Abstract
Bacillus amyloliquefaciens strain SQR9, isolated from the cucumber rhizosphere, suppresses the growth of Fusarium oxysporum in the cucumber rhizosphere and protects the host plant from pathogen invasion through efficient root colonization. In the Gram-positive bacterium Bacillus, the response regulator DegU regulates genetic competence, swarming motility, biofilm formation, complex colony architecture, and protease production. In this study, we report that stepwise phosphorylation of DegU in B. amyloliquefaciens SQR9 can influence biocontrol activity by coordinating multicellular behavior and regulating the synthesis of antibiotics. Results from in vitro and in situ experiments and quantitative PCR (qPCR) studies demonstrate the following: (i) that the lowest level of phosphorylated DegU (DegU∼P) (the degQ mutation) impairs complex colony architecture, biofilm formation, colonization activities, and biocontrol efficiency of Fusarium wilt disease but increases the production of macrolactin and bacillaene, and (ii) that increasing the level of DegU∼P by degQ and degSU overexpression significantly improves complex colony architecture, biofilm formation, colonization activities, production of the antibiotics bacillomycin D and difficidin, and efficiency of biocontrol of Fusarium wilt disease. The results offer a new strategy to enhance the biocontrol efficacy of Bacillus amyloliquefaciens SQR9.
INTRODUCTION
Knowledge of rhizosphere ecology and its implications for plant physiology have dramatically changed traditional crop management practices, especially with regard to plant nutrition and plant defense mechanisms (1). Today, it is widely accepted that certain strains of rhizosphere bacteria, referred to as biocontrol agents (BCAs), suppress soilborne pathogens and stimulate plant growth. The main mechanisms by which BCAs suppress pathogens are antibiosis, competition for niches and nutrients, plant growth promotion, leading to more robust plants, and induction of systemic resistance in plants. Most BCAs are applied through soil. Thus, rhizosphere competence is considered a prerequisite for an effective BCA (2).
In natural environments, microorganisms usually exist in multicellular aggregates associated with solid surfaces that are generally described as biofilms (3). In Bacillus, some genes, including yvcA and the yqxM-sipW-tasA operon, are essential for this process. YvcA is a putative membrane-bound lipoprotein required for complex colony development in strain NCIB 3610 but not for pellicle formation in strain ATCC 6051. TasA is a protein component of the extracellular matrix, and its correct localization depends on the activities of both the SipW and YqxM proteins. Transcription of the yqxM operon is inhibited by SinR through direct promoter binding, an interaction that may in turn be disrupted by its antagonist, SinI. The pleiotropic regulator AbrB represses transcription of the yqxM operon via indirect and direct mechanisms (4).
Bacillus species. strains are used for the production of a wide range of antibiotics, such as polymyxins (B. polymyxa), which destroy membrane integrity, as well as edeines (B. brevis), which inhibit the formation of the initiation complex on the 30S ribosomal subunit (5). Furthermore, more lipopeptides and polyketides were identified in B. amyloliquefaciens (6). Little is known about the regulatory mechanisms that control the expression of antibiotics produced by Bacillus species strains.
Two-component signal transduction systems are the major family of signaling proteins by which bacteria sense and respond to changes in the environment. These include a sensor kinase that is autophosphorylated in response to a signal, followed by the subsequent transfer of the phosphate group to a two-domain response regulator (7). The phosphorylated response regulator often functions as a transcription factor. DegS/DegU is one of 10 well-characterized systems that regulate several postexponential processes, such as competence for DNA uptake, bacterial motility, and degradative enzyme synthesis. Phosphorylated DegU (DegU∼P) activates the expression of more than 170 genes, including aprE (encoding alkaline protease), nprE (encoding neutral protease), bpr (encoding bacillopeptidase), and sacB (encoding levansucrase) (8). Moreover, DegU∼P was found to probably be involved in poly-γ-glutamic acid (γ-PGA) production (7). A model is proposed to describe DegU's function as a rheostat regulating swarming motility, complex colony architecture, and extracellular protease production as a function of the DegU∼P concentration (9). The function of the DegSU two-component system in biofilm formation is poorly understood. This is partly due to the fact that analysis of DegSU has been carried out using a laboratory strain, ATCC 6051, in which expression of DegU-regulated genes is more complex than that in undomesticated strains.
The transfer of the phosphate moiety from DegS to DegU is enhanced in the presence of DegQ, a small protein consisting of 46 amino acids. Some laboratory isolates of Bacillus subtilis contain a point mutation within the promoter of degQ that is associated with a reduced level of DegQ synthesis; this in turn reduces the level of DegU∼P in the cell (10). The actions of DegU and DegQ are strongly interconnected. DegU∼P binds to the direct-repeat sequence of sacB and enhances its expression. Although B. subtilis 168 carries the degQ gene in its chromosome, this degQ gene is expressed only at a low level. A previous study reported that a single-base substitution within the −10 region of the degQ promoter (degQ36 mutation) allows overexpression of the intact degQ gene, and this leads to increased expression of sacB (9). It has also been reported that increased expression of the pleiotropic regulator DegQ in B. subtilis 168 enhances antibiotic production. Interestingly, most of the wild-type Bacillus isolates that produce peptide antibiotics show significantly elevated degQ expression compared to that of the domesticated lab strain B. subtilis 168 (11).
B. amyloliquefaciens SQR9, which was isolated from the cucumber (Cucumis sativus Linn.) rhizosphere, suppressed the growth of Fusarium oxysporum, the causative agent of vascular wilt of cucumber (12). SQR9 produces three families of lipopeptides: bacillomycin D, fengycins, and surfactins. Furthermore, a previous study indicated that bacillomycin D acts both as a signal for its biofilm formation and as the weapon of B. amyloliquefaciens SQR9 for suppressing F. oxysporum and other fungal and bacterial pathogens (13). The disruption of the abrB gene in SQR9 can improve biofilm formation and root colonization (14). In the present study, we constructed three derivatives of B. amyloliquefaciens SQR9 with different levels of DegU phosphorylation. Our results strongly suggest that altering the phosphorylation level of DegU in B. amyloliquefaciens SQR9 influences the biocontrol activity of soilborne Fusarium wilt disease by coordinating multicellular behavior and regulating the synthesis of bacillomycin D.
MATERIALS AND METHODS
Strains and culture conditions.
The strains and plasmids used in this study are described in Table 1. E. coli TOP10 was used as the host for all plasmids. B. amyloliquefaciens strain SQR9 (CGMCC accession no. 5808; China General Microbiology Culture Collection Center) was used throughout this study. Fusarium oxysporum CIPP1012 (ACCC accession no. 30220; Agricultural Culture Collection of China) was used as the pathogen and was obtained from the Institute of Vegetables and Flowers, Chinese Academy of Agricultural Sciences (Beijing, China). The pathogenic Ralstonia solanacearum strain QLRs-1115 was previously isolated from a wilted tomato plant (15).
TABLE 1.
Microorganisms and plasmids used in this study
| Strain or plasmid | Description | Reference or source |
|---|---|---|
| Fungus | ||
| F. oxysporum f. sp. cucumerinum J. H. Owen CIPP1012 | Laboratory stock (13) | |
| Bacterial strain | ||
| E. coli TOP10 | F− mcrA Δ(mrr-hsdRMS-mcrBC) ψ80 lacZΔM15 ΔlacX74 nupG recA1 araD139Δ(ara-leu)7697 galE15 galK16 rpsL (Strr) endA1λ− | Invitrogen (Shanghai, China) |
| R. solanacearum QLRs-1115 | Laboratory stock | |
| B. amyloliquefaciens SQR9 | Wild type | 13 |
| B. amyloliquefaciens SQR9-gfp | GFP-labeled B. amyloliquefaciens SQR9, Kanr | 12 |
| SQR9M4 | B. amyloliquefaciens SQR9 ΔdegQ::cm | This study |
| SQR9M4-gfp | GFP-labeled B. amyloliquefaciens SQR9ΔdegQ::cm, Kanr | This study |
| SQR9M4Q | B. amyloliquefaciens SQR9M4 with pUBCQ (Cmr Kanr) | This study |
| SQR9Q | B. amyloliquefaciens SQR9 with pUBCQ (Kanr) | This study |
| SQR9Q-gfp | B. amyloliquefaciens SQR9 with pUBCQ and pHAPII (Kanr) | This study |
| SQR9SUQ | B. amyloliquefaciens SQR9 with pUBCSUQ (Kanr) | This study |
| SQR9SUQ-gfp | B. amyloliquefaciens SQR9 with pUBCSUQ and pHAPII (Kanr) | This study |
| Plasmids | ||
| pMD 19-T | Ampr; MCS | TaKaRa (Dalian, China) |
| pNW33n | Cmr; MCS | 37 |
| pUBC19 | Ampr Kmr; B. subtilis-E. coli shuttle vector | 12 |
| pUBCQ | pUBC19 containing degQ | This study |
| pUBCSUQ | pUBC19 containing P43-degQ and degSU | This study |
| pHAPII | pUBC19 containing gfp fused to HAPII promoter | 12 |
| p43NMK | Ampr Kmr; B. subtilis expression vector, mpd expression cassette | 38 |
Growth conditions.
Escherichia coli strains were routinely grown in LB medium (10 g NaCl, 5 g yeast extract, and 10 g tryptone per liter). B. amyloliquefaciens strains were routinely grown in LB medium and, where appropriate, in MSgg medium (5 mM potassium phosphate, 100 mM morpholinepropanesulfonic acid [MOPS], pH 7, 2 mM MgCl2, 700 μM CaCl2, 50 μM MnCl2, 50 μM FeCl3, 1 μM ZnCl2, 2 mM; thiamine, 0.5% glycerol, 0.5% glutamate, 50 μg ml−1 tryptophan, 50 μg ml−1 phenylalanine, and 50 μg ml−1 threonine). For antibiotic production and liquid chromatography-mass spectrometry (LC-MS) characterization, B. amyloliquefaciens strains were grown in Landy medium (16). E. coli strains were grown at 37°C, and B. amyloliquefaciens strains were grown at 30°C. The antibiotics were used, as required, at the following concentrations: ampicillin, 100 μg ml−1; chloramphenicol, 5 μg ml−1; kanamycin, 30 μg ml−1.
Construction of the fused fragments for allelic exchange.
To construct the fragment used to delete the 0.1-kb region in the degQ gene of B. amyloliquefaciens SQR9, a 0.9-kb fragment upstream of the 0.1-kb region in degQ and a 1.1-kb fragment downstream of this region were amplified with primer sets p1/p2 and p3/p4 (see Table S1 in the supplemental material), respectively. The 1.- kb chloramphenicol resistance (Cm) gene was amplified from plasmid pNW33n with primer set P5/P6 (see Table S1), which overlapped with the upstream fragments and the downstream fragments of the 0.1-kb region in the degQ gene.
All PCR products were gel purified using an AxyPrep DNA gel purification and extraction kit (Axygen, Hangzhou, China). In the first step of the overlapping PCR, the mixture included 12 μl water, 5 μl PrimeSTAR buffer (5×), 2 μl dinucleoside triphosphate (dNTP) mix (2.5 mM [each]), 2 μl (20 ng) front fragment, 2 μl (20 ng) back fragment, 1.5 μl (15 ng) Cmr fragment, and 0.5 μl PrimeSTAR HS DNA polymerase. The PCR program was as follows: denaturing at 98°C for 2 min; 20 cycles of 98°C for 10 s, annealing at 55°C for 15 s, and extension at 72°C for 1.5 min. For the second step of overlapping PCR, the mixture included 32 μl water, 10 μl PrimeSTAR buffer, 4 μl dNTP mix, 1 μl forward primer of the front fragment, 1 μl reverse primer of the back fragment, 1 μl unpurified product from the first-step PCR, and 1 μl PrimeSTAR HS DNA polymerase. The PCR program was as follows: denaturing at 98°C for 2 min; 35 cycles of 98°C for 10 s, annealing at 54°C for 15 s, and extension at 72°C for 3.5 min. Through the overlapping PCR, the fused fragment was obtained and used to delete the 0.1-kb region in the degQ gene of B. amyloliquefaciens SQR9.
Disruption of the degQ gene in B. amyloliquefaciens SQR9.
The fused fragment was individually transformed into the competent cells of B. amyloliquefaciens SQR9, and the transformants were selected on LB agar plates containing 5 μg ml−1 chloramphenicol. The desired mutants with a deletion encompassing the 0.1-kb region in the degQ gene were verified by PCR using the primer sets p7/p8 (see Table S1 in the supplemental material) and confirmed by Southern blotting experiments (data not shown); the degQ disruption mutant of SQR9 was designated SQR9M4.
Complementation of disrupted degQ gene.
Plasmid pUBCQ was constructed to complement degQ disruption in SQR9M4. The degQ gene (including its promoter), bordered by PstI and HindIII sites, was amplified from SQR9 genomic DNA using primers p9/p10 (see Table S1 in the supplemental material). After purification and digestion with the corresponding restriction enzymes, the fragments were cloned into the B. amyloliquefaciens-E. coli shuttle vector pUBC19 (see Fig. S1). The plasmid was sequenced to test whether PCR-generated mutations were present. Plasmid pUBCQ was transformed into strain SQR9M4 to obtain the degU gene complemented strain.
Construction of high-DegU∼P-level B. amyloliquefaciens SQR9 strains.
Plasmid pUBCQ was also transformed into strain SQR9 to obtain the high-level-DegU∼P strain and designated B. amyloliquefaciens SQR9Q. Plasmid pUBCSUQ was used to construct a strain with the highest level of DegU∼P. The degQ gene was placed under the control of the P43 promoter, and we introduced the degSU genes into this plasmid. The P43 promoter was amplified from plasmid pP43NMK with primer set p11/p12 (see Table S1 in the supplemental material), and degQ and degSU (including three promoters of the degSU operon) were amplified from SQR9 genomic DNA using primers p13/p14 and p15/p16 (see Table S1), respectively. Subsequently, the three fragments were fused by PCR with primer set p11/p16 (see Table S1). After purification and digestion with the corresponding restriction nuclease, the fragments were cloned into the B. amyloliquefaciens-E. coli shuttle vector pUBC19 to get plasmid pUBCSUQ (see Fig. S1). Plasmid pUBCSUQ was sequenced to test whether PCR-generated mutations were present and, if negative, transformed into B. amyloliquefaciens SQR9 to obtain strain B. amyloliquefaciens SQR9SUQ.
Purification by reverse-phase (RP) HPLC.
For the purification of macrolactin, high-pressure liquid chromatography (HPLC) was performed using a HPLC 1200 device (1200 series; Agilent, Santa Clara, CA) to purify the active substance. A 20-μl sample, pretreated using an XAD-16 column, was injected into the HPLC column (Eclipse XDB-C18, 4.6 by 250 mm, 5 μm; Agilent, Santa Clara, CA). The temperature of the column was maintained at 20°C throughout the experiment. The purification was performed using a solvent containing 60% A (0.1% [vol/vol] CH3COOH) and 40% B (CH3CN) at a flow rate of 0.6 ml/min. To produce a stable baseline, the column was equilibrated using 60% A and 40% B solvent. An UV detector was used to detect peaks at 230 nm.
For the purification of bacillomycin D, fengycin, and surfactin, 10-μl sample was injected into a HPLC column, 9.4 mm by 150 mm (Agilent Technologies, Santa Clara, CA, USA). The temperature was kept at 35°C during the experiment. The flow rate was 0.84 ml/min using gradient elution with the detecting wavelength of 210 nm. Mobile phase A and mobile phase B were acetonitrile and 0.1% acetic acid in water, respectively. The elution conditions were 0 to 9 min of 60 to 93% (vol/vol) A, 40 to 7% (vol/vol) B, and 9 to 20 min of 93% A, 7% B.
For the purification of difficidin and bacillaene, 10-μl samples were injected onto a, HPLC column (Eclipse XDB-C18, 4.6 by 250 mm, 5 μm; Agilent, Santa Clara, CA). The temperature was kept at 30°C during the experiment. The run was performed with a flow rate of 0.75 ml/min and a gradient of solvents A (0.1% [vol/vol] HCOOH) and B (CH3CN), which reached 100% B after 20 min. A concentration of 100% CH3CN-HCOOH was held for a further 2 min. To equilibrate the column, it was treated with 5% CH3CN-HCOOH for 3 min. A UV detector was used to detect peaks at 230 nm.
A fraction collector (Analyt FC, G1364C; Agilent, Santa Clara, CA) was used to collect the pure compounds, and the fractions were collected using the time and peaks mode. The injections were performed repeatedly to collect a sufficient quantity of secondary metabolite. The fractions were lyophilized, and the residues were dissolved in 500 μl of methanol for mass spectrometry analysis.
MS analysis.
Samples were collected from an HPLC fraction collector, and the molecular mass and molecular formula of each secondary metabolite substance were determined using a liquid chromatography/electrospray ionization-mass spectrometry (LC/ESI-MS) system (1200 series and ESI-MS, 6410 Triple Quad LC/MS; Agilent, Santa Clara, CA) with a C18 column (50 by 2.1 mm, 1.8 μm) at a flow rate of 0.4 ml/min. The mobile phase was the same as the purification by HPLC section. For MS analysis, the electrospray needle was operated at a spray voltage of 4.5 KV. The capillary temperature was 300°C. The MS analysis was done by electrospray ionization in positive-ion mode, and the mass spectra were acquired in an m/z range of 50 to 1,200 at a scan rate of 500 atomic mass units (amu)/s.
Biofilm assay of B. amyloliquefaciens SQR9 and its derivative strains.
The biofilm assay was carried out using a modified version of the microtiter plate assay as described by Hamon et al. (17). Briefly, B. amyloliquefaciens cells were grown in 24-well polyvinylchloride (PVC) microtiter plates (Fisher Scientific, Shanghai, China) at 30°C in biofilm growth medium (MSgg medium) (18). The inoculum for the microtiter plates was obtained by growing the cells in MSgg medium under aeration to mid-exponential phase and subsequently diluting the cells to an optical density at 600 nm (OD600) value of 0.01 in fresh biofilm growth medium. Samples of 100 μl of the diluted cells were then placed in each well of a 24-well PVC microtiter plate. The microtiter plates were incubated while stationary. At various time points during the incubation of B. amyloliquefaciens strains in the microtiter plates, the presence of adhered cells was monitored by staining with crystal violet (CV) as follows. Growth medium and nonadherent cells were removed from the microtiter plate wells, which were subsequently rinsed with washing buffer (0.15 M ammonium sulfate, 100 mM potassium phosphate, pH 7, 34 mM sodium citrate, and 1 mM MgSO4). Biofilm cells were stained with 1% CV in washing buffer at room temperature for 20 min. Excess CV was subsequently removed, and the wells were rinsed with water. The CV that had stained the cells was then solubilized in 200 μl of 80% ethanol–20% acetone. Biofilm formation was quantified by measuring the OD570 value of each well using a Bio-Rad model 550 plate reader (17).
Transcription analysis of genes involved in biofilm formation and antibiotic synthesis.
Total RNA was isolated from B. amyloliquefaciens strains (SQR9, SQR9M4, SQR9Q, and SQR9SUQ) grown in MSgg and Landy medium. The cells were harvested by centrifugation at 4°C for 10 min at 5,000 × g. Total RNA samples were extracted using an RNAiso Plus kit (TaKaRa, Dalian, China) according to the manufacturer's protocol. RNA was reversely transcribed into cDNA in a 20-μl reverse transcription system (TaKaRa, Dalian, China) according to the manufacturer's instructions.
Transcription levels of yqxM, yvcA, bamD (in the bacillomycin D operon), sftA (in the surfactin operon), dfnA (in the difficidin operon), baeB (in the bacillaene operon), mlnA (in the macrolactin operon), and fenA (in the fengycin operon) of SQR9M4, SQR9Q, and SQR9SUQ relative to those for SQR9 were measured by reverse transcription-quantitative PCR (RT-qPCR) using a SYBR Premix Ex Taq (Perfect Real Time) kit (TaKaRa, Dalian, China). The sequences of primers used to amplify these genes are listed in Table S1 in the supplemental material. The recA gene was used as an internal control. Reactions were carried out on an ABI 7500 system (ABI, USA) under the followed condition: cDNA was denatured for 10 s at 95°C, followed by 40 cycles consisting of 5 s at 95°C and 34 s at 60°C. The method of 2−△△CT was used to analyze the real-time PCR data (19).
Microscopy.
In the hydroponics system, the root samples were monitored at 6 days after inoculation. The collected roots were cut into 1 to 2 cm in length, put on microscope slides, and visualized using a confocal laser scanning microscope (CLSM) (model TCS SP2; Leica, Heidelberg, Germany) with excitation wavelengths of 488 nm. Emitted light in the range of 500 to 600 nm was collected for green fluorescent protein (GFP) visualization. Images were obtained using Leica confocal software, version 2.61.
Root colonization by B. amyloliquefaciens SQR9 and its derivative strains in hydroponic culture.
The shuttle plasmid pHAPII was used to introduce plasmid-borne GFP genes into B. amyloliquefaciens SQR9 and its derivative strains by electroporation (20). To study root colonization, B. amyloliquefaciens strains were labeled with GFP to enable the monitoring. Colonies of B. amyloliquefaciens strains (SQR9-gfp, SQR9M4-gfp, SQR9Q-gfp, and SQR9SUQ-gfp) were inoculated into 50 ml of LB broth with appropriate antibiotics, and the cultures were incubated at 30°C until reaching stationary phase. The cells were washed twice in M8 buffer (22 mM Na2HPO4, 22 mM KH2PO4, and 100 mM NaCl, pH 7) and resuspended in 500 ml M8 buffer prior to use. Axenic prepared cucumber seedlings were soaked in 500 ml bacterial suspension for 30 min at 30°C. Then, the seedlings were aseptically transplanted to containers with 200 ml of sterile 1/2 MS culture medium (21). The plants were incubated in a growth chamber at 28°C with a 16-h light regimen. Ten repeats were performed for each strain.
To monitor B. amyloliquefaciens strains colonized on cucumber seedling roots, root samples were taken at 0, 2, 4, 6, 8, and 15 days after inoculation; 0.2 g of roots were homogenized in 1.8 ml of PBS buffer using a mortar and pestle until a fine homogenate was obtained. The homogenates were serially diluted and plated on LB plates with appropriate antibiotics. After culture at 30°C for 2 days, the bacterial colonies were examined using fluorescence microscopy (Olympus DP71), and those emitting green fluorescence were counted.
Pot experimental design.
The trial was conducted from 7 July to 15 September 2012 in the greenhouse of Nanjing Agricultural University. The soils used for the pot experiments were collected from a field with a history of cucumber cultivation. The field is located in Nanjing, Jiangsu Province, China, and the soil had the following properties: pH 5.4; organic matter, 23.2 g kg−1; available N, 159.2 mg kg−1; available Pm 138.9 mg kg−1; available K, 272. mg kg−1; total N, 1.9 g kg−1; total P, 1.9 g kg−1; and total K, 16.4 g kg−1.
Seeds of cucumber, Jinchun no. 4, were surface disinfected in 2% sodium hypochlorite for 3 min, rinsed three times in sterile distilled water, and subsequently allowed to germinate in 9-cm petri dishes covered with sterile wet filter paper at 30°C. After germinating, the seeds were planted in seedling trays. After 2 weeks, the seedlings with two true leaves were transplanted into larger pots (16 cm in diameter by 15.5 cm high) with 1 kg of soil. Five treatments in the pot experiment were designed as follows. CK (control), SQR9, SQR9M4, SQR9Q, and SQR9SUQ were used. All strains were individually mixed into the soil: FOC (Fusarium oxysporum f. sp. cucumerinum) at 105 spores g−1 soil and B. amyloliquefaciens strains (SQR9, SQR9M4, SQR9Q, and SQR9SUQ) at 108 CFU g−1; the treatment without B. amyloliquefaciens strains was the control. Each treatment was replicated 30 times, which included three blocks in a completely randomized design (10 plants for each block). The seedlings were incubated in a growth chamber at 30°C under a 16-h light regimen and irrigated with 1/2 Hoagland medium.
The disease index was recorded for each individual plant from 6 days to 9 days after FOC inoculation and expressed on a scale of 0 to 4 as follows: 0, the entire plant was healthy; 1, <25% of leaves were wilted; 2, 25% to 50% of leaves were wilted; 3, 50% to 75% of leaves were wilted; 4, 75% to 100% of leaves were wilted. The disease index for each treatment was calculated using the following formula (22): disease index = [∑(rating × number of plants rated)/(total number of plants × highest rating)] ×100.
Statistical analysis.
The data were statistically analyzed using analysis of variance, followed by Fisher's least-significant-difference test (P < 0.05) using SPSS software (SPSS Inc., Chicago, IL, USA).
RESULTS
Phosphorylation of DegU is essential for both complex colony architecture and biofilm formation.
To investigate the effect of the level of DegU∼P on the biocontrol efficiency, B. amyloliquefaciens SQR9 derivatives with different levels of DegU∼P were constructed (Fig. 1). It has been shown previously that degQ enhances the phospho transfer from phospho-DegS to DegU (10). The lowest-DegU∼P-level strain was constructed through disruption of the degQ gene. The intermediate-DegU∼P-level strain, SQR9Q, was constructed through the overexpression of the degQ gene, and the highest-DegU∼P-level strain, SQR9SUQ, was constructed by overexpressing all degQ, degS, and degU genes.
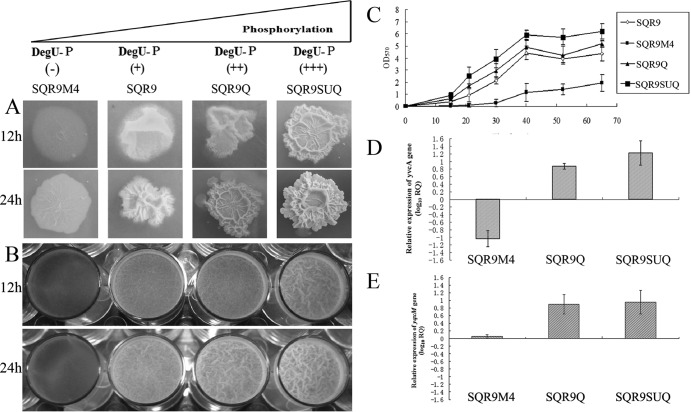
FIG 1.
Colony morphology and biofilm formation activity of B. amyloliquefaciens strains as a function of DegU phosphorylation. (A) Complex colony morphology of strains SQR9M4, the wild-type strain, SQR9Q, and SQR9SUQ. (B) Microtiter plate assay of biofilm formation by different B. amyloliquefaciens SQR9 strains. (C) OD570 of solubilized crystal violet from the microtiter plate assay over time for different B. amyloliquefaciens SQR9 strains. (D) Transcriptional levels of yvcA for different B. amyloliquefaciens SQR9 strains relative to that for wild-type B. amyloliquefaciens SQR9, evaluated by relative quantification PCR. (E) Transcriptional levels of yqxM for different B. amyloliquefaciens SQR9 strains relative to that for wild-type B. amyloliquefaciens SQR9, evaluated by qPCR. The experiments were carried out with pellicle obtained from microtiter plates when cells were grown in MSgg medium for 24 h. The B. amyloliquefaciens SQR9 recA gene was used as an internal reference gene. Bars represent standard deviations of data from three biological replicates.
DegU∼P is required to regulate complex colony architecture. The results in Fig. 1A show that in solid medium, the wild type was found to produce colonies with complex architectural features. SQR9M4 showed impaired complex colony architecture: upon closer examination of the degQ deletion mutant, it was apparent that the central aerial structures observed on colonies formed by the wild-type strain were not present and the colony remained flattened against the agar surface. Additionally, the degQ deletion mutant did not form the fruiting body-like structures that could be observed for the wild-type strain (Fig. 1A). The wild-type phenotype was restored when the disrupted degQ gene was complemented through expression of degQ in a plasmid see (Fig. S2 in the supplemental material). On the other hand, strains SQR9Q and SQR9SUQ demonstrated the extended formation of aerial colony architecture on agar plates compared with results for the wild-type strain, SQR9 (Fig. 1A).
The four strains showed indistinctive growth curves (see Fig. S3), but in liquid medium, the degQ mutant formed thin and fragile biofilm compared with wild-type strain SQR9 (Fig. 1B). In contrast, the strain SQR9SUQ in high level of DegU∼P formed robust rugose biofilms. Quantitative analysis of the biofilm biomass indicated that the degQ mutant exhibited significantly lower levels of biofilm biomass. At all time points tested, the degQ mutant exhibited a level of CV staining that did not exceed the OD570 value of 2. This is significantly below the level of CV staining observed with wild-type cells, which typically ranged from an OD570 of 3.5 to 4.5 at the 60-h time point. Moreover, it is worth mentioning that the level of CV staining of SQR9SUQ was 1.5-fold higher than that observed for the wild-type strain (Fig. 1C).
Transcription of genes involved in biofilm formation was evaluated. As shown in Fig. 1D and E, the highest transcription levels of yqxM and yvcA were detected for SQR9SUQ, followed by SQR9Q. The transcripts of yvcA for SQR9M4 were on average 1 order of magnitude below those for SQR9.
High phosphorylation of DegU increased rhizosphere colonization.
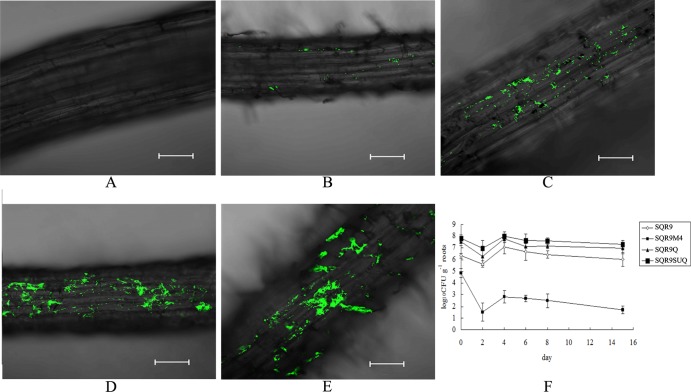
To further test whether B. amyloliquefaciens SQR9 and its derivative strains differ in their abilities to form biofilm on a natural matrix surface, their degree of colonization on roots of cucumber seedlings was monitored. After 6 days post-cocultivation of cucumber roots and green fluorescent protein (GFP)-labeled B. amyloliquefaciens strains (SQR9-gfp, SQR9M4-gfp, SQR9Q-gfp, and SQR9SUQ-gfp) in a hydroponic system, roots were viewed by confocal scanning laser microscopy (CSLM). A segment of the primary root, located around 2 to 4 cm distant from the root tip, was found to be heavily colonized by SQRSUQ, which formed microcolonies on the surface of the outer epidermis cells of the primary root (Fig. 2). However, in the same position, only few and small regions of the roots were colonized by the degQ mutant cells, and significantly reduced biofilm formation was observed compared with that for other strains (Fig. 2). Cucumber seedlings grown in sterile soil with B. amyloliquefaciens strains showed similar root colonization (data not shown), suggesting that increasing levels of DegU∼P can improve rhizosphere colonization of B. amyloliquefaciens SQR9.
FIG 2.
Colonization capabilities of B. amyloliquefaciens strains depend on DegU phosphorylation. CLSM micrographs of cucumber roots colonized by GFP-tagged B. amyloliquefaciens SQR9 strains. (A) Control; (B) SQR9M4-gfp; (C) SQR9-gfp; (D) SQR9Q-gfp; (E) SQR9SUQ-gfp; (F) populations of different B. amyloliquefaciens strains colonizing cucumber seedling roots. The data are expressed as log10 CFU per gram of fresh cucumber root.
In the hydroponic system, B. amyloliquefaciens (SQR9-gfp, SQR9M4-gfp, SQR9Q-gfp, and SQR9SUQ-gfp) on cucumber roots was enumerated 2, 4, 6, 8, and 15 days after inoculation by isolation, serial dilution, plating, and fluorescence microscopy, as previously described. At all time points investigated, the population of the degQ mutant was significantly decreased compared to those of the other strains (Fig. 2F).
Characterization of nonribosomal lipopeptides and polyketides produced by B. amyloliquefaciens SQR9 through HPLC/ESI-MS.
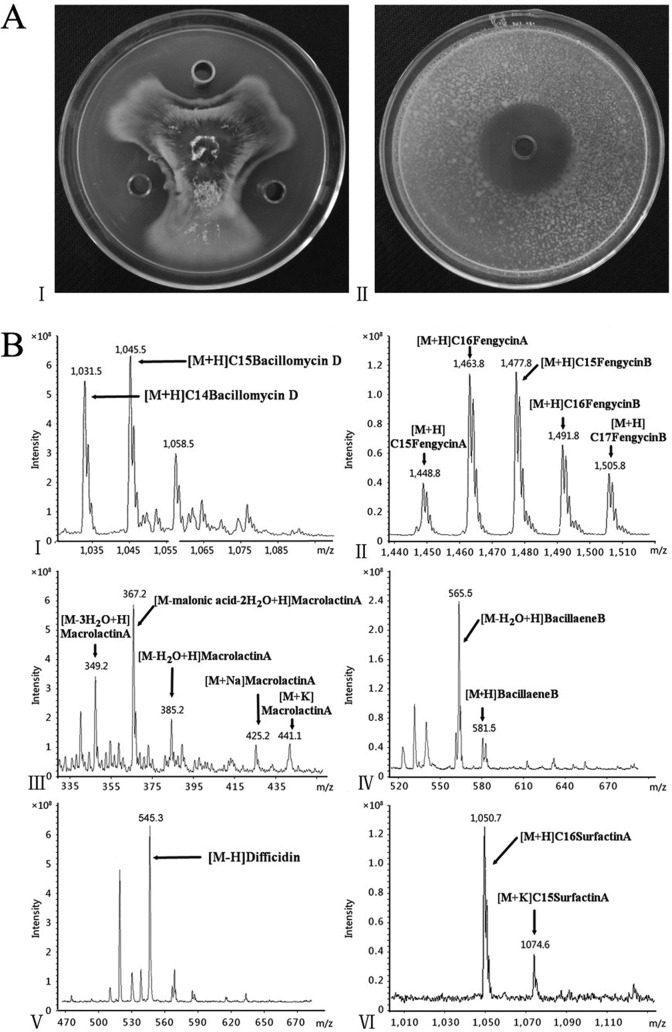
B. amyloliquefaciens SQR9 exhibited efficient biocontrol activity against the cucumber wilting pathogen Fusarium oxysporum and a broad range of other fungal and bacterial pathogens (Fig. 3A). Genomic analysis (unpublished) of B. amyloliquefaciens SQR9 indicated that more than 8% of the genome is devoted to the synthesis of various lipopeptides and polyketides (see Table S2 in the supplemental material). To investigate the effects of the various DegU∼P levels on their production, the potential lipopeptide and polyketide antibiotics predicted by genomic analysis were verified through HPLC/ESI-MS (see Fig. S4). Three cyclic lipopeptides are produced by B. amyloliquefaciens SQR9: surfactin, fengycin, and bacillomycin D. Antibiotics were identified based on the comparisons of these MS data with fractions previously identified by MS analysis for other B. amyloliquefaciens strains (see Table S3) (23). Three antibacterial polyketides, bacillaene, difficidin, and macrolactin, were identified by MS (Fig. 3B). Their mass numbers are summarized in Table S3. In positive mode, bacillaene variants with molecular masses [M + H]+ = 581.5 and 583.5, as well as their dehydrated species ([M + H-18]+ = 563.5 and 565.5), were detected (24). Difficidin and its oxidized form, oxydifficidin, appeared mainly as their alkali ion adducts, whereas the intensity of the mass peaks of their protonated forms at m/z 545.3 and 561.3 are very low. Oxydifficidin bears a hydroxyl group at position 5 of the difficidin ring system, showing mass signals 16 mass units higher than those of the main metabolite. For macrolactin A, the mass spectra in Fig. 3B exhibited m/z ratios of 349.2, 367.2, and 385.2, respectively, and [M + Na]+ and [M + K]+ ions were also found under the positive-ion-mode condition (25).
FIG 3.
LC-MS spectra of antibiotics produced by SQR9 after 36 h of cultivation in Landy medium. (A) Antagonistic activities against F. oxysporum (I) and Ralstonia solanacearum (II) of the supernatant of B. amyloliquefaciens SQR9. (B) LC-MS analysis of general antibiotics produced by B. amyloliquefaciens SQR9: bacillomycin D (I), fengycin (II), macrolactin A (III), bacillaene B (IV), difficidin (V), and surfactin A (VI).
Phosphorylation of DegU influences the production of antibiotics.
The transcription of the bamD, sftA, dfnA, baeB, mlnA, and fenA genes was investigated by real-time qPCR in B. amyloliquefaciens strains SQR9, SQR9M4, SQR9Q, and SQR9SUQ. At 12-h time points, compared with SQR9, bamD and dfnA transcription in strain SQR9SUQ was increased by 5- and 7-fold, respectively. However, compared with results for SQR9, a high level of DegU∼P (in strain SQR9SUQ) inhibited mlnA and fenA transcription by 4 and 5 times, respectively. The low level of DegU∼P in strain SQR9M4 stimulated the transcription of sftA, baeB, mlnA, and fenA 4-, 6-, 4-, and 17-fold, respectively, compared with results for SQR9. At 36-h time points, the stationary phase of SQR9 growth, which is very important for secondary metabolite production, transcription of bamD and dfnA in strain SQRSUQ increased significantly, with average transcription levels 10 and 6 times higher, respectively, than those for SQR9 (Fig. 4).
FIG 4.
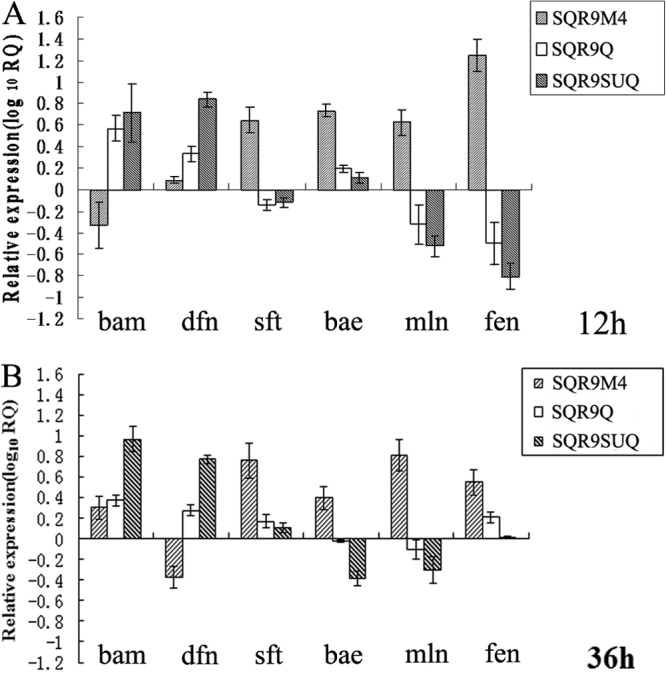
Transcriptional levels of bamD (bam). dfnA (dfn), sftA (sft), baeB (bae), mlnA (mln), and fenA (fen) for different B. amyloliquefaciens SQR9 strains relative to those for wild-type B. amyloliquefaciens SQR9, evaluated by RT-qPCR. The experiments were carried out with B. amyloliquefaciens cells grown in Landy medium for 12 h (A) and 36 h (B). The B. amyloliquefaciens SQR9 recA gene was used as an internal reference gene. Bars represent standard deviations of data from three biological replicates.
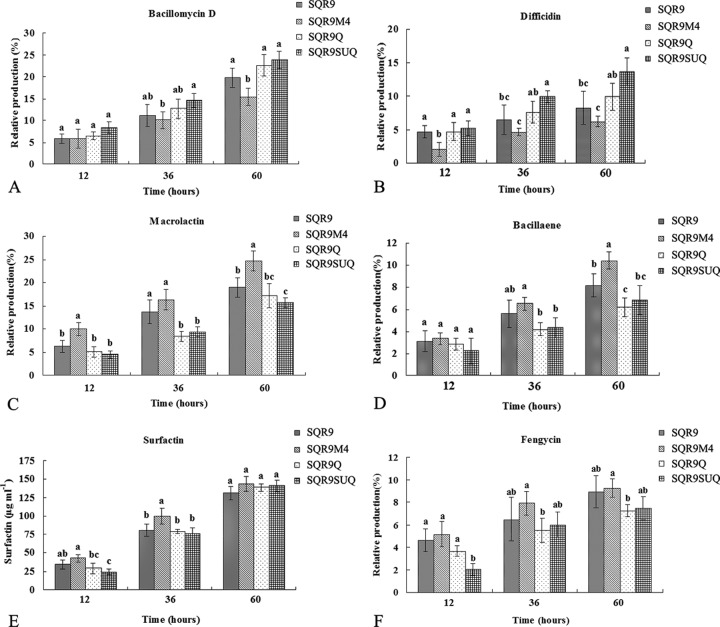
Using HPLC conditions developed in the present study (see Materials and Methods), the production of bacillomycin D, fengycin, surfactin, macrolactin, difficidin, and bacillaene in liquid cultures of B. amyloliquefaciens strains SQR9, SQR9M4, SQR9Q, and SQR9SUQ was analyzed. Two methods for measuring the antibiotics in different strains were used: the external standard method (for antibiotics for which commercial standards are available) and the area normalization method (for antibiotics for which commercial standards are not available) (26, 27). For surfactin (Sigma, USA), two methods were used at the same time to test the accuracy of the area normalization method, and these yielded similar results (Fig. 5E; see also Fig. S5B in the supplemental material). As shown in Fig. 5, the production of bacillomycin D and difficidin by SQRSUQ was significantly increased in comparison with that for SQR9M4. In contrast, the production levels of macrolactin and bacillaene for SQRSUQ were significantly decreased in comparison with those for SQR9M4. There was no significant difference in the production of surfactin and fengycin among the strains with different levels of DegU∼P.
FIG 5.
Production of bacillomycin D (A), difficidin (B), macrolactin (C), bacillaene (D), surfactin (E), and fengycin (F) for different B. amyloliquefaciens strains relative to that for wild-type B. amyloliquefaciens SQR9, evaluated by HPLC.
Increasing levels of DegU∼P in B. amyloliquefaciens SQR9 improve biocontrol activity against cucumber wilt disease.
Both antibiotic production and root colonization by biocontrol agents are critical for effective control of soilborne pathogens, since elevated DegU∼P in B. amyloliquefaciens SQR9 increases its root colonization and bacillomycin D production. Considering that bacillomycin D is the major antibiotic with which B. amyloliquefaciens SQR9 suppresses Fusarium oxysporum, the cucumber wilt disease pathogen (13), we predicted that B. amyloliquefaciens SQR9 strains with a high level of DegU∼P would perform better in biocontrol against Fusarium oxysporum. To test this hypothesis, biocontrol assays were conducted with the cucumber/Fusarium pathosystem to compare the abilities of the different B. amyloliquefaciens SQR9 strains in disease control. As shown in Fig. 6, there were significant differences among the five treatments. The disease incidence of SQR9M4 treatment averaged 66.7%. Treatment with SQR9 showed an average disease incidence of 43.3%. Treatment with strain SQR9SUQ demonstrated a disease incidence of 23%, which was significantly higher than those with the wild-type strain and with strain SQR9M4.
FIG 6.
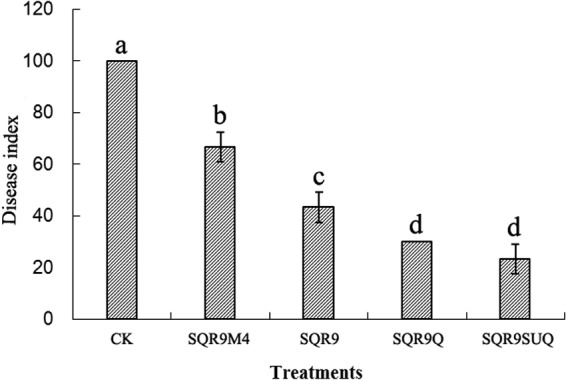
Effects of different treatments on the incidence of Fusarium wilt. Bars with different letters indicated statistical differences among the four treatments by Duncan's test (P < 0.05).
DISCUSSION
The root surface and surrounding rhizosphere are significant carbon sinks (28). Thus, along root surfaces, there are various suitable nutrient-rich niches attracting a great diversity of microorganisms, including phytopathogens (29). Competition for these nutrients and niches is a fundamental mechanism by which BCAs protect plants from phytopathogens. Root colonization by rhizosphere bacteria is linked with biofilm formation (30, 31). The transcription factor DegU, the response regulator of the DegSU two-component system, has a key role in regulating multicellular behavior (9). Previous study demonstrated that the regulatory control exerted by DegU is more flexible than a simple molecular switch and has the capacity to integrate physiological responses along a gradient of DegU phosphorylation. Recently, Verhamme et al. (9) demonstrated that gradual phosphorylation of DegU coordinates multicellular behavior, by using different approaches (7). Their data indicated that swarming motility is activated by very low levels of DegU∼P, and complex colony architecture is activated when the levels of DegU∼P are increased. DegQ is a small pleiotropic regulatory protein which controls the expression of degradative enzymes, intracellular proteases, and several secreted enzymes (32). It has been shown previously that overexpression of DegQ enhances transcription of DegU-regulated genes in the laboratory strain 168 (33). The degQ36 mutation is a C-to-T transition in the degQ promoter which makes the promoter stronger and thus stimulates transcription of DegU-regulated genes in the laboratory strain (10). In the present study, we constructed three mutant strains of B. amyloliquefaciens SQR9 with various levels of DegU phosphorylation. Results from in vitro (Fig. 1), root in situ (Fig. 2), and qPCR (Fig. 1) studies demonstrate that complex colony architecture and biofilm formation are activated when the levels of DegU∼P are increased. By labeling the B. amyloliquefaciens strains, we observed that SQR9SUQ has a significantly higher colonization efficiency than the wild-type and degQ mutant strains. As far as we know, this is the first report in which it is demonstrated that increased levels of DegU phosphorylation can improve colonization efficiency on the root surface.
Phosphorylation is a common posttranslational modification of proteins involved in transmitting intracellular signals. Current methods for detecting phosphorylation have a number of limitations (34). Therefore, we monitored the expression of DegU-regulated genes to detect the levels of DegU phosphorylation in different strains. yvcA is a gene that is directly regulated by DegU∼P (9). The transcription of yvcA demonstrated that the levels of DegU∼P in the four strains were gradually increased. Also, we found similar results when we tested ycdA and aprE, genes that can be directly bound by DegU (data not shown). For the B. subtilis mutant strain 3610, high levels of DegU∼P inhibit complex colony architecture: Verhamme et al. used an allele of degU that encodes a histidine-to-leucine mutation at amino acid 12 (H12L), encoding a DegU protein that exhibits a 7-fold-higher rate of dephosphorylation than the wild-type protein (9). In our case, we overexpressed only the degQ gene, and the level of DegU∼P was lower than that in the B. subtilis mutant strain 3610.
An increasing number of DegU-regulated genes were identified by transcriptome and microarray analyses (8). However, the relationship between DegU∼P and antibiotic production is not clear at present. Recently, DegQ was shown to enhance the production of the peptide antibiotics plipastatin and iturin A (35, 36). However, the effects of DegQ are only indirect and are mediated via DegU, since DegQ shares no homology with typical transcriptional regulators, i.e., DNA-binding proteins, and DegQ overexpression cannot complement for the loss of DegU in terms of bacillomycin D synthesis. In the present study, the production of bacillomycin D and difficidin was significantly increased when we increased the levels of DegU∼P, whereas the production of macrolactin and bacillaene was significantly decreased. In a previous study, a series of in vivo and in vitro data indicated that DegU binds to Pbmy, the promoter of bacillomycin D, and directly activates its expression (37). Moreover, DegU∼P is more suitable for optimal promoter binding and activation. To our knowledge, this is the first report which indicates that different levels of DegU∼P can influence the production of difficidin, macrolactin, and bacillaene. The mechanism of how DegU∼P regulates the production of antibiotics is currently unknown.
In conclusion, the results reported here indicate that gradual phosphorylation of DegU in B. amyloliquefaciens SQR9 can influence biocontrol activity by coordinating multicellular behavior and also regulating the synthesis of bacillomycin D. Increasing the levels of DegU∼P in B. amyloliquefaciens SQR9 can improve biocontrol activity. In natural environments, to respond to various conditions and incorporate input from various environmental signals, the colonization mechanism of B. amyloliquefaciens SQR9 may be complex and controlled by multiple regulators. An important challenge for the future will be to determine the relationship between such regulators and signals that control biofilm formation and colonization.
Supplementary Material
ACKNOWLEDGMENTS
This research was financially supported by the National Natural Science Foundation of China (31330069) and the Chinese Ministry of Science and Technology (2011BAD11B03 and 2013AA102802); R.Z. and Q.S. were also supported by the 111 Project (B12009) and the Priority Academic Program Development (PAPD) of Jiangsu Higher Education Institutions.
We thank E. (Ben) J. J. Lugtenberg of Leiden University for his critical review and for polishing the language of the manuscript.
We have no conflict of interest to declare.
Footnotes
Published ahead of print 28 February 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.03943-13.
REFERENCES
- 1.Lugtenberg BJJ, Chin-A-Woeng TFC, Bloemberg GV. 2002. Microbe-plant interactions: principles and mechanisms. Antonie Van Leeuwenhoek 81:373–383. 10.1023/A:1020596903142 [DOI] [PubMed] [Google Scholar]
- 2.Chet I, Chernin L. 2003. Biocontrol, microbial agents in soil. Encyclopedia of environmental microbiology. John Wiley & Sons, Inc., Hoboken, NJ [Google Scholar]
- 3.Ram RJ, VerBerkmoes NC, Thelen MP, Tyson GW, Baker BJ, Blake RC, Shah M, Hettich RL, Banfield JF. 2005. Community proteomics of a natural microbial biofilm. Science 308:1915–1920. 10.1126/science.1109070 [DOI] [PubMed] [Google Scholar]
- 4.Vlamakis H, Chai Y, Beauregard P, Losick R, Kolter R. 2013. Sticking together: building a biofilm the Bacillus subtilis way. Nat. Rev. Microbiol. 11:157–168. 10.1038/nrmicro2960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hancock RE. 1997. Peptide antibiotics. Lancet 349:418–422. 10.1016/S0140-6736(97)80051-7 [DOI] [PubMed] [Google Scholar]
- 6.Chen XH, Koumoutsi A, Scholz R, Eisenreich A, Schneider K, Heinemeyer I, Morgenstern B, Voss B, Hess WR, Reva O, Junge H, Voigt B, Jungblut PR, Vater J, Süssmuth R, Liesegang H, Strittmatter A, Gottschalk G, Borriss R. 2007. Comparative analysis of the complete genome sequence of the plant growth-promoting bacterium Bacillus amyloliquefaciens FZB42. Nat. Biotechnol. 25:1007–1014. 10.1038/nbt1325 [DOI] [PubMed] [Google Scholar]
- 7.Murray EJ, Kiley TB, Stanley-Wall NR. 2009. A pivotal role for the response regulator DegU in controlling multicellular behaviour. Microbiology 155:1–8. 10.1099/mic.0.023903-0 [DOI] [PubMed] [Google Scholar]
- 8.Ogura M, Yamaguchi H, Yoshida K, Fujita Y, Tanaka T. 2001. DNA microarray analysis of Bacillus subtilis DegU, ComA and PhoP regulons: an approach to comprehensive analysis of B. subtilis two-component regulatory systems. Nucleic Acids Res. 29:3804–3813. 10.1093/nar/29.18.3804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Verhamme DT, Kiley TB, Stanley-Wall NR. 2007. DegU co-ordinates multicellular behaviour exhibited by Bacillus subtilis. Mol. Microbiol. 65:554–568. 10.1111/j.1365-2958.2007.05810.x [DOI] [PubMed] [Google Scholar]
- 10.Kobayashi K. 2007. Gradual activation of the response regulator DegU controls serial expression of genes for flagellum formation and biofilm formation in Bacillus subtilis. Mol. Microbiol. 66:395–409. 10.1111/j.1365-2958.2007.05923.x [DOI] [PubMed] [Google Scholar]
- 11.Tsuge K, Ano T, Hirai M, Nakamura Y, Shoda M. 1999. The genes degQ, pps, and lpa-8 (sfp) are responsible for conversion of Bacillus subtilis 168 to plipastatin production. Antimicrob. Agents Chemother. 43:2183–2192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cao Y, Zhang Z, Ling N, Yuan Y, Zheng X, Shen B, Shen Q. 2011. Bacillus subtilis SQR 9 can control Fusarium wilt in cucumber by colonizing plant roots. Biol. Fertil. Soils 47:495–506. 10.1007/s00374-011-0556-2 [DOI] [Google Scholar]
- 13.Xu Z, Shao J, Li B, Yan X, Shen Q, Zhang R. 2013. Contribution of bacillomycin D in Bacillus amyloliquefaciens SQR9 to antifungal activity and biofilm formation. Appl. Environ. Microbiol. 79:808–815. 10.1128/AEM.02645-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Weng J, Wang Y, Li J, Shen Q, Zhang R. 2013. Enhanced root colonization and biocontrol activity of Bacillus amyloliquefaciens SQR9 by abrB gene disruption. Appl. Microbiol. Biotechnol. 97:8823–8830. 10.1007/s00253-012-4572-4 [DOI] [PubMed] [Google Scholar]
- 15.Tan S, Dong Y, Liao H, Huang J, Song S, Xu Y, Shen Q. 2013. Antagonistic bacterium Bacillus amyloliquefaciens induces resistance and controls the bacterial wilt of tomato. Pest Manag. Sci. 69:1245–1252. 10.1002/ps.3491 [DOI] [PubMed] [Google Scholar]
- 16.Landy M, Warren GH, Rosenman SB, Coli LG. 1967. Bacillomycin D, an antibiotic from Bacillus subtilis active against pathogenic fungi. Proc. Soc. Exp. Biol. Med. 67:539–541 [DOI] [PubMed] [Google Scholar]
- 17.Hamon MA, Lazazzera BA. 2001. The sporulation transcription factor Spo0A is required for biofilm development in Bacillus subtilis. Mol. Microbiol. 42:1199–1209. 10.1046/j.1365-2958.2001.02709.x [DOI] [PubMed] [Google Scholar]
- 18.Kearns DB, Chu F, Branda SS, Kolter R, Losick R. 2005. A master regulator for biofilm formation by Bacillus subtilis. Mol. Microbiol. 55:739–749. 10.1111/j.1365-2958.2004.04440.x [DOI] [PubMed] [Google Scholar]
- 19.Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25:402–408. 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- 20.Zhang N, Wu K, He X, Li S, Zhang Z, Shen B, Yang X, Zhang R, Huang Q, Shen Q. 2011. A new bioorganic fertilizer can effectively control banana wilt by strong colonization with Bacillus subtilis N11. Plant Soil 344:87–97. 10.1007/s11104-011-0729-7 [DOI] [Google Scholar]
- 21.Murashige T, Skoog F. 1962. A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol. Plant 15:473–497. 10.1111/j.1399-3054.1962.tb08052.x [DOI] [Google Scholar]
- 22.Peltier AJ, Grau CR. 2008. The influence of light on relationships between sclerotinia stem rot of soybean in field and controlled environments. Plant Dis. 92:1510–1514. 10.1094/PDIS-92-11-1510 [DOI] [PubMed] [Google Scholar]
- 23.Koumoutsi A, Chen X-H, Henne A, Liesegang H, Hitzeroth G, Franke P, Vater J, Borriss R. 2004. Structural and functional characterization of gene clusters directing nonribosomal synthesis of bioactive cyclic lipopeptides in Bacillus amyloliquefaciens strain FZB42. J. Bacteriol. 186:1084–1096. 10.1128/JB.186.4.1084-1096.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen X-H, Vater J, Piel J, Franke P, Scholz R, Schneider K, Koumoutsi A, Hitzeroth G, Grammel N, Strittmatter AW, Gottschalk G, Süssmuth RD, Borriss R. 2006. Structural and functional characterization of three polyketide synthase gene clusters in Bacillus amyloliquefaciens FZB 42. J. Bacteriol. 188:4024–4036. 10.1128/JB.00052-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yuan J, Li B, Zhang N, Waseem R, Shen Q, Huang Q. 2012. Production of bacillomycin- and macrolactin-type antibiotics by Bacillus amyloliquefaciens NJN-6 for suppressing soilborne plant pathogens. J. Agric. Food Chem. 60:2976–2981. 10.1021/jf204868z [DOI] [PubMed] [Google Scholar]
- 26.Coutrim MX, Nakamura LA, Collins CH. 1993. Quantification of 2,4-dinitrophenyl-hydrazones of low molecular mass aldehydes and ketones using HPLC. Chromatographia 37:185–190. 10.1007/BF02275859 [DOI] [Google Scholar]
- 27.Bais HP, Fall R, Vivanco JM. 2004. Biocontrol of Bacillus subtilis against infection of Arabidopsis roots by Pseudomonas syringae is facilitated by biofilm formation and surfactin production. Plant Physiol. 134:307–319. 10.1104/pp.103.028712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Compant S, Duffy B, Nowak J, Clément C, Barka EA. 2005. Use of plant growth-promoting bacteria for biocontrol of plant diseases: principles, mechanisms of action, and future prospects. Appl. Environ. Microbiol. 71:4951–4959. 10.1128/AEM.71.9.4951-4959.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nelson EB. 2004. Microbial dynamics and interactions in the spermosphere. Annu. Rev. Phytopathol. 42:271–309. 10.1146/annurev.phyto.42.121603.131041 [DOI] [PubMed] [Google Scholar]
- 30.Lugtenberg BJJ, Kravchenko LV, Simons M. 1999. Tomato seed and root exudate sugars: composition, utilization by Pseudomonas biocontrol strains and role in rhizosphere colonization. Environ. Microbiol. 1:439–446. 10.1046/j.1462-2920.1999.00054.x [DOI] [PubMed] [Google Scholar]
- 31.Bloemberg GV, Lugtenberg BJ. 2001. Molecular basis of plant growth promotion and biocontrol by rhizobacteria. Curr. Opin. Plant Biol. 4:343–350. 10.1016/S1369-5266(00)00183-7 [DOI] [PubMed] [Google Scholar]
- 32.Mäder U, Antelmann H, Buder T, Dahl M, Hecker M, Homuth G. 2002. Bacillus subtilis functional genomics: genome-wide analysis of the DegS-DegU regulon by transcriptomics and proteomics. Mol. Genet. Genomics 268:455–467. 10.1007/s00438-002-0774-2 [DOI] [PubMed] [Google Scholar]
- 33.Dahl MK, Msadek T, Kunst F, Rapoport G. 1992. The phosphorylation state of the DegU response regulator acts as a molecular switch allowing either degradative enzyme synthesis or expression of genetic competence in Bacillus subtilis. J. Biol. Chem. 267:14509–14514 [PubMed] [Google Scholar]
- 34.Anderson JC, Peck SC. 2008. A simple and rapid technique for detecting protein phosphorylation using one-dimensional isoelectric focusing gels and immunoblot analysis. Plant J. 55:881–885. 10.1111/j.1365-313X.2008.03550.x [DOI] [PubMed] [Google Scholar]
- 35.Borgmeier C, Biedendieck R, Hoffmann K, Jahn D, Meinhardt F. 2011. Transcriptome profiling of degU expression reveals unexpected regulatory patterns in Bacillus megaterium and discloses new targets for optimizing expression. Appl. Microbiol. Biotechnol. 92:583–596. 10.1007/s00253-011-3575-x [DOI] [PubMed] [Google Scholar]
- 36.Tsuge K, Inoue S, Ano T, Itaya M, Shoda M. 2005. Horizontal transfer of Iturin A operon, itu, to Bacillus subtilis 168 and conversion into an Iturin A producer. Antimicrob. Agents Chemother. 49:4641–4648. 10.1128/AAC.49.11.4641-4648.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Koumoutsi A, Chen X-H, Vater J, Borriss R. 2007. DegU and YczE positively regulate the synthesis of bacillomycin D by Bacillus amyloliquefaciens strain FZB42. Appl. Environ. Microbiol. 73:6953–6964. 10.1128/AEM.00565-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang X-Z, Cui Z, Hong Q, Li S. 2005. High-level expression and secretion of methyl parathion hydrolase in Bacillus subtilis WB800. Appl. Environ. Microbiol. 71:4101–4103. 10.1128/AEM.71.7.4101-4103.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.