Abstract
Understanding the interactions of plant-parasitic nematodes with antagonistic soil microbes could provide opportunities for novel crop protection strategies. Three arable soils were investigated for their suppressiveness against the root knot nematode Meloidogyne hapla. For all three soils, M. hapla developed significantly fewer galls, egg masses, and eggs on tomato plants in unsterilized than in sterilized infested soil. Egg numbers were reduced by up to 93%. This suggested suppression by soil microbial communities. The soils significantly differed in the composition of microbial communities and in the suppressiveness to M. hapla. To identify microorganisms interacting with M. hapla in soil, second-stage juveniles (J2) baited in the test soil were cultivation independently analyzed for attached microbes. PCR-denaturing gradient gel electrophoresis of fungal ITS or 16S rRNA genes of bacteria and bacterial groups from nematode and soil samples was performed, and DNA sequences from J2-associated bands were determined. The fingerprints showed many species that were abundant on J2 but not in the surrounding soil, especially in fungal profiles. Fungi associated with J2 from all three soils were related to the genera Davidiella and Rhizophydium, while the genera Eurotium, Ganoderma, and Cylindrocarpon were specific for the most suppressive soil. Among the 20 highly abundant operational taxonomic units of bacteria specific for J2 in suppressive soil, six were closely related to infectious species such as Shigella spp., whereas the most abundant were Malikia spinosa and Rothia amarae, as determined by 16S rRNA amplicon pyrosequencing. In conclusion, a diverse microflora specifically adhered to J2 of M. hapla in soil and presumably affected female fecundity.
INTRODUCTION
Root knot nematodes (Meloidogyne spp.) are among the most damaging pathogens of many crops worldwide and are important pests in Europe (1). Chemical nematicides are costly and restricted due to their adverse impact on the environment and human health, whereas cultural control or host plant resistance are often not practical or not available (2). Alternative management strategies could include biological control methods. Microbial pathogens or antagonists of root knot nematodes have high potential for nematode suppression. Many fungal or bacterial isolates have been found that antagonize root knot nematodes either directly by toxins, enzymatically, parasitically, or indirectly by inducing host plant resistance (3). Indigenous microbial communities of arable soils were occasionally reported to suppress root knot nematodes (4–7). Soils that suppress Meloidogyne spp. are of interest for identifying antagonistic microorganisms and the mechanisms that regulate nematode population densities. Understanding the ecological factors that enable these antagonists to persist, compete, and function may improve the basis for integrated management strategies. Cultivation-independent approaches were used in several studies to analyze the diversity of bacteria or fungi associated with the plant-parasitic nematode genera Bursaphelenchus (8), Heterodera (9–11), or Rotylenchulus (12). Papert et al. (13) showed by PCR-denaturing gradient gel electrophoresis (DGGE) of 16S rRNA genes that the bacterial colonization of egg masses of Meloidogyne fallax differed from the rhizoplane community. An rRNA sequence most similar to that of the egg-parasitizing fungus Pochonia chlamydosporia was frequently detected in egg masses of Meloidogyne incognita that derived from a suppressive soil (4).
Root knot nematodes spend the majority of their life protected inside the root. After hatching, second-stage juveniles (J2) of root knot nematodes migrate through soil to penetrate host roots. During this searching, they are most exposed to soil microbes. Root knot nematodes do not ingest microorganisms, and their cuticle is the main barrier against microbes. The collagen matrix of the cuticle is covered by a continuously shed and renewed surface coat mainly composed of highly glycosylated proteins, which likely is involved in evading host immune defense and microbial attack (14). Attachment of microbes to the J2 cuticle while dwelling through soil may result in the transport of microbes to roots, endophytic colonization, coinfection of roots, or the defense response of the plant triggered by microbe-associated molecular pattern. Attached microbes may also directly inhibit or infect J2 or later colonize eggs of nematodes (15). Despite its potential ecological importance, the microbiome associated with J2 of root knot nematodes has not yet been analyzed by cultivation-independent methods.
In the present study, three arable soils were investigated for their suppressiveness against the root knot nematode Meloidogyne hapla. The bacteria and fungi attached to J2 incubated in these soils were analyzed based on their 16S rRNA genes or internal transcribed spacer (ITS), respectively, and compared to the microbial communities of the bulk soil. The objectives were (i) to test whether a specific subset of soil microbes attaches to J2 of M. hapla, (ii) to test whether attached species differ between soils of varying suppressive potential, and (iii) to identify bacteria and fungi that putatively interact with J2 of M. hapla.
MATERIALS AND METHODS
Soils.
Soils were obtained from three different locations in Germany and included a Luvic-Phaeozem with medium clayey silt and 17.2% clay (loess loam, pH 7.3, organic carbon content [Corg] = 1.8%) from a field of the plant breeder KWS Saat AG in Klein Wanzleben (Kw), a Gleyic-Fluvisol with heavy sandy loam and 27.5% clay (alluvial loam, pH 6.7, Corg = 1.8%) from a lettuce field in Golzow (Go), and an Arenic-Luvisol with less silty sand and 5.5% clay (diluvial sand, pH 6.1, Corg = 0.9%) from a field in Grossbeeren (Gb). These soils were selected because of a low abundance of M. hapla despite the presence of suitable environmental conditions and susceptible plants. The soils were previously characterized in detail (16), and data on microbial communities were available. Soil samples were collected from eight plots within each field. Each sample consisted of ∼3 kg composed of 12 soil cores taken from the top 30 cm. All samples were kept in polyethylene bags and stored at 4°C until further processing.
Greenhouse assay for soil suppressiveness.
The suppressiveness against M. hapla of the microbial communities in the three soils was determined by comparing the reproduction of inoculated J2 on tomato plants in natural and sterilized soil. Native soil without inoculated J2 served as control for putative indigenous root knot nematodes. Thus, each of the eight replicate soil samples of each soil was divided into three portions for the three treatments. The portion for the J2 inoculation into sterilized soil was autoclaved at 134°C for 10 min to kill indigenous microbes, followed by a 20-min dry cycle. Each portion of the soil samples was separately mixed with steamed loamy sand at a ratio of 1:1 to improve physical soil properties for greenhouse culture and placed in 1.2-kg portions in 15-cm-diameter pots. Two-week-old seedlings of Solanum lycopersicum ‘Moneymaker' were transplanted into the pots. One week after transplanting, 1,600 freshly hatched J2 of M. hapla were inoculated into each pot, except the control for putative indigenous root knot nematodes. The J2 were inoculated by transferring 1 ml of a suspension with 200 J2 ml−1 into each of eight holes at the periphery of the pot (7 cm from stem base, 2 cm deep), so that the J2 could interact with soil microbes before penetrating tomato roots. The pots were arranged in a randomized block design, so that in total 72 pots (8 replicate blocks × 3 soils × 3 treatments) were maintained in the greenhouse at 20 ± 2°C at ambient light. Plants were watered and fertilized as needed. Two months after inoculation, root systems were washed free of adhering soil and weighted. Egg masses attached to the roots were stained with 0.4% cochenille red solution (Brauns-Heitmann, Warburg, Germany) for 15 min. Galls and egg masses were counted. Roots were vigorously shaken for 3 min in 2% chlorine to free the eggs from the gelatinous matrices. The suspension was poured through a 250-μm-aperture sieve to remove roots. Eggs were collected on a 20-μm-pore-size sieve and counted.
Soil baiting with J2 and DNA extraction.
To analyze the microorganisms attaching to J2 when they move through soil, J2 were inoculated in each soil and extracted after exposure to the microbial communities in the three soils. Four replicate tubes per soil type with 2,000 inoculated J2 in 50 g of soil were kept at 20 ± 2°C in the dark for 7 days. The soil moisture was adjusted to 15%. J2 were extracted from the soil by centrifugal flotation with MgSO4 solution (17), collected on 25-μm-aperture sieves, and transferred with sterile water into petri dishes. Under the stereomicroscope, 100 J2 from each replicate, which were morphologically identified as root knot nematodes, were captured by using a needle. DNA from J2 with adhering microorganisms was extracted by using a FastPrep FP120 bead-beating system (MP Biomedicals, Santa Ana, CA) for 30 s at high speed, a FastDNA Spin kit for soil (MP Biomedicals), and the Geneclean spin kit (MP Biomedicals) for further purification. In parallel, total soil DNA was extracted from 0.5 g of bulk soil of each tube by the same method for comparison of the microbial communities from nematode samples to those of the surrounding soil.
PCR-DGGE of fungal ITS and bacterial 16S rRNA gene fragments.
PCR amplifications of fungal ITS and of 16S rRNA genes of bacteria or bacterial groups from total DNA of soil and J2 samples and separation of the PCR products in DGGE were performed as previously described (18). In brief, bacterial 16S rRNA gene fragments were amplified either directly from total DNA using the primer pair F984GC/R1378 or via PCR with primers that were designed to target the bacterial groups Alphaproteobacteria, Betaproteobacteria, Pseudomonas, Actinobacteriales, Enterobacteriaceae, or Bacillus (all primer sequences are shown in Table S1 in the supplemental material). The fungal ITS fragments were amplified using a nested PCR approach with primer pairs ITS1F/ITS4 and ITS1FGC/ITS2. DGGE was done by using the PhorU2 system (Ingeny, Goes, Netherlands) as previously described (18).
Analysis of ribosomal sequences of microbes attached to J2.
For the DGGE fingerprints of bacterial groups and fungal ITS fragments that showed nematode-specific bands, PCR products were cloned and sequenced to identify the corresponding microbial species by sequence comparison to the GenBank entries. For Alphaproteobacteria and Pseudomonas, PCR products obtained with the primer pair F984GC/R1378 were used; for Bacillus, products produced with the primer pair BacF/R1378 were used; for fungal profiles, products of the primer pair ITS1FGC/ITS2 were used (see Table S1 in the supplemental material). PCR products were cloned using the vector pGEM-T and Escherichia coli JM109 high-efficiency competent cells (Promega, Madison, WI). Based on the PCR-DGGE analyses, cloned amplicons corresponding in electrophoretic mobility to nematode-specific bands were sequenced (Macrogen, Amsterdam, Netherlands).
Barcoded amplicon pyrosequencing was used to analyze 16S rRNA genes of total J2-associated bacteria. PCR with the universal bacterial primers F27/R1494 was performed as previously described (19). The products were purified with a Minelute PCR purification kit (Qiagen, Hilden, Germany) and used as target to amplify the V3-V4 region of 16S rRNA genes with fusion primers containing the Roche-454 A and B Titanium sequencing adapters, an eight-base barcode sequence in adaptor A, and specific sequences V3F/V4R targeting the ribosomal region. Library preparation and sequencing were done on a 454 Genome Sequencer FLX platform according to standard 454 protocols (Roche-454 Life Sciences, Branford, CT) by Biocant (Cantanhede, Portugal). Pyrosequencing data were evaluated according to the method of Ding et al. (20). Briefly, sequences matching the barcode and primer were selected for blastn searches in the database SILVA 115 SSU Ref (21) and a subset of that containing the strains with the species name. Chimera were truncated, barcodes and primers were removed, and sequences shorter than 200 bp were discarded. Multiple alignments and operational taxonomic unit (OTU) assignment (>97% similarity) were performed using the software package Mothur v1.14.0 (22). OTUs were regarded as specific for J2 that comprised >1% of all sequences of J2 samples and that were not detected in soil or had at least 100 times higher relative abundance on J2 compared to soil.
Statistical analysis.
For the greenhouse experiment, the numbers of galls, egg masses, eggs per gram of root, and eggs per egg mass after propagation of inoculated J2 were compared between pots with native and sterilized soil for each soil type. The data were log transformed and a linear model with soil, treatment, and soil·treatment as fixed effects and block as a random effect was applied (see Table S2 in the supplemental material). For pairwise comparisons between soil types the Tukey-Kramer adjustment was applied.
Sequence accession numbers.
Sequences for DGGE bands were deposited in GenBank under accession no. KF225704 to KF225718 and KF257370 to KF257399. Pyrosequencing data were deposited at the NCBI Sequence Read Archive under study accession number SRP029944.
RESULTS
Microbes of the three soils reduced progeny of M. hapla to different extent.
To assess the suppressive effect of the microbial soil communities on M. hapla, the nematode propagation on tomato was compared between sterilized and native soils. Significantly fewer galls, egg masses, eggs, and a reduced rate of fecundity (eggs per egg mass) were found on roots from native soils than in sterilized soils 8 weeks after J2 inoculation (P < 0.001, ANOVA with soil origin and sterilization as fixed effects, see Table S2). Also soil origin had a significant effect on nematode counts and fecundity (P < 0.015), except for egg masses (P = 0.055). In nonsterilized soil Kw the lowest numbers of galls, egg masses, eggs, and eggs per egg mass were found compared to soils Go and Gb (Table 1). The number of eggs was reduced by 93% in native soil Kw compared to the sterilized control and was significantly lower than for the other soils, suggesting that the microbial community of soil Kw had a more suppressive effect. The reduction in galls and egg masses for soil Kw was less pronounced than egg reduction (58 and 68%, respectively). The least suppressive soil Go had significantly more galls, egg masses, and eggs in the nonsterilized treatment than soil Kw (Table 1), with significantly lower reductions compared to the sterilized control (30, 38, and 63%, respectively). In contrast to the native soils, in sterilized soils the numbers of galls and egg masses were highly similar between soils. Egg numbers and fecundity in sterilized soils were fewest for Go and highest for Gb, whereas sterilized soil Kw did not show the lowest counts among the soils, as seen for the soils with indigenous microbial communities (Table 1). This suggested a minor role of the physicochemical soil differences compared to biotic factors. In control pots without J2 inoculation, indigenous root knot nematodes developed only five galls on one tomato plant in soil Kw, which was too low to confound nematode counts of the inoculated nonsterilized pots (data not shown).
TABLE 1.
Effect of soil biota on fertility of M. hapla on tomato plants in three infested soils
| Parameter | Soil treatment | Mean log10 (no. g−1 root fresh wt) ± SDa |
||
|---|---|---|---|---|
| Soil Kw | Soil Go | Soil Gb | ||
| Galls | Sterilized | 1.53 ± 0.18A | 1.57 ± 0.21A | 1.54 ± 0.11A |
| Nonsterilized | 1.09 ± 0.33A | 1.45 ± 0.06B | 1.17 ± 0.19A | |
| Egg masses | Sterilized | 1.47 ± 0.17A | 1.49 ± 0.20A | 1.45 ± 0.11A |
| Nonsterilized | 0.86 ± 0.44A | 1.28 ± 0.13B | 0.91 ± 0.39AB | |
| Eggs | Sterilized | 4.48 ± 0.08AB | 4.45 ± 0.14A | 4.58 ± 0.12B |
| Nonsterilized | 3.31 ± 0.19A | 3.95 ± 0.27B | 3.86 ± 0.21B | |
| Fecundity (eggs/egg mass) | Sterilized | 3.01 ± 0.13AB | 2.96 ± 0.07A | 3.13 ± 0.10B |
| Nonsterilized | 2.45 ± 0.35A | 2.67 ± 0.24AB | 2.95 ± 0.41B | |
Values are means of eight replicate root systems. Different letters within a row indicate a significant difference between means for either sterilized or native soils (P < 0.05, Tukey-Kramer adjustment).
Fungal attachment to M. hapla in soil.
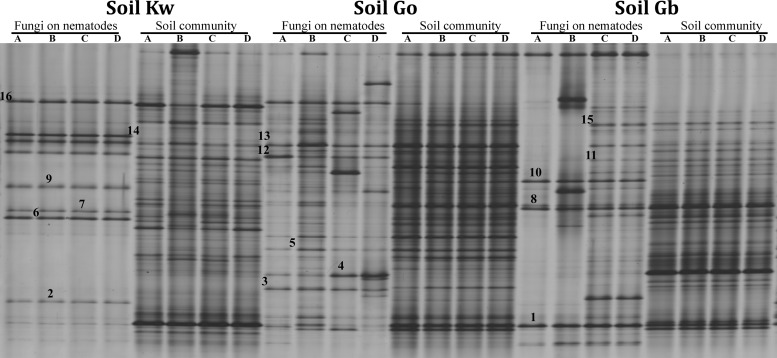
The fungi sticking to J2, which were extracted from the three soils and washed, were analyzed by PCR-DGGE of fungal ITS fragments. ITS profiles of DNA from J2 showed 20 (for soil Kw) to 40 (for soil Gb) clearly visible bands, while profiles of fungal soil communities were much more complex (Fig. 1). Several fungal ITS types were abundant in all replicate DNA samples from J2 of one or more soils but not in the surrounding soil, suggesting specific attachment to the J2 in soil (Fig. 1, bands 2, 3, 4, 6, 9, 11, 13, and 15). Some of the fungal ITS types associated with J2 were also abundant in soil, but the relative band intensity within the profile was higher for the J2 samples than for soil, which indicated an enrichment on J2 (Fig. 1, bands 1, 5, 7, 8, 10, 12, and 14). The most reproducible patterns were detected on J2 from replicates of the most suppressive soil Kw, evidencing the most specific fungal attachment compared to those from the other two soils. The DNA sequences of ITS types were determined to identify fungal species that potentially interacted with the J2 in soil. The sequences corresponded to fungal ITS of eight genera of Ascomycota, five genera of Basidiomycota, Rhizopodium (Chytridiomycota), and Mortierella (Fungi incertae sedis) (Table 2). Bands 9 and 15, of which the DNA was most closely related to the genera Davidiella and Rhizophydium, respectively, were associated with J2 from all three soils, even though they were mostly below the detection limit in the soil fungal communities. Some bands were common on both nematodes and soil samples in the three soils, such as bands 1, 12, and 14, which corresponded to Malassezia restricta, Mortierella sp., and Ascomycete sp., respectively (Table 2). Eight of the ITS types associated with J2 were soil type specific, four of which were only detected on J2 (Table 2, bands 3, 4, 6, and 13), while the other four were obtained from both J2 and soil samples (Table 2, bands 5, 7, 8, and 10). The sequences of these bands exhibited 98 to 100% similarity to known sequences of fungal species in GenBank (Table 2). Furthermore, two of the attached ITS types seemed to be specific for J2 samples in two of the three soils (Table 2, bands 2 and 11). The ITS type of band 2 was found in J2 samples from the two most suppressive soils, Kw and Gb, and corresponded to Aspergillus penicillioides (99.7% identities). In contrast to J2 from soils Go and Gb, J2 extracted from the most suppressive soil Kw were specifically associated with ITS types closely related to Eurotium sp., Ganoderma applanatum, and Cylindrocarpon olidum (Table 2, bands 6, 7, and 13).
FIG 1.
DGGE profiles of fungal ITS fragments amplified from DNA of M. hapla J2 from three arable soils and from total soil DNA. Fungal ITS types are marked that were enriched in nematode samples and characterized by sequencing (Table 2). A, B, C, and D refer to replicate soil baiting assays for each soil.
TABLE 2.
Identification and frequency of the dominant nematode-specific DGGE bands
| DGGE type and band no. | Closest GenBank match (organism, GenBank no.)a | % identity | No. of samples where band was found |
|||||
|---|---|---|---|---|---|---|---|---|
| Nematodes |
Soil |
|||||||
| Kw | Go | Gb | Kw | Go | Gb | |||
| Fungus DGGE | ||||||||
| 1 | Malassezia restricta, EU400587 | 98.7 | 4 | 4 | 4 | 4 | 4 | 4 |
| 2 | Aspergillus penicillioides, GU017496 | 99.6 | 4 | 0 | 2 | 0 | 0 | 0 |
| 3 | Cryptococcus pseudolongus, AB105353 | 100 | 0 | 4 | 0 | 0 | 0 | 0 |
| 4 | Chaetomium globosum, JX501299 | 98.2 | 0 | 4 | 0 | 0 | 0 | 0 |
| 5 | Arthopyreniaceae, FJ439584 | 100 | 0 | 4 | 0 | 0 | 4 | 0 |
| 6 | Eurotium sp., AM901702 | 100 | 4 | 0 | 0 | 0 | 0 | 0 |
| 7 | Ganoderma applanatum, JX501311 | 99.6 | 4 | 0 | 0 | 2 | 0 | 0 |
| 8 | Cladosporinum cladosporioides, AJ300335 | 100 | 0 | 0 | 4 | 0 | 0 | 4 |
| 9 | Davidiella sp., JX164064 | 99.6 | 4 | 4 | 4 | 0 | 0 | 0 |
| 10 | Cryptococcus sp., JX164076 | 100.0 | 0 | 0 | 4 | 0 | 4 | 4 |
| 11 | Trichosporonales, EF060720 | 98.3 | 4 | 4 | 0 | 0 | 0 | 0 |
| 12 | Mortierella sp., JF439489 | 99.6 | 4 | 4 | 2 | 4 | 4 | 4 |
| 13 | Cylindrocarpon olidum, GU198183 | 99.0 | 4 | 0 | 0 | 0 | 0 | 0 |
| 14 | Ascomycete, AM410609 | 99.2 | 4 | 4 | 4 | 4 | 4 | 4 |
| 15 | Rhizophydium sp., DQ485617 | 98.7 | 4 | 4 | 2 | 0 | 0 | 0 |
| Bacillus DGGE | ||||||||
| 1 | Bradyrhizobium pachyrhizi, NR_043037 | 97.9 | 0 | 3 | 0 | 0 | 0 | 0 |
| 2 | Sphingomonas insulae, NR_044187 | 99.4 | 1 | 1 | 3 | 0 | 0 | 0 |
| 3 | Staphylococcus epidermidis, NR_036904 | 100 | 4 | 4 | 4 | 0 | 0 | 0 |
| 4 | Staphylococcus epidermidis, NR_036904 | 99.6 | 4 | 4 | 4 | 0 | 0 | 0 |
| 5 | Micrococcus endophyticus, NR_044365 | 98.6 | 3 | 3 | 4 | 0 | 0 | 0 |
| 6 | Bacillus megaterium, NR_043401 | 99.7 | 4 | 4 | 4 | 0 | 0 | 0 |
| 7 | Micrococcus luteus, NR_037113 | 99.2 | 4 | 4 | 4 | 4 | 4 | 4 |
| 8 | Propionibacterium acnes, NR_040847 | 100 | 4 | 4 | 4 | 4 | 4 | 4 |
| 9 | Methylobacterium rhodesianum, NR_041028 | 97.2 | 2 | 1 | 3 | 0 | 0 | 0 |
| 10 | Streptococcus thermophilus, NR_074827 | 100 | 0 | 0 | 3 | 0 | 0 | 0 |
| Alphaproteobacterium DGGE | ||||||||
| 1 | Solirubrobacter soli, NR_041365 | 99.8 | 2 | 3 | 1 | 3 | 3 | 0 |
| 2 | Janthinobacterium lividum, NR_026365 | 99.8 | 1 | 0 | 3 | 0 | 0 | 0 |
| 3 | Rhizobium phaseoli, NR_044112 | 99.8 | 4 | 4 | 4 | 0 | 0 | 0 |
| 4 | Pedomicrobium australicum, NR_026337 | 96.0 | 1 | 3 | 3 | 4 | 4 | 4 |
| 5 | Ochrobactrum anthropi, NR_074243 | 99.5 | 4 | 3 | 2 | 2 | 4 | 0 |
| 6 | Hyphomonadaceae, NR_041967 | 91.0 | 3 | 2 | 3 | 4 | 4 | 4 |
| 7 | Nitrospira moscoviensis, NR_029287 | 96.3 | 2 | 3 | 0 | 0 | 0 | 0 |
| 8 | Rhodobiaceae, NR_042626 | 92.8 | 2 | 2 | 2 | 0 | 0 | 0 |
| 9 | Devosia chinhatensis, NR_044214 | 96.6 | 0 | 3 | 2 | 4 | 4 | 4 |
| 10 | Kaistia soli, NR_044302 | 96.0 | 0 | 2 | 3 | 0 | 0 | 0 |
| 11 | Magnetospirillum gryphiswaldense, NR_027605 | 96.3 | 1 | 3 | 1 | 0 | 0 | 0 |
| 12 | Bosea eneae, NR_028798 | 95.5 | 4 | 4 | 4 | 0 | 0 | 0 |
| 13 | Rhodobacter blasticus, NR_043735 | 96.3 | 4 | 4 | 4 | 4 | 4 | 4 |
| Pseudomonas DGGE | ||||||||
| 1 | Pseudomonas asplenii, NR_040802 | 99.5 | 0 | 3 | 0 | 0 | 0 | 0 |
| 2 | Pseudomonas tuomuerensis, NR_043990 | 99.1 | 2 | 3 | 2 | 4 | 4 | 4 |
| 3 | Pseudomonas koreensis, NR_025228 | 100 | 3 | 0 | 0 | 0 | 0 | 0 |
| 4 | Pseudomonas jessenii, NR_024918 | 99.3 | 1 | 3 | 3 | 0 | 4 | 3 |
| 5 | Pseudomonas jessenii, NR_024918 | 99.1 | 1 | 1 | 3 | 0 | 0 | 4 |
| 6 | Pseudomonas koreensis, NR_025228 | 99.8 | 3 | 0 | 1 | 0 | 0 | 0 |
| 7 | Pseudomonas taetrolens, NR_036909 | 98.9 | 4 | 4 | 4 | 4 | 4 | 4 |
Details for the BLASTN results and taxonomy are given in the supplemental material. (T), type strain.
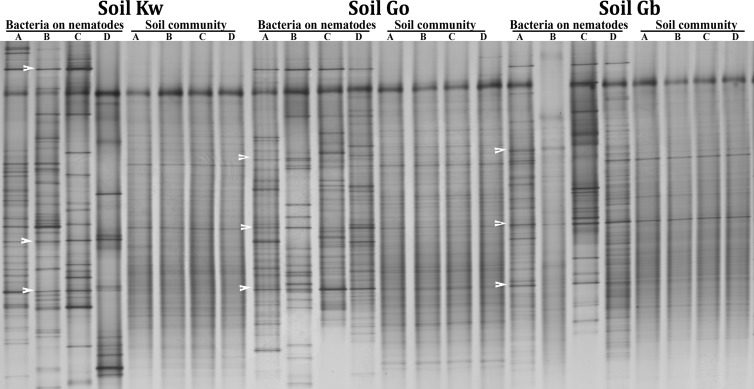
Bacterial attachment to M. hapla in soil.
The bacteria associated with J2 in the three soils were analyzed by PCR-DGGE and 454-pyrosequencing of 16S rRNA genes. DGGE profiles of DNA from J2 showed fewer and more intense bands than those from directly extracted soil DNA, indicating that only a subset of the species in soil were present on the J2 (Fig. 2). The bacterial communities differed among the three soils, as did the communities on the J2 from the three soils. Some bacteria seemed to be attached to the nematodes in all soils. The bacterial community associated with J2 displayed a higher degree of variability than the fungal community structure. In the most suppressive soil, Kw, J2 were most frequently colonized with some highly abundant but variable species, whereas the patterns associated with J2 from the other two soils were more consistent.
FIG 2.
DGGE profiles of bacterial 16S rRNA genes amplified from DNA of M. hapla J2from three arable soils and from total soil DNA. A, B, C, and D refer to replicate soil baiting assays for each soil.
Some bacterial groups that were suspected to interact with root knot nematodes were investigated by DGGE fingerprinting using group-specific 16S rRNA gene primers for Actinobacteriales, Alphaproteobacteria, Betaproteobacteria, Bacillus, Enterobacteriaceae, and Pseudomonas. The fingerprints were highly variable among replicate J2 samples (see Fig. S1 in the supplemental material). Nematode-specific bands representing attachment to J2 in the three soils were mainly detected in DGGE fingerprints generated with primers, which were designed to preferentially target 16S rRNA genes of Alphaproteobacteria, Bacillus, and Pseudomonas. Bacterial 16S rRNA genes amplified based on the selective specificity of primer BacF were most clearly enriched in J2 samples (Table 2). Among them, four intense bands were detected in most J2 samples from all soils (Table 2; see also Fig. S1A, bands 3 to 6, in the supplemental material), of which the sequences belonged to the genera Staphylococcus, Micrococcus, and Bacillus (Table 2). The majority of cloned 16S rRNA genes amplified based on the specificity of primer F203α belonged to the Alphaproteobacteria (Table 2). Despite the high variability of these bacteria from nematode samples, a few bands were dominant on most J2 from the three soils (Table 2; see Fig. S1B in the supplemental material), which were related to Rhizobium phaseoli (99.8% identities) or Bosea sp., respectively. Bacteria from J2 samples that were much more abundant for the most suppressive soil Kw were not apparent, but more intense bands were related to sequences of the actinobacterial species Solirubrobacter soli, and the alphaproteobacterial species Ochrobactrum anthropi and Anderseniella sp. (Table 2). In Pseudomonas-specific DGGE fingerprints, bands related to P. koreensis were most clearly associated with J2 from soil Kw (Table 2, bands 3, 6; see also Fig. S1D in the supplemental material). Other pseudomonads that were relatively more abundant in J2 samples than in the soil samples were similar to P. asplenii, P. tuomuerensis, P. jessenii, or P. taetrolens. DGGE fingerprints from 16S rRNA genes of Actinobacteriales, Betaproteobacteria, and Enterobacteriaceae showed high variability among replicate J2 samples, so that bacteria specifically attached to the nematodes were hardly distinguishable from randomly attached bacteria (see Fig. S1C, E, and F in the supplemental material).
Bacteria on J2 based on 16S rRNA gene amplicon pyrosequencing.
Bacterial 16S rRNA gene sequences from nematode and soil samples were determined by barcoded amplicon pyrosequencing. A total of 22,347 sequences from 12 nematode samples were obtained and analyzed together with sequences from all three bulk soils. The sequences were grouped, based on 97% identity, into 12,425 OTU, of which 87% were unique to soil samples, 9% had a higher relative abundance on J2 than in soil, and 6% were unique to J2 samples. Thus, the diversity of bacterial OTU associated with the J2 in soil was strongly reduced compared to soil. The overlap of abundant OTU between J2 and soil samples was low. The 24 OTU that were most abundant in nematode samples (>1%) but not detected in soil or that were at least 100 times higher in relative abundance on J2 than in soil are shown in Table 3. They mainly belonged to the Alpha-, Beta-, and Gammaproteobacteria, Firmicutes, and Actinobacteria. Nineteen of the OTU had >99% sequence identity with strains of well-studied species, nine of which are associated with infectious diseases (Streptococcus salivarius, Peptoniphilus gorbachii, Mycoplasma wenyonii, Brucella sp., Paracoccus yeei, Neisseria mucosa, Shigella flexneri, Acinetobacter schindleri, and Acinetobacter johnsonii). In the most suppressive soil, Kw, J2 were especially associated with 18 OTU, of which the most abundant OTU were related to the species Rothia amarae, Malikia spinosa, Shigella spp., Janthinobacterium lividum, Geobacillus stearothermophilus, and Pseudomonas kilonensis. Three of the OTU, which were mainly detected on J2 from soil Kw but also on J2 from soil Gb, were closely related to yet-uncultured bacteria of the Gemmatimonadetes, Deltaproteobacteria, or Rhodospirillaceae, respectively.
TABLE 3.
OTU of bacteria that were highly enriched on soil-derived J2 of M. hapla compared to the bacterial community in soil, based on 16S rRNA gene amplicon pyrosequencing
| Most similar cultured species or environmental sequence of the OTU specific for J2 (GenBank accession no., % identity)a | No. of sequences |
||
|---|---|---|---|
| J2 from Kw | J2 from Gb | J2 from Go | |
| Micrococcus yunnanensis (KC469953, 100) | 9 | 21 | 612 |
| Rothia amarae (T) (AY043359, 100) | 835 | 0 | 0 |
| Geobacillus stearothermophilus (T) (AB021196, 99.2) | 394 | 74 | 0 |
| Streptococcus salivarius (T) (AY188352, 100) | 0 | 651 | 0 |
| Anaerococcus octavius (T) (Y07841, 99.2) | 91 | 4 | 177 |
| Peptoniphilus gorbachii (T) (DQ911241, 100) | 118 | 0 | 28 |
| Clostridium disporicum (T) (Y18176, 99.6) | 202 | 3 | 0 |
| Mycoplasma wenyonii (CP003703, 99.7) | 110 | 1 | 3 |
| Uncultured Gemmatimonas in rhizosphere (EU159980, 98.9) | 101 | 1 | 0 |
| Uncultured deltaproteobacterium (HE613616, 100) | 96 | 3 | 0 |
| Ochrobactrum sp./Brucella sp. (AJ242584/AY594216, 99.8) | 147 | 17 | 0 |
| Hirschia maritima (T) (FM202386, 96.0) | 128 | 0 | 0 |
| Haematobacter missouriensis (T) (DQ342315, 100) | 222 | 0 | 0 |
| Paracoccus yeei (T) (AY014173, 100) | 161 | 0 | 0 |
| Uncultured Rhodospirillaceae (GQ263062, 100) | 261 | 5 | 0 |
| Malikia spinosa (AB077038, 98.5) | 962 | 0 | 48 |
| Janthinobacterium lividum (T) (Y08846, 99.8) | 480 | 13 | 0 |
| Neisseria mucosa (HG005351, 99.8) | 104 | 0 | 0 |
| Vogesella indigofera (AB021385, 99.2) | 0 | 421 | 0 |
| Shigella flexneri/S. fergusonii (T) (X96963/AF530475, 100) | 518 | 0 | 109 |
| Acinetobacter schindleri (T) (AJ278311, 99.6) | 0 | 76 | 305 |
| Acinetobacter johnsonii (X81663, 100) | 0 | 229 | 67 |
| Enhydrobacter aerosaccus (T) (AJ550856, 100) | 172 | 3 | 67 |
| Pseudomonas kilonensis (T) (NR_028929, 100) | 281 | 9 | 0 |
| Total sequences | 7,647 | 8,664 | 6,164 |
That is, OTU that comprised >1% of sequences from all J2 or only from Kw-J2 samples and that were not detected in soil or had at least a 100-fold higher relative abundance on J2 compared to soil. Sequence data from soil were obtained from NCBI sequence read archive study accession number SRP029944. Details for the BLASTN results and taxonomy are given in the supplemental material. (T), type strain.
DISCUSSION
This study has revealed by cultivation-independent techniques that diverse microbial communities attached to J2 of M. hapla when they were moving through soil. Several fungal and bacterial types were abundant on J2 but not in the surrounding soil, while other types detectable in soil were highly enriched on J2 relative to other soil microbes. This suggested a specific attachment of these microbes to the cuticle surface of J2. Evidence is gathering that species-specific characteristics of cuticle and surface coat determine microbial attachment to J2 and that the highly glycosylated mucins of the surface coat play a role in specificity (14). Bacterial adhesion changes with genetically determined modification of the complex carbohydrates of the surface coat (23, 24). The Gram-positive obligate parasites of root knot nematodes, Pasteuria spp., are highly host specific in endospore attachment to the cuticle. Thus far, only a few examples for nonparasitic attachment of bacteria or fungi to the cuticle of plant-parasitic nematodes have been described (25, 26), and images of the J2 surface by scanning electron microscopy indicated a rather low abundance of microorganisms with the exception of highly specialized parasites (27). Also, we found evidence for a rather low number of microbes on the cuticle, evidenced by high variation between microbial DGGE fingerprints from J2, and low amounts of direct PCR products from DNA of J2 samples. The importance of the surface coat of the nematode cuticle in the recognition by nematode parasites has been recognized, but studies have focused on highly specialized nematode parasites (28) and more recently on potential human pathogens (29).
In our study, soil suppressiveness to M. hapla was most likely caused by indigenous soil microbes since it was not observed in sterilized controls. In addition, differences in suppressiveness between the three soils investigated corresponded to differences in microbial soil communities and J2 attached microbes, while progenies of M. hapla in the sterilized soils were rather similar or did not correlate with the differences in the soils with indigenous microbial communities. However, some fungi and bacteria were found attached to J2 from all three soils, which therefore have not severely contributed to the differences in suppressiveness between the soils. It cannot be ruled out that some of these common microbes were already associated with the inoculated J2. In previous studies, sensitivity to pasteurization or biocide treatment also provided evidence of the biological nature of soil suppressiveness to plant-parasitic nematodes (4, 30).
For all three soils, the reduction in the numbers of egg masses and eggs was more pronounced than the effect on galling. This observation suggested a mode of action directed against nematode reproduction rather than against J2 vitality or the initial infection by juveniles. We surmised that reduction of reproduction was mediated by microbial attachment to juveniles in soil while searching for host plant roots. This attachment may have resulted in the transport of microbes into the root to the location of egg development. Although no indication of the presence of known parasites became evident, this mode of action points to the involvement of antagonists that get attached to J2 in soil and then reduce the fecundity in females of the target nematode, as reported for Pasteuria penetrans, or egg-parasitic fungi (31, 32). Accordingly, a baiting assay similar to the one we used had been successful in searching for egg parasites of root knot nematodes (33). Transport of cuticle-attached microbes, which are not egg parasites, to the host plant of the nematode has been shown for the phytopathogenic fungus Dilophospora alopecuri adhering to the J2 cuticle of Anguina funesta (34). Other attached microbes may establish as endophytes. Specific endophytes were observed to significantly reduce the progeny of root knot nematodes, probably by indirect mechanisms based on endophyte-plant interactions rather than directly by nematicidal activity (35).
In our study by cultivation-independent methods, we identified bacteria and fungi associated with J2 in soils with different levels of suppressiveness against M. hapla. Two fungi were found on J2 from all tested soils that have been reported as attachments to the nematode surface. A fungus of the genus Rhizophydium was previously reported as attachment to Criconemoides sp. (36), and fungi related to Malassezia restricta have been found in association with the soil nematodes Malenchus sp. and Tylolaimophorus typicus (37). In our study, a fungus related to Cylindrocarpon olidum was only abundant on J2 from the most suppressive soil Kw. Isolates of this genus were shown to reduce the number of galls of M. javanica on tomato roots (38) or to inhibit egg hatch of Meloidogyne spp. by metabolites (39). Cladosporinum cladosporioides, which was only associated with J2 from the Gb soil, was previously found to be associated with Meloidogyne sp. females (40) and with Rotylenchulus reniformis vermiform stages and eggs (12).
Genera or species of the bacterial attachments to J2 from the three soils were also found in association with different plant-parasitic nematodes in previous studies (8, 9, 41, 42). J2 from the most suppressive soil Kw were often associated with OTU similar to species that were reported to be involved in infectious diseases (Mycoplasma wenyonii, Peptoniphilus gorbachii, Brucella sp., Paracoccus yeei, Neisseria mucosa, and Shigella flexneri). These OTU may have in common with their pathogenic relatives that they efficiently attach to tissue surfaces as part of their lifestyle and thereby become enriched on the cuticle of J2. Other J2-enriched OTU were related to soil bacteria such as Rothia amarae, Malikia spinosa, Janthinobacterium lividum, Geobacillus stearothermophilus, or Pseudomonas kilonensis. These bacteria might antagonize M. hapla after cuticle attachment but have not yet been found associated with root knot nematodes. This can be explained by the bias of cultivation approaches which were used in most previous investigations. In a study on the bacterial community associated with cysts of Heterodera glycines, fewer than 5% of the bacteria could be cultured, and there was limited resemblance of the dominant species detected by DGGE analysis and the plating method (9).
In conclusion, a diverse microflora specifically adhered to J2 of M. hapla in soil, which might lead to colonization of eggs and play a role in nematode suppression. Several bacteria and fungi from soil enriched on the baiting J2 extracted from soil reportedly possess some nematicidal properties against plant parasitic nematodes. These should be evaluated for their potential as biocontrol agents. The sequence tags of these microbes could be useful to develop targeted cultivation methods for these species, for cultivation-independent study of the in situ interaction with M. hapla, and to survey their population increase in response to soil treatments. Management of arable soils to increase the abundance of antagonistic bacteria and fungi could become a substantial part in nematode control.
Supplementary Material
ACKNOWLEDGMENTS
This study was funded by a grant to M.A. from the Egyptian government. The Norwegian Research Council provided support (project 1110411), together with the Foundation for Research Levy on Agricultural Products, the Agricultural Agreement Research Fund (project 199604/I99), and the Norwegian Food Safety Authority.
We thank E. Woldt for excellent technical assistance, R. Grosch (IGZ, Grossbeeren, Germany) and W. Joachim (KWS Saat AG, Klein Wanzleben, Germany) for discussions and access to fields, S. Schreiter for soil sampling, and G.-C. Ding for help with the pyrosequencing data.
Footnotes
Published ahead of print 14 February 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.03905-13.
REFERENCES
- 1.Wesemael WML, Viaene N, Moens M. 2011. Root knot nematodes (Meloidogyne spp.) in Europe. Nematology 13:3–16. 10.1163/138855410X526831 [DOI] [Google Scholar]
- 2.Nyczepir AP, Thomas SH. 2009. Current and future management strategies in intensive crop production systems, p 412–443 In Perry RN, Moens M, Starr JL. (ed), Root knot nematodes. CAB International, Wallingford, United Kingdom [Google Scholar]
- 3.Hallmann J, Davies KG, Sikora R. 2009. Biological control using microbial pathogens, endophytes and antagonists, p 380–411 In Perry RN, Moens M, Starr JL. (ed), Root knot nematodes. CAB International, Wallingford, United Kingdom [Google Scholar]
- 4.Bent E, Loffredo A, McKenry MV, Becker JO, Borneman J. 2008. Detection and investigation of soil biological activity against Meloidogyne incognita. J. Nematol. 40:109–118 http://journals.fcla.edu/jon/article/view/67776 [PMC free article] [PubMed] [Google Scholar]
- 5.Orion D, Kritzman G, Meyer SLF, Erbe EF, Chitwood DJ. 2001. A role of the gelatinous matrix in the resistance of root knot nematode (Meloidogyne spp.) eggs to microorganisms. J. Nematol. 33:203–207 http://journals.fcla.edu/jon/article/view/67251 [PMC free article] [PubMed] [Google Scholar]
- 6.Pyrowolakis A, Westphal A, Sikora RA, Becker JO. 2002. Identification of root knot nematode suppressive soils. Appl. Soil Ecol. 19:51–56. 10.1016/S0929-1393(01)00170-6 [DOI] [Google Scholar]
- 7.Stirling GR, Mankau R. 1978. Parasitism of Meloidogyne eggs by a new fungal parasite. J. Nematol. 10:236–240 http://journals.fcla.edu/jon/article/view/65088 [PMC free article] [PubMed] [Google Scholar]
- 8.Tian XL, Cheng XY, Mao ZC, Chen GH, Yang JR, Xie BY. 2011. Composition of bacterial communities associated with a plant-parasitic nematode Bursaphelenchus mucronatus. Curr. Microbiol. 62:117–125. 10.1007/s00284-010-9681-7 [DOI] [PubMed] [Google Scholar]
- 9.Nour SM, Lawrence JR, Zhu H, Swerhone GDW, Welsh M, Welacky TW, Topp E. 2003. Bacteria associated with cysts of the soybean cyst nematode (Heterodera glycines). Appl. Environ. Microbiol. 69:607–615. 10.1128/AEM.69.1.607-615.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yin B, Valinsky L, Gao XB, Becker JO, Borneman J. 2003. Bacterial rRNA genes associated with soil suppressiveness against the plant-parasitic nematode Heterodera schachtii. Appl. Environ. Microbiol. 69:1573–1580. 10.1128/AEM.69.3.1573-1580.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yin B, Valinsky L, Gao X, Becker JO, Borneman J. 2003. Identification of fungal rDNA associated with soil suppressiveness against Heterodera schachtii using oligonucleotide fingerprinting. Phytopathology 93:1006–1013. 10.1094/PHYTO.2003.93.8.1006 [DOI] [PubMed] [Google Scholar]
- 12.Castillo JD, Lawrence KS, Morgan-Jones G, Ramirez CA. 2010. Identification of fungi associated with Rotylenchulus reniformis. J. Nematol. 42:313–318 http://journals.fcla.edu/jon/article/view/78286 [PMC free article] [PubMed] [Google Scholar]
- 13.Papert A, Kok CJ, van Elsas JD. 2004. Physiological and DNA fingerprinting of the bacterial community of Meloidogyne fallax egg masses. Soil Biol. Biochem. 36:1843–1849. 10.1016/j.soilbio.2004.04.038 [DOI] [Google Scholar]
- 14.Davies KG, Curtis RHC. 2011. Cuticle surface coat of plant-parasitic nematodes. Annu. Rev. Phytopathol. 49:135–156. 10.1146/annurev-phyto-121310-111406 [DOI] [PubMed] [Google Scholar]
- 15.Stirling GR. 1991. Biological control of nematodes: progress, problems and prospects. CAB International, Wallingford, United Kingdom [Google Scholar]
- 16.Rühlmann J. 2006. The box plot experiment in Grossbeeren after six rotations: effect of fertilization on crop yield. Arch. Agron. Soil Sci. 52:313–319. 10.1080/03650340600638701 [DOI] [Google Scholar]
- 17.Hooper DJ, Hallmann J, Subbotin S. 2005. Methods for extraction, processing and detection of plant and soil nematodes, p 53–86 In Luc M, Sikora RA, Bridge J. (ed), Plant parasitic nematodes in subtropical and tropical agriculture, 2nd ed. CAB International, Wallingford, United Kingdom [Google Scholar]
- 18.Weinert N, Meincke R, Gottwald C, Heuer H, Gomes NC, Schloter M, Berg G, Smalla K. 2009. Rhizosphere communities of genetically modified zeaxanthin-accumulating potato plants and their parent cultivar differ less than those of different potato cultivars. Appl. Environ. Microbiol. 75:3859–3865. 10.1128/AEM.00414-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Heuer H, Kopmann C, Binh CTT, Top Smalla EMK. 2009. Spreading antibiotic resistance through spread manure: characteristics of a novel plasmid type with low %G+C content. Environ. Microbiol. 11:937–949. 10.1111/j.1462-2920.2008.01819.x [DOI] [PubMed] [Google Scholar]
- 20.Ding GC, Heuer H, Smalla K. 2012. Dynamics of bacterial communities in two unpolluted soils after spiking with phenanthrene: soil type specific and common responders. Front. Microbiol. 3:290. 10.3389/fmicb.2012.00290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, Peplies J, Glöckner FO. 2013. The SILVA rRNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 41:D590–D596. 10.1093/nar/gks1219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ, Sahl JW, Stres B, Thallinger GG, Van Horn DJ, Weber CF. 2009. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 75:7537–7541. 10.1128/AEM.01541-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gravato-Nobre MJ, Stroud D, O'Rourke D, Darby C, Hodgkin J. 2011. Glycosylation genes expressed in seam cells determine complex surface properties and bacterial adhesion to the cuticle of Caenorhabditis elegans. Genetics 187:141–155. 10.1534/genetics.110.122002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Davies KG, Rowe JA, Williamson VM. 2008. Inter- and intra-specific cuticle variation between amphimictic and parthenogenetic species of root knot nematode (Meloidogyne spp.) as revealed by a bacterial parasite (Pasteuria penetrans). Int. J. Parasitol. 38:851–859. 10.1016/j.ijpara.2007.11.007 [DOI] [PubMed] [Google Scholar]
- 25.Bird AF. 2004. Surface adhesion to nematodes and its consequences, p 295–329 In Chen ZX, Chen SY, Dickson DW. (ed), Nematology: advances and perspectives, vol 1 CAB International, Wallingford, United Kingdom [Google Scholar]
- 26.Hallmann J, Quadt-Hallmann A, Rodriguez-Kabana R, Kloepper JW. 1998. Interactions between Meloidogyne incognita and endophytic bacteria in cotton and cucumber. Soil Biol. Biochem. 30:925–937. 10.1016/S0038-0717(97)00183-1 [DOI] [Google Scholar]
- 27.Sayre RM, Wergin WP. 1977. Bacterial parasite of a plant nematode: morphology and ultrastructure. J. Bacteriol. 129:1091–1101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Curtis RHC, Jones JT, Davies KG, Sharon E, Spiegel Y. 2011. Plant nematode surfaces, p 115–144 In Davies KG, Spiegel Y. (ed), Biological control of plant-parasitic nematodes: building coherence between microbial ecology and molecular mechanisms. Springer, New York, NY [Google Scholar]
- 29.Maghodia AB, Spiegel Y, Sela S. 2008. Interactions between Escherichia coli and the plant-parasitic nematode Meloidogyne javanica. J. Appl. Microbiol. 105:1810–1816. 10.1111/j.1365-2672.2008.03945.x [DOI] [PubMed] [Google Scholar]
- 30.Westphal A. 2005. Detection and description of soils with specific nematode suppressiveness. J. Nematol. 37:121–132 http://journals.fcla.edu/jon/article/view/67544 [PMC free article] [PubMed] [Google Scholar]
- 31.Weibelzahl-Fulton E, Dickson DW, Whitty EB. 1996. Suppression of Meloidogyne incognita and M. javanica by Pasteuria penetrans in field soil. J. Nematol. 28:43–49 http://journals.fcla.edu/jon/article/view/66791 [PMC free article] [PubMed] [Google Scholar]
- 32.Kerry B. 1988. Fungal parasites of cyst nematodes. Agr. Ecosyst. Environ. 24:293–305. 10.1016/0167-8809(88)90073-4 [DOI] [Google Scholar]
- 33.Stirling GR, White AM. 1982. Distribution of a parasite of root knot nematodes in South-Australian vineyards. Plant Dis. 66:52–53. 10.1094/PD-66-52 [DOI] [Google Scholar]
- 34.Atanasoff D. 1925. The Dilophospora desease of cereals. Phytopathology 15:11–40 [Google Scholar]
- 35.Sikora RA, Pocasangre L, Felde A, Niere B, Vu TT, Dababat AA. 2008. Mutualistic endophytic fungi and in-planta suppressiveness to plant parasitic nematodes. Biol. Control. 46:15–23. 10.1016/j.biocontrol.2008.02.011 [DOI] [Google Scholar]
- 36.Esser RP, Schubert TS. 1983. Fungi that utilize zoospores to parasitize nematodes. Nematol. Circular 101:1–4 [Google Scholar]
- 37.Renker C, Alphei J, Buscot F. 2003. Soil nematodes associated with the mammal pathogenic fungal genus Malassezia (Basidiomycota: Ustilaginomycetes) in Central European forests. Biol. Fertil. Soils 37:70–72. 10.1007/s00374-002-0556-3 [DOI] [Google Scholar]
- 38.Freitas LG, Ferraz S, Muchovej JJ. 1995. Effectiveness of different isolates of Paecilomyces lilacinus and an isolate of Cylindrocarpon destructans on the control of Meloidogyne javanica. Nematropica 25:109–115 http://journals.fcla.edu/nematropica/article/view/64128 [Google Scholar]
- 39.Meyer SLF, Huettel RN, Liu XZ, Humber RA, Juba J, Nitao JK. 2004. Activity of fungal culture filtrates against soybean cyst nematode and root knot nematode egg hatch and juvenile motility. Nematology 6:23–32. 10.1163/156854104323072883 [DOI] [Google Scholar]
- 40.Amer-Zareen Imran AM, Zaki MJ. 2000. Fungal parasites of root knot nematodes. Pakistan J. Biol. Sci. 3:478–480. 10.3923/pjbs.2000.478.480 [DOI] [Google Scholar]
- 41.Proença DN, Francisco R, Santos CV, Lopes A, Fonseca L, Abrantes IMO, Morais PV. 2010. Diversity of bacteria associated with Bursaphelenchus xylophilus and other nematodes isolated from Pinus pinaster trees with pine wilt disease. PLoS One 5:e15191. 10.1371/journal.pone.0015191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stirling GR, Wachtel MF. 1980. Mass-production of Bacillus penetrans for the biological-control of root knot nematodes. Nematologica 26:308–312. 10.1163/187529280X00260 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.