Abstract
The generation of NADPH by malic enzyme (ME) was postulated to be a rate-limiting step during fatty acid synthesis in oleaginous fungi, based primarily on the results from research focusing on ME in Mucor circinelloides. This hypothesis is challenged by a recent study showing that leucine metabolism, rather than ME, is critical for fatty acid synthesis in M. circinelloides. To clarify this, the gene encoding ME isoform E from Mortierella alpina was homologously expressed. ME overexpression increased the fatty acid content by 30% compared to that for a control. Our results suggest that ME may not be the sole rate-limiting enzyme, but does play a role, during fatty acid synthesis in oleaginous fungi.
INTRODUCTION
Oleaginous fungi, such as the commercial production species Mortierella alpina and Mucor circinelloides, can accumulate fatty acids to more than 20% of their cell dry weight (1). However, the mechanism of fatty acid biosynthesis in these organisms is still not fully understood. The carbon flux pathway and provision of NADPH are two major events during fatty acid synthesis. NADPH is particularly important as the sole source of reducing power during fatty acid synthesis in oleaginous fungi (2–5).
The NADPH-generating enzyme malic enzyme (ME) (NADP+ dependent; EC 1.1.1.40), catalyzing the decarboxylation of malate to pyruvate (malate + NADP+ = pyruvate + CO2 + NADPH), was speculated to play a pivotal role during fatty acid synthesis in oleaginous fungi (3, 5). When ME activity was inhibited, the cell fatty acid content of M. circinelloides decreased from 24% to 2% (6). Moreover, in the M. circinelloides strain R7B, overexpression of the ME genes from M. alpina and M. circinelloides produced a 2.5-fold increase in fatty acid content (7). These results led to the conclusion that a rate-limiting step in fatty acid biosynthesis is the generation of NADPH by ME (7). However, a recent study revealed that leucine auxotrophy caused a 2.5-fold decrease in cell fatty acid content and that leuA gene expression restored its level in M. circinelloides strain R7B. ME overexpression, however, did not alter the fatty acid content despite a significant increase in ME activity (8). It was thus proposed that the leucine metabolic pathway, by participating in acetyl coenzyme A (acetyl-CoA) generation, may be critical during fatty acid synthesis in M. circinelloides (8, 9).
M. circinelloides R7B, generated by random mutagenesis (10), may contain other, unknown mutations in addition to that in the leuA gene, which can complicate the interpretation of the above-mentioned results. Therefore, a recipient strain with a known genetic mutation(s) and stable fatty acid synthesis characteristics will be advantageous for future research. To this end, a uracil auxotroph of M. alpina ATCC 32222 was generated via homologous recombination. The M. alpina ATCC 32222 cytosolic ME (isoform E)-encoding gene (11), named malE1, was cloned and homologously overexpressed in order to evaluate the role of ME in fatty acid synthesis.
MATERIALS AND METHODS
Strains and growth conditions.
Wild-type M. alpina (ATCC 32222; DDBJ/EMBL/GenBank accession no. ADAG00000000 [first version, ADAG01000000]) was cultured on potato dextrose agar (PDA) medium. M. alpina uracil-auxotrophic strains were maintained on GY medium, consisting of 30 g/liter glucose, 5 g/liter yeast extract, 2 g/liter KNO3, 1 g/liter NaH2PO4 and 0.3 g/liter MgSO4 · 7H2O, supplemented with 5-fluoroorotic acid (5-FOA) (0.5 mg/ml) and uracil (0.05 mg/ml). Escherichia coli strain TOP 10 was used for plasmid construction and cultivated at 37°C on LB agar plates containing 100 μg/ml kanamycin. Agrobacterium tumefaciens C58C1, which was kindly provided by Yasuyuki Kubo (Kyoto Prefectural University, Japan), was used as a transfer DNA (T-DNA) donor for fungal transformation. YEP medium, consisting of 10 g/liter tryptone, 10 g/liter yeast extract, and 5 g/liter NaCl, was used for Agrobacterium tumefaciens C58C1 cultivation at 28°C. GY medium was used for extracting M. alpina genomic DNA; when this medium was used for positive selection of uracil auxotrophs, 5-FOA (1 mg/ml) and uracil (0.05 mg/ml) were added. The compositions of synthetic complete (SC) medium, minimal medium (MM), and induction medium (IM) were described before (12, 13). For the fatty acid analysis, M. alpina was grown at 28°C in Kendrick medium (14), consisting of 50 g/liter glucose, 2.0 g/liter diammonium tartrate, 7.0 g/liter KH2PO4, 2.0 g/liter Na2HPO4, 1.5 g/liter MgSO4 · 7H2O, 1.5 g/liter yeast extract, 0.1 g/liter CaCl2 · 2H2O, 8 mg/liter FeCl3 · 6H2O, 1 mg/liter ZnSO4 · 7H2O, 0.1 mg/liter CuSO4 · 5H2O, 0.1 mg/liter Co(NO3)2 · 6H2O, and 0.1 mg/liter MnSO4 · 5H2O, pH 6.0. The incubation protocols were as previously described (15). The proliferative-phase cultures were inoculated at 10% (vol/vol) into a 2-liter fermentor containing 1.5 liters of Kendrick medium. Fermentors were held at 28°C, with stirring at 500 rpm with an aeration rate of 0.5 vol/vol/min (vvm), and the initial pH was set at 6.0.
Construction of the T-DNA binary vector.
Total RNA was extracted with TRIzol reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer's instructions. Reverse transcription (RT) was performed using the PrimeScript RT reagent kit (TaKaRa, Otsu, Shiga, Japan) following the manufacturer's instructions. The primers used in this study are summarized in Table S1 in the supplemental material. The endonucleases used in this study were purchased from New England BioLabs (Beverly, MA, USA). The malE1 gene was amplified from cDNA with primer pair malE1F/malE1R and subcloned into pGEM-T Easy vector (Promega, Madison, WI, USA), followed by sequence analysis on an ABI Prism 3730. The T-DNA binary vector pBIG2RHPH2 (16) was used as the backbone. A multiple-cloning site (MCS) was amplified with the primer pair MCS-F/MCS-R from pBluescript II SK(+). The MCS fragments were doubly digested with NheI and MunI and then introduced into pBIG2RHPH2 doubly digested with XbaI and EcoRI. The resulting plasmid was named pBIG4. In order to generate M. alpina ATCC 32222 uracil-auxotrophic strains, the orotate phosphoribosyl transferase (OPRTase) (EC.2.4.2.10)-encoding gene ura5, which is essential for uracil synthesis, was disrupted by double homologous recombination. As illustrated in Fig. S1 in the supplemental material, the left and right arms of the ura5 gene were amplified from the wild-type M. alpina genomic DNA with primers P1/P2 (for the ura5 left arm, URA5-L) and P3/P4 (for the ura5 right arm, URA5-R), respectively. The 2.8-kb Δura5 fragments were constructed by fusion PCR with equimolar amounts of URA5-L/URA5-R PCR fragments as templates and P1/P4 as primers. The resulting fragment was purified and digested with EcoRI and KpnI, followed by ligation into pBIG4 treated with the same restriction enzymes. The resulting plasmid was named pBIG4-Δura5. As illustrated in Fig. S2 in the supplemental material, primer pair HisproD1F/TrpCtR (12) was used to amplify the hygromycin B phosphotransferase gene (HPH) fragment from pD4 (17). The resulting fragments were doubly digested with XbaI and EcoRI and introduced into pET28a(+) to construct pET28a-HPHs. As described before (18), the hpt gene was replaced by the ura5 gene, which was amplified with primer pair URA5F/URA5R. The resulting plasmid was named pET28a-ura5s. The ura5s cassette was gel purified from XbaI- and EcoRI-digested pET28a-ura5s. The ura5s cassette was introduced into pBIG2RHPH2 to replace the HPH cassette by XbaI and EcoRI digestion and ligation. The resulting plasmid was designated pBIG2-ura5s. The malE1 gene, which was PCR amplified from cDNA, was digested with BspHI and BamHI and introduced into pET28a-HPHs to replace the hpt gene. The resulting plasmid, named pET28a-malE1, was then digested with SpeI and XbaI. The malE1 expression cassette was introduced into the XbaI site of pBIG2-ura5s to form pBIG2-ura5s-malE1.
ATMT.
Agrobacterium tumefaciens-mediated transformation (ATMT) was performed with a modification of a previously described protocol (12, 19). M. alpina spores were harvested from 2-week cultures growing on GY agar medium, which was supplemented with 0.05 g/liter uracil for uracil-auxotrophic strains. A. tumefaciens C58C1 was electrotransformed with the corresponding binary vector. A. tumefaciens transformants were isolated on YEP agar plates supplemented with 100 μg/ml kanamycin and 100 μg/ml rifampin. Positive transformants were confirmed by PCR. A single colony of A. tumefaciens C58C1 harboring the appropriate binary vector was inoculated into 20 ml of liquid MM containing 100 μg/ml kanamycin and 100 μg/ml rifampin and cultured at 28°C with shaking at 200 rpm for 48 h. Bacterial cells were harvested by centrifugation at 4,000 × g for 5 min at room temperature and the supernatant was removed. The cells were washed by gently resuspending pellets in 5 ml of fresh liquid IM, centrifuging, and removing the supernatant. The washed cells were diluted with IM to an optical density at 600 nm (OD600) of 0.3 to 0.5 and then preincubated for 8 to 12 h at 28°C with shaking at 200 rpm to an OD600 of 0.8 to 1.2. If cells were at a higher density, they were diluted to an appropriate OD600 with IM. One hundred microliters of the induced cell suspension was mixed with an equal volume of M. alpina spore suspension (108/ml) (19) and then spread onto nitrocellulose or cellophane membranes, which were placed on a solid cocultivation medium (IM containing 0.9 g/liter instead of 1.8 g/liter glucose). The plates were incubated at 23°C for 24 to 72 h in a temperature-controlled dark incubator. The membranes were transferred to uracil-free SC plates supplemented with 50 μg/ml cefotaxime and 50 μg/ml spectinomycin to inhibit the growth of bacteria. When nitrocellulose membranes were used, 0.03% Nile blue A was added to help detect fungal colonies against the natural color of the membranes. The plates were incubated at 25 to 30°C until colonies appeared. The transformed candidates were transferred to uracil-free SC agar plates, and this was repeated three times to obtain stable transformants. For wild-type M. alpina ura5 gene disruption, the cocultivated membranes were transferred to GY plates supplemented with 0.05 mg/ml uracil, 50 μg/ml cefotaxime, and 50 μg/ml spectinomycin. After an extended incubation time of at least 2 weeks, spores were gently scraped off the surface with a sterile loop. The spore suspensions were filtrated through a 40-μm cell strainer and spread on GY plates supplemented with 50 μg/ml cefotaxime, 50 μg/ml spectinomycin, and 1 mg/ml 5-FOA, a chemical used for positive selection of auxotrophic mutants. The plates were incubated at 25 to 30°C in a temperature-controlled dark incubator. When visible colonies appeared, the mycelium was transferred to fresh GY plates supplemented with 1 mg/ml 5-FOA, followed by a series of subcultures (≥3) to obtain stable strains. These stable 5-FOA-resistant strains were maintained as candidate uracil-auxotrophic M. alpina strains for further identification. All experiments were carried out in triplicate.
Genomic DNA preparation.
M. alpina strains were cultivated in GY liquid medium at 28°C with shaking at 200 rpm for 4 days. M. alpina genomic DNA was extracted as described previously (15).
Confirmation of the existence of T-DNA in the genome.
The existence of integrated T-DNA in genomic DNA was identified by PCR. The PCR cycling conditions were 95°C for 10 min, followed by 30 cycles of amplification at 95°C for 30 s, 55°C for 30 s, and 72°C for 3 min and a final extension step of 72°C for 10 min. Two pairs of promoter- and terminator-specific primers (HisproF1/TrpCR1 and HisproF2/TrpCR2; see Table S1 in the supplemental material) were designed to confirm whether transformation was successful or not.
RT-qPCR analysis.
The primer pairs used for reverse transcription-quantitative PCR (RT-qPCR) (malE1RTF/malE1RTR and 18SRTF/18SRTR) are shown in Table S1 in the supplemental material. Total RNA was isolated from M. alpina strains and reverse transcribed with the PrimeScript RT reagent kit (TaKaRa, Otsu, Shiga, Japan) according to the manufacturer's instructions. RT-qPCR was performed on an ABI Prism 7900 sequence detection system (Applied Biosystems, Foster City, CA) with Power SYBR green PCR master mix (Applied Biosystems, Foster City, CA) according to the manufacturer's instructions. Reaction mixtures were composed of 10 μl of SYBR green PCR master mix, 0.5 μl of each primer pair, 8 μl of distilled water, and 1 μl of DNA template or distilled water as a no-template control. The PCR cycling conditions were 50°C for 2 min, 95°C for 10 min, and 40 cycles of amplification at 95°C for 15 s and 60°C for 30 s. The expression of the internal control gene (18S rRNA) was used as the normalization standard for gene expression.
Protein analysis.
The protein analysis was performed essentially as described before (20). For Western blot analysis, two SDS-polyacrylamide gels with 10 μg cellular protein extract per lane were used, one for Western blot analysis and the other for Coomassie blue staining to verify the loading quantity. Polyvinylidene difluoride (PVDF) membranes (Millipore, Billerica, MA, USA) were employed instead of nitrocellulose membranes. A specific rabbit antibody raised against M. alpina malic enzyme 1 (ME1), encoded by the malE1 gene, was prepared by Sangon (Shanghai China) and used to probe membranes, followed by horseradish peroxidase (HRP)-conjugated goat anti-rabbit IgG (Santa Cruz, California, USA).
ME activity determination.
Mycelia collected from filtrates were frozen and ground in liquid nitrogen and then suspended in extraction buffer (7). The suspension was centrifuged at 10,000 × g for 10 min at 4°C. The supernatant was used for the determination of protein concentration using the Bradford method. ME activity was detected using continuous spectrophotometric assays at 340 nm at 30°C as described before (6). Reaction mixtures contained 80 mM KH2PO4/KOH (pH 7.5), 3 mM MgCl2, 0.6 mM NADP+, 25 mM l-malate, and approximately 0.1 mg/ml protein, and 1 unit of enzyme activity was defined as the amount of enzyme required to produce 1 nmol NADPH per min.
NADP and NADPH quantification.
About 100 mg (fresh weight) of mycelia was collected from different samples and stored in liquid nitrogen until further analysis. NADP and NADPH levels were determined using the NADP/NADPH quantification colorimetric kit (Biovision, CA, USA) according to the manufacturer's instructions.
Fatty acid methyl ester (FAME) analysis.
For fatty acid analysis, approximately 50 mg of mycelia (dry weight) was used for each lipid extraction. Fatty acids were extracted and methyl esterified as described previously (15). Fatty acid profiles were analyzed as their methyl esters by gas chromatography (GC-2010; Shimadzu Co., Kyoto, Japan) with a DB-Waxetr column (30 m by 0.32 mm; film thickness, 0.25 μm). The temperature program was as follows: 120°C for 3 min, ramp to 190°C at 5°C per min, ramp to 220°C at 4°C per min, and hold for 20 min. Nitrogen was the carrier gas at a constant flow of 3 ml per min.
Cell dry weight determination.
Biomass was harvested by filtration and then washed twice with distilled water and frozen in liquid nitrogen. The cell dry weight was determined gravimetrically after lyophilization.
Determination of glucose and ammonium concentrations.
The glucose concentration in the culture medium was determined using a glucose oxidase test kit (Rongsheng Biotech, Shanghai, China) according to the manufacturer's instructions, with a lower detection limit of 0.4 mg/liter. The ammonium concentration was calculated by the indophenol method (21) with ammonium sulfate as an ammonia standard; the lower detection limit of ammonium is 0.2 mg/liter. Five milliliters each of diluted solution 1 (10 g/liter phenol and 0.05 g/liter sodium nitroprusside) and diluted solution 2 (5 g sodium hydroxide and 0.42 g sodium hypochlorite) was mixed with 1 ml sample. Spectrophotometric determination of the solution was carried out at 625 nm after incubation at 55°C for 3 min.
RESULTS
Isolation and identification of M. alpina uracil-auxotrophic strains.
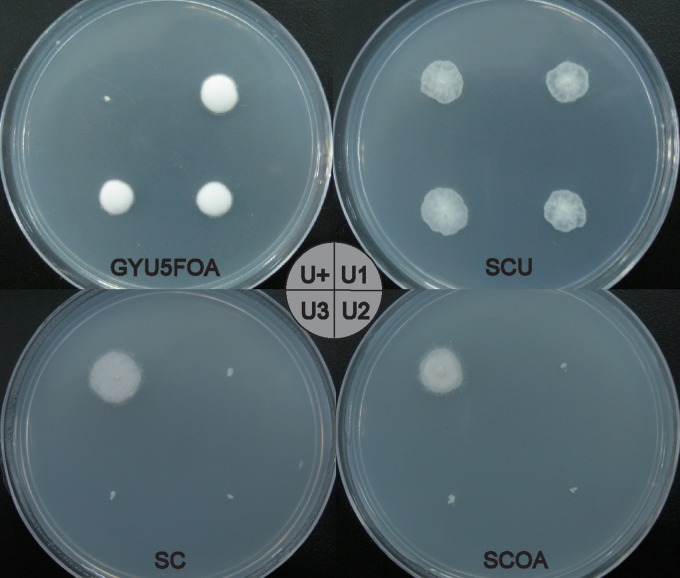
The in situ spore generation strategy was applied in the cocultivation step of ATMT, because the existence of prototrophic nuclei in the polynuclear and nonseptum mycelium of M. alpina is lethal in the presence of 5-FOA, in spite of the existence of a uracil-auxotrophic nucleus. After transferring three generations by spores in the presence of 5-FOA, 9 strains exhibited stable 5-FOA resistance. Sequence analysis with the ABI Prism 3730 showed the expected absence of an 18-bp sequence (positions 213 to 230) encoding a putative conserved domain of the active site (22) in the ura5 gene in all stable 5-FOA-resistant strains. The colony morphology of these uracil-auxotrophic strains was indistinguishable from that of the wild-type strain. These strains could grow on SC medium only when 0.05 mg/ml uracil was added; they could not grow on SC medium supplemented with 0.5 mg/ml orotic acid instead of uracil (Fig. 1). This suggested that the function of the ura5 gene had been completely abolished (23). No apparent differences in malE1 expression level, ME1 protein level, specific activity, or fatty acid content were observed between the M. alpina prototrophic strain and three of the uracil-auxotrophic strains (named MAU1, MAU2, and MAU3) (Fig. 2). MAU1 (CCFM 501) was used as the recipient strain for the following study.
FIG 1.
Growth of M. alpina ATCC 32222 and its uracil auxotrophs on different media. The mycelium of each strain (U+, M. alpina wild type; U1, MAU1; U2, MAU2; U3, MAU3) was transferred to each plate and incubated at 28°C for 3 days. GYU5FOA, GY medium supplemented with 0.05 g/liter uracil and 1 mg/ml 5-FOA; SC, SC medium; SCU, SC medium supplemented with 0.05 g/liter uracil; SCOA, SC medium supplemented with 0.5 mg/ml orotic acid. The circle in the middle indicates the position of each strain on the plate.
FIG 2.
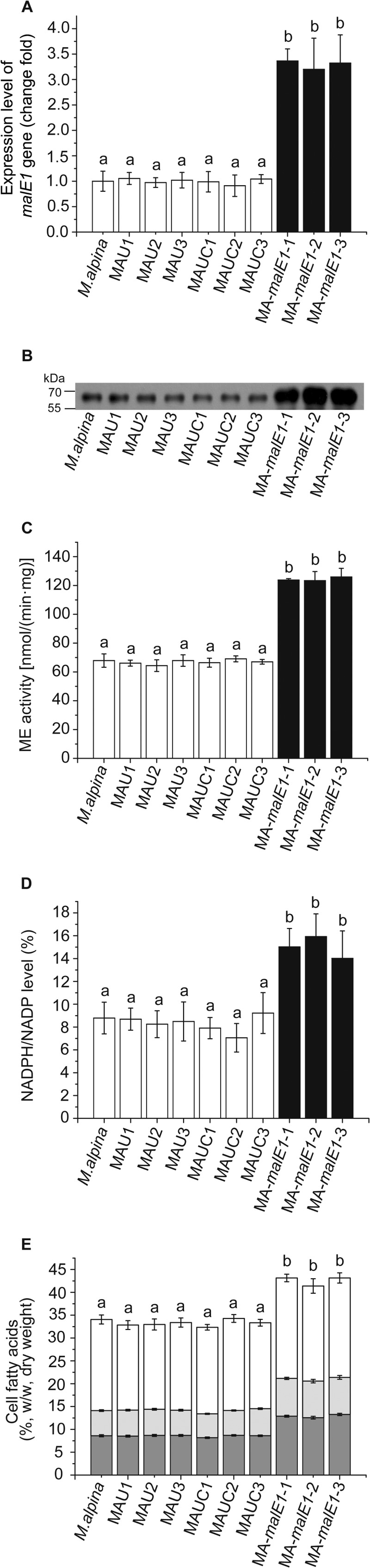
malE1 expression level (A), Western blot analysis of ME1 (B), specific activity (C), NADPH level (D), and cell fatty acid composition (E) in the prototrophic strain ( M. alpina), auxotrophic strains (MAU1, MAU2, and MAU3), complementary-phenotype strains (MAUC1, MAUC2, and MAUC3) and malE1-overexpressing strains (MA-malE1-1, MA-malE1-2, and MA-malE1-3). Samples were taken from cultures of each strain growing in Kendrick medium at an initial pH of 6.0 in a stirred 2-liter fermentor with direct aeration for 144 h at 28°C. In panels A, C, and D, the open bars represent the nonoverexpressing strains and the black bars represent the malE1-overexpressing strains. In panel E, the dark gray bars represent AA, the light gray bars represent other ω6 PUFAs (linoleic acid, γ-linolenic acid, and dihomo-γ-linolenic acid), and the open bars represent other fatty acids. Three independent experiments were performed, and error bars represent standard deviations. Data with different letters are significantly different (P < 0.05).
Homologous expression of ME.
The malE1 gene was homologously overexpressed in the auxotrophic MAU1 strain derived from M. alpina ATCC 32222.
(i) Establishment of an M. alpina ATCC 32222 transformation system.
By using the uracil-auxotrophic strain (MAU1) and binary vector pBIG2-ura5s, an M. alpina ATCC 32222 ATMT system was constructed. Twenty-three putative transformants were randomly picked, and their stability was checked by transferring them on GY plates containing 5-FOA and on SC uracil-free plates (18). After being subcultured for three generations, 15 transformants which could not grow on GY plates supplied with 5-FOA but could grow on SC uracil-free plates were recognized as mitotically stable transformants. On average, 65% of the total colonies yielded stable transformants in triplicate experiments. About 31 positive colonies were obtained from one membrane, and 20 of them were stable transformants. About 108 spores cocultivated on 10 membranes were prepared at once. The transformation frequency was around 200 transformants/108 spores, which is lower than the frequency which was described before (12). This may be due to the different host strain and nonoptimized cocultivation conditions (24).
The presence of integrated T-DNA in the genomic DNA isolated from transformed strains was confirmed by PCR using two pairs of promoter- and terminator-specific primers (HisproF1/TrpCR1 and HisproF2/TrpCR2). As shown in Fig. S3 in the supplemental material, all the pBIG2ura5s-transformed complementary-phenotype strains (MAUC1, MAUC2, and MAUC3) showed an 818-bp or 861-bp fragment, in contrast to the negative controls (M. alpina and MAU1). There was no evidence of multiple-copy insertion in the transformants (see Fig. S3 in the supplemental material); this may due to a high rate (94%) of single-copy insertion in ATMT (12). RT-qPCR, Western blotting, and specific activity analyses of ME revealed the same levels of transcription, translation, ME activity, and NADPH in these stable transformants (Fig. 2A, B, C, and D), suggesting that the endogenous expression of ME was not affected. The fatty acid contents and compositions in three randomly selected mitotically stable transformants (MAUC1, MAUC2, and MAUC3) showed no significant difference from those of the wild-type strain (Fig. 2E). Our construction of the transformation system for M. alpina may be applicable to other fungi with defined genome sequences.
(ii) Characterization of the malE1 transformed strains.
The malE1-carrying DNA fragment (1,752 bp) was directly amplified from cDNA with primer pair malE1F/malE1R. Analysis of the malE1 coding sequence showed 84.54% identity with the mce2 coding sequence (see Fig. S4 in the supplemental material) (7). Binary vector pBIG2-ura5s-malE1 (see Fig. S2 in the supplemental material) was constructed for homologous expression of the malE1 gene in MAU1. The presence of T-DNA fragments containing a ura5 gene expression cassette and a malE1 gene overexpression cassette in genomic DNA was identified by PCR. Besides the 818-bp and 861-bp fragments, the presence of 1,916-bp and 1,959-bp products identified the presence of the malE1 overexpression cassette in the genome (see Fig. S3 in the supplemental material).
The expression of the malE1 gene in three randomly selected malE1-overexpressing strains (MA-malE1-1, MA-malE1-2, and MA-malE1-3) was analyzed after culturing them for 144 h. The results revealed an approximately 3-fold-higher expression level of the malE1 gene in all malE1-overexpressing strains than in the other strains analyzed (Fig. 2A). Western blot analysis illustrated consistent translation levels of the malE1 gene in M. alpina prototrophic, uracil-auxotrophic, and complementary-phenotype strains. All three malE1-overexpressing strains exhibited higher ME protein levels (Fig. 2B; see S5A in the supplemental material). The malic enzyme activity exhibited the same pattern: lower ME activity in all the malE1-nonoverexpressing strains, around 50 nmol/(min · mg), and higher ME activity in all the overexpressing strains, around 100 nmol/(min · mg) (Fig. 2C). Cell NADPH levels in malE1-overexpressing strains were significantly higher than those in other strains (Fig. 2D).
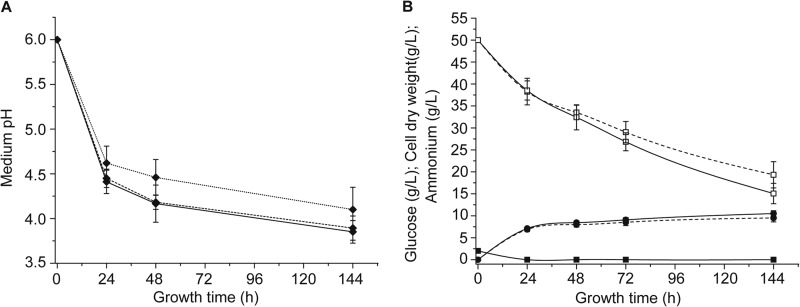
The control strain selected for time course experiments was MAU1, because it is the recipient strain for the transformation and has the same fatty acid content and composition as the M. alpina prototrophic strain and complementary-phenotype strains (Fig. 2E). Samples of both strains taken at different cultivation times (24, 48, 72, and 144 h) were analyzed. The results revealed that the expression of the malE1 gene in MA-malE1-1 was 1.5- to 3-fold higher than that in MAU1 (Fig. 3A). Expression of the malE1 gene in the control strain displayed a decreasing slope over time, in contrast with a more stable trend in the MA-malE1-1 strain. We note that the expression level of the malE1 gene in strain MA-malE1-1 seemed not to be maintained at a stable high level but decreased slightly, which was different from results described for M. circinelloides (8). This may be because the homologous promoter (His550 promoter) used in this work was regulated at the transcriptional level, which was confirmed by our transcriptome analysis (data not shown); alternately, this may indicate regulation of endogenous ME1 at the RNA level by small RNAs (25). Western blot analysis revealed a higher ME1 protein level in MA-malE1-1 than in MAU1 (Fig. 3B; see Fig. S5B in the supplemental material). The specific activity of ME showed a steady decrease after a peak at 48 h in both strains (Fig. 3C), which was the same trend as described before when M. alpina was cultured under oxygen-limited conditions (26). In the present study, without pH control, cultures under oxygen-limited (in sterilized bottles) and aerobic (in fermentors) conditions exhibited similar medium pH curves (Fig. 4A), along with the same ME activity trends. This indicated that the changes in ME activity may also be related to medium pH. NADPH levels in both strains exhibited the same variations with ME activity (Fig. 3D; see Fig. S6A in the supplemental material). Cell growth of MA-malE1-1 and MAU1 was also compared. The growth patterns of two strains were similar: mycelia grew rapidly during the first 24 h, and there was a higher glucose consumption rate; after the nitrogen was exhausted, cells stopped growing to accumulate fatty acids, and there was a relative lower glucose consumption rate (Fig. 4B).
FIG 3.
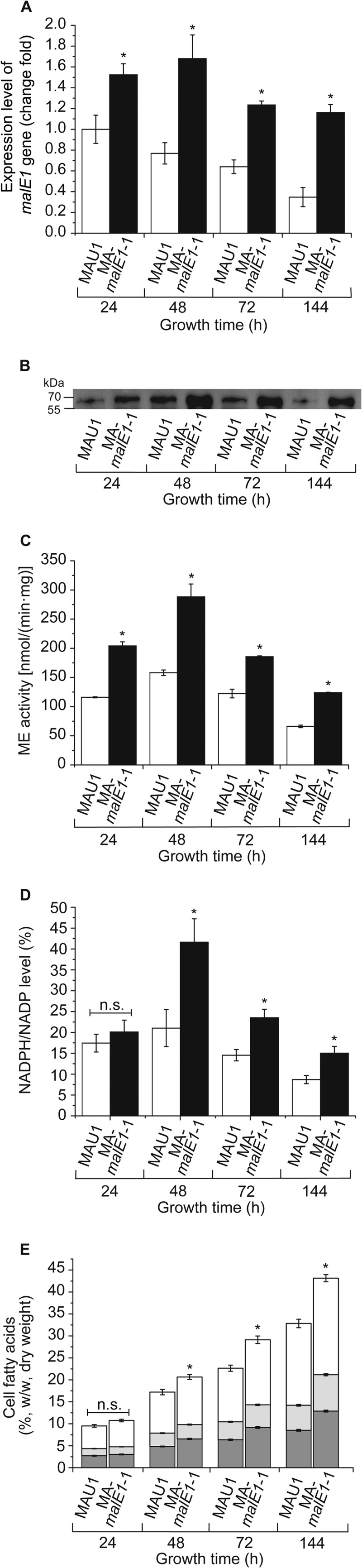
malE1 expression level (A), Western blot analysis of ME1 (B), specific activity (C), NADPH level (D), and cell fatty acid composition (E) in the control strain MAU1 and overexpressing strain MA-malE1-1. Samples were taken at the indicated times from cultures of both strains growing in Kendrick medium at an initial pH of 6.0 in a stirred 2-liter fermentor with direct aeration for up to 144 h at 28°C. In panels A, C, and D, the open bars represent the control strain MAU1 and the black bars represent the malE1 overexpression strain MA-malE1-1. In panel E, the dark gray bars represent AA, the light gray bars represent other ω6 PUFAs (linoleic acid, γ-linolenic acid, and dihomo-γ-linolenic acid), and the open bars represent other fatty acids. Three independent experiments were performed, and error bars represent standard deviations. n.s., not significant. *, P < 0.05 compared with the control.
FIG 4.
Growth of M. alpina strains in Kendrick medium. (A) Medium pH of MAU1 (dashed line) and overexpressing strain MA-malE1-1 (solid line) in Kendrick medium at an initial pH of 6.0 in a 2-liter fermentor at 28°C with direct aeration. The dotted line represents the medium pH measured over time in M. alpina culture medium from 500-ml stirred bottles containing 100 ml Kendrick medium. (B) Growth of control strain MAU1 (dashed lines) and overexpressing strain MA-malE1-1 (solid lines) in Kendrick medium at an initial pH of 6.0 in a 2-liter fermentor at 28°C with direct aeration. Cell dry weight (closed circles), ammonium tartrate concentration (closed squares), glucose concentration (open squares) are shown. Three independent experiments were performed and error bars represent standard deviations.
Fatty acid synthesis in M. alpina strains.
Overexpression of the malE1 gene led to an elevated fatty acid content of approximately 43% of cell dry weight at 144 h, compared to 33% in nonoverexpressing strains. The fatty acid yield of MA-malE1-1 was 0.129 ± 0.011 g/g glucose, which was higher than the 0.102 ± 0.006 g/g glucose for MAU1 (three independent experiments were performed). Increased ME activity also resulted in an increase in total ω6 polyunsaturated fatty acid (PUFA) content (from 42% to 50% of total fatty acids) and arachidonic acid (AA) content (from 25% to 30% of total fatty acids). The improved ω6 PUFA content suggested that ME was also generating NADPH for fatty acid desaturase (14). In time course experiments, the difference in fatty acid content between MA-malE1-1 and MAU1 became more and more significant during the fatty acid accumulation phase (after nitrogen exhaustion), from 10% to 30% (Fig. 3E), indicating that overexpression of ME caused a relatively higher fatty acid synthesis rate. When M. alpina was cultured at pH 4.2 in a fully controlled fermentor for 144 h, the fatty acid content reached only 33% of cell dry weight, compared to 38% at pH 6.0 (see Fig. S6B in the supplemental material), the same trends as described for M. circinelloides (27) and Trichoderma reesei (28).
DISCUSSION
Production of fatty acids in microorganisms is of great interest, especially for fatty acids of significant commercial value such as AA, eicosapentaenoic acid (EPA), and docosahexaenoic acid (DHA). NADPH is known to be an essential element during fatty acid synthesis in oleaginous fungi; however, whether ME plays a pivotal role is still controversial (3, 7, 8). In the present study, the M. alpina uracil-auxotrophic strains were different from M. circinelloides R7B used in the previous studies (7). The auxotrophic strain MAU1 with a defined genetic background, generated by homologous recombination, may be a more appropriate experimental recipient than auxotrophic strains obtained by random chemical mutagenesis.
In M. alpina, malE1 overexpression increased the fatty acid content by only 30% compared to that in the control, despite a 2-fold increase in ME activity. This may be because the increased ME activity did not lead to an appropriate elevated supplement of NADPH for fatty acid synthesis; alternatively, the NADPH generated by ME may be utilized by other metabolic pathways. It would appear that the role of M. alpina ME during fatty acid synthesis may not be as pivotal as postulated before (3, 7). The study of ME in filamentous fungi is far behind that of ME in plants, where ME is known to be involved in a variety of metabolic pathways, such as photosynthesis, protein and lipid biosynthesis, cellular pH regulation, and defense responses (29–31).
The carbon metabolic pathway may be another factor that affects the performance of ME in malE1-overexpressing strains. Recently, acetyl-CoA generated from the endogenous leucine metabolic pathway was postulated to be another rate-limiting step during fatty acid synthesis in M. circinelloides (8), indicating that the amino acid metabolism may intersect with fatty acid synthesis as an acetyl-CoA resource. Likewise, in M. alpina, phenylalanine and tyrosine were suggested to be metabolized by the phenylalanine-hydroxylating system to contribute NADPH and acetyl-CoA for lipid metabolism (32). The supplement of acetyl-CoA might play a more important role during fatty acid synthesis in oleaginous fungi than previously speculated (7). In the oleaginous yeast Yarrowia lipolytica, genetic engineering aimed at constructing a strain for industrial production of EPA led to estimated fatty acid contents of 30% to 56.5% (33), suggesting that the carbon flux of the fatty acid biosynthetic pathway not only may determine the composition but also may be important for the content of fatty acids in oleaginous fungi.
Recently, a study in Aspergillus oryzae revealed that nitrogen starvation promotes intracellular malic acid biosynthesis (34). With its large expansion of low-pH secretory hydrolytic enzymes (35), A. oryzae is becoming a promising species for the industrial production of malic acid. In M. alpina, under nitrogen starvation, the increase of ME activity could be recognized as a response to the accumulation of intracellular malic acid, which was inhibited by the decrease of the medium pH. However, a lower medium pH condition, even though it corresponded with a higher ME activity, is unsuitable for the accumulation of fatty acids in M. alpina. Interestingly, higher ME activity was associated with a lower fatty acid content, which was also observed in M. circinelloides (36); this may confirm our results about the role of ME during fatty acid synthesis in oleaginous fungi.
Combined with previous studies in M. circinelloides (8), our results suggest that ME activity in M. alpina may not be the sole rate-limiting step but that it does play a role in fatty acid synthesis and that the biosynthesis of fatty acids in oleaginous fungi is influenced by multiple steps during the process.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the National Natural Science Foundation of China (21276108 and 31271812), the Program for New Century Excellent Talents (NCET-13-0831), the National High Technology Research and Development Program of China (2011AA100905 and 2012AA022105C), the National Science Fund for Distinguished Young Scholars (31125021), the National Basic Research Program 973 of China (2012CB720802), the 111 project B07029, and the Fundamental Research Funds for the Central Universities (JUSRP51320B).
Footnotes
Published ahead of print 14 February 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.00140-14.
REFERENCES
- 1.Thorpe R, Ratledge C. 1972. Fatty acid distribution in triglycerides of yeasts grown on glucose or n-alkanes. J. Gen. Microbiol. 72:151–163. 10.1099/00221287-72-1-151 [DOI] [Google Scholar]
- 2.Ratledge C, Wynn JP. 2002. The biochemistry and molecular biology of lipid accumulation in oleaginous microorganisms. Adv. Appl. Microbiol. 51:1–51. 10.1016/S0065-2164(02)51000-5 [DOI] [PubMed] [Google Scholar]
- 3.Ratledge C. 2004. Fatty acid biosynthesis in microorganisms being used for single cell oil production. Biochimie 86:807–815. 10.1016/j.biochi.2004.09.017 [DOI] [PubMed] [Google Scholar]
- 4.Evans CT, Ratledge C. 1985. Possible regulatory roles of ATP: citrate lyase, malic enzyme, and AMP deaminase in lipid accumulation by Rhodosporidium toruloides CBS 14. Can. J. Microbiol. 31:1000–1005. 10.1139/m85-189 [DOI] [Google Scholar]
- 5.Wynn JP, bin Abdul Hamid A, Ratledge C. 1999. The role of malic enzyme in the regulation of lipid accumulation in filamentous fungi. Microbiology 145:1911–1917. 10.1099/13500872-145-8-1911 [DOI] [PubMed] [Google Scholar]
- 6.Wynn JP, Kendrick A, Ratledge C. 1997. Sesamol as an inhibitor of growth and lipid metabolism in Mucor circinelloides via its action on malic enzyme. Lipids 32:605–610. 10.1007/s11745-997-0077-1 [DOI] [PubMed] [Google Scholar]
- 7.Zhang Y, Adams IP, Ratledge C. 2007. Malic enzyme: the controlling activity for lipid production? Overexpression of malic enzyme in Mucor circinelloides leads to a 2.5-fold increase in lipid accumulation. Microbiology 153:2013–2025. 10.1099/mic.0.2006/002683-0 [DOI] [PubMed] [Google Scholar]
- 8.Rodríguez-Frómeta RA, Gutiérrez A, Torres-Martínez S, Garre V. 2013. Malic enzyme activity is not the only bottleneck for lipid accumulation in the oleaginous fungus Mucor circinelloides. Appl. Microbiol. Biotechnol. 97:3063–3072. 10.1007/s00253-012-4432-2 [DOI] [PubMed] [Google Scholar]
- 9.Kohlhaw GB. 2003. Leucine biosynthesis in fungi: entering metabolism through the back door. Microbiol. Mol. Biol. Rev. 67:1–15. 10.1128/MMBR.67.1.1-15.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Roncero MIG. 1984. Enrichment method for the isolation of auxotrophic mutants of Mucor using the polyene antibiotic N-glycosyl-polifungin. Carlsberg Res. Commun. 49:685–690. 10.1007/BF02907499 [DOI] [Google Scholar]
- 11.Zhang H, Zhang L, Chen H, Chen YQ, Ratledge C, Song Y, Chen W. 2013. Regulatory properties of malic enzyme in the oleaginous yeast, Yarrowia lipolytica, and its non-involvement in lipid accumulation. Biotechnol. Lett. 35:2091–2098. 10.1007/s10529-013-1302-7 [DOI] [PubMed] [Google Scholar]
- 12.Ando A, Sumida Y, Negoro H, Suroto DA, Ogawa J, Sakuradani E, Shimizu S. 2009. Establishment of Agrobacterium tumefaciens-mediated transformation of an oleaginous fungus, Mortierella alpina 1S-4, and its application for eicosapentaenoic acid producer breeding. Appl. Environ. Microbiol. 75:5529–5535. 10.1128/AEM.00648-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Takeno S, Sakuradani E, Murata S, Inohara-Ochiai M, Kawashima H, Ashikari T, Shimizu S. 2004. Cloning and sequencing of the ura3 and ura5 genes, and isolation and characterization of uracil auxotrophs of the fungus Mortierella alpina 1S-4. Biosci. Biotechnol. Biochem. 68:277–285. 10.1271/bbb.68.277 [DOI] [PubMed] [Google Scholar]
- 14.Kendrick A, Ratledge C. 1992. Desaturation of polyunsaturated fatty acids in Mucor circinelloides and the involvement of a novel membrane-bound malic enzyme. Eur. J. Biochem. 209:667–673. 10.1111/j.1432-1033.1992.tb17334.x [DOI] [PubMed] [Google Scholar]
- 15.Wang L, Chen W, Feng Y, Ren Y, Gu Z, Chen H, Wang H, Thomas MJ, Zhang B, Berquin IM, Li Y, Wu J, Zhang H, Song Y, Liu X, Norris JS, Wang S, Du P, Shen J, Wang N, Yang Y, Wang W, Feng L, Ratledge C, Chen YQ. 2011. Genome characterization of the oleaginous fungus Mortierella alpina. PLoS One 6:e28319. 10.1371/journal.pone.0028319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tsuji G, Fujii S, Fujihara N, Hirose C, Tsuge S, Shiraishi T, Kubo Y. 2003. Agrobacterium tumefaciens-mediated transformation for random insertional mutagenesis in Colletotrichum lagenarium. J. Gen. Plant Pathol. 69:230–239. 10.1007/s10327-003-0040-4 [DOI] [Google Scholar]
- 17.Mackenzie DA, Wongwathanarat P, Carter AT, Archer DB. 2000. Isolation and use of a homologous histone H4 promoter and a ribosomal DNA region in a transformation vector for the oil-producing fungus Mortierella alpina. Appl. Environ. Microbiol. 66:4655–4661. 10.1128/AEM.66.11.4655-4661.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Takeno S, Sakuradani E, Murata S, Inohara-Ochiai M, Kawashima H, Ashikari T, Shimizu S. 2004. Establishment of an overall transformation system for an oil-producing filamentous fungus, Mortierella alpina 1S-4. Appl. Microbiol. Biot. 65:419–425. 10.1007/s00253-004-1622-6 [DOI] [PubMed] [Google Scholar]
- 19.Michielse CB, Hooykaas PJ, van den Hondel CA, Ram AF. 2008. Agrobacterium-mediated transformation of the filamentous fungus Aspergillus awamori. Nat. Protoc. 3:1671–1678. 10.1038/nprot.2008.154 [DOI] [PubMed] [Google Scholar]
- 20.Chen H, Gu Z, Zhang H, Wang M, Chen W, Lowther WT, Chen YQ. 2013. Expression and purification of integral membrane fatty acid desaturases. PLoS One 8:e58139. 10.1371/journal.pone.0058139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chaney AL, Marbach EP. 1962. Modified reagents for determination of urea and ammonia. Clin. Chem. 8:130–132 [PubMed] [Google Scholar]
- 22.Marchler-Bauer A, Lu S, Anderson JB, Chitsaz F, Derbyshire MK, DeWeese-Scott C, Fong JH, Geer LY, Geer RC, Gonzales NR. 2011. CDD: a Conserved Domain Database for the functional annotation of proteins. Nucleic Acids Res. 39:D225–D229. 10.1093/nar/gkq1189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yamagishi K, Kimura T, Oita S, Sugiura T, Hirai H. 2007. Transformation by complementation of a uracil auxotroph of the hyper lignin-degrading basidiomycete Phanerochaete sordida YK-624. Appl. Microbiol. Biotechnol. 76:1079–1091. 10.1007/s00253-007-1093-7 [DOI] [PubMed] [Google Scholar]
- 24.Michielse CB, Hooykaas PJ, van den Hondel CA, Ram AF. 2005. Agrobacterium-mediated transformation as a tool for functional genomics in fungi. Curr. Genet. 48:1–17. 10.1007/s00294-005-0578-0 [DOI] [PubMed] [Google Scholar]
- 25.Lee H-C, Li L, Gu W, Xue Z, Crosthwaite SK, Pertsemlidis A, Lewis ZA, Freitag M, Selker EU, Mello CC. 2010. Diverse pathways generate microRNA-like RNAs and Dicer-independent small interfering RNAs in fungi. Mol. Cell 38:803–814. 10.1016/j.molcel.2010.04.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang Y, Ratledge C. 2008. Multiple isoforms of malic enzyme in the oleaginous fungus, Mortierella alpina. Mycol. Res. 112:725–730. 10.1016/j.mycres.2008.01.003 [DOI] [PubMed] [Google Scholar]
- 27.Xia C, Zhang J, Zhang W, Hu B. 2011. A new cultivation method for microbial oil production: cell pelletization and lipid accumulation by Mucor circinelloides. Biotechnol. Biofuels 4:15. 10.1186/1754-6834-4-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brown D, Hasan M, Lepe-Casillas M, Thornton A. 1990. Effect of temperature and pH on lipid accumulation by Trichoderma reesei. Appl. Microbiol. Biot. 34:335–339 [Google Scholar]
- 29.Drincovich MaF, Casati P, Andreo CS. 2001. NADP-malic enzyme from plants: a ubiquitous enzyme involved in different metabolic pathways. FEBS Lett. 490:1–6. 10.1016/S0014-5793(00)02331-0 [DOI] [PubMed] [Google Scholar]
- 30.Lai LB, Tausta SL, Nelson TM. 2002. Differential regulation of transcripts encoding cytosolic NADP-malic enzyme in C3 and C4 Flaveria species. Plant Physiol. 128:140–149. 10.1104/pp.010449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Parker D, Beckmann M, Zubair H, Enot DP, Caracuel-Rios Z, Overy DP, Snowdon S, Talbot NJ, Draper J. 2009. Metabolomic analysis reveals a common pattern of metabolic re-programming during invasion of three host plant species by Magnaporthe grisea. Plant J. 59:723–737. 10.1111/j.1365-313X.2009.03912.x [DOI] [PubMed] [Google Scholar]
- 32.Wang H, Chen H, Hao G, Yang B, Feng Y, Wang Y, Feng L, Zhao J, Song Y, Zhang H. 2013. Role of the phenylalanine-hydroxylating system in aromatic substance degradation and lipid metabolism in the oleaginous fungus Mortierella alpina. Appl. Environ. Microbiol. 79:3225–3233. 10.1128/AEM.00238-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xue Z, Sharpe PL, Hong S-P, Yadav NS, Xie D, Short DR, Damude HG, Rupert RA, Seip JE, Wang J. 2013. Production of omega-3 eicosapentaenoic acid by metabolic engineering of Yarrowia lipolytica. Nat. Biotechnol. 31:734–740. 10.1038/nbt.2622 [DOI] [PubMed] [Google Scholar]
- 34.Knuf C, Nookaew I, Brown SH, McCulloch M, Berry A, Nielsen J. 2013. Investigation of malic acid production in Aspergillus oryzae under nitrogen starvation conditions. Appl. Environ. Microbiol. 79:6050–6058. 10.1128/AEM.01445-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Machida M, Asai K, Sano M, Tanaka T, Kumagai T, Terai G, Kusumoto K-I, Arima T, Akita O, Kashiwagi Y. 2005. Genome sequencing and analysis of Aspergillus oryzae. Nature 438:1157–1161. 10.1038/nature04300 [DOI] [PubMed] [Google Scholar]
- 36.Song Y, Wynn JP, Li Y, Grantham D, Ratledge C. 2001. A pre-genetic study of the isoforms of malic enzyme associated with lipid accumulation in Mucor circinelloides. Microbiology 147:1507–1515 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.