Abstract
Deinococcus radiodurans RNA ligase (DraRnl) is the founding member of a family of end-joining enzymes encoded by diverse microbes and viruses. DraRnl ligates 3′-OH, 5′-PO4 nicks in double-stranded nucleic acids in which the nick 3′-OH end is RNA. Here we gauge the effects of 3′-OH and 5′-PO4 base mispairs and damaged base lesions on the rate of nick sealing. DraRnl is indifferent to the identity of the 3′-OH nucleobase, provided that it is correctly paired. With 3′-OH mispairs, the DraRnl sealing rate varies widely, with G-T and A-C mispairs being the best substrates and G-G, G-A, and A-A mispairs being the worst. DraRnl accepts 3′ A–8-oxoguanine (oxoG) to be correctly paired, while it discriminates against U-oxoG and G-oxoG mispairs. DraRnl displays high activity and low fidelity in sealing 3′-OH ends opposite an 8-oxoadenine lesion. It prefers 3′-OH adenosine when sealing opposite an abasic template site. With 5′-PO4 mispairs, DraRnl seals a 5′ T-G mispair as well as it does a 5′ C-G pair; in most other respects, the ligation fidelity at 5′ mispairs is similar to that at 3′ mispairs. DraRnl accepts a 5′ A-oxoG end to be correctly paired, yet it is more tolerant of 5′ T-oxoG and 5′ G-oxoG mispairs than the equivalent configurations on the 3′ side of the nick. At 5′ nucleobase-abasic site nicks, DraRnl prefers to ligate when the nucleobase is a purine. The biochemical properties of DraRnl are compatible with its participation in the templated repair of RNA damage or in the sealing of filled DNA gaps that have a 3′ ribopatch.
INTRODUCTION
Ionizing radiation (IR) damages nucleic acids via reactive hydroxyl radicals, generated by the radiolysis of water, that chemically alter the nucleobases and sugar-phosphate backbone. Closely spaced lesions on opposing strands of duplex DNA can lead to lethal double-strand breaks. Cells and viruses deploy multiple enzymatic pathways to recognize and repair radiation-damaged DNA. The final step in the repair cascade is the restoration of the phosphodiester backbone by DNA ligase, an enzyme that seals 3′-OH and 5′-PO4 ends (1, 2).
RNA is also susceptible to base and backbone damage by IR and oxidative stress. Hydroxyl radicals and the bleomycin class of anticancer drugs cleave RNA in vitro and in vivo (3–6). Oxidative damage triggered by exposure of cells to hydrogen peroxide generates 8-oxopurine nucleobases in RNA, principally, 8-oxoguanine (oxoG) (7). Accumulation of oxoG in cellular RNA is a feature of human neurodegenerative syndromes, such as Alzheimer's disease, parkinsonism, and amyotrophic lateral sclerosis (8, 9). It is envisioned that damaged RNAs are degraded (9–11). However, it may be advantageous at times for cells and viruses to repair broken RNAs, rather than simply resynthesize them, in order to rapidly restore the necessary templates and machinery for protein synthesis. Enzymes capable of healing and sealing broken RNA ends are widely prevalent in bacterial, archaeal, and eukaryal taxa and are also encoded by bacterial and eukaryal viruses (12–25). It has been suggested that RNA repair is a general mechanism to circumvent or recover from damage inflicted by site-specific ribotoxin endoribonucleases that trigger cell death or arrest cell growth (26–29). It is not yet clear whether RNA repair is also a mechanism to evade radiation-induced damage to the RNA backbone.
RNA ligases are the central agents of RNA repair. Classic ATP-dependent RNA ligases seal broken RNAs with 5′-PO4 and 3′-OH ends. Three distinct nucleotidyl transfer steps define this reaction: (i) RNA ligase reacts with ATP to generate a covalent enzyme–(lysyl-Nζ)-AMP intermediate, (ii) AMP is transferred to the 5′-PO4 end of the RNA to form 5′-adenylated RNA (AppRNA), and (iii) ligase catalyzes the attack of the 3′-OH end on the AppRNA intermediate to form a 3′-5′ phosphodiester and release AMP (30). An alternative sealing pathway is catalyzed by the RtcB family of RNA ligases, whereby broken RNAs with 3′-PO4 and 5′-OH ends are joined in a GTP-dependent reaction via covalent enzyme–(histidinyl-Nε)-GMP and RNA3′ ppG intermediates (31–33).
The radiation-resistant bacterium Deinococcus radiodurans encodes an ATP-dependent RNA ligase, DraRnl, with a distinctive biochemical specificity and domain architecture (34, 35). DraRnl is a monomeric 342-amino-acid protein composed of an adenylyltransferase domain (common to all classic polynucleotide ligases) and a signature N-terminal module, a putative OB fold. DraRnl seals 3′-OH, 5′-PO4 nicks in duplex RNAs, and it requires Mn2+ for activity. DraRnl has a strict requirement for RNA in the 3′-OH strand of the nick but is nonselective in vitro for DNA versus RNA in the 5′-PO4 strand or the template strand. The adenylyltransferase domain of DraRnl is closely related to that of other RNA ligases that seal duplex nicks, specifically, bacteriophage T4 RNA ligase 2 (Rnl2) and the RNA-editing ligases (RELs) of kinetoplastid protozoa Trypanosoma and Leishmania (36, 37).
Although nick-sealing RNA ligases are found in diverse taxa and viruses, their biological functions have been established only for the RELs, which are agents of mitochondrial mRNA editing (38–40). In this process, guide RNA (gRNA) templates anneal to the pre-mRNA via canonical Watson-Crick interactions and also G-U pairs to form a pre-mRNA–gRNA duplex with a bulge at the editing site. The pre-mRNA–gRNA duplex is endonucleolytically cleaved in the pre-mRNA strand to form a 3′-OH, 5′-PO4 break. When the extrahelical segment is in the gRNA, cleavage creates a gapped duplex, to which a uridylyltransferase enzyme adds UMP nucleotides to the 3′-OH break end, across from purines in the gRNA, until the gap is filled. When the unpaired bulge is on the pre-mRNA strand, cleavage generates a 3′-OH flap from which UMPs are resected until the 3′-OH strand can pair as a duplex nick with the gRNA. In both contexts, the RELs seal the resulting nicks. Iterative gRNA-directed editing of the pre-mRNA at multiple sites eventually establishes the proper translational reading frame. We can speculate that the oxidative environment of the kinetoplast mitochondria might damage gRNA templates, by converting purines to 8-oxopurines. Oxidized gRNAs could engender mutagenic RNA editing if nucleotides are misincorporated opposite 8-oxopurines in the gRNA during gap filling and the mispaired 3′-OH N-oxopurine nicks are sealed by RELs. Recent studies of bacteriophage T4 Rnl2, a homolog of the RELs, highlight its ability to seal, in high yield, nicks with mispaired 3′-OH ends, including 3′ A-oxoG ends (41).
Nothing is known as yet about the biological functions of DraRnl, which exemplifies a distinct ligase clade. Therefore, we sought insights from in vitro studies of DraRnl's catalytic repertoire and substrate preferences to suggest plausible roles in vivo. Because the Deinococcus operon that encodes DraRnl is transcriptionally upregulated after exposure of bacteria to ionizing radiation (42), we naturally invoked a role for the ligase activity in nucleic acid repair, presumably in a pathway involving nicked duplexes with 3′-OH RNA ends. In the study described here, we investigated the fidelity of DraRnl in nick ligation and its ability to contend with damaged base lesions at the nick. We interrogated the effect of base pair identity at the nick 3′-OH and 5′-PO4 ends and the impact of all base mispairs on nick sealing. We also gauged the effects of abasic sites and oxopurines in the template strand opposite the nick 3′-OH and 5′-PO4 nucleotides.
MATERIALS AND METHODS
Recombinant DraRnl.
Plasmid pET28b-His10Smt3-DraRnl encodes DraRnl fused to an N-terminal His10Smt3 domain under the transcriptional control of a bacteriophage T7 RNA polymerase promoter. Cultures (8 liters) of Escherichia coli BL21(DE3)/pET28b-His10Smt3-DraRnl cells were grown at 37°C in terrific broth medium supplemented with 30 μg/ml kanamycin until the A600 reached 0.6. The cultures were chilled on ice for 30 min, adjusted to 0.25 mM IPTG (isopropyl-β-d-thiogalactopyranoside) and 2% (vol/vol) ethanol, and then incubated for 17 h at 17°C with continuous shaking. Cells were harvested by centrifugation, and the pellets were stored at −20°C. All subsequent procedures were performed at 4°C. Thawed cells were resuspended in 250 ml of buffer A (50 mM Tris-HCl, pH 8.0, 0.5 M NaCl, 20 mM imidazole) containing 10% sucrose and then supplemented with lysozyme (1 mg/ml), phenylmethylsulfonyl fluoride (0.1 M), four protease inhibitor tablets (cOmplete, Mini, EDTA free; Roche), and Triton X-100 (0.1%). After incubation for 1 h, the lysate was sonicated to reduce viscosity. Insoluble material was removed by centrifugation at 14,000 rpm for 45 min, and the soluble extract was applied to an 8-ml column of His60 Ni Superflow resin (Clontech). The column was serially washed with buffers containing 20 mM imidazole, 50 mM imidazole, and then 3 M KCl. Bound material was then eluted stepwise with 100 mM, 200 mM, and 500 mM imidazole in 50 mM Tris-HCl (pH 8.0), 0.5 M NaCl. The elution profile was monitored by SDS-PAGE. The His10Smt3-DraRnl protein was primarily recovered in the 200 mM imidazole fraction. The N-terminal His10Smt3 tag was removed by cleavage with Ulp1 (an Smt3-specific protease; 1:100 ratio of Ulp1 to His10Smt3-DraRnl) during overnight dialysis against 50 mM Tris-HCl (pH 8.0), 250 mM NaCl, 10 mM imidazole. The dialysate was applied to a 3.5-ml His60 Ni column equilibrated with 50 mM Tris-HCl (pH 8.0), 250 mM NaCl, 10 mM imidazole. Tag-free DraRnl was recovered in the flowthrough. The DraRnl preparation was concentrated by centrifugal ultrafiltration (10,000-molecular-weight cutoff; Amicon) and then applied to a 120-ml S200 gel filtration column (GE Healthcare) equilibrated with 50 mM Tris-HCl (pH 8.0), 500 mM NaCl, 5 mM dithiothreitol (DTT), 2 mM EDTA, 10% glycerol. The peak DraRnl fractions were pooled, concentrated to 1.5 mg/ml by centrifugal ultrafiltration, and stored at −80°C. The protein concentration was determined with Bio-Rad dye-binding reagent using bovine serum albumin as a standard.
Singly nicked duplex substrates.
The substrates employed were singly nicked duplexes composed of an RNAOH strand and a 5′-phosphate DNA (pDNA) strand (labeled with 32P at the nick 5′-PO4) annealed to a 36-mer DNA template strand. RNA oligonucleotides were purchased from Dharmacon and deprotected according to the manufacturer's instructions. DNA oligonucleotides, including those with chemical modifications (8-oxoguanine, 8-oxoadenine [oxoA], or tetrahydrofuran [THF] abasic nucleosides), were purchased from Eurofins MWG Operon. The 5′ 32P-labeled DNA oligonucleotides were prepared by label transfer from [γ-32P]ATP catalyzed by bacteriophage T4 polynucleotide kinase and then purified free of ATP by electrophoresis through a nondenaturing 18% polyacrylamide gel. To form the nicked duplexes, the radiolabeled pDNA strand, RNAOH strand, and DNA template strand were annealed at a 1:5:2 molar ratio in 150 mM NaCl, 10 mM Tris-HCl (pH 8.0), 1 mM EDTA by incubation for 10 min at 65°C, 15 min at 37°C, and then 30 min at 22°C.
Nick-sealing assay.
Reaction mixtures (10 μl) containing 50 mM Tris-acetate, pH 6.5, 5 mM DTT, 10 mM MnCl2, 100 nM (1 pmol) 5′ 32P-labeled nicked duplex substrate, and 1 μM (10 pmol) DraRnl were incubated at 22°C. The reactions were initiated by the addition of DraRnl and quenched after 0.5, 1, 2, 5, 10, and 15 min by adding 15 μl of 80% formamide, 100 mM EDTA, 0.025% xylene cyanol, 0.025% bromophenol blue. The samples were heated for 5 min at 95°C and then analyzed by electrophoresis through an 18% polyacrylamide gel containing 8 M urea in 45 mM Tris-borate, 1 mM EDTA, which separated the 32P-labeled RNApDNA product from the pDNA strand. The extents of ligation (RNApDNA/[RNApDNA + pDNA]) were quantified by scanning the gel with a Fujix BAS2500 imager.
RESULTS
Sealing of nicked duplexes with correctly paired 3′-OH and 5′-PO4 ends.
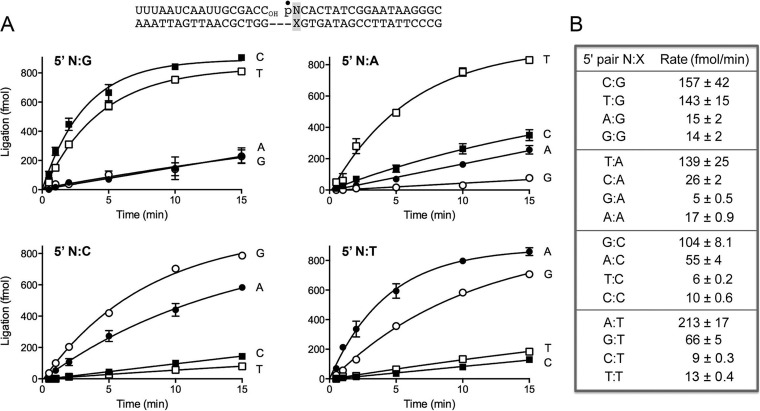
We analyzed the rate and extent of nick sealing by DraRnl under conditions of enzyme excess, varying the identity of the nucleobase (N)-X base pair (where X indicates any nucleobase) at the nick 3′-OH end as U-A, A-T, G-C, or C-G. The substrates employed were singly nicked duplexes composed of an 18-mer RNAOH strand and an 18-mer pDNA strand (labeled with 32P at the nick 5′-PO4) annealed to a 36-mer DNA template strand (Fig. 1A). The nicked duplexes (100 nM) were reacted with a 10-fold molar excess of DraRnl (1 μM) in the presence of 10 mM Mn2+ and 0.2 mM ATP. The reactions were quenched with EDTA and formamide after 0.5, 1, 2, 5, 10, and 15 min, and the products were analyzed by urea-PAGE, which separated the radiolabeled 18-mer pDNA strand and the sealed 36-mer RNApDNA strand. The kinetic profiles are shown in Fig. 1A, where each datum is the average of at least three separate time course experiments ± standard error of the mean (SEM). The sealed products accumulated steadily with time, attaining an endpoint at 10 to 15 min and with 80 to 85% of the substrate being ligated. The initial rates of sealing of the 3′ C-G, U-A, G-C, and A-T nicks, calculated by linear regression in Prism software, were 198, 173, 212, and 195 fmol/min, respectively (Fig. 1B). We conclude that DraRnl does not kinetically discriminate between paired 3′-OH ends with different 3′-OH nucleobases under the reaction conditions employed.
FIG 1.
Ligation of nicked duplexes with paired and mispaired 3′-OH ends. (A) The structure of the nicked duplex substrate is shown at the top, with the 5′ 32P label at the nick denoted by a closed circle. The 3′-OH strand is all RNA. The 3′ N-X base pair at the nick (highlighted in the shaded box) is variable. Ligase reactions were performed as described in Materials and Methods. The yields of the 36-mer RNApDNA ligation product are plotted as a function of time; the experiments are presented in sets of four according to the identity of the X nucleobase, with the N nucleobase (specified to the right of each curve) being the variable within the set. Each datum is the average of at least 3 separate experiments ± SEM. In this and all subsequent figures, 1,000 fmol represents 100% ligation of the input 32P-labeled strand. (B) The initial rates of sealing of the indicated 3′ N-X nicks were calculated by linear regression in Prism.
To vary the identity of the 5′-PO4 N-X base pair at the nick, a 17-mer RNAOH strand and one of four 19-mer pDNA strands with a 5′ 32P-labeled G, A, T, or C nucleotide were annealed to 36-mer DNA template strands to form the nicked duplexes shown in Fig. 2A (which, except for the 1-nucleotide leftward shift in the position of the nick, were identical in nucleobase sequence to the substrate used in Fig. 1). Whereas the yields of sealed product did not differ according to the identity of the 5′-PO4 base pair (Fig. 2A), the initial rates of sealing of the 5′-PO4 C-G, U-A, G-C, and A-T nicks were 157, 139, 104, and 213 fmol/min, respectively (Fig. 2B). Thus, the rate varies within a 2-fold range according to the identity of the paired 5′-PO4 nucleobase.
FIG 2.
Ligation of nicked duplexes with paired and mispaired 5′-PO4 ends. (A) The structure of the nicked duplex substrate is shown at the top, with the 5′ 32P label at the nick denoted by a closed circle. The 3′-OH strand is all RNA. The 5′ N-X base pair at the nick (highlighted in the shaded box) is variable. Ligase reactions were performed as described in Materials and Methods. The yields of the sealed 36-mer RNApDNA product are plotted as a function of time; the experiments are presented in sets of four according to the identity of the X nucleobase, with the N nucleobase (specified to the right of each curve) being the variable within the set. Each datum is the average of at least 3 separate experiments ± SEM. (B) The initial rates of sealing of the indicated 5′ N-X nicks were calculated by linear regression in Prism.
Effect of 3′-OH base mispairs on the rate of nick sealing.
The fidelity of strand joining refers to the degree to which ligases discriminate in the sealing of substrates with paired versus mispaired bases at the nick. Here we evaluated the fidelity of DraRnl, focusing first on the effects of all 3′-OH base mispairs on the rate of nick sealing under conditions of enzyme excess. The kinetic profiles are shown in Fig. 1A, and the initial rates are compiled in Fig. 1B and grouped as N-G, N-A, N-C, and N-T subsets, to allow easy comparison of the correct versus mispaired N-X nicks for each X template base. We found that DraRnl was especially sensitive to 3′ N-purine mispairs. The rates of sealing of 3′ C-A, G-A, and A-A nicks were 19-fold, 87-fold, and 87-fold lower, respectively, than the rate of ligation of the correctly paired 3′ U-A nick (Fig. 1B). The rates of joining of mispaired 3′ U-G, A-G, and G-G nicks were 10-fold, 20-fold, and 66-fold lower than the rate of sealing of the correctly paired 3′ C-G nick.
The 3′ purine-pyrimidine mispaired ends 3′ A-C and 3′ G-T were the best of the mispaired substrates for DraRnl, being sealed, respectively, at one-fifth and one-third the rates of the correctly paired 3′ G-C and A-T nicks (Fig. 1A and B). The 3′ pyrimidine-pyrimidine mispairs were poorly tolerated. The mispaired U-C and C-C nicks were sealed 16-fold and 30-fold slower, respectively, than the paired 3′ G-C substrate. The mispaired 3′ C-T and U-T ends were ligated 10-fold and 22-fold slower, respectively, than the paired 3′ A-T end (Fig. 1B). These results indicate that DraRnl fidelity in sealing nicks with 3′ mispairs varies over an ∼30-fold range. Within the series of 3′ N-purine and 3′ N-pyrimidine mispaired nicks, the U-G and G-T wobble pairs were sealed best by DraRnl, a noteworthy trend given the common occurrence of wobble pairs in RNA duplexes (43).
Effect of 5′-PO4 base mispairs on the rate of nick sealing.
We next turned our attention to the fidelity of DraRnl in sealing nicks with 5′-PO4 mispairs (Fig. 2A). The kinetic profiles are shown in Fig. 2A, and the initial rates are listed in Fig. 1B, again grouped as N-G, N-A, N-C and N-T subsets. The effects of the 5′-PO4 mispairs on ligation rates were generally concordant with the mispair effects seen at the 3′-OH end, with a few notable exceptions. For example, the 5′ T-G wobble pair end was sealed at virtually the same rate as the 5′ C-G end (143 versus 157 fmol/min); in contrast, the 3′ U-G wobble pair slowed ligation 10-fold. The T-G wobble was the best substrate among all the 5′ N-X mispair configurations (followed by the reciprocal G-T wobble end, which was sealed at 66 fmol/min). The rates of joining of mispaired 5′ A-G and G-G nicks were 10-fold and 11-fold lower, respectively, than the rates of sealing of the correctly paired 3′ C-G nick. The 5′ C-A, G-A, and A-A nicks were ligated 5-fold, 28-fold, and 8-fold slower, respectively, than the correctly paired 5′ T-A nick (Fig. 2B).
Among the 5′ N-pyrimidine mispairs, the 5′ A-C and G-T ends were the best substrates, being sealed, respectively, at one-half and one-third the rates for the correctly paired 5′ G-C and A-T nicks (Fig. 1A and B). The 5′ pyrimidine-pyrimidine mispairs were poorly ligated. The 5′ T-C and C-C ends were sealed 17-fold and 10-fold slower, respectively, than the paired 5′ G-C end. The 5′ T-T and C-T nicks were repaired 16-fold and 23-fold slower, respectively, than the paired 5′ A-T end (Fig. 2B).
Taken together, the 3′ and 5′ mispair effects highlight a distinctive property of DraRnl compared to nick-sealing DNA ligases. DraRnl displays similar patterns of fidelity on the 3′-OH and 5′-PO4 sides of the nick. This is best exemplified by the very similar rates at which the equivalent 3′-OH N-T and 5′-PO4 N-T nicks are sealed by DraRnl (compare the bottom panels in Fig. 1B and 2B). In contrast, DNA ligases are generally biased in their fidelity of mispair sealing, being highly sensitive to 3′-OH mispairs and comparatively tolerant of 5′-PO4 mispairs (44–54).
Effect of 8-oxopurines in the template strand at the nick 3′-OH end.
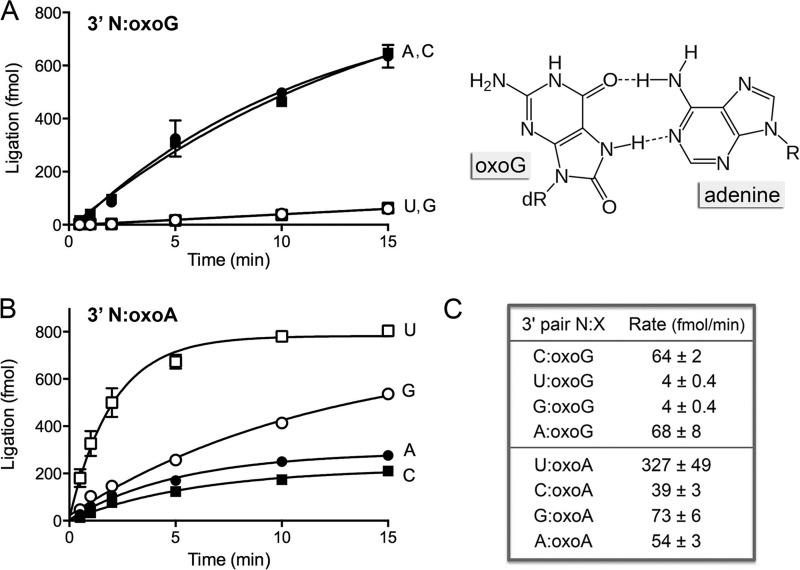
Oxidative damage to purine nucleobases generates 8-oxoguanine (oxoG) and 8-oxoadenine (oxoA). The effects and metabolism of oxoG in DNA have been studied in depth because of its mutagenic properties, which stem from the tendency of oxoG in the syn conformation to mispair with adenine (Fig. 3). This pairing prompts DNA polymerases to embed A-oxoG mispairs, which can ultimately lead to G → T transversions. RNA is also prone to oxidative base damage. The frequency of oxoG lesions in total RNA is reported to be higher than that in total DNA, and several studies have correlated elevated RNA oxoG levels with human neurodegenerative diseases (7–10). Kinetoplastid mRNA editing occurs in the oxidative milieu of the mitochondrion, where the gRNA purine-tract template for U insertion might acquire oxopurine lesions that could affect the efficiency and fidelity of RNA gap filling and thereby impact the efficiency and outcome of the ligation reaction performed by RELs. Were ligases to seal mispairs at 8-oxopurines in the template strand, this would be promutagenic and might result in misediting of kinetoplastid mRNAs. There have been reports of oxopurine effects on bacteriophage T4 and human DNA ligases (55, 56) and on bacteriophage T4 Rnl2 (41). Here we investigated the effect of oxopurine lesions in the template strand opposite the nick termini on the kinetics of nick sealing by DraRnl.
FIG 3.
Effect of 8-oxopurines in the template strand at the nick 3′-OH end. (A) The substrates for sealing were versions of the nicked duplex shown in Fig. 1A, in which the X nucleobase of the 3′-OH N-X end was either oxoG (A) or oxoA (B). The chemical structure of the A-oxoG pair with oxoG in the syn conformation is shown at the right in panel A; hydrogen-bonding contacts are indicated by dashed lines. dR, deoxyribose. Ligase reactions were performed as described in Materials and Methods. The yields of the sealed 36-mer RNApDNA product are plotted as a function of time; the experiments are presented in sets of four, with the N nucleobase (specified to the right of each curve) being the variable within the set. Each datum is the average of at least 3 separate experiments ± SEM. (C) The initial rates of sealing of the indicated 3′ N-oxopurine nicks were calculated by linear regression in Prism.
We first addressed the kinetics of sealing of nicks with all combinations of 3′-OH N-oxoG and N-oxoA base pairs (Fig. 3), with an eye toward comparing the rates of sealing these damaged substrates to the rates for the corresponding undamaged N-G and N-A nicks. The rate of 3′ C-oxoG nick ligation (64 fmol/min) was one-third the rate of sealing a 3′ C-G nick (198 fmol/min). On the other hand, the 3′ U-oxoA nick was ligated twice as fast as the 3′ U-A nick (327 fmol/min versus 173 fmol/min). The reasons for the disparate impact of the 8-oxo atom in the context of a 3′ C-oxoG pair versus a 3′ U-oxoA pair are elusive. Nonetheless, the instructive finding was that the rate of sealing of the 3′ A-oxoG nick (68 fmol/min) was virtually equivalent to that of the 3′ C-oxoG nick (Fig. 3A and C) and was 7-fold faster than the rate of sealing of an unmodified 3′ A-G mispaired nick (10 fmol/min; Fig. 1B). Thus, DraRnl readily accepted the 3′ base pair between syn oxoG and adenine as a correct pair. In contrast, the 3′ G-oxoG and U-oxoG ends were treated as mispairs and were sealed 16-fold more slowly (4 fmol/min) than the C-oxoG end (Fig. 3A and C).
The salutary effect of the oxoA template nucleotide across from the nick 3′-OH extended to the 3′ C-oxoA, G-oxoA, and A-oxoA mispaired nicks, for which the initial rates of nick sealing (39, 73, and 54 fmol/min, respectively; Fig. 3B and C) were much higher than those seen with the unmodified 3′ C-A, G-A, and A-A mispairs (Fig. 1B). Thus, the effect of the template oxoA is to relax the fidelity of DraRnl at a 3′ N-A mispair.
Effect of 8-oxopurines in the template strand at the nick 5′-PO4 end.
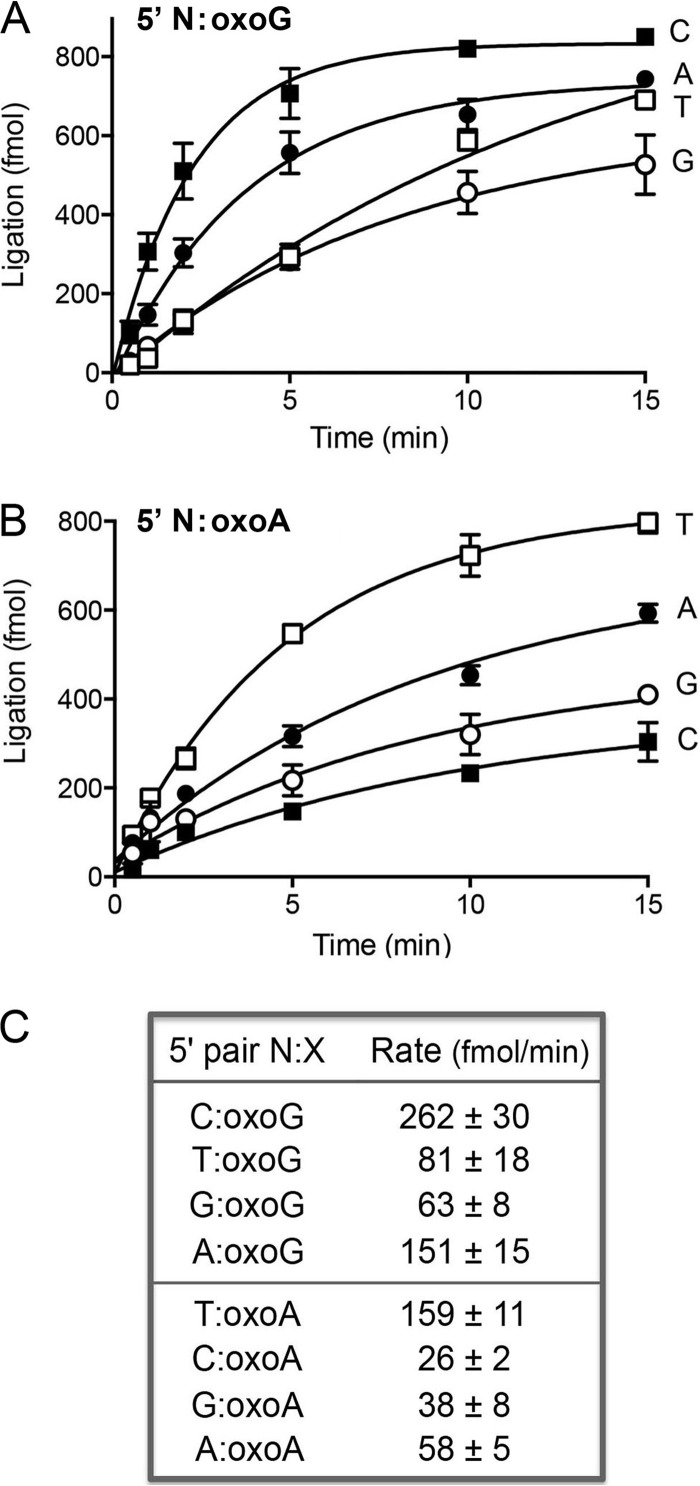
We proceeded to test the effects of template strand oxopurines opposite the 5′-PO4 nucleotide of the nick (Fig. 4). Of note, the rate of sealing of the modified 5′ C-oxoG paired end (262 fmol/min) was 1.7-fold higher than the rate of ligation of an unmodified 5′ C-G pair (157 fmol/min). Thus, a C-oxoG pair exerted a very different influence on ligation kinetics when situated on the 3′-OH side of the nick (a 3-fold rate decrement) than when situated on the 5′-PO4 side (a modest rate increase). The rate of sealing of a 5′ A-oxoG end (151 fmol/min; Fig. 4) was 10-fold higher than that of an unmodified A-G nick (15 fmol/min; Fig. 2) and ∼60% as high as that of a 5′ C-oxoG end, signifying that DraRnl sees the 5′ base pair between syn oxoG and adenine as a correct pair. Whereas the rate of sealing of the 5′ T-oxoG nick (81 fmol/min; Fig. 4) was ∼60% of that of the unmodified 5′ T-G wobble pair (143 fmol/min), the 5′ G-oxoG nick was ligated 4.5-fold faster (63 fmol/min) than the unmodified G-G nick (14 fmol/min). These results highlight a trend whereby a template oxoG opposite the nick 5′-PO4 nucleotide allows more promiscuous sealing by DraRnl.
FIG 4.
Effect of 8-oxopurines in the template strand at the nick 5′-PO4 end. (A) The substrates for sealing were versions of the nicked duplex shown in Fig. 2A in which the X nucleobase of the 5′-PO4 N-X end was either oxoG (A) or oxoA (B). Ligase reactions were performed as described in Materials and Methods. The yields of sealed 36-mer RNApDNA are plotted as a function of time; the experiments are presented in sets of four, with the N nucleobase (specified to the right of each curve) being the variable within the set. Each datum is the average of at least 3 separate experiments ± SEM. (C) The initial rates of sealing of the indicated 5′ N-oxopurine nicks were calculated by linear regression in Prism.
DraRnl sealed a 5′ T-oxoA nick at much the same rate as it repaired an unmodified 5′ T-A end (159 versus 139 fmol/min). The 5′ C-oxoA end was sealed at the same rate as a 5′ C-A end (26 fmol/min). However, the rates of sealing of 5′ G-oxoA (38 fmol/min) and 5′ A-oxoA (58 fmol/min) ends were 8-fold and 3-fold higher, respectively, than those of the unmodified G-A and A-A mispairs. The effect of the 8-oxo atom is to lower the fidelity of DraRnl at a 5′ purine-A mispair.
Sealing opposite abasic sites in the template strand.
Abasic sites are among the most common DNA lesions and present a threat to genome integrity by causing DNA and RNA polymerases to stall, slip, or misincorporate at an abasic site in the template strand. Abasic sites in RNA are generated by potent ribotoxins with N-glycosidase activity (66). How ligases deal with abasic sites is an open question. Here we examined the ability of DraRnl to seal nicked duplexes with THF abasic sites in the template strand opposite the 3′-OH ribonucleotide (Fig. 5A) and 5′-PO4 deoxynucleotide (Fig. 5B) at the nick. All N-abasic configurations were tested. By comparing the ligation rates for N-abasic nicks to those for the correctly paired N-X substrates and N-X mispairs, we aimed to distinguish, if applicable, the effects of having no base pairing to the template at the nick from the effects of mispairing.
FIG 5.
Effect of an abasic site opposite the nick 3′-OH or 5′-PO4 nucleotide. (A) The substrates for sealing were versions of the nicked duplex in Fig. 1A in which the X position of the 3′-OH N-X end was a THF abasic site (A) or variants of the nicked duplex in Fig. 2A in which the X position of the 5′-PO4 N-X end was an abasic site (B). Ligase reactions were performed as described in Materials and Methods. The yields of sealed 36-mer RNApDNA are plotted as a function of time; the N nucleobase is specified to the right of each curve. The initial rates of sealing of the indicated 3′ and 5′ N-abasic site nicks were calculated by linear regression in Prism.
DraRnl sealed the 3′ A-abasic nick at a rate of 18 fmol/min (Fig. 5A), which was 11-fold lower than the rate of ligation of a 3′ A-T paired end (198 fmol/min). The 3′ A-abasic end was a better substrate than the 3′ A-A and A-G mispairs but less effective than the 3′ A-C mispair (Fig. 1B). DraRnl was extremely feeble in repairing 3′ G-abasic (1 fmol/min), 3′ C-abasic (3 fmol/min), and 3′ U-abasic (2 fmol/min) ends (Fig. 5A). The magnitude of the 3′ G-abasic defect was similar to that seen for the 3′ G-G and 3′ G-A mispairs (Fig. 1B). The 3′ C-abasic and U-abasic ends were less effective substrates than any of the 3′ C-N and 3′ U-N mispairs (Fig. 1B).
Among the 5′-PO4 N-abasic nicks, DraRnl was the most proficient at sealing 5′ A-abasic (38 fmol/min) and 5′ G-abasic (17 fmol/min) ends (Fig. 5B), albeit it did so 6-fold slower than it sealed the respective 5′ A-T and 5′ G-C paired ends. The 5′ A-abasic lesion was sealed about 2-fold better than the 5′ A-G and A-A mispairs but 70% as well as the A-C mispair (Fig. 2B). DraRnl handled the 5′ G-abasic nick better than the 5′ G-A mispair, about the same as the 5′ G-G mispair, and less well than the 5′ G-T mispair (Fig. 2B). The 5′ C-abasic and 5′ T-abasic ends were very poor substrates (3 and 4 fmol/min, respectively).
Taken together, these experiments underscore a preference for adenine as the nucleobase when DraRnl seals at a nick with an abasic lesion across from the 3′-OH or 5′-PO4 terminus. The obedience by DrRnl of an A rule for ligating opposite an abasic lesion at the nick 3′-OH end dovetails nicely with the preference of many DNA and RNA polymerases for A incorporation opposite an abasic site in the DNA template (57–59).
DISCUSSION
DraRnl is the founding member of a family of nick-sealing RNA ligase enzymes composed of a covalent nucleotidyltransferase domain (common to all classic polynucleotide ligases) linked to a signature N-terminal domain predicted to be an RNA-binding OB fold (34). When last reported on in 2007, the phylogenetic distribution of DraRnl homologs was sparse (35). An updated search for DraRnl family members now shows them to be encoded by diverse microbial taxa and viruses. We find DraRnl homologs in 35 bacterial genera representing 10 different phyla: Actinobacteria, Bacteroidetes, Chloroflexi, Cyanobacteria, Deinococcus-Thermus, Firmicutes, Fusobacteria, Planctomycetes, Proteobacteria, and Spirochaetes (see Table S1 in the supplemental material). Bacteriophages that prey on Aeromonas, Caulobacter, Mycobacterium, and Sinorhizobium also encode homologs of DraRnl. We identified a lone archaeal DraRnl homolog in Methanobrevibacter ruminantium. Of note is the presence of DraRnl homologs in many unicellular eukarya, including the amoebae Dictyostelium and Polysphondylium, the amoeboflagellate Naegleria, and 13 genera of fungi (see Table S1 in the supplemental material).
The biological functions of DraRnl and its many homologs are uncharted. Here, we sought clues from in vitro studies to home in on plausible substrates and invoke roles in vivo. The substrate specificity of DraRnl, whereby it seals nicks in duplex nucleic acids in which the 3′-OH end is RNA (34), is compatible with its participation in the repair of radiation-induced RNA damage, yet the contributions of RNA damage to bacterial sensitivity to IR are uncharted, as are the broken RNAs that might be subject to templated repair by DraRnl.
An alternative scenario is that DraRnl's sealing activity is deployed for DNA repair, especially in gap repair and nonhomologous end-joining (NHEJ) events in which ribonucleotides are added to a DNA 3′-OH end during gap filling and cross-break priming by repair polymerases. Synthesis of a ribopatch during DNA break repair is potentially advantageous as a stopgap measure in nondividing cells that have limited deoxynucleoside triphosphate (dNTP) pools. Indeed, the bacterial NHEJ polymerase (POL) LigD-POL is more adept at short templated additions and gap filling with ribonucleoside triphosphates than with dNTPs and prefers to add ribonucleotides across from an abasic template lesion (60, 61). The eukaryal NHEJ polymerase μ is also proficient at gap filling with ribonucleotides (62–64). After such a ribopatch fill-in, the repair ligase is presented with a 3′-OH, 5′-PO4 nick in which the 5′-PO4 strand and the template strand are DNA and the 3′-OH end is RNA, this being the model substrate presently employed for our analyses of DraRnl. Prior studies had shown that DraRnl is more effective at ligating a 3′-OH RNA strand and a 5′-PO4 DNA strand than it is in sealing a nick in which the 3′-OH and 5′-PO4 strands are both RNA (34). The efficiency of ligation depends on the number of ribonucleotides at the 3′-OH end, whereby DraRnl is unable to seal a DNA nick with a single 3′-OH ribonucleotide but is progressively more active at nicks with 3′-OH diribonucleotide and triribonucleotide ends (34). Thus, the biochemical properties of DraRnl are consistent with a role in nick sealing after synthesis of a ribopatch.
Here, we found that DraRnl was indifferent to the identity of the 3′-OH nucleobase, as long as it was correctly paired. We examined the fidelity of ligation at mispair ends and sites of template strand damage, in light of the propensity of repair polymerases to generate mismatches, especially when confronted with template lesions. With the 3′-OH mispairs (which we suspect are more commonly encountered by ligases during gap repair than 5′-PO4 mispairs), the DraRnl sealing rate varied widely in a hierarchical fashion, with G-T and A-C mispairs being the best substrates and the G-G, G-A, and A-A mispairs being the worst. It is noteworthy that when the 3′-OH mispaired nicks were recently used to interrogate the fidelity of bacteriophage T4 Rnl2, the 3′ A-C and G-T configurations were the ones that were best tolerated by Rnl2 (41). Bacteriophage T4 Rnl2 and DraRnl exemplify distinct families of nick-sealing enzymes that require a 3′-OH RNA strand (34, 65). Rnl2 and DraRnl rely for nucleic acid recognition on structurally unrelated domains appended to the C and N termini of the core nucleotidyltransferase domain, respectively (30, 34–36). The similar responses of DraRnl and Rnl2 to 3′-OH mispaired nicks, notwithstanding the fact that Rnl2 is inherently much faster at nick sealing than DraRnl under conditions of enzyme excess (41), suggest that the two families of nick-sealing enzymes recognize common features of their substrates that are affected by distortion of the nick 3′ terminus.
We found that the responses of DraRnl to oxoG and abasic lesions in the template strand across from the nick 3′-OH end are in tune with the promutagenic properties of DNA and RNA polymerases that copy over these lesions to generate A-oxoG and A-abasic configurations (57–59). First, DraRnl accepts the A-oxoG pair as correct, while it discriminates against 3′ U-oxoG and G-oxoG mispairs. Bacteriophage T4 Rnl2 displays similar properties in this regard (41). Second, DraRnl obeys the A rule in its preference for sealing when a 3′-OH A is opposite an abasic template site. An interesting feature of DraRnl highlighted by the present study is its relatively high activity and infidelity in sealing nicks with 3′-OH ends opposite a template oxoA lesion (compared with its response to an oxoG lesion). The net effect of these properties is that DraRnl can embed mutations during either ribopatched DNA repair or repair by a templated RNA repair pathway analogous to kinetoplastid RNA editing.
For completeness and comparison, we surveyed the effects of 5′-PO4 mispairs on the rate of nick sealing by DraRnl. Duplex RNA structures are often interspersed with single-nucleotide mispairs that, if incised on the 5′ side of the mispair, would be potential substrates for repair by DraRnl. The finding that DraRnl seals a nick with a 5′ T-G mispair as well as it handles a 5′ C-G paired end again seems well tuned to the common occurrence of U-G wobble pairs in duplex RNAs. In most other respects, the fidelity of DraRnl at 5′ mispairs is similar to its behavior at 3′ mispairs; i.e., the 5′ G-T and A-C mispairs are its next best substrates. As noted above, DraRnl differs from many nick-sealing DNA ligases that evince much higher fidelity at mispairs on the 3′-OH side of the nick than on the 5′-PO4 side (44–54).
With respect to oxoG template lesions opposite the 5′-PO4 end, DraRnl treated the A-oxoG configuration as correctly paired, yet it was more tolerant of the 5′ T-oxoG and 5′ G-oxoG mispairs than the equivalent configuration on the 3′ side of the nick. The sealing of 5′ N-oxoA nicks was generally similar to that of the 3′ N-oxoA series. Whereas DraRnl adopts the A rule in sealing a 5′-PO4 end across from an abasic template site, the enzyme also accepted a G-abasic site lesion at the nick 5′ end.
In conclusion, our studies of DraRnl add to an emerging picture of the substrate preferences of nick-sealing RNA ligases and their responses to nucleobase damage.
Supplementary Material
ACKNOWLEDGMENTS
This research was supported by NIH grants GM42498 (to S.S.) and F32-ES22914 (to B.J.S.).
S.S. is an American Cancer Society research professor.
Footnotes
Published ahead of print 14 February 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.00020-14.
REFERENCES
- 1.Ellenberger T, Tomkinson AE. 2008. Eukaryotic DNA ligases: structural and functional insights. Annu. Rev. Biochem. 77:313–338. 10.1146/annurev.biochem.77.061306.123941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shuman S. 2009. DNA ligases: progress and prospects. J. Biol. Chem. 284:17365–17369. 10.1074/jbc.R900017200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Culver GM, Noller HF. 2000. Directed hydroxyl radical probing of RNA from iron(II) tethered to proteins in ribonucleoprotein complexes. Methods Enzymol. 318:461–475. 10.1016/S0076-6879(00)18070-X [DOI] [PubMed] [Google Scholar]
- 4.Joseph S, Noller HF. 2000. Directed hydroxyl radical probing using Fe (II) tethered to RNA. Methods Enzymol. 318:175–190. 10.1016/S0076-6879(00)18052-8 [DOI] [PubMed] [Google Scholar]
- 5.Hecht SM. 1994. RNA degradation by bleomycin, a naturally occurring bioconjugate. Bioconjug. Chem. 5:513–526. 10.1021/bc00030a006 [DOI] [PubMed] [Google Scholar]
- 6.Abraham AT, Lin JJ, Newtown DL, Rybak S, Hecht SM. 2003. RNA cleavage and inhibition of protein synthesis by bleomycin. Chem. Biol. 10:45–52. 10.1016/S1074-5521(02)00306-X [DOI] [PubMed] [Google Scholar]
- 7.Liu M, Gong X, Alluri RK, Wu J, Sablo T, Li Z. 2012. Characterization of RNA damage under oxidative stress in Escherichia coli. Biol. Chem. 393:123–132. 10.1515/hsz-2011-0247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nunomura A, Moreira PI, Castellani RJ, Lee HG, Zhu X, Smith MA, Perry G. 2012. Oxidative damage to RNA in aging and neurodegenerative disorders. Neurotox. Res. 22:231–248. 10.1007/s12640-012-9331-x [DOI] [PubMed] [Google Scholar]
- 9.Wurtmann EJ, Wolin SL. 2009. RNA under attack: cellular handling of RNA damage. Crit. Rev. Biochem. Mol. Biol. 44:34–49. 10.1080/10409230802594043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li Z, Wu J, DeLeo CJ. 2006. RNA damage and surveillance under oxidative stress. IUBMB Life 58:581–588. 10.1080/15216540600946456 [DOI] [PubMed] [Google Scholar]
- 11.Wu J, Jiang Z, Liu M, Gong X, Wu S, Burns CM, Li Z. 2009. Polynucleotide phosphorylase protects Escherichia coli against oxidative stress. Biochemistry 48:2012–2020. 10.1021/bi801752p [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Silber R, Malathi VG, Hurwitz J. 1972. Purification and properties of bacteriophage T4-induced RNA ligase. Proc. Natl. Acad. Sci. U. S. A. 69:3009–3013. 10.1073/pnas.69.10.3009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang LK, Lima CD, Shuman S. 2002. Structure and mechanism of T4 polynucleotide kinase: an RNA repair enzyme. EMBO J. 21:3873–3880. 10.1093/emboj/cdf397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Das U, Shuman S. 2013. Mechanism of RNA 2′,3′-cyclic phosphate end-healing by T4 polynucleotide kinase-phosphatase. Nucleic Acids Res. 41:355–365. 10.1093/nar/gks977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.El Omari K, Ren J, Bird LE, Bona MK, Klarmann G, LeGrice SFJ, Stammers DK. 2006. Molecular architecture and ligand recognition determinants for T4 RNA ligase. J. Biol. Chem. 281:1573–1579. 10.1074/jbc.M509658200 [DOI] [PubMed] [Google Scholar]
- 16.Wang LK, Nandakumar J, Schwer B, Shuman S. 2007. The C-terminal domain of T4 RNA ligase 1 confers specificity for tRNA repair. RNA 13:1235–1244. 10.1261/rna.591807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Martins A, Shuman S. 2004. Characterization of a baculovirus enzyme with RNA ligase, polynucleotide 5′ kinase and polynucleotide 3′ phosphatase activities. J. Biol. Chem. 279:18220–18231. 10.1074/jbc.M313386200 [DOI] [PubMed] [Google Scholar]
- 18.Ho CK, Shuman S. 2002. Bacteriophage T4 RNA ligase 2 (gp24.1) exemplifies a family of RNA ligases found in all phylogenetic domains. Proc. Natl. Acad. Sci. U. S. A. 99:12709–12714. 10.1073/pnas.192184699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brooks MA, Meslet-Cladiére L, Graille M, Kuhn J, Blondeau K, Myllykallio H, van Tilbeurgh H. 2008. The structure of an archaeal homodimeric ligase which has RNA circularization activity. Protein Sci. 17:1336–1345. 10.1110/ps.035493.108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Martins A, Shuman S. 2005. An end-healing enzyme from Clostridium thermocellum with 5′ kinase, 2′,3′ phosphatase, and adenylyltransferase activities. RNA 11:1271–1280. 10.1261/rna.2690505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang LK, Das U, Smith P, Shuman S. 2012. Structure and mechanism of the polynucleotide kinase component of the bacterial Pnkp-Hen1 RNA repair system. RNA 18:2277–2286. 10.1261/rna.036061.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Smith P, Wang LK, Nair PA, Shuman S. 2012. The adenylyltransferase domain of bacterial Pnkp defines a unique RNA ligase family. Proc. Natl. Acad. Sci. U. S. A. 109:2296–2301. 10.1073/pnas.1116827109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tanaka N, Shuman S. 2011. RtcB is the RNA ligase component of an Escherichia coli RNA repair operon. J. Biol. Chem. 286:7727–7731. 10.1074/jbc.C111.219022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sawaya R, Schwer B, Shuman S. 2003. Genetic and biochemical analysis of the functional domains of yeast tRNA ligase. J. Biol. Chem. 278:43928–43938. 10.1074/jbc.M307839200 [DOI] [PubMed] [Google Scholar]
- 25.Remus BS, Shuman S. 2013. A kinetic framework for tRNA ligase and enforcement of a 2′-phosphate requirement for ligation highlights the design logic of an RNA repair machine. RNA 19:659–669. 10.1261/rna.038406.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Amitsur M, Levitz R, Kaufman G. 1987. Bacteriophage T4 anticodon nuclease, polynucleotide kinase, and RNA ligase reprocess the host lysine tRNA. EMBO J. 6:2499–2503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schwer B, Sawaya R, Ho CK, Shuman S. 2004. Portability and fidelity of RNA-repair systems. Proc. Natl. Acad. Sci. U. S. A. 101:2788–2793. 10.1073/pnas.0305859101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nandakumar J, Schwer B, Schaffrath R, Shuman S. 2008. RNA repair: an antidote to cytotoxic eukaryal RNA damage. Mol. Cell 31:278–286. 10.1016/j.molcel.2008.05.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chan CM, Zhou C, Huang RH. 2009. Reconstituting bacterial RNA repair and modification in vitro. Science 326:247. 10.1126/science.1179480 [DOI] [PubMed] [Google Scholar]
- 30.Nandakumar J, Shuman S, Lima CD. 2006. RNA ligase structures reveal the basis for RNA specificity and conformational changes that drive ligation forward. Cell 127:71–84. 10.1016/j.cell.2006.08.038 [DOI] [PubMed] [Google Scholar]
- 31.Tanaka N, Chakravarty AK, Maughan B, Shuman S. 2011. A novel mechanism of RNA repair by RtcB via sequential 2′,3′-cyclic phosphodiesterase and 3′-phosphate/5′-hydroxyl ligation reactions. J. Biol. Chem. 286:43134–43143. 10.1074/jbc.M111.302133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chakravarty AK, Subbotin R, Chait BT, Shuman S. 2012. RNA ligase RtcB splices 3′-phosphate and 5′-OH ends via covalent RtcB-(histidinyl)-GMP and polynucleotide-(3′)pp(5′)G intermediates. Proc. Natl. Acad. Sci. U. S. A. 109:6072–6077. 10.1073/pnas.1201207109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Desai KK, Bingman CA, Phillips GN, Raines RT. 2013. Structures of the noncanonical RNA ligase RtcB reveal the mechanism of histidine guanylylation. Biochemistry 52:2518–2525. 10.1021/bi4002375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Martins A, Shuman S. 2004. An RNA ligase from Deinococcus radiodurans. J. Biol. Chem. 279:50654–50661. 10.1074/jbc.M407657200 [DOI] [PubMed] [Google Scholar]
- 35.Raymond A, Shuman S. 2007. Deinococcus radiodurans RNA ligase exemplifies a novel ligase clade with a distinctive N-terminal module that is important for 5′-PO4 nick sealing and ligase adenylylation but dispensable for phosphodiester formation at an adenylylated nick. Nucleic Acids Res. 35:839–849. 10.1093/nar/gkl1090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ho CK, Wang LK, Lima CD, Shuman S. 2004. Structure and mechanism of RNA ligase. Structure 12:327–339. 10.1016/j.str.2004.01.011 [DOI] [PubMed] [Google Scholar]
- 37.Deng J, Schnaufer A, Salavati R, Stuart KD, Hol WG. 2004. High resolution crystal structure of a key editosome enzyme from Trypanosoma brucei: RNA editing ligase 1. J. Mol. Biol. 343:601–613. 10.1016/j.jmb.2004.08.041 [DOI] [PubMed] [Google Scholar]
- 38.Schnaufer A, Panigrahi AK, Panicucci B, Igo RP, Salavati R, Stuart K. 2001. An RNA ligase essential for RNA editing and survival of the bloodstream form of Trypanosoma brucei. Science 291:2159–2162. 10.1126/science.1058655 [DOI] [PubMed] [Google Scholar]
- 39.Blanc V, Alfonzo JD, Aphasizhev R, Simpson L. 1999. The mitochondrial RNA ligase from Leishmania tarentolae can join RNA molecules bridged by a complementary RNA. J. Biol. Chem. 274:24289–24296. 10.1074/jbc.274.34.24289 [DOI] [PubMed] [Google Scholar]
- 40.Aphasizhev R, Aphasizheva I. 2011. Uridine insertion/deletion editing in trypanosomes: a playground for RNA-guided information transfer. Wiley Interdiscip. Rev. RNA 2:669–685. 10.1002/wrna.82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chauleau M, Shuman S. 2013. Kinetic mechanism of nick sealing by T4 RNA ligase 2 and effects of 3′-OH base mispairs and damaged base lesions. RNA 19:1840–1847. 10.1261/rna.041731.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu Y, Zhou J, Omelchenko MV, Beliaev AS, Venkateswaran A, Stair J, Wu L, Thompson DK, Xu D, Rogozin IB, Gaidamakova EK, Zhai M, Makarova KS, Koonin EV, Daly MJ. 2003. Transcriptome dynamics of Deinococcus radiodurans recovering from ionizing radiation. Proc. Natl. Acad. Sci. U. S. A. 100:4191–4196. 10.1073/pnas.0630387100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Varani G, McClain WH. 2000. The G·U wobble base pair. EMBO Rep. 1:18–23. 10.1093/embo-reports/kvd001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Barany F. 1991. Genetic disease detection and DNA amplification using cloned thermostable ligase. Proc. Natl. Acad. Sci. U. S. A. 88:189–193. 10.1073/pnas.88.1.189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Luo J, Bergstrom DE, Barany F. 1996. Improving the fidelity of Thermus thermophilus DNA ligase. Nucleic Acids Res. 24:3071–3078. 10.1093/nar/24.15.3071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tong J, Barany F, Cao W. 2000. Ligation reaction specificities of an NAD+-dependent DNA ligase from the hyperthermophile Aquifex aeolicus. Nucleic Acids Res. 28:1447–1454. 10.1093/nar/28.6.1447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tomkinson AE, Tappe NJ, Friedberg EC. 1992. DNA ligase I from Saccharomyces cerevisiae: physical and biochemical characterization of the CDC9 gene product. Biochemistry 31:11762–11771. 10.1021/bi00162a013 [DOI] [PubMed] [Google Scholar]
- 48.Husain I, Tomkinson AE, Burkhart WA, Moyer MB, Ramos W, Mackey ZB, Besterman JM, Chen J. 1995. Purification and characterization of DNA ligase III from bovine testes: homology with DNA ligase II and vaccinia DNA ligase. J. Biol. Chem. 270:9683–9690. 10.1074/jbc.270.16.9683 [DOI] [PubMed] [Google Scholar]
- 49.Bhagwat AA, Sanderson RJ, Lindahl T. 1999. Delayed DNA joining at 3′ mismatches by human DNA ligases. Nucleic Acids Res. 27:4028–4033. 10.1093/nar/27.20.4028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shuman S. 1995. Vaccinia virus DNA ligase: specificity, fidelity, and inhibition. Biochemistry 34:16138–16147. 10.1021/bi00049a029 [DOI] [PubMed] [Google Scholar]
- 51.Sriskanda V, Shuman S. 1998. Specificity and fidelity of strand joining by Chlorella virus DNA ligase. Nucleic Acids Res. 26:3536–3541. 10.1093/nar/26.15.3536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nakatani M, Ezaki S, Atomi H, Imanaka T. 2002. Substrate recognition and fidelity of strand joining by an archaeal DNA ligase. Eur. J. Biochem. 269:650–656. 10.1046/j.0014-2956.2001.02695.x [DOI] [PubMed] [Google Scholar]
- 53.Lamarche BJ, Showalter AK, Tsai MD. 2005. An error-prone viral DNA ligase. Biochemistry 44:8408–8417. 10.1021/bi047706g [DOI] [PubMed] [Google Scholar]
- 54.Wang Y, Lamarche BJ, Tsai MD. 2007. Human DNA ligase IV and the ligase IV/XRCC4 complex: analysis of nick ligation fidelity. Biochemistry 46:4962–4976. 10.1021/bi0621516 [DOI] [PubMed] [Google Scholar]
- 55.Hashimoto K, Tominaga Y, Nakabeppu Y, Moriya M. 2004. Futile short-patch DNA base excision repair of adenine:8-oxoguanine mispair. Nucleic Acids Res. 32:5928–5934. 10.1093/nar/gkh909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhao X, Muller JG, Halasyam M, David SS, Burrows CJ. 2007. In vitro ligation of oligodeoxynucleotide containing C8-oxidized purine lesions using bacteriophage T4 DNA ligase. Biochemistry 46:3734–3744. 10.1021/bi062214k [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Obeid S, Blatter N, Kranaster R, Schnur A, Diederichs K, Welte W, Marx A. 2010. Replication through an abasic DNA lesion: structural basis for adenine selectivity. EMBO J. 29:1738–1747. 10.1038/emboj.2010.64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhou W, Doetsch PW. 1993. Effects of abasic sites and DNA single-strand breaks on prokaryotic RNA polymerases. Proc. Natl. Acad. Sci. U. S. A. 90:6601–6605. 10.1073/pnas.90.14.6601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chen YH, Bogenhagen DF. 1993. Effects of DNA lesions on transcription elongation by T7 RNA polymerase. J. Biol. Chem. 268:5849–5855 [PubMed] [Google Scholar]
- 60.Yakovleva L, Shuman S. 2006. Nucleotide misincorporation, 3′-mismatch extension, and responses to abasic sites and DNA adducts by the polymerase component of bacterial DNA ligase D. J. Biol. Chem. 281:25026–25040. 10.1074/jbc.M603302200 [DOI] [PubMed] [Google Scholar]
- 61.Zhu H, Shuman S. 2010. Gap filling activities of Pseudomonas LigD polymerase and functional interactions of LigD with the DNA end-binding Ku protein. J. Biol. Chem. 285:4815–4825. 10.1074/jbc.M109.073874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ruiz JF, Juarez R, Garcia-Diaz M, Terrados G, Picher AJ, Gonzalez-Barrera S, Fernandez de Henestrosa AR, Blanco L. 2003. Lack of sugar discrimination by human Pol mu requires a single glycine residue. Nucleic Acids Res. 31:4441–4449. 10.1093/nar/gkg637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Nick McElhinny SA, Ramsden DA. 2003. Polymerase mu is a DNA-directed DNA/RNA polymerase. Mol. Cell. Biol. 23:2309–2315. 10.1128/MCB.23.7.2309-2315.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Martin MJ, Garcia-Ortiz MV, Esteban V, Blanco L. 2013. Ribonucleotides and manganese ions improve non-homologous end joining by human Polμ. Nucleic Acids Res. 41:2428–2436. 10.1093/nar/gks1444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Nandakumar J, Shuman S. 2004. How an RNA ligase discriminates RNA versus DNA damage. Mol. Cell 16:211–221. 10.1016/j.molcel.2004.09.022 [DOI] [PubMed] [Google Scholar]
- 66.Walsh MJ, Dodd JE, Hautbergue GM. 2013. Ribosome-inactivating proteins: potent poisons and molecular tools. Virulence 4:774–784. 10.4161/viru.26399 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.