inflammation-induced diseases (21). Breastfed infants are leaner than formula-fed counterparts (22, 23) and display lower incidences of obesity, diabetes, and inflammatory bowel diseases later in life (24), and a link between diet, infant gut microbiota, obesity development, and inflammatory pathways has thus been suggested (25).
The discovery of the existence of so-called gut enterotypes in adult human subjects has received considerable attention, although final consensus on the number and characteristics of the enterotypes has not yet been achieved (26, 27). In this context, it has recently been proposed by us that adults can be grouped into two distinct groups based on the levels of Bacteroides spp. and particularly Prevotella spp., which are bimodally distributed in adult subjects and remain stable over a period of at least 6 months, and that these two genera can therefore serve as markers for the corresponding enterotypes (28). Each adult individual is estimated to harbor approximately 160 different high-abundance species, many of which are expected to be shared between individuals but present at very different levels in different individuals (29). Looking at the total pool of species present, earlier studies have suggested that as little as 1% may be shared with another individual (7, 30). Therefore, some redundancy of colonic bacterial processing would be expected to exist, and it has been demonstrated that within the intestinal metagenome, the phylogenetic variation is much more pronounced than the variation in functional capacities of the ecosystem (31). While interdependence and recognizable patterns of gut bacterial colonization in the intestinal ecosystem exist (32–35) and are known to be influenced, e.g., by dietary differences (6), only a few studies have combined the longitudinal development of representative bacterial taxa with parallel development of growth, body composition, and nutritional parameters in early life.
The aims of the present study were to describe patterns of microbial establishment during the first 3 years of life and to identify putative correlations of such patterns with dietary habits and physiological parameters, particularly focusing on development of body weight. We used the previously published quantitative PCR (qPCR)-based gut low-density array (GULDA) (36) to determine relative abundances and interbacterial relations of 31 different bacterial 16S rRNA gene targets, representing different phylogenetic levels, in fecal samples from a longitudinal cohort study of approximately 300 healthy Danish infants sampled at 9, 18, and 36 months after birth. Results were correlated with measures of growth, body composition, and nutritional records obtained for the same children, in order to reveal possible associations with the microbial composition and temporal development. Additionally, we performed the first comprehensive analysis of establishment of gut enterotypes (26, 27) in early life, using the Prevotella/Bacteroides ratio as a proxy for the enterotypes driven by the abundance of these genera.
MATERIALS AND METHODS
SKOT cohort.
The present study is based on data and samples collected during an observational cohort study of approximately 300 apparently healthy Danish singleton term infants. The cohort, titled SKOT, based on a Danish abbreviation, was followed for a period of 3 years with visits at 9, 18, and 36 months of age, and several papers based on the cohort have been published (37–42). The study protocol was approved by the Committees on Biomedical Research Ethics for the Capital Region of Denmark (H-KF-2007-0003). Fecal samples and information on birth mode and gender, measurements of growth and body composition, food questionnaires, and background interviews were collected at the visits during the study. The numbers of participants completing the 9-, 18-, and 36-month visits were 311, 290, and 264, respectively.
DNA extraction and qPCR analysis.
Total community DNA was extracted from a total of 698 fecal samples on the Maxwell 16 system using the Maxwell 16 DNA Tissue DNA purification kit (Promega Biotech AB, Sweden). The DNA concentrations were determined fluorometrically (Qubit dsDNA HS assay; Invitrogen) and adjusted to 1 ng/μl prior to use as the template in qPCR. The qPCR analysis was performed using the GULDA platform previously described (36). Briefly, each 384-well PCR plate accommodated simultaneous analysis of four DNA samples to determine the relative abundance of 31 bacterial selected 16S rRNA gene targets (Table 1) representing different phylogenetic levels. A universal bacterial primer set was included as the reference gene. All qPCRs were performed in duplicate in transparent 384-well MicroAmp optical reaction plates (Applied Biosystems) sealed with MicroAmp optical adhesive film on an ABI Prism 7900HT system (Applied Biosystems, Naerum, Denmark). Following the thermocycling program, the raw fluorescence data recorded by the SDS software were exported to the LinRegPCR program (43, 44). This software was used to perform baseline correction and calculate the mean PCR efficiency per amplicon group. This was used to calculate the initial quantities N0 (arbitrary fluorescence units) for each amplicon using the formula N0 = threshold/(EffCTmean), where Effmean denotes the mean PCR efficiency per amplicon, threshold is the optimal “cutoff” in the exponential region, and CT is the cycle number, where each sample exceeds this threshold. The relative abundances of the 31 specific amplicon groups were obtained by normalization to the N0 value obtained for the universal bacterial amplicon group determined in the same array (see Fig. S1 in the supplemental material).
TABLE 1.
16S rRNA gene targets included on gut low-density array (GULDA)a
| Amplicon ID | Phylum | Class/family/genus | Species/group/familyb |
|---|---|---|---|
| U1 | Universal (all phyla) | Universal | Universal |
| F1b | Firmicutes | All | All |
| F2 | Firmicutes | Lactobacillus | spp. |
| F3 | Firmicutes | Lactobacillus | L. plantarum |
| F4 | Firmicutes | Lactobacillus | L. acidophilus |
| F5 | Firmicutes | Clostridium | C. butyricum |
| F6 | Firmicutes | Clostridia | Cluster IV (C. leptum group) |
| F7 | Firmicutes | Clostridia | Cluster XIVa (C. coccoides-Eubacterium rectale group) |
| F8 | Firmicutes | Eubacterium | E. hallii |
| F9 | Firmicutes | Roseburia | spp. |
| F10 | Firmicutes | Enterococcus | spp. |
| B1 | Bacteroidetes | All | All |
| B2 | Bacteroidetes | Bacteroides-Prevotella | spp. |
| B3 | Bacteroidetes | Bacteroides | spp. |
| B4 | Bacteroidetes | Bacteroides | B. fragilis group |
| B5 | Bacteroidetes | Bacteroides | B. vulgatus |
| B6 | Bacteroidetes | Bacteroides | B. thetaiotaomicron |
| B7 | Bacteroidetes | Bacteroides | B. eggerthii |
| B8 | Bacteroidetes | Bacteroides | B. distasonis |
| B9 | Bacteroidetes | Prevotella | spp. |
| B10 | Bacteroidetes | Alistipes | spp. |
| A1b | Actinobacteria | Bifidobacterium | spp. |
| A2 | Actinobacteria | Bifidobacterium | B. bifidum |
| A3 | Actinobacteria | Bifidobacterium | B. adolescentis |
| A4 | Actinobacteria | Bifidobacterium | B. catenulatum/pseudocatenulatum |
| A5 | Actinobacteria | Bifidobacterium | B. longum |
| A6 | Actinobacteria | Bifidobacterium | B. breve |
| P1 | Proteobacteria | Enterobacteriaceae | Family |
| P2 | Proteobacteria | Escherichia | E. coli |
| P3 | Proteobacteria | Desulfovibrio | spp. |
| V1 | Verrucomicrobia | Akkermansia | A. muciniphila |
| E1 | Euryarchaeota | Methanobrevibacter | M. smithii |
Adapted from reference 36.
spp., multiple species.
A universal limit of detection (LODU) of 10−5 (N0, specific/N0, universal) was applied to the normalized N0 values due to qPCR analysis limitations. LODU was set to this value based on previous results using the GULDA setup and roughly corresponds to target CT values above 30 cycles. All normalized N0 values equal to or above LODU were included in the analysis, while samples below LODU were set to 0.5 LODU. Results were calculated as the arithmetic mean of normalized N0 values of the two technical repeats. Differences between CT values of technical replicates were typically less than 0.5. For samples where qPCR was successful for only one of the replicates, this value was used. Water as the template was used as the negative control. For 40 samples, no PCR amplification was detectable using the universal bacterial primer, and thus, the final number of fecal DNA samples for gut microbiota analysis was 658 representing 218, 232, and 208 at 9, 18, and 36 months, respectively. For 132 subjects, samples from all three time points were obtained. Note that a small subset of the data (obtained from 6 infants at two time points) has previously been published in order to illustrate the applicability of the PCR-based GULDA platform (36).
Parameters of nutrition, growth, and body composition.
Information on duration of breastfeeding and level of iron supplementation was obtained from background interviews (see Table S1 in the supplemental material). Parameters of infant diet (see Table S2) were estimated from parent-completed precoded dietary records over seven consecutive days (45). Dietary intake was calculated with GIES software (version 1.000d; The National Food Institute, DTU Food, Soborg, Denmark). Based on anthropometric measures described previously (39, 42), age- and gender-specific Z scores at birth and 9, 18, and 36 months (see Table S3) were calculated with the WHO Anthro software (Department of Nutrition, World Health Organization, Geneva, Switzerland). At 3 years, body composition was estimated by both dual-energy X-ray absorptiometry (DXA) and bioelectrical impedance analysis (BIA). BIA is a simple method for measuring body composition (46), and whole-body resistance, reactance, and impedance were measured using a single-frequency (50-kHz) tetrapolar BIA (Quantum III; RJL Systems, Michigan, USA) between right hand and right foot. Whole-body DXA scans were performed in a subgroup of the SKOT children (n = 101) with a Lunar Prodigy Advance densitometer (GE Healthcare, Madison, WI, USA) using the software enCore, version 12.30 (procedure described in detail by Jensen et al. [39]). Here, DXA fat-free mass (FFM) and fat mass (FM), resistance index (height2/resistance), and FFM and FM predicted from BIA were all used as measures for body composition at 3 years.
Assessment of changes in the gut microbial composition.
The normalized N0 values obtained for each bacterial taxon were log10 transformed and used as input for multivariate principal component analysis (PCA) using LATENTIX version 2.11 (Latent5 Aps, Frederiksberg, Denmark). Univariate statistical analysis was performed using the GraphPad Prism software (version 5.03; GraphPad Software Inc., La Jolla, CA). Only individuals for whom samples at all three examinations were available were included. Specific primer results, which never exceeded LODU at either 9, 18, or 36 months for a given individual, were excluded from further analysis. Consequently, the number of individuals for analysis of each bacterial taxon ranged from n = 25 to n = 132. Fold changes (FC) for specific gene targets were calculated as the pairwise (log2) ratio of normalized, but not log10-transformed, abundances at 9, 18, and 36 months, giving three ratios: 18 months/9 months, 36 months/9 months, and 36 months/18 months. Mean and corresponding standard error of the mean (SEM) values were calculated, and a one-sample t test was performed to test if the fold changes differed significantly from zero. The Wilcoxon signed-rank-sum test was performed as an alternative when data were not normally distributed. Correction for multiple testing (47) was performed for the 90 comparisons (3 × 30) presented in Fig. 2. Three statistical significance levels were employed: P < 0.05, P < 0.01, and P < 0.001. Spearman correlations R and corresponding P values between bacterial fold changes from 9 to 18 months and from 18 to 36 months, respectively, and changes in parameters of growth and body composition and nutritional parameters (see Tables S1, S2, and S3 in the supplemental material) were calculated using GraphPad (Table 2). The Mann-Whitney test was used to compare effects of continued or terminated breastfeeding at the 9-month examination on the relative abundances of all bacteria at 9, 18, and 36 months, respectively.
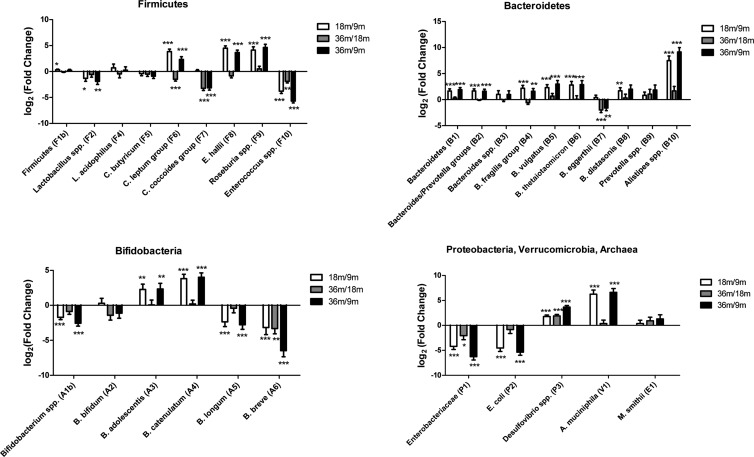
FIG 2.
Progressive development of the gut microbial composition. Log2(fold changes) of microbial 16S rRNA gene targets occurring from 9 to 18 months (white) and 18 to 36 months (gray) and cumulative values from 9 to 36 months (black). Statistical significance of one-sided t tests: *, P < 0.05; **, P < 0.01; ***, P < 0.001. Data were corrected for multiple testing, using a maximal false discovery rate of 5%.
TABLE 2.
Spearman correlation analysis of the relative differences occurring in BMI from 9 to 18 months, 18 to 36 months, and 9 to 36 months (ΔBMI) with corresponding bacterial fold changes in the same perioda
| Taxon | n | 9–18 mo |
18–36 mo |
9–36 mo |
|||
|---|---|---|---|---|---|---|---|
| P | R | P | R | P | R | ||
| Firmicutes (F1b) | 132 | 0.02* | 0.20 | NS | NS | ||
| C. leptum group (F6) | 132 | 0.02* | 0.21 | NS | NS | ||
| E. hallii (F8) | 131 | 0.03* | 0.19 | NS | NS | ||
| Enterobacteriaceae (P1) | 57 | 0.16 | −0.19 | 0.03* | −0.28 | NS | |
| M. smithii (E1) | 25 | 0.04* | −0.42 | NS | NS | ||
Since fold change and Δ calculations require valid measurements for each included individual at all three time points, the number of individuals (n) was different for each bacterial target. R designates the Spearman correlation coefficient. Only bacterial taxa with P values below 0.05 (*) are included in the table. NS, not significant (P > 0.05).
Spearman correlations at 9, 18, and 36 months.
For the time-independent analysis, all valid samples for each of the three time points, 9, 18, and 36 months, were included (n > 200 in each age group). Pairwise correlations between all measured SKOT parameters, including the microbiota, at all three time points were performed using GraphPad. Spearman correlations were applied for all pairwise correlations. False discovery rates (FDR) were calculated by a classical one-stage method (47).
The ratios between relative abundances of the Prevotella (B9) and Bacteroides (B3) targets were calculated as a proxy for the corresponding Prevotella- or Bacteroides-driven gut enterotypes (26). The logged relative abundances of Bacteroides spp. and Prevotella spp., frequency distributions of Bacteroides spp. and Prevotella spp., and corresponding ratio (P/B) were calculated for n = 69, 84, and 130 individuals, at 9, 18, and 36 months, respectively. A Kernel density plot was fitted to all histograms using the Kernel add-in package for Microsoft Excel. Characterization of the frequency distributions as uni- or bimodal was tested with a dip test, calculated by the R package diptest (R package version 0.75-4, based on Fortran and S-plus from Dario Ringach, New York University).
Samples from the n = 79 individuals giving qPCR results for both B3 (Bacteroides spp.) and B9 (Prevotella spp.) at both 18 and 36 months were stratified as either high- or low-P/B enterotypes and investigated for their putative cooccurrence with specifically high or low levels of physiological parameters (nutrition, growth, or body composition). Finally, the fold change from 18 to 36 months using the same stratification was correlated with the longitudinal development of all parameters of nutrition, growth, and body composition from 18 to 36 months.
RESULTS AND DISCUSSION
Development of the gut microbiota.
Although a quite extensive amount of literature on the possible factors involved in microbiota development in early life exists (for reviews, see references 48 and 49), these studies typically focus on microbial colonization immediately after birth (50, 51), during weaning at 4 to 6 months (3, 52), or up to 1 year (7) and, in a single recent study, 2 years of age (53). To our knowledge, no previous studies including numbers of participants as high as those in the present study have focused on the development occurring in the microbiota between infancy and 3 years of age.
We observed a clear change in the microbiota during this period, in particular from age 9 to 18 months (Fig. 1). The 9-month samples appeared to cluster less closely together than the later samples and were characterized by more lactic acid bacteria and enterobacteria than seen for samples taken at ages 18 and 36 months. The two other age groups comprised a higher number of different microbes, including both Firmicutes and Bacteroidetes. In line with this, the majority of specific changes in abundances of given bacterial taxa occurred between 9 and 18 months (Fig. 2; see also Fig. S2 in the supplemental material). We observed a consistent and significant increase of several species within the Bacteroidetes phylum, which is consistent with reported findings seen after introduction of complementary feeding (6, 54). Additionally, we observed a significant decrease in the relative abundance of Bifidobacterium spp. (FC, −2.56; P < 0.001). Within these, Bifidobacterium longum (FC, −2.77; P < 0.001) and Bifidobacterium breve (FC, −6.47; P < 0.001) were observed to decrease, while Bifidobacterium adolescentis (FC, 2.33; P < 0.01) and Bifidobacterium catenulatum (FC, 4.00; P < 0.001) increased during the period from 9 to 36 months of age, indicating that during early childhood, the conditions in the gut and/or the diet changes in ways that favor the latter species of bifidobacteria later in life. For example, breast milk is known to contain bifidogenic human milk oligosaccharides (HMOs), which are atypical carbohydrates, resistant to enzymatic hydrolysis in the upper gastrointestinal tract (55, 56). B. longum, Bifidobacterium bifidum, and B. breve are particularly abundant in breastfed children (57) and known to be highly proficient in capturing and utilizing HMOs as their sole carbon source, while B. adolescentis is unable to degrade these oligosaccharides (58).
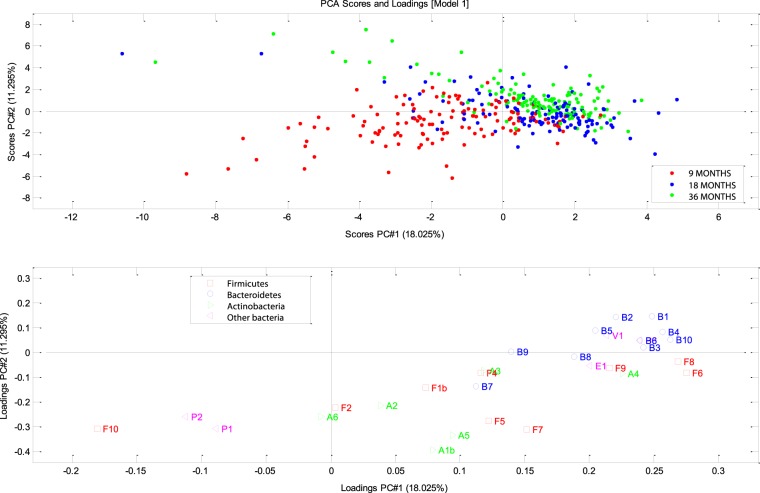
FIG 1.
Principal component analysis (PCA) of the GULDA microbiota. Upper plot: scores (individuals); lower plot: loadings (bacterial 16S rRNA gene targets). This figure shows the two primary principal components, PC1 and PC2, which explain 18.02% and 11.29% of data variation, respectively. Bacterial targets primarily associated with the lower left quadrant and thus relatively highly abundant in the 9-month samples were Enterococcus spp. (F10), Enterobacteriaceae (P1), and Escherichia coli (P2) and, to a lesser extent, B. breve (A6) and Lactobacillus spp. (F2). Bacterial targets appearing in higher abundances in the 36-month samples were the Bacteroidetes (B1), the Bacteroides-Prevotella group (B2), Bacteroides spp. (B3), B. fragilis group (B4), B. vulgatus (B5), Bacteroides thetaiotaomicron (B6), Alistipes spp. (B10), A. muciniphila (V1), and Desulfovibrio spp. (P3). Complete explanations for all labels are given in Table 1.
Lactobacillus spp. (FC, −1.33; P < 0.05) and Enterobacteriaceae (FC, −4.21; P < 0.001) were found to decrease between 9 and 18 months (Fig. 2), while an increase was observed for the butyrate-producing taxa Clostridium leptum group (FC, 2.32; P < 0.001), E. hallii (FC, 3.65; P < 0.001), and Roseburia spp. (FC, 4.62; P < 0.001) from age 9 to 36 months. This is in accordance with previous findings seen at cessation of breastfeeding and introduction of formula feeding and/or cow's milk (1, 2, 5, 24, 54, 59–61). Conversely, the butyrate-producing Clostridium coccoides group (FC, −3.14; P < 0.001) was seen to be reduced between 9 and 18 months. A previous report (62), based on a cross-sectional study of 40 children, indicates that the C. coccoides group increases until 6 months of age and thereafter remains at a stable level.
The fact that we observed significant changes still occurring from 18 to 36 months (Fig. 1 and 2) suggests that convergence toward adult-like stability, characterized by high levels of Firmicutes and Bacteroidetes and smaller fractions of Actinobacteria, Proteobacteria, and Verrucomicrobia (63–65), was still occurring during this period. This is in line with a recent large cross-sectional study of humans from different age groups showing that bacterial communities of the gut evolve toward adult-like configurations during the first 3 years of life (66) but contradicts older reports proposing that full stability is reached already at 12 months (6, 7, 67).
Correlations between relative abundance of bacterial groups and physiological parameters.
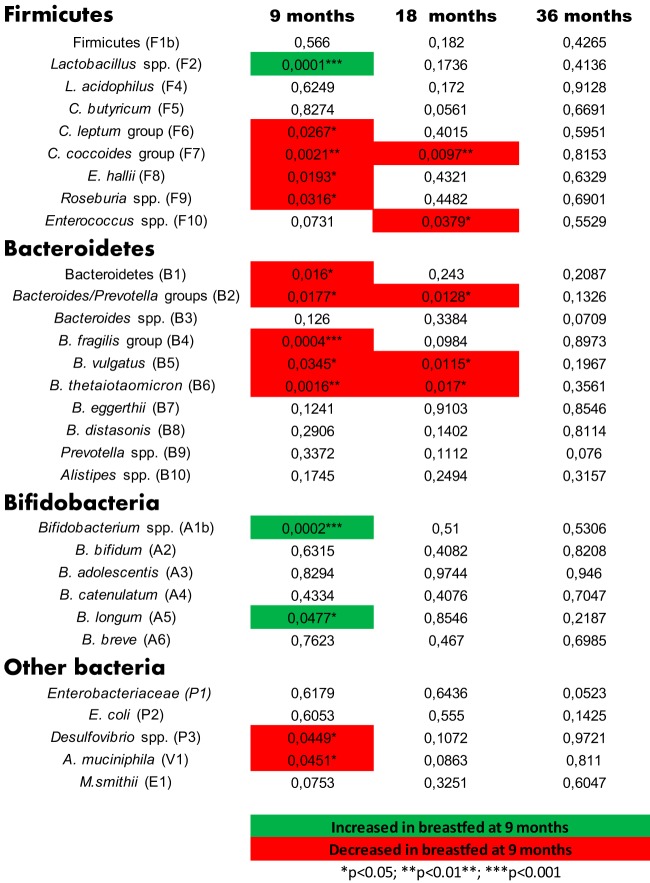
In agreement with previous studies from other researchers (58), continued breastfeeding at 9 months was associated positively with high relative abundances of Lactobacillus spp., Bifidobacterium spp., and B. longum at 9 months (Fig. 3). Additionally, compared to infants no longer breastfed at 9 months, infants still breastfed at 9 months had lower levels of a number of butyrate-producing taxa, including C. leptum group, C. coccoides group, E. hallii, and Roseburia spp. Breastfeeding at 9 months was also associated with lower levels of Desulfovibrio spp. and Akkermansia muciniphila, as well as of the Bacteroidetes phylum and several taxa therein. For the C. coccoides group and some of the Bacteroides species, the differences were still present after 18 months, while at 36 months the breastfeeding history no longer influenced the microbiota (Fig. 3). Breastfeeding has been shown to significantly reduce the risk of overweight/obesity later in childhood as well as in adult life (68); however, conflicting reports of the role of breastfeeding in obesity also exist (69). We speculate that the observation that continued breastfeeding at 9 months delays progression of specific bacterial taxa may be of relevance for development and later-life health.
FIG 3.
Effect of breastfeeding on infant gut microbiota. P values of Mann-Whitney statistical tests addressing differences between relative bacterial abundances at 9, 18, and 36 months, dependent on whether or not the infants were still breastfed at the 9-month examination. Green indicates an increase in children breastfed at 9 months, and red indicates a corresponding decrease.
We observed only very few correlations between the abundances of specific gut bacteria and the physiological parameters measured (see Fig. S3 in the supplemental material). Significant associations with P < 0.001 and false discovery rates (q) below 0.08 were observed at 9 months, where duration of breastfeeding (breast milk days, indicating the number of days with either partial or exclusive breastfeeding as estimated by the mothers) was shown to correlate positively with Lactobacillus and Bifidobacterium targets. Less significantly (P < 0.01, q < 0.32), negative associations with duration of breastfeeding were seen for C. leptum group, E. hallii, Roseburia spp., Bacteroides/Prevotella groups, Bacteroides fragilis, Bacteroides vulgatus, Desulfovibrio spp., and A. muciniphila.
Supporting these observations, many of the opposite trends for correlations were observed between these bacterial targets and the intake of infant formula, including a negative correlation (P < 0.001, q < 0.08) with the abundance of Lactobacillus spp. Additionally, the data reflected that duration of breast milk consumption (breast milk days) was negatively associated with the overall energy intake, as previously reported (38). We have previously shown that the breastfed infants had lower body mass indexes (BMIs) at both 9 and 18 months (41). No other significant correlations were found between the gut microbiota and nutritional parameters, or measures of growth and body composition (including DXA and BIA examinations at 36 months), gender, or birth mode (vaginal versus caesarean) at any of the three time points (data not shown). Previous reports on correlations between BMI and gut microbiota composition in young children exist (70, 71); however, these were observed in cross-sectional cohort studies where sampling was focused on high versus normal BMI, while the infants included in the present study constituted a younger and leaner population, with only 8% classified as overweight (72).
Exploiting the longitudinal observations from the present study, we found significant (P < 0.05) positive correlations between increase of body mass index (ΔBMI) and the increase of the Firmicutes phylum, the C. leptum group, and E. hallii (belonging to the C. coccoides group) between 9 and 18 months (Table 2). Additionally, increases in Methanobrevibacter smithii were negatively correlated with ΔBMI from 9 to 18 months, while reductions in Enterobacteriaceae were associated with higher ΔBMI from 18 to 36 months. Similar results were obtained for ΔBMI-for-age Z score (ΔBAZ) and Δweight-for-length Z score (ΔWFL) but not for changes in Z scores for weight for age (WAZ), length for age (HAZ), subscapular skinfold for age (SSZ), or triceps skinfold for age (TSZ) (data not shown). No other significant correlations between changes in body compositional measures and changes in nutritional parameters and/or bacterial targets were observed. Although excessive weight gain during the first 6 months after birth has been shown to be particularly predictive of later obesity (73–76), we found no significant characteristics in the microbiota after 9 months, which corresponded to changes in BAZ or WAZ between birth and 9 months (data not shown).
The clostridial targets selected for this study represent colonic butyrate-producing bacteria, assisting in the conversion of polysaccharides to monosaccharides and short-chain fatty acids (SCFA) constituting energy for the host (77, 78). The development of abundances of the selected taxa C. butyricum, C. leptum group, C. coccoides group, E. hallii, and Roseburia spp. (the two last taxa belonging to the C. coccoides group) was very different for the different targets (Fig. 2). Significant differences in carbohydrate metabolism and butyrate production between different clostridia have been demonstrated to be of relevance to the pathogenesis of obesity (79). It remains to be established whether certain dietary compounds, arguably containing high concentrations of specific complex polysaccharides, are specifically subjected to catabolism by the C. leptum group, E. hallii, and Roseburia spp. between 9 and 18 months, as suggested by the data (Fig. 2).
Correlation between bacterial groups at 9, 18, and 36 months.
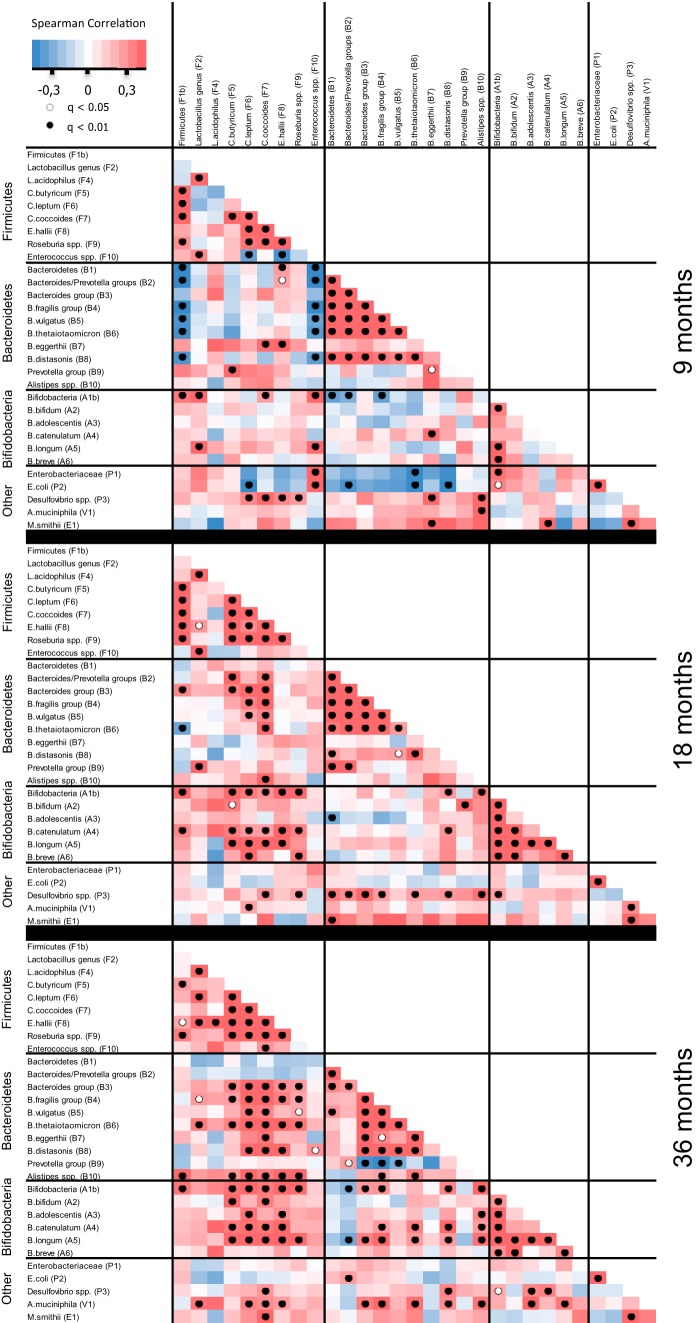
Correlations between the abundances of the bacterial 16S rRNA gene targets were investigated for each of the three age groups (Fig. 4). Although there were more differences between the 9-month pattern and the two later groups, differences between 18 and 36 months were also observed.
FIG 4.
Spearman pairwise correlation map of measured bacterial 16S rRNA gene targets at 9, 18, and 36 months. Because each time point was analyzed separately, the included number of individuals was >200 at each of the three samplings. The color gradient denotes Spearman R values. Dots indicate significant correlations, corrected for false discovery rates (q).
At 9 months, we found coabundance (positive Spearman correlations) between the butyrate-producing Firmicutes C. leptum group, C. coccoides group, and Clostridium butyricum. Similarly, coabundance was seen for many of the Bacteroides species, with the notable exception of Bacteroides eggerthii. With this specific exception, a high abundance of Firmicutes was also clearly associated with a low abundance of Bacteroides species. While the other Bacteroides species were increasing in abundance during the study, B. eggerthii was reduced (Fig. 2). The reverse cooccurrence of B. eggerthii and the other Bacteroides species was no longer as significant at 36 months (Fig. 4), indicating that later in life, the environment in the gut no longer represses this organism more than the other Bacteroides organisms. Additionally, the clear association between a high abundance of Firmicutes and enterococci and a low abundance of Bacteroides spp. seen at 9 months has disappeared at 18 and 36 months, indicating that it is particularly in infancy that these groups are mutually exclusive of each other.
Bacteroides spp. and Clostridiales spp. have previously been seen to cocluster in the so-called enterotype driven by the abundance of Bacteroides spp. in healthy human adults (26). This is in agreement with our data from age 18 months and becomes even clearer at 36 months; however, at 9 months we found no coabundance of these groups (Fig. 4). It seems plausible that the development of this coabundance is a logical consequence of adaptation to a Western-type diet after weaning. However, analysis of possible correlations between bacterial targets and the investigated nutritional parameters (see Table S2 in the supplemental material) did not result in statistically significant cooccurrences (data not shown).
We found it noteworthy that at 9 months, when Bifidobacterium spp. in general were most abundant (Fig. 2), there was a clear cooccurrence of particularly B. longum with the other lactic acid-producing taxa Lactobacillus spp. and Enterococcus spp.; however, no cooccurrence between specific species of Bifidobacterium was seen (Fig. 4). This pattern was reversed at 18 and 36 months, when the cooccurrence with lactobacilli and enterococci was no longer present, while cooccurrence of the specific Bifidobacterium species B. longum with B. bifidum, B. adolescentis, and B. catenulatum, and further of B. bifidum with B. catenulatum and B. breve was evident. Interestingly, these cooccurrences existed independently of the fact that the average abundance of certain Bifidobacterium species was increased during the experimental period, while others were reduced as discussed above (Fig. 2).
Enterotype development in the infant gut.
Lately, the existence of three distinct enterotypes, driven by the abundance of Bacteroides spp., Prevotella spp., and Ruminococcus spp., respectively, has been given particular attention (26). While evidence is mounting in support of the distinction between the Bacteroides- and Prevotella-driven groupings, the existence of the third group, driven by the abundance of Ruminococcus spp., is not as clearly supported (80). In the present study, we used the relative abundance between Prevotella spp. and Bacteroides spp., measured as the Prevotella/Bacteroides ratio (P/B), as a proxy for the enterotypes driven by these two genera as proposed by Arumugam et al. (26). It is important to note that we do not mean to propose that the enterotypes are characterized solely by the abundance of these taxa but merely that their abundance can be used as a marker for more complex differences characterizing these two types of intestinal bacterial communities. For the first time, this approach allowed addressing the establishment of enterotypes during infancy.
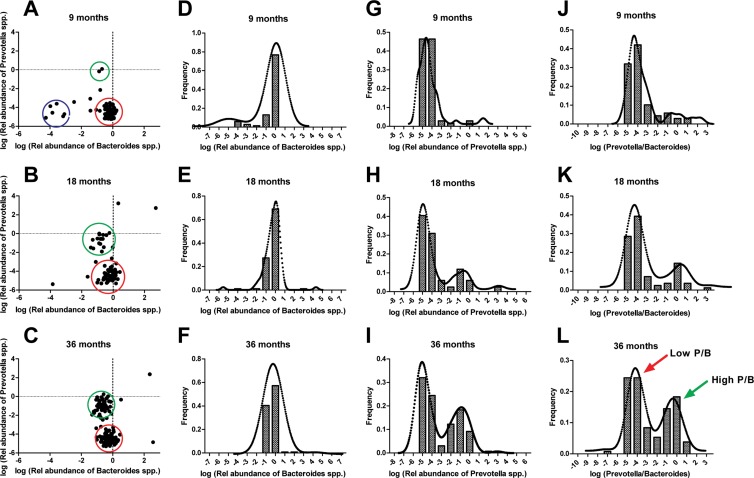
In agreement with the existence of enterotypes, we observed a negative correlation between Prevotella spp. and Bacteroides spp. at 36 months of age but not at 9 or 18 months (Fig. 4). The relative abundances of both Prevotella spp. and Bacteroides spp. are below detection limits at birth, given the absence of these bacteria in the prenatal environment of the maternal uterus (48). We propose that Bacteroides spp. colonize better than Prevotella spp. between birth and 9 months (9), as almost all individuals were characterized by a low P/B at 9 months of age (Fig. 5A). At 18 months, a smaller subset of individuals established a higher P/B (Fig. 5B), a pattern which was even more pronounced after 36 months, where two distinct groups appeared (Fig. 5C). We observed a unimodal distribution of Bacteroides abundances at all three sampling points (Fig. 5D to F), while an increasingly bimodal pattern of Prevotella abundances (Fig. 5G to I) developed from 9 to 36 months. The ratio of logged P/B values similarly showed an increasingly bimodal pattern with age, with high-P/B samples characterized by a logged ratio above a level of −2 (P/B > 0.01) and low-P/B samples below −2 (P/B < 0.01) (Fig. 5J and K). When tested statistically for the presence of bimodal distribution, there was significance at the 36-month Prevotella abundance (P = 0.004) (Fig. 5I) and an even clearer bimodality for the 36-month P/B (P = 6.6E−5) (Fig. 5L), but not at the earlier time points. Consequently, development of the two enterotypes starts between 18 and 36 months and is driven by changes in Prevotella spp., rather than Bacteroides spp. However, stratification of samples into P/B types at either 18 or 36 months did not reveal any additional correlations between bacterial 16S rRNA gene targets and host physiological phenotypes.
FIG 5.
Enterotype defined as P/B development from 9 to 36 months. Relative abundances of log(Bacteroides spp.) and log(Prevotella spp.) show a distinct development from 9 to 36 months (A to C), moving from low relative abundances of both groups (blue circle) to a Bacteroides-prevalent microbiota (red circle) at 9 months. From 9 to 36 months, an increasing subgroup of Prevotella-prevalent samples appear (green circle), indicating segregation of specific individuals from a Bacteroides-driven into a Prevotella-driven enterotype. This progressive development is also evident from the corresponding histograms of frequency distributions of log(Bacteroides spp.) (D to F) and log(Prevotella spp.) (G to I) abundance, and particularly from the distributions of the logged P/B (J to L). The dotted curve in panels D to L shows a Kernel density plot, which is a modification of the histogram patterns, supporting the underlying statistical distributions found. Panels A, D, G, and J (9 months) represent 69 individuals; panels B, E, H, and K (18 months) represent 84 individuals; and panels C, F, I, and L (36 months) represent 130 individuals, as only individuals with relative abundances of Bacteroides spp. and Prevotella spp. exceeding the detection limit were included.
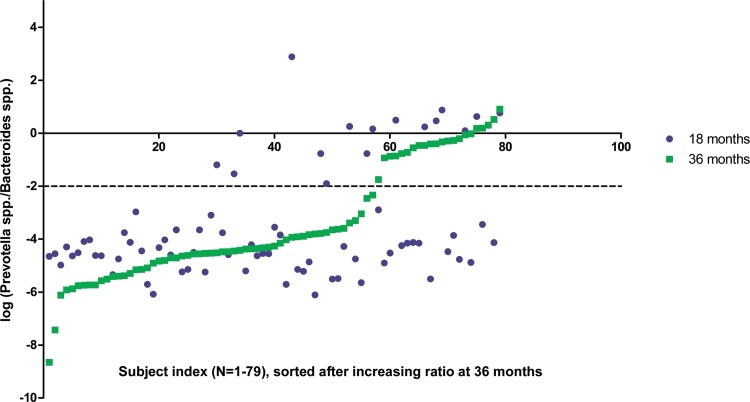
Out of the 79 individuals for whom the P/B could be calculated for both 18 and 36 months, 70% remained in the same P/B group between 18 and 36 months, while 18% and 11%, respectively, shifted their enterotypes from low P/B to high P/B or vice versa (Fig. 6). Previous studies have shown that although the adult gut microbiota of any individual is quite resilient to major perturbations, enterotypes may shift in a few individuals when measured over longer periods of time (81, 82) and by long-term dietary intervention schemes (80). In the present study, we did not identify any correlation of enterotype with diet or BMI. No data on antibiotic treatment were collected for the cohort; however, since antibiotic treatment is known to induce major changes in the composition of the infant gut microbiota (60, 83, 84), it cannot be excluded that antibiotic usage prior to 18- or 36-month samplings influenced the observed shifts. Nevertheless, our results (Fig. 6) strongly support the notion that between 18 and 36 months, the P/B enterotype is still not as stably established as reported in adults (28). In light of recent findings showing that enterotypes driven by Bacteroides spp. and Prevotella spp. affect risk markers for atherosclerosis (85) and that gut microbiome composition correlates with obesity and metabolic markers in adults (86), we propose that the establishment of microbiota during infancy may affect health status in adult life.
FIG 6.
Changes in P/B occurring between age 18 and 36 months The 79 individuals giving qPCR results for both 18 (blue) and 36 (green) months were sorted after increasing logged P/B at 36 months. Samples above the dotted line belong to the high-P/B group (Prevotella-driven enterotype), while samples below this line belong to the low-P/B group (Bacteroides-driven enterotype). Forty-eight of 79 individuals remained in the low-P/B group, while 8/79 individuals remained in the high-P/B group from age 18 to age 36 months. Fourteen of 79 and 9/79 individuals shifted from low to high P/B, or from high to low P/B, respectively.
Concluding remarks.
We have studied the establishment of intestinal microbiota in a large cohort of Danish infants and analyzed the microbial data in relation to a vast amount of dietary and physiological measures. We demonstrate significant differences in microbiota composition between infants either breastfed or no longer breastfed at 9 months but additionally show that the effects of breastfeeding on the microbiota are no longer prevalent at age 36 months. Positive correlations between increases in BMI, C. leptum group, and E. hallii were observed from 9 to 18 months, indicating that these butyrate-producing groups may contribute importantly to host energy harvest. Additionally, we show for the first time that human enterotypes, expressed as a bimodal distribution of the Prevotella/Bacteroides ratio, start being established between 18 and 36 months of age. In this period, where we observe an ongoing development of the microbiota toward an adult-like composition, enterotypes are still more susceptible to shifting than previously seen for adults.
Considering the increasing evidence supporting a key role of gut microbiota composition in human health, the presented data constitute an important new body of knowledge on microbiota development during infancy, which is likely to constitute a window where the microbiota can be more significantly influenced by intervention. In this context, the current development in next-generation sequencing is expected to contribute importantly to our understanding of human microbiome establishment during the coming years.
Supplementary Material
ACKNOWLEDGMENTS
We thank the Danish Council for Independent Research for financial support (grant no. 10-093725/FTP). The SKOT study was supported by the Danish Directorate for Food, Fisheries and Agribusiness (grant no. 3304-FSE-06-0503).
We thank Bodil Madsen and Vivian Julia Anker for excellent technical assistance with qPCR and DNA purification, respectively. Additionally, we thank Inge Tetens, Ellen Trolle, Ulla Gondolf, and Majken Ege for collecting the nutritional data and Louise Beltoft Borup Andersen for providing the processed nutritional data, while Line Rieck Schmidt, Sara Dyhrberg, and Louise Nissen-Schmidt are acknowledged for additional experimental assistance.
Footnotes
Published ahead of print 28 February 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.00342-14.
REFERENCES
- 1.Adlerberth I, Wold AE. 2009. Establishment of the gut microbiota in Western infants. Acta Paediatr. 98:229–238. 10.1111/j.1651-2227.2008.01060.x [DOI] [PubMed] [Google Scholar]
- 2.Fallani M, Young D, Scott J, Norin E, Amarri S, Adam R, Aguilera M, Khanna S, Gil A, Edwards CA, Dore J. 2010. Intestinal microbiota of 6-week-old infants across Europe: geographic influence beyond delivery mode, breast-feeding, and antibiotics. J. Pediatr. Gastroenterol. Nutr. 51:77–84. 10.1097/MPG.0b013e3181d1b11e [DOI] [PubMed] [Google Scholar]
- 3.Fallani M, Amarri S, Uusijarvi A, Adam R, Khanna S, Aguilera M, Gil A, Vieites JM, Norin E, Young D, Scott JA, Dore J, Edwards CA. 2011. Determinants of the human infant intestinal microbiota after the introduction of first complementary foods in infant samples from five European centres. Microbiology 157:1385–1392. 10.1099/mic.0.042143-0 [DOI] [PubMed] [Google Scholar]
- 4.Harmsen HJ, Wildeboer-Veloo AC, Raangs GC, Wagendorp AA, Klijn N, Bindels JG, Welling GW. 2000. Analysis of intestinal flora development in breast-fed and formula-fed infants by using molecular identification and detection methods. J. Pediatr. Gastroenterol. Nutr. 30:61–67. 10.1097/00005176-200001000-00019 [DOI] [PubMed] [Google Scholar]
- 5.Roger LC, Costabile A, Holland DT, Hoyles L, McCartney AL. 2010. Examination of faecal Bifidobacterium populations in breast- and formula-fed infants during the first 18 months of life. Microbiology 156:3329–3341. 10.1099/mic.0.043224-0 [DOI] [PubMed] [Google Scholar]
- 6.Koenig JE, Spor A, Scalfone N, Fricker AD, Stombaugh J, Knight R, Angenent LT, Ley RE. 2011. Succession of microbial consortia in the developing infant gut microbiome. Proc. Natl. Acad. Sci. U. S. A. 108(Suppl 1):4578–4585. 10.1073/pnas.1000081107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Palmer C, Bik EM, DiGiulio DB, Relman DA, Brown PO. 2007. Development of the human infant intestinal microbiota. PLoS Biol. 5:e177. 10.1371/journal.pbio.0050177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Spor A, Koren O, Ley R. 2011. Unravelling the effects of the environment and host genotype on the gut microbiome. Nat. Rev. Microbiol. 9:279–290. 10.1038/nrmicro2540 [DOI] [PubMed] [Google Scholar]
- 9.Jost T, Lacroix C, Braegger CP, Chassard C. 2012. New insights in gut microbiota establishment in healthy breast fed neonates. PLoS One 7:e44595. 10.1371/journal.pone.0044595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jost T, Lacroix C, Braegger CP, Rochat F, Chassard C. 23 August 2013. Vertical mother-neonate transfer of maternal gut bacteria via breastfeeding. Environ. Microbiol. 10.1111/1462-2920.12238 [DOI] [PubMed] [Google Scholar]
- 11.Mackie RI, Sghir A, Gaskins HR. 1999. Developmental microbial ecology of the neonatal gastrointestinal tract. Am. J. Clin. Nutr. 69:1035S–1045S [DOI] [PubMed] [Google Scholar]
- 12.Mandar R, Mikelsaar M. 1996. Transmission of mother's microflora to the newborn at birth. Biol. Neonate 69:30–35. 10.1159/000244275 [DOI] [PubMed] [Google Scholar]
- 13.Sekirov I, Russell SL, Antunes LC, Finlay BB. 2010. Gut microbiota in health and disease. Physiol. Rev. 90:859–904. 10.1152/physrev.00045.2009 [DOI] [PubMed] [Google Scholar]
- 14.Kinross JM, von Roon AC, Holmes E, Darzi A, Nicholson JK. 2008. The human gut microbiome: implications for future health care. Curr. Gastroenterol. Rep. 10:396–403. 10.1007/s11894-008-0075-y [DOI] [PubMed] [Google Scholar]
- 15.Othman M, Aguero R, Lin HC. 2008. Alterations in intestinal microbial flora and human disease. Curr. Opin. Gastroenterol. 24:11–16. 10.1097/MOG.0b013e3282f2b0d7 [DOI] [PubMed] [Google Scholar]
- 16.Lau DC, Douketis JD, Morrison KM, Hramiak IM, Sharma AM, Ur E. 2007. 2006 Canadian clinical practice guidelines on the management and prevention of obesity in adults and children. CMAJ 176:S1–S13. 10.1503/cmaj.061409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Willers SM, Brunekreef B, Smit HA, van der Beek EM, Gehring U, de Jongste C, Kerkhof M, Koppelman GH, Wijga AH. 2012. BMI development of normal weight and overweight children in the PIAMA study. PLoS One 7:e39517. 10.1371/journal.pone.0039517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI. 2006. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature 444:1027–1031. 10.1038/nature05414 [DOI] [PubMed] [Google Scholar]
- 19.Vael C, Desager K. 2009. The importance of the development of the intestinal microbiota in infancy. Curr. Opin. Pediatr. 21:794–800. 10.1097/MOP.0b013e328332351b [DOI] [PubMed] [Google Scholar]
- 20.Heilbronn LK, Campbell LV. 2008. Adipose tissue macrophages, low grade inflammation and insulin resistance in human obesity. Curr. Pharm. Des. 14:1225–1230. 10.2174/138161208784246153 [DOI] [PubMed] [Google Scholar]
- 21.Tremaroli V, Backhed F. 2012. Functional interactions between the gut microbiota and host metabolism. Nature 489:242–249. 10.1038/nature11552 [DOI] [PubMed] [Google Scholar]
- 22.Gibbs BG, Forste R. 2 April 2013. Socioeconomic status, infant feeding practices and early childhood obesity. Pediatr. Obes. 10.1111/j.2047-6310.2013.00155.x [DOI] [PubMed] [Google Scholar]
- 23.Owen CG, Martin RM, Whincup PH, Davey-Smith G, Gillman MW, Cook DG. 2005. The effect of breastfeeding on mean body mass index throughout life: a quantitative review of published and unpublished observational evidence. Am. J. Clin. Nutr. 82:1298–1307 [DOI] [PubMed] [Google Scholar]
- 24.Le Huerou-Luron I, Blat S, Boudry G. 2010. Breast- v. formula-feeding: impacts on the digestive tract and immediate and long-term health effects. Nutr. Res. Rev. 23:23–36. 10.1017/S0954422410000065 [DOI] [PubMed] [Google Scholar]
- 25.Sanz Y, Santacruz A, De Palma G. 2008. Insights into the roles of gut microbes in obesity. Interdiscip. Perspect. Infect. Dis. 2008:829101. 10.1155/2008/829101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Arumugam M, Raes J, Pelletier E, Le Paslier D, Yamada T, Mende DR, Fernandes GR, Tap J, Bruls T, Batto JM, Bertalan M, Borruel N, Casellas F, Fernandez L, Gautier L, Hansen T, Hattori M, Hayashi T, Kleerebezem M, Kurokawa K, Leclerc M, Levenez F, Manichanh C, Nielsen HB, Nielsen T, Pons N, Poulain J, Qin J, Sicheritz-Ponten T, Tims S, Torrents D, Ugarte E, Zoetendal EG, Wang J, Guarner F, Pedersen O, de Vos WM, Brunak S, Dore J, Antolin M, Artiguenave F, Blottiere HM, Almeida M, Brechot C, Cara C, Chervaux C, Cultrone A, Delorme C, Denariaz G, Dervyn R, Foerstner KU, Friss C, van de Guchte M, Guedon E, Haimet F, Huber W, van Hylckama-Vlieg J, Jamet A, Juste C, Kaci G, Knol J, Lakhdari O, Layec S, Le Roux K, Maguin E, Merieux A, Melo MR, M'rini C, Muller J, Oozeer R, Parkhill J, Renault P, Rescigno M, Sanchez N, Sunagawa S, Torrejon A, Turner K, Vandemeulebrouck G, Varela E, Winogradsky Y, Zeller G, Weissenbach J, Ehrlich SD, Bork P. 2011. Enterotypes of the human gut microbiome. Nature 473:174–180. 10.1038/nature09944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Koren O, Knights D, Gonzalez A, Waldron L, Segata N, Knight R, Huttenhower C, Ley RE. 2013. A guide to enterotypes across the human body: meta-analysis of microbial community structures in human microbiome datasets. PLoS Comput. Biol. 9:e1002863. 10.1371/journal.pcbi.1002863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Roager HM, Licht TR, Poulsen SK, Larsen TM, Bahl MI. 2014. Microbial enterotypes, inferred by the Prevotella-to-Bacteroides ratio, remain stable during a 6-month randomized controlled diet intervention with the new Nordic diet. Appl. Environ. Microbiol. 80:1142–1149. 10.1128/AEM.03549-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Qin J, Li R, Raes J, Arumugam M, Burgdorf KS, Manichanh C, Nielsen T, Pons N, Levenez F, Yamada T, Mende DR, Li J, Xu J, Li S, Li D, Cao J, Wang B, Liang H, Zheng H, Xie Y, Tap J, Lepage P, Bertalan M, Batto JM, Hansen T, Le Paslier D, Linneberg A, Nielsen HB, Pelletier E, Renault P, Sicheritz-Ponten T, Turner K, Zhu H, Yu C, Li S, Jian M, Zhou Y, Li Y, Zhang X, Li S, Qin N, Yang H, Wang J, Brunak S, Dore J, Guarner F, Kristiansen K, Pedersen O, Parkhill J, Weissenbach J, Bork P, Ehrlich SD, Wang J. 2010. A human gut microbial gene catalogue established by metagenomic sequencing. Nature 464:59–65. 10.1038/nature08821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Eckburg PB, Bik EM, Bernstein CN, Purdom E, Dethlefsen L, Sargent M, Gill SR, Nelson KE, Relman DA. 2005. Diversity of the human intestinal microbial flora. Science 308:1635–1638. 10.1126/science.1110591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huttenhower C, Haley EM, Hibbs MA, Dumeaux V, Barrett DR, Coller HA, Troyanskaya OG. 2009. Exploring the human genome with functional maps. Genome Res. 19:1093–1106. 10.1101/gr.082214.108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Faust K, Sathirapongsasuti JF, Izard J, Segata N, Gevers D, Raes J, Huttenhower C. 2012. Microbial co-occurrence relationships in the human microbiome. PLoS Comput. Biol. 8:e1002606. 10.1371/journal.pcbi.1002606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Freilich S, Kreimer A, Meilijson I, Gophna U, Sharan R, Ruppin E. 2010. The large-scale organization of the bacterial network of ecological co-occurrence interactions. Nucleic Acids Res. 38:3857–3868. 10.1093/nar/gkq118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Konopka A. 2009. What is microbial community ecology? ISME J. 3:1223–1230. 10.1038/ismej.2009.88 [DOI] [PubMed] [Google Scholar]
- 35.Trosvik P, Stenseth NC, Rudi K. 2010. Convergent temporal dynamics of the human infant gut microbiota. ISME J. 4:151–158. 10.1038/ismej.2009.96 [DOI] [PubMed] [Google Scholar]
- 36.Bergstrom A, Licht TR, Wilcks A, Andersen JB, Schmidt LR, Gronlund HA, Vigsnaes LK, Michaelsen KF, Bahl MI. 2012. Introducing GUt low-density array (GULDA): a validated approach for qPCR-based intestinal microbial community analysis. FEMS Microbiol. Lett. 337:38–47. 10.1111/1574-6968.12004 [DOI] [PubMed] [Google Scholar]
- 37.Engel S, Tronhjem KM, Hellgren LI, Michaelsen KF, Lauritzen L. 2013. Docosahexaenoic acid status at 9 months is inversely associated with communicative skills in 3-year-old girls. Matern. Child Nutr. 9:499–510. 10.1111/j.1740-8709.2012.00411.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gondolf UH, Tetens I, Michaelsen KF, Trolle E. 2012. Dietary habits of partly breast-fed and completely weaned infants at 9 months of age. Public Health Nutr. 15:578–586. 10.1017/S1368980011003247 [DOI] [PubMed] [Google Scholar]
- 39.Jensen SM, Molgaard C, Ejlerskov KT, Christensen LB, Michaelsen KF, Briend A. 20 November 2012. Validity of anthropometric measurements to assess body composition, including muscle mass, in 3-year-old children from the SKOT cohort. Matern. Child Nutr. 10.1111/mcn.12013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Klingenberg L, Christensen LB, Hjorth MF, Zangenberg S, Chaput JP, Sjodin A, Molgaard C, Michaelsen KF. 2013. No relation between sleep duration and adiposity indicators in 9–36 months old children: the SKOT cohort. Pediatr. Obes. 8:e14–e18. 10.1111/j.2047-6310.2012.00109.x [DOI] [PubMed] [Google Scholar]
- 41.Madsen AL, Larnkjaer A, Molgaard C, Michaelsen KF. 2011. IGF-I and IGFBP-3 in healthy 9 month old infants from the SKOT cohort: breastfeeding, diet, and later obesity. Growth Horm. IGF Res. 21:199–204. 10.1016/j.ghir.2011.05.003 [DOI] [PubMed] [Google Scholar]
- 42.Madsen AL, Schack-Nielsen L, Larnkjaer A, Molgaard C, Michaelsen KF. 2010. Determinants of blood glucose and insulin in healthy 9-month-old term Danish infants; the SKOT cohort. Diabet. Med. 27:1350–1357. 10.1111/j.1464-5491.2010.03134.x [DOI] [PubMed] [Google Scholar]
- 43.Ramakers C, Ruijter JM, Deprez RH, Moorman AF. 2003. Assumption-free analysis of quantitative real-time polymerase chain reaction (PCR) data. Neurosci. Lett. 339:62–66. 10.1016/S0304-3940(02)01423-4 [DOI] [PubMed] [Google Scholar]
- 44.Ruijter JM, Ramakers C, Hoogaars WM, Karlen Y, Bakker O, van den Hoff MJ, Moorman AF. 2009. Amplification efficiency: linking baseline and bias in the analysis of quantitative PCR data. Nucleic Acids Res. 37:e45. 10.1093/nar/gkp045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gondolf UH, Tetens I, Hills AP, Michaelsen KF, Trolle E. 2012. Validation of a pre-coded food record for infants and young children. Eur. J. Clin. Nutr. 66:91–96. 10.1038/ejcn.2011.133 [DOI] [PubMed] [Google Scholar]
- 46.Houtkooper LB, Lohman TG, Going SB, Howell WH. 1996. Why bioelectrical impedance analysis should be used for estimating adiposity. Am. J. Clin. Nutr. 64(3 Suppl):436S–448S [DOI] [PubMed] [Google Scholar]
- 47.Benjamini Y, Hochberg Y. 1995. Controlling the false discovery rate—a practical and powerful approach to multiple testing. J. R. Stat. Soc. Series B Stat. Methodol. 57:289–300 [Google Scholar]
- 48.Scholtens PA, Oozeer R, Martin R, Amor KB, Knol J. 2012. The early settlers: intestinal microbiology in early life. Annu. Rev. Food Sci. Technol. 3:425–447. 10.1146/annurev-food-022811-101120 [DOI] [PubMed] [Google Scholar]
- 49.Fouhy F, Ross RP, Fitzgerald GF, Stanton C, Cotter PD. 2012. Composition of the early intestinal microbiota: knowledge, knowledge gaps and the use of high-throughput sequencing to address these gaps. Gut Microbes 3:203–220. 10.4161/gmic.20169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Biasucci G, Rubini M, Riboni S, Morelli L, Bessi E, Retetangos C. 2010. Mode of delivery affects the bacterial community in the newborn gut. Early Hum. Dev. 86(Suppl 1):13–15. 10.1016/j.earlhumdev.2010.01.004 [DOI] [PubMed] [Google Scholar]
- 51.Fanaro S, Chierici R, Guerrini P, Vigi V. 2003. Intestinal microflora in early infancy: composition and development. Acta Paediatr. Suppl. 91:48–55. 10.1111/j.1651-2227.2003.tb00646.x [DOI] [PubMed] [Google Scholar]
- 52.Mitsou EK, Kirtzalidou E, Oikonomou I, Liosis G, Kyriacou A. 2008. Fecal microflora of Greek healthy neonates. Anaerobe 14:94–101. 10.1016/j.anaerobe.2007.11.002 [DOI] [PubMed] [Google Scholar]
- 53.Avershina E, Storro O, Oien T, Johnsen R, Pope P, Rudi K. 2014. Major faecal microbiota shifts in composition and diversity with age in a geographically restricted cohort of mothers and their children. FEMS Microbiol. Ecol. 87:280–290. 10.1111/1574-6941.12223 [DOI] [PubMed] [Google Scholar]
- 54.Edwards CA, Parrett AM. 2002. Intestinal flora during the first months of life: new perspectives. Br. J. Nutr. 88(Suppl 1):S11–S18. 10.1079/BJN2002625 [DOI] [PubMed] [Google Scholar]
- 55.Engfer MB, Stahl B, Finke B, Sawatzki G, Daniel H. 2000. Human milk oligosaccharides are resistant to enzymatic hydrolysis in the upper gastrointestinal tract. Am. J. Clin. Nutr. 71:1589–1596 [DOI] [PubMed] [Google Scholar]
- 56.Gnoth MJ, Kunz C, Kinne-Saffran E, Rudloff S. 2000. Human milk oligosaccharides are minimally digested in vitro. J. Nutr. 130:3014–3020 [DOI] [PubMed] [Google Scholar]
- 57.Matsuki T, Watanabe K, Tanaka R. 2003. Genus- and species-specific PCR primers for the detection and identification of bifidobacteria. Curr. Issues Intest. Microbiol. 4:61–69 [PubMed] [Google Scholar]
- 58.Sela DA, Mills DA. 2010. Nursing our microbiota: molecular linkages between bifidobacteria and milk oligosaccharides. Trends Microbiol. 18:298–307. 10.1016/j.tim.2010.03.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bezirtzoglou E, Tsiotsias A, Welling GW. 2011. Microbiota profile in feces of breast- and formula-fed newborns by using fluorescence in situ hybridization (FISH). Anaerobe 17:478–482. 10.1016/j.anaerobe.2011.03.009 [DOI] [PubMed] [Google Scholar]
- 60.Penders J, Thijs C, Vink C, Stelma FF, Snijders B, Kummeling I, van den Brandt PA, Stobberingh EE. 2006. Factors influencing the composition of the intestinal microbiota in early infancy. Pediatrics 118:511–521. 10.1542/peds.2005-2824 [DOI] [PubMed] [Google Scholar]
- 61.Stark PL, Lee A. 1982. The microbial ecology of the large bowel of breast-fed and formula-fed infants during the first year of life. J. Med. Microbiol. 15:189–203. 10.1099/00222615-15-2-189 [DOI] [PubMed] [Google Scholar]
- 62.Hopkins MJ, Macfarlane GT, Furrie E, Fite A, Macfarlane S. 2005. Characterisation of intestinal bacteria in infant stools using real-time PCR and northern hybridisation analyses. FEMS Microbiol. Ecol. 54:77–85. 10.1016/j.femsec.2005.03.001 [DOI] [PubMed] [Google Scholar]
- 63.Andersson AF, Lindberg M, Jakobsson H, Backhed F, Nyren P, Engstrand L. 2008. Comparative analysis of human gut microbiota by barcoded pyrosequencing. PLoS One 3:e2836. 10.1371/journal.pone.0002836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ley RE, Turnbaugh PJ, Klein S, Gordon JI. 2006. Microbial ecology: human gut microbes associated with obesity. Nature 444:1022–1023. 10.1038/4441022a [DOI] [PubMed] [Google Scholar]
- 65.Tap J, Mondot S, Levenez F, Pelletier E, Caron C, Furet JP, Ugarte E, Munoz-Tamayo R, Paslier DL, Nalin R, Dore J, Leclerc M. 2009. Towards the human intestinal microbiota phylogenetic core. Environ. Microbiol. 11:2574–2584. 10.1111/j.1462-2920.2009.01982.x [DOI] [PubMed] [Google Scholar]
- 66.Yatsunenko T, Rey FE, Manary MJ, Trehan I, Dominguez-Bello MG, Contreras M, Magris M, Hidalgo G, Baldassano RN, Anokhin AP, Heath AC, Warner B, Reeder J, Kuczynski J, Caporaso JG, Lozupone CA, Lauber C, Clemente JC, Knights D, Knight R, Gordon JI. 2012. Human gut microbiome viewed across age and geography. Nature 486:222–227. 10.1038/nature11053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Favier CF, Vaughan EE, de Vos WM, Akkermans AD. 2002. Molecular monitoring of succession of bacterial communities in human neonates. Appl. Environ. Microbiol. 68:219–226. 10.1128/AEM.68.1.219-226.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Harder T, Bergmann R, Kallischnigg G, Plagemann A. 2005. Duration of breastfeeding and risk of overweight: a meta-analysis. Am. J. Epidemiol. 162:397–403. 10.1093/aje/kwi222 [DOI] [PubMed] [Google Scholar]
- 69.Lefebvre CM, John RM. 12 July 2013. The effect of breastfeeding on childhood overweight and obesity: a systematic review of the literature. J. Am. Assoc. Nurse Pract. 10.1002/2327-6924.12036 [DOI] [PubMed] [Google Scholar]
- 70.Kalliomaki M, Collado MC, Salminen S, Isolauri E. 2008. Early differences in fecal microbiota composition in children may predict overweight. Am. J. Clin. Nutr. 87:534–538 [DOI] [PubMed] [Google Scholar]
- 71.Karlsson CL, Onnerfalt J, Xu J, Molin G, Ahrne S, Thorngren-Jerneck K. 2012. The microbiota of the gut in preschool children with normal and excessive body weight. Obesity (Silver Spring) 20:2257–2261. 10.1038/oby.2012.110 [DOI] [PubMed] [Google Scholar]
- 72.Cole TJ, Bellizzi MC, Flegal KM, Dietz WH. 2000. Establishing a standard definition for child overweight and obesity worldwide: international survey. BMJ 320:1240–1243. 10.1136/bmj.320.7244.1240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chomtho S, Wells JC, Williams JE, Davies PS, Lucas A, Fewtrell MS. 2008. Infant growth and later body composition: evidence from the 4-component model. Am. J. Clin. Nutr. 87:1776–1784 [DOI] [PubMed] [Google Scholar]
- 74.Fewtrell MS. 2007. Session 6: infant nutrition: future research developments in Europe EARNEST, the early nutrition programming project: EARly Nutrition programming—long-term Efficacy and Safety Trials and integrated epidemiological, genetic, animal, consumer and economic research. Proc. Nutr. Soc. 66:435–441. 10.1017/S0029665107005708 [DOI] [PubMed] [Google Scholar]
- 75.Ong KK, Ahmed ML, Emmett PM, Preece MA, Dunger DB. 2000. Association between postnatal catch-up growth and obesity in childhood: prospective cohort study. BMJ 320:967–971. 10.1136/bmj.320.7240.967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Young BE, Johnson SL, Krebs NF. 2012. Biological determinants linking infant weight gain and child obesity: current knowledge and future directions. Adv. Nutr. 3:675–686. 10.3945/an.112.002238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Louis P, Scott KP, Duncan SH, Flint HJ. 2007. Understanding the effects of diet on bacterial metabolism in the large intestine. J. Appl. Microbiol. 102:1197–1208. 10.1111/j.1365-2672.2007.03322.x [DOI] [PubMed] [Google Scholar]
- 78.Pryde SE, Duncan SH, Hold GL, Stewart CS, Flint HJ. 2002. The microbiology of butyrate formation in the human colon. FEMS Microbiol. Lett. 217:133–139. 10.1111/j.1574-6968.2002.tb11467.x [DOI] [PubMed] [Google Scholar]
- 79.Duncan SH, Belenguer A, Holtrop G, Johnstone AM, Flint HJ, Lobley GE. 2007. Reduced dietary intake of carbohydrates by obese subjects results in decreased concentrations of butyrate and butyrate-producing bacteria in feces. Appl. Environ. Microbiol. 73:1073–1078. 10.1128/AEM.02340-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wu GD, Chen J, Hoffmann C, Bittinger K, Chen YY, Keilbaugh SA, Bewtra M, Knights D, Walters WA, Knight R, Sinha R, Gilroy E, Gupta K, Baldassano R, Nessel L, Li H, Bushman FD, Lewis JD. 2011. Linking long-term dietary patterns with gut microbial enterotypes. Science 334:105–108. 10.1126/science.1208344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Moeller AH, Degnan PH, Pusey AE, Wilson ML, Hahn BH, Ochman H. 2012. Chimpanzees and humans harbour compositionally similar gut enterotypes. Nat. Commun. 3:1179. 10.1038/ncomms2159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Rajilic-Stojanovic M, Heilig HG, Tims S, Zoetendal EG, de Vos WM. 15 October 2012. Long-term monitoring of the human intestinal microbiota composition. Environ. Microbiol. 10.1111/1462-2920.12023 [DOI] [PubMed] [Google Scholar]
- 83.Adlerberth I. 2008. Factors influencing the establishment of the intestinal microbiota in infancy. Nestle Nutr. Workshop Ser. Pediatr. Program 62:13–29. 10.1159/000146245 [DOI] [PubMed] [Google Scholar]
- 84.Reinhardt C, Reigstad CS, Backhed F. 2009. Intestinal microbiota during infancy and its implications for obesity. J. Pediatr. Gastroenterol. Nutr. 48:249–256. 10.1097/MPG.0b013e318183187c [DOI] [PubMed] [Google Scholar]
- 85.Koeth RA, Wang Z, Levison BS, Buffa JA, Org E, Sheehy BT, Britt EB, Fu X, Wu Y, Li L, Smith JD, DiDonato JA, Chen J, Li H, Wu GD, Lewis JD, Warrier M, Brown JM, Krauss RM, Tang WH, Bushman FD, Lusis AJ, Hazen SL. 2013. Intestinal microbiota metabolism of L-carnitine, a nutrient in red meat, promotes atherosclerosis. Nat. Med. 19:576–585. 10.1038/nm.3145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Le Chatelier E, Nielsen T, Qin J, Prifti E, Hildebrand F, Falony G, Almeida M, Arumugam M, Batto JM, Kennedy S, Leonard P, Li J, Burgdorf K, Grarup N, Jorgensen T, Brandslund I, Nielsen HB, Juncker AS, Bertalan M, Levenez F, Pons N, Rasmussen S, Sunagawa S, Tap J, Tims S, Zoetendal EG, Brunak S, Clement K, Dore J, Kleerebezem M, Kristiansen K, Renault P, Sicheritz-Ponten T, de Vos WM, Zucker JD, Raes J, Hansen T, Bork P, Wang J, Ehrlich SD, Pedersen O. 2013. Richness of human gut microbiome correlates with metabolic markers. Nature 500:541–546. 10.1038/nature12506 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.