Abstract
The length of the flagellar hook is controlled by the soluble protein FliK. FliK is structurally divided into two halves with distinct functions; the N-terminal half determines hook length, while the C-terminal half switches the secretion substrate specificity, consequently terminating hook elongation. FliK properly achieves both functions only when it is secreted. In a previous paper, we showed that a temperature-sensitive flgE mutant of Salmonella enterica serovar Typhimurium, SJW2219, produced basal bodies with short hooks (average length, 25 nm) at 37°C. In this study, we show that the mutant cells grown at 37°C secrete FliK but not flagellin (FliC), indicating that FliK is abortively secreted into the medium when the hook is shorter than 30 nm. In contrast, FliK unfailingly switches the gate modes when the hook is longer than 30 nm. Taking the FliC, FliK, and FlgM secretion patterns into account, we conclude that FliK determines the minimal length of the hook. We will discuss how FliK detects the critical switching point of the secretion gate.
INTRODUCTION
The bacterial flagellum is composed mainly of three structures, the basal body, the external filament, and the hook that connects the basal body and the filament. The hook is a curved tube that is composed of many copies of the hook protein FlgE (1, 2). In the pathway of flagellar morphogenesis, hook assembly starts after completion of the basal body, which consists of the rod and ring structures. FliC (flagellin) assembly to form the filament starts only after completion of the hook (3). The soluble protein FliK somehow senses the completion of hook elongation. In this process, FliK plays two roles by monitoring hook length and switching the substrate specificity of the export apparatus so that FlgE export ceases and flagellin (FliC) export begins. In the latter role, FliK is responsible for switching the secretion mode from hook elongation to filament elongation (4, 5). The average length of the hook is controlled at about 55 nm in wild-type Salmonella strains (6).
The role of FliK in determining hook length led to a conventional model in which FliK acts as a physical ruler (7). However, this model could not fully explain the broad distribution of hook lengths. This problem was partly overcome by Erhardt et al. (8), who demonstrated that the probability of switching the specificity of the export apparatus is an function of increasing hook length. They combined experimental data with mathematical analysis to show that FliK secretion immediately triggered the specificity switch in hooks greater than the length of the FliK ruler. Here we present direct evidence that when FliK is secreted from long hooks, it always switches the secretion gate for FliC, supporting their model.
In this study, we employed a temperature-sensitive (TS) flgE mutant of Salmonella enterica serovar Typhimurium, SJW2219, which has a T149N FlgE point mutation (9). At the nonpermissive temperature (37°C), the mutant cells produced many basal bodies with short hooks, whose length was 25 nm, on average. When the cells were grown at 30°C, the hook length distribution was the same as that of wild-type cells but the rate of elongation was significantly lower (10).
We constructed fliK and fliC mutants, observed their flagella by electron microscopy, and analyzed the proteins secreted from the mutants by SDS-PAGE. We found that a considerable amount of FliK but little FliC was secreted from immature hooks, indicating that FliK is secreted when the hook is short but does not cause switching of the export apparatus. We also showed that when FliK was secreted from long hooks, it always switched the secretion gate for FliC. Therefore, we concluded that FliK does not determine the final hook length but rather determines the minimal hook length.
MATERIALS AND METHODS
Strains and growth conditions.
The S. enterica serovar Typhimurium strains used in this study are listed in Table 1. Cells were cultured in LB medium (1% [wt/vol] tryptone, 0.5% [wt/vol] yeast extract, 1% [wt/vol] NaCl) or TY medium (1% [wt/vol] tryptone, 0.5% [wt/vol] yeast extract) at either 30 or 37°C.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Genotype | Source or reference |
|---|---|---|
| Strains | ||
| SJW1103 | Wild type | S. Yamaguchi |
| SJW2219 | flgE2219 (T149N) | 9 |
| UA001 | flgE2219 (T149N) fliK6125 [FliK95′-YscP (217-381)-′96FliK] | This study |
| UA002 | flgE2219 (T149N)/pTrc99AFF4-fliC | This study |
| SJW2536 | ΔfliC | S. Yamaguchi |
| SJW108 | ΔfliK | S. Yamaguchi |
| TH16783 | ΔaraBAD7606::fliK+ ΔfliK6140 | M. Erhardt |
| TH18945 | flgM5794::FRT flgE2219 (T149N) fljBenx fljBenxvh2 | K. Hughes |
| Plasmids | ||
| pTH8256a | FliK95::YscP 217-381 | 13 |
| pAH25 | pTrc99AFF4-fliC | 16 |
Plasmid pTH8256 was used to construct UA001 by recombination.
Hook length measurement.
To measure hook length, intact flagella were isolated and the hook basal bodies were prepared as previously described (10).
SDS-PAGE.
Proteins secreted into the medium were analyzed by SDS-PAGE as previously described (11, 12). Briefly, cells were grown at 30 or 37°C until they reached an optical density at 600 nm (OD600) of 1.0, at which point cells were harvested for analysis. The cell pellet and the culture supernatant were separated by differential centrifugation. The proteins secreted into the culture, which were in the supernatant, were precipitated with 7% trichloroacetic acid (TCA). The precipitated proteins were then dissolved in SDS sample buffer and boiled for 3 min. Fractions were subjected to SDS-PAGE. The amount of sample applied to SDS gels was normalized by the number of cells in the sample as determined by OD600 measurement. SDS-PAGE was carried out with a minigel kit (ATTO). The acrylamide concentration of the gel was 12.5%. Gels were stained with Coomassie brilliant blue (CBB).
Western blot analysis.
Samples were subjected to SDS-PAGE, and protein bands were transferred to polyvinylidene fluoride membranes. Polyclonal anti-FliC or anti-FliK antibodies were used for Western blot analysis.
Electron microscopy.
Samples were prepared as previously described (10). Briefly, samples were negatively stained with 1 or 2% (wt/vol) phosphotungstic acid (pH 7.0) and observed with a JEM-1200EXII electron microscope (JEOL, Tokyo, Japan). Micrographs were taken at an accelerating voltage of 80 kV.
Preparation of osmotically shocked cells.
Cells collected from 5 ml of culture were suspended in 100 μl of a sucrose solution (20% [wt/vol] sucrose, 5 mM EDTA, pH 7). After 10 min of incubation on ice, the suspension was abruptly diluted with 1.9 ml prechilled water containing 5 mM EDTA. Intact cells were removed by low-speed centrifugation, and the osmotically shocked cell products that remained in the supernatant were collected by high-speed centrifugation (2,000 × g, 20 min). Samples were negatively stained with phosphotungstic acid and observed by electron microscopy (12).
RESULTS
Hook length control in a hook mutant.
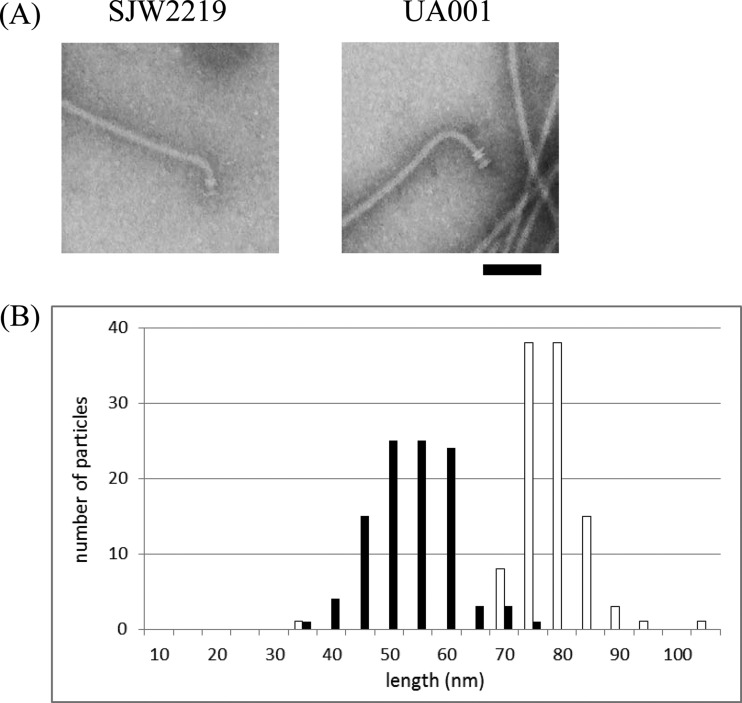
TS flgE mutant strain SJW2219 produces normal flagella at 30°C (Fig. 1A, left panel). The average length of the hooks in intact flagella is 55 nm (n = 101) (Fig. 1B, filled bars), which is the same as that observed in wild-type flagella. However, the hook became shorter (45 nm) after the removal filaments by acid treatment. We showed that the interaction between the hook and hook-associated protein 1 (HAP1) FlgK was weak in this mutant. In conclusion, we assumed that the 10-nm difference between intact and acid-treated hooks was due to the removal of FlgK from the hook (10). In order to confirm that FliK controls hook length even in this hook mutant, we constructed a mutant with a modified FliK molecular length.
FIG 1.
(A) Electron micrographs of flagellar basal structures from strains UA001 (right) and SJW2219 (left). Intact flagella were isolated from cells grown in TY medium at 30°C as previously described (10). The hook length of UA001 is significantly greater than that of SJW2219. Samples were negatively stained with 2% phosphotungstic acid (pH 7). Bar, 100 nm. (B) Hook length distribution of intact flagella isolated from strains UA001 (empty bars) and SJW2219 (filled bars) grown at 30°C. The number of particles counted was 105 for UA001 and 101 for SJW2219.
We introduced a modified fliK gene lengthened by insertion of a foreign gene fragment into SJW2219 and named the construct UA001 (Table 1). UA001 cells produced a FliK variant 570 amino acids long. At 30°C, these cells produced correspondingly longer hooks (Fig. 1A, right panel) with an average length of 81 nm and a standard deviation of 7 nm (n = 105) (Fig. 1B, empty bars). This distribution of hook length is the same as that of a mutant carrying the same lengthened fliK gene (UA001) in a wild-type flgE background (13). Therefore, we conclude that FliK controls hook length despite the slow elongation of the hook in SJW2219 cells.
FliC is not produced by SJW2219 grown at 37°C.
TS mutant SJW2219 cells rarely formed intact flagella at 37°C (Fig. 2A). Accordingly, they secreted negligible amounts of flagellin (FliC). Why was the fliC gene exported by a hook mutant when it was grown at 30°C but not when it was grown at 37°C? It is known that mutant cells with a flgE deletion (thus hookless) and cells with a fliK deletion (thus polyhooked) do not produce flagellin. The reason is the presence of an anti-sigma factor, FlgM, which inhibits the expression of late genes, including fliC, flgK, and flgL (14). When the export apparatus is switched, FlgM is secreted and thus the block to late gene expression is removed. In both mutants, however, the anti-sigma factor FlgM is not secreted from the cell and continues to suppress the expression of late genes, including fliC. It seems likely that FlgM is not secreted by short-hook mutant cells. In the absence of FlgM in the cytoplasm, the sigma factor FliA is free to act and thus allows the late flagellar genes to be transcribed (15). Thus, even the mutant cells should produce FliC in the absence of FlgM. To test this possibility, a flgM deletion mutation was introduced into SJW2219, resulting in strain TH18945 (Table 1).
FIG 2.
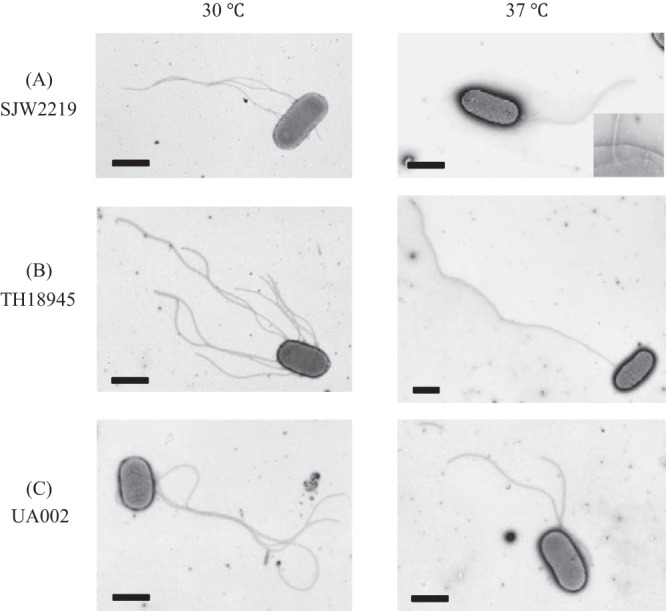
Electron micrographs of SJW2219 (A), TH18945 (B), and UA002 (C) cells grown in TY medium at either 30 or 37°C. In an enlarged image of the SJW2219 flagellar basal structures in the membrane, both intact and immature basal structures are visible (A, inset). Cells were negatively stained with 1% phosphotungstic acid (pH 7). Bars, 1 μm.
SJW2219 and TH18945 cells were grown in TY medium at either 30 or 37°C, and samples were analyzed by electron microscopy (Fig. 2A and B). Flagella in TH18945 cells were more numerous or longer than those from SJW2219 at either growth temperature (Fig. 2B). It should be noted that when TH18945 cells were grown at 37°C, only a small fraction of them had intact flagella (6%) but their filaments were obviously much longer than those attached to SJW2219 cells grown at 37°C (Fig. 2B, right panel), thus confirming that an increased amount of FliC was secreted and assembled as expected.
We analyzed the secreted and intracellular proteins from SJW2219 and TH18945 cells by SDS-PAGE, followed by Western blotting. TH18945 cells grown at 30°C held an amount of FliC similar to that of SJW2219 cells (Fig. 3A, lanes 5 and 7), but the former secreted more FliC into the medium than the latter (lanes 1 and 3). On the other hand, the cellular level of FliC was low in both TH18945 and SJW2219 cells grown at 37°C, albeit FliC levels remained higher in TH18945 than in SJW2219 (Fig. 3A and B, lanes 6 and 8). These results are consistent with our EM observations (Fig. 2B). TH18945 cells secreted little FliC, i.e., amounts similar to those secreted by SJW2219 cells at 37°C (Fig. 3A, lanes 2 and 4). These data suggest that FliC production in TH18945 cells grown at 37°C is inhibited in a FlgM-independent manner, possibly because of increased degradation by cellular proteases (see Discussion). In contrast, as much of both FlgE and FliK was produced and secreted by TH18945 cells at either temperature as by SJW2219 cells, as detected by Western blot assays (Fig. 3C and D). By comparison of band densities in CBB-stained and immunostained gels, we now know the position of each protein in CBB-stained gels (11, 12). Since the amounts of proteins are more quantitatively estimated in CBB-stained gels than in immunostained gels, we will use only CBB-stained gels here.
FIG 3.
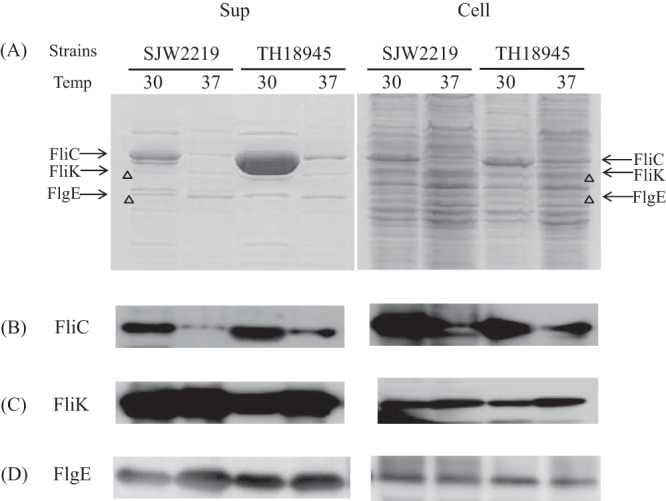
(A) SDS-PAGE pattern of proteins secreted into the culture medium (Sup) by SJW2219 and TH18945 cells (Cell). Cells were grown in TY medium either at 30 or 37°C. Secreted proteins were recovered from the cultures by pelleting the cells and precipitating the supernatant with TCA (Sup). Whole cells were collected from the pellets (Cell). The gels were stained with CBB. The acrylamide concentration of the gel was 12.5%. The proteins secreted by SJW2219 cells grown at a temperature (Temp) 30°C are shown in lane 1, while those of cells incubated at 37°C are shown in lane 2. Secreted proteins from TH18945 cells grown at 30°C are shown in lane 3, and those from the same cells grown at 37°C are in lane 4. Proteins from the pellets of SJW2219 cells grown at 30°C are in lane 5, and those from the same cells grown at 37°C are in lane 6. Proteins from pellets of TH18945 cells grown at 30°C are in lane 7, and those from the same cells grown at 37°C are in lane 8. (B) Western blot analysis of the supernatant and cellular fractions of SJW2219 and TH18945 cells with anti-FliC polyclonal antibodies at a 1:20,000 dilution. Cells were grown at 30 and 37°C to an OD600 of 1.0. Samples in lanes are the same order as those in panel A. (C) Detection of intracellular and extracellular FliK with polyclonal anti-FliK antibodies (1:20,000 dilution). Samples in lanes are in the same order as those in panel A. (D) Detection of intracellular and extracellular FlgE with polyclonal anti-FlgE antibodies (1:20,000 dilution). Samples in lanes are in the same order as those in panel A.
FliC does not pass the secretion gate of SJW2219 at 37°C.
SJW2219 cells did not secrete FliC at 37°C, presumably because of the inhibition of fliC gene expression by anti-sigma factor FlgM. flgM deletion mutant strain TH18945 grown at 37°C did not secrete FliC, because the intracellular FliC level was too low. We wondered if FliC overproduced in SJW2219 cells would be secreted. To test this, we introduced FliC-overproducing plasmid pAH25 (16) into SJW2219 and named the resulting strain UA002 (Table 1).
UA002 cells were grown in TY medium at 30 or 37°C and observed by electron microscopy. fliC gene expression was activated by the addition of isopropyl-β-d-thiogalactopyranoside (IPTG). The UA002 flagella became a little longer after activation at both temperatures, confirming that an increased amount of FliC was secreted in a manner similar to that in TH18945 cells (Fig. 2C).
The secreted and intracellular levels of FliC and FliK in SJW2219 and UA002 cells were examined as described above. On the basis of the density of the 58-kDa band of FliC in the SDS gels, the secreted and intracellular levels of FliC increased after activation in UA002 cells (Fig. 4, Sup). Although the intracellular level of FliC was the same at both temperatures, the amount of FliC secreted was much smaller at 37°C than at 30°C. This indicates that FliC was not secreted by UA002 cells at 37°C, at least not significantly more than the amount that leaks from a few intact flagella. When FliC was overproduced in TH18945 or UA002 cells at 37°C, the amounts of FliC in the cells stayed at the same level as in SJW2219 cells. Since FliC overproduction occurred as expected at 30°C, it should have occurred at 37°C. We assume that this low level of FliC is probably because of the negative feedback that suppressed fliC expression. In conclusion, given that the number of basal bodies is the same at both temperatures, we argue that FliC cannot pass the secretion gate in SJW2219 cells grown at 37°C.
FIG 4.
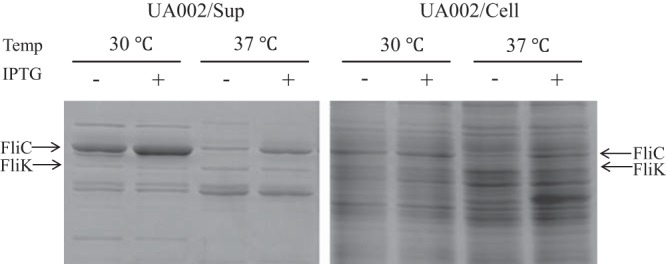
SDS-PAGE of proteins secreted into the culture medium (Sup) by SJW2219 and UA002 cells (Cell). Cells were grown in TY medium at a temperature (Temp) of either 30 or 37°C. Secreted proteins were recovered and examined as described in the legend to Fig. 3. Supernatants from SJW2219 are in lanes 1 and 3 (grown at 30 and 37°C, respectively), and those from UA002 are in lanes 2 and 4 (grown at 30 and 37°C, respectively). Proteins from whole SJW2219 cells are in lanes 5 and 7, and those from whole UA002 cells are in lanes 6 and 8 (grown at 30 and 37°C, respectively).
In contrast, the band density of FliK at 42 kDa secreted from activated UA002 cells was the same as that secreted from nonactivated UA002 cells or from SJW2219 cells grown at either temperature (Fig. 4). This indicates that FliK failed to switch the substrate specificity of the export apparatus and was thus abortively secreted from the short hooks in SJW2219 cells at 37°C.
FliK fails to switch substrate specificity in cells with short hooks.
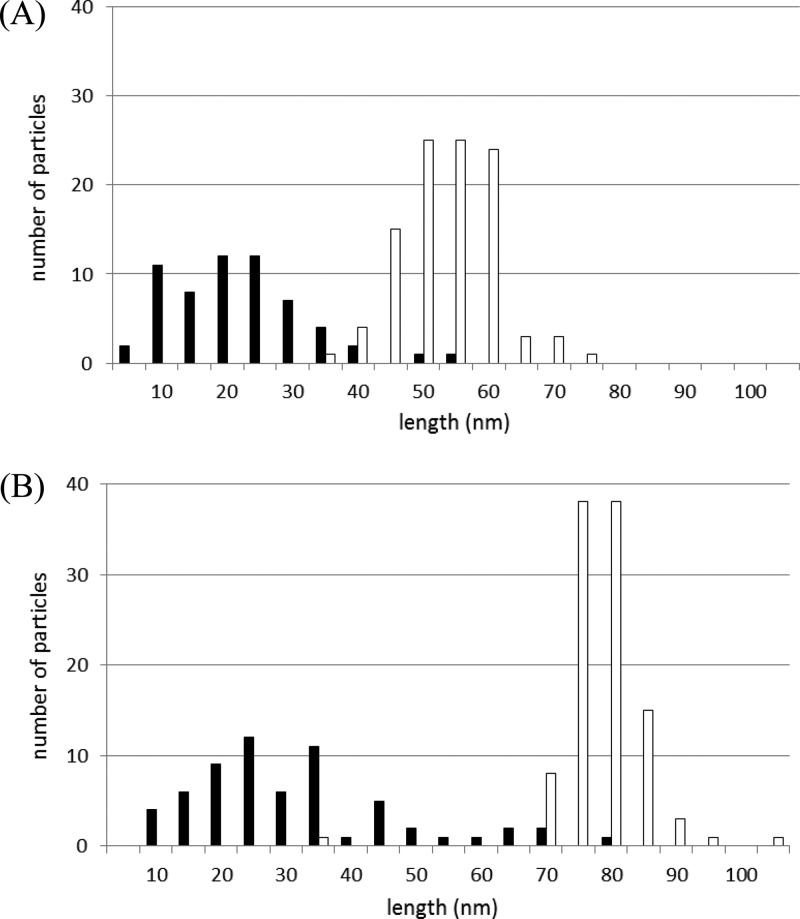
In a previous paper (10), we showed that SJW2219 cells produce not only intact flagella but also intermediate structures at any temperature (Fig. 2A, right panel, inlet). The basal bodies with short hooks occupied one-fourth of the population of flagellar particles at both 30 and 37°C. The average length of the short hooks was ca. 25 nm (10). We observed that the hooks with filaments attached were longer than ca. 20 nm in the wild type (data not shown). Since termination of hook growth is a probabilistic process, we asked whether there were filaments attached to hooks that were shorter than 20 nm.
We examined hooks isolated from SJW2219 cells grown at 30°C and separated the hook lengths in intact flagella from those in intermediate structures. Two peaks appeared in the length distribution diagram (Fig. 5A). The average hook length was 24 ± 10 nm (n = 61) in intermediate structures (filled bars) and 56 ± 7 nm (n = 103) in intact flagella (empty bars). It should be noted that hooks with and without filaments attached are both found in the region between the two peaks at 24 and 56 nm. Thus, there must be a critical hook length in this intermediate region at which FliK secretion is coupled with substrate specificity switching and filament growth is initiated. If this is the case, the average hook length of intermediate structures in UA001 cells should be greater than 24 nm. We again examined hook lengths in UA001 cells.
FIG 5.
Histograms of strain SJW2219 (A) and UA001 (B) hook lengths. The average hook length of intact particles (empty bars) of strain UA001 was 81 ± 7 nm (n = 105), whereas that of immature particles (filled bars) was 34 ± 16 nm (n = 63).
Flagellar structures were isolated from UA001 cells grown at 37°C as described above. There were many intermediate hook basal body structures similar to those observed in SJW2219. We measured the lengths of the hooks in intact flagella (hooks with filaments attached) and intermediates (hooks without filaments). The distribution of hook lengths is bimodal (Fig. 5B). The average hook length was 81 ± 7 nm (n = 105) in intact flagella (empty bars), whereas it was 34 ± 16 nm (n = 63) in intermediates (filled bars). The region in which hooks with and without filaments are mixed is shifted to the greater lengths; that is, it lies between 34 and 81 nm (Fig. 5B), indicating that the minimal hook length needed for filament assembly is greater.
FliK always switches substrate specificity in cells with longer hooks.
As we have seen, FliK does not switch substrate specificity when the hook is short even though FliK is being secreted. Does FliK therefore always switch substrate specificity when the hook is long? Is FliK exported through long hooks in the same manner as it is in short hooks, and does it still switch export specificity? To address these questions, we employed strain TH16783, which is a fliK deletion mutant carrying a plasmid with an arabinose-inducible fliK gene (Table 1).
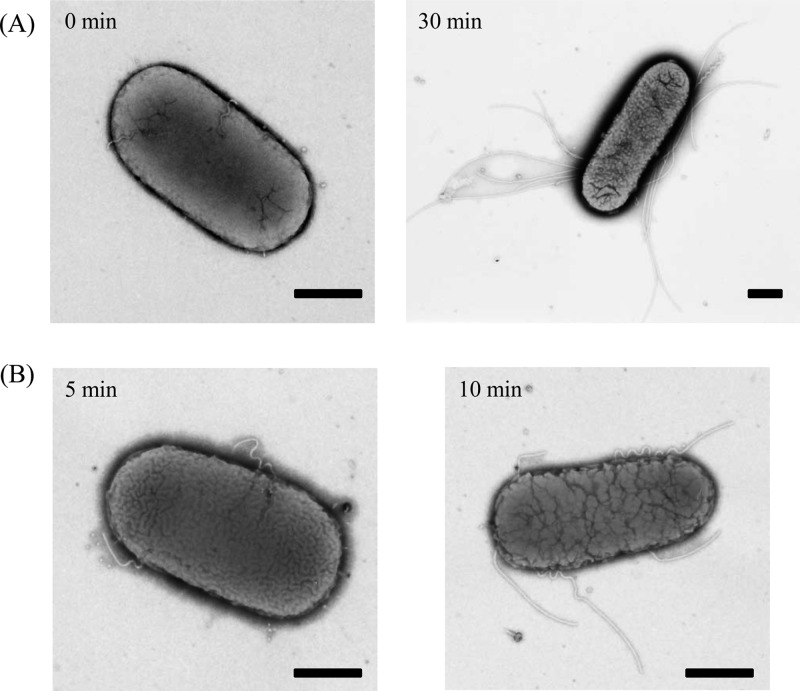
TH16783 cells produced polyhooks when arabinose was absent from the medium (Fig. 6A, left panel). Upon the addition of arabinose, the fliK gene was expressed and TH16783 cells produced normal flagella after 30 min (Fig. 6A, right panel). Why and when did polyhooks disappear? We found that short filaments were attached to the tips of polyhooks after 5 min of arabinose induction. The number of polyhook filaments gradually increased as time passed (Fig. 6B). At longer times, these polyhook filaments seemed to be diluted out by intact flagella newly produced in the presence of FliK. This indicates that immediately after FliK is produced in a polyhook mutant, FliK switches the export gate, allowing the export of FliC. Therefore, we conclude that FliK switches substrate export specificity when the hook is long.
FIG 6.
Electron micrographs of TH16783 cells at 0 and 30 min (A) or 5 and 10 min (B) after the addition of arabinose. Cells were negatively stained with 1% phosphotungstic acid (pH 7). Bars, 500 nm.
FlgM is not secreted by SJW2219 cells at 37°C.
FlgM secretion is as important as FliK secretion in the completion of the flagellum. We examined the onset of FlgM secretion by analyzing the amount of FlgM secreted into the medium by various mutants. FlgM is a small molecule of 97 amino acids and runs near the moving front in SDS gels, where no other bands are observed (Fig. 7A). After a 30-min incubation, a considerable amount of FlgM was secreted from wild-type cells. As much FlgM was secreted by fliC deletion mutant SJW2536 as by the wild type, while only a little FlgM was secreted by fliK deletion mutant SJW108 (Fig. 7B). FlgM was secreted by SJW2219 cells at 30°C but not at 37°C (Fig. 7C), agreeing with the observations mentioned above.
FIG 7.
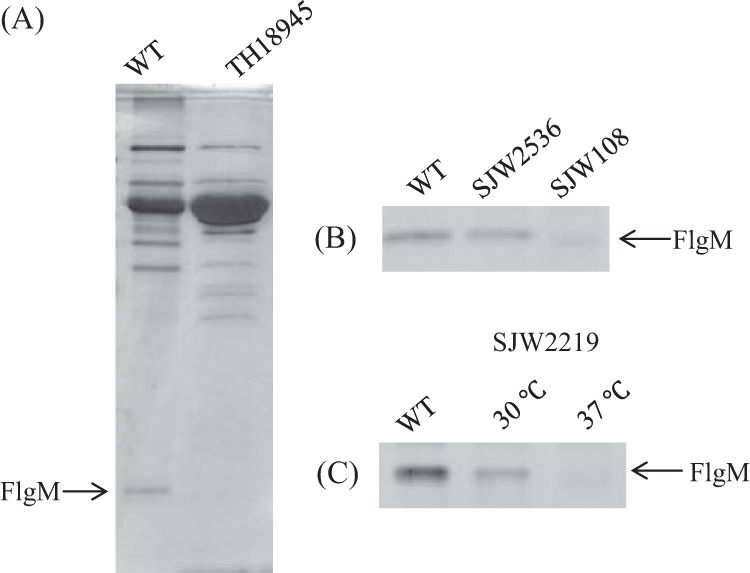
SDS-PAGE pattern of FlgM secreted into the culture medium by SJW2219 cells. (A) Patterns of FlgM secretion by the wild type (WT) and flgM deletion mutant TH18945. (B) Patterns of FlgM secretion by the wild type, fliC deletion mutant SJW2536, and fliK deletion mutant SJW108. (C) Patterns of FlgM secretion by wild-type SJW2219 grown at 30 and 37°C.
Taking all of the data obtained into account, we can add FliK and FlgM to the conventional secretion model; FliK belongs to the early proteins, while FlgM belongs to the late proteins. FliK is secreted without switching gate specificity until the hook reaches a certain length. When FliK switches substrate specificity, the secretion gate shuts off the early proteins, including FliK itself, and allows the late proteins, including FlgM, to pass.
In conclusion, the secretion gate changes export specificity from rod-hook proteins to flagellin-HAP proteins at a certain point during hook elongation. Let us call the rod-hook secretion state the early phase and the filament class secretion state the late phase. In the early phase, FliK is secreted but FlgM is not. On the contrary, in the late phase, FlgM is secreted but FliK is not. The two phases are switched by FliK, but other substrate proteins are passively secreted through the gate according to the phase to which they belong (Fig. 8). Therefore, FliK determines the minimal length of the hook that can support filament assembly. This minimal hook length must be intimately related to the function of the hook as a universal joint.
FIG 8.
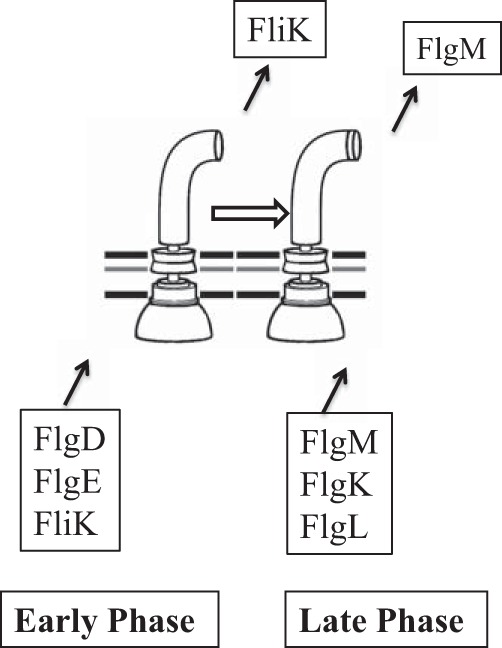
Diagram of the two secretion substrate specificity phases that are switched from the early phase to the late phase by FliK. Secreted proteins of the rod-hook class (represented by FlgD, FlgE, and FliK) are secreted through the early-phase gate, while flagellin-HAP proteins (represented by FlgM, FlgK, and FlgL) are secreted through the late-phase gate. FliK and FlgM are regulatory proteins, while the other secreted substrates are structural. At a certain point in hook elongation, FliK switches substrate specificity from the early phase to the late phase, presumably through a productive interaction with the C terminus of FliK and the FlhB component of the secretion apparatus in the cytoplasmic membrane.
DISCUSSION
Flagellar filament assembly starts after completion of the hook. To ensure that the hook reaches an appropriate length to act as a universal joint, we argue that FliK must measure the length of the hook as it is elongated. Is this so? The rate of wild-type hook elongation has not been determined, presumably because elongation happens too quickly to observed. In contrast, mutant SJW2219 hooks are short because of a low rate of polymerization of the mutant hook protein, which gave us an opportunity to quantitatively monitor the switching of the secretion gate by FliK. In fact, we found that a considerable amount of FliK was secreted from short hooks into the medium.
By analysis of patterns of FliK and FliC secretion from various mutants, we concluded that FliK does not measure the final hook length, but instead it appears to determine the minimum length of the hook. Late gene products, including FlgM, are constantly passed through the secretion gate of the late phase. How does FliK determine the minimal hook length? Erhardt et al. (8) suggested that the speed of FliK passage through the gate might determine the minimal length. They argue that when the hook is short, FliK passes the gate too fast to interact but when the hook is long, FliK passes the gate slowly enough to interact with the switching mechanism. This idea solves the logical problem that a single molecule of FliK plays two roles, measuring and switching to determine hook length. In their theory, FliK plays only one role; FliK does not measure hook length but determines it. Our data strongly support and augment their idea.
ACKNOWLEDGMENTS
We thank Satoshi Inaba, Saori Higaki, and Yuya Tokumaru for their technical help and Tatsuya Yamazaki for illustration. We also thank Kelly Hughes for kindly providing TH strains, Georges Dreyfus for invaluable discussion, and David DeRosier for revising the manuscript.
Footnotes
Published ahead of print 21 February 2014
REFERENCES
- 1.Macnab RM. 1996. Flagella and motility, p. 123–145 In Neidhardt FC, Curtiss R, III, Ingraham JL, Lin ECC, Low KB, Magasanik B, Reznikoff WS, Riley M, Schaechter M, Umbarger HE. (ed), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. ASM Press, Washington, DC [Google Scholar]
- 2.Aizawa S-I. 2009. Flagella, p. 393–403 In Schaechter M. (ed), Encyclopedia of microbiology. Elsevier, Oxford, United Kingdom [Google Scholar]
- 3.Aizawa S-I. 1996. Flagellar assembly in Salmonella typhimurium. Mol. Microbiol. 19:1–5. 10.1046/j.1365-2958.1996.344874.x [DOI] [PubMed] [Google Scholar]
- 4.Hughes KT. 2012. Flagellar hook length is controlled by a secreted molecular ruler. J. Bacteriol. 194:4793–4796. 10.1128/JB.00343-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aizawa S-I. 2012. Mystery of FliK in length control of the flagellar hook. J. Bacteriol. 194:4798–4800. 10.1128/JB.06239-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hirano T, Yamaguchi S, Oosawa K, Aizawa SI. 1994. Roles of FliK and FlhB in determination of flagellar hook length in Salmonella typhimurium. J. Bacteriol. 176:5439–5449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Minamino T, Saijo-Hamano Y, Furukawa Y, González-Pedrajo B, Macnab RM, Namba K. 2004. Domain organization and function of Salmonella FliK, a flagellar hook length control protein. J. Mol. Biol. 341:491–502. 10.1016/j.jmb.2004.06.012 [DOI] [PubMed] [Google Scholar]
- 8.Erhardt M, Singer HM, Wee DH, Keener JP, Hughes KT. 2011. An infrequent molecular ruler controls flagellar hook length in Salmonella enterica. EMBO J. 30:2948–2961. 10.1038/emboj.2011.185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Moriya N, Minamino T, Hughes KT, Macnab RM, Namba K. 2006. The type III flagellar export specificity switch is dependent on FliK ruler and a molecular clock. J. Mol. Biol. 359:466–477. 10.1016/j.jmb.2006.03.025 [DOI] [PubMed] [Google Scholar]
- 10.Uchida K, Dono K, Aizawa S-I. 2013. Length control of the flagellar hook in a temperature-sensitive flgE mutant of Salmonella enterica serovar Typhimurium. J. Bacteriol. 195:3590–3595. 10.1128/JB.00302-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Komoriya K, Shibano N, Higano T, Azuma N, Yamaguchi S, Aizawa SI. 1999. Flagellar proteins and type III-exported virulence factors are the predominant proteins secreted into the culture media of Salmonella typhimurium. Mol. Microbiol. 34:767–779. 10.1046/j.1365-2958.1999.01639.x [DOI] [PubMed] [Google Scholar]
- 12.Mizusaki H, Takaya A, Yamamoto T, Aizawa SI. 2008. Signal pathway in the salt-activated expression of the Salmonella pathogenicity island 1 type III secretion system in Salmonella enterica serovar Typhimurium. J. Bacteriol. 190:4624–4631. 10.1128/JB.01957-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shibata S, Takahashi N, Chevance FF, Karlinsey JE, Hughes KT, Aizawa SI. 2007. FliK regulates flagellar hook length as an internal ruler. Mol. Microbiol. 64:1404–1415. 10.1111/j.1365-2958.2007.05750.x [DOI] [PubMed] [Google Scholar]
- 14.Chilcott GS, Hughes KT. 1998. The type III secretion determinants of the flagellar anti-transcription factor, FlgM, extend from the amino-terminus into the anti-sigma 28 domain. Mol. Microbiol. 30:1029–1040. 10.1046/j.1365-2958.1998.01131.x [DOI] [PubMed] [Google Scholar]
- 15.Kutsukake K, Iino T. 1994. Role of the FliA-FlgM regulatory system on the transcriptional control of the flagellar regulon and flagellar formation in Salmonella typhimurium. J. Bacteriol. 176:3598–3605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Inaba S, Hashimoto M, Jyot J, Aizawa S-I. 2013. Exchangeability of the flagellin (FliC) and the cap protein (FliD) among different species in flagellar assembly. Biopolymers 99:63–72. 10.1002/bip.22141 [DOI] [PubMed] [Google Scholar]