Abstract
The reactive nature of heme enables its use as an enzymatic cofactor while rendering excess heme toxic. The importance of heme detoxification machinery is highlighted by the presence of various types of these homeostatic systems in Gram-positive and Gram-negative microorganisms. A number of pathogens possess orthologs of the HssRS/HrtAB heme detoxification system, underscoring a potential role this system plays in the survival of bacteria in heme-rich environments such as the vertebrate host. In this work, we sought to determine the role of this system in protection against metalloporphyrin heme analogues identified by previous studies as antimicrobial agents. Our findings demonstrate that only toxic metalloporphyrins maximally activate expression of the Staphylococcus aureus heme detoxification system, suggesting that the sensing mechanism of HssRS might require a component of the associated toxicity rather than or in addition to the metalloporphyrin itself. We further establish that only a subset of toxic metalloporphyrins elicit the oxidative damage previously shown to be a significant component of heme toxicity whereas all toxic noniron metalloporphyrins inhibit bacterial respiration. Finally, we demonstrate that, despite the fact that toxic metalloporphyrin treatment induces expression of S. aureus heme detoxification machinery, the HrtAB heme export pump is unable to detoxify most of these molecules. The ineffectiveness of HrtAB against toxic heme analogues provides an explanation for their increased antimicrobial activity relative to heme. Additionally, these studies define the specificity of HssRS/HrtAB, which may provide future insight into the biochemical mechanisms of these systems.
INTRODUCTION
Staphylococcus aureus is a Gram-positive coccus that asymptomatically colonizes the skin and nostrils of nearly one-third of the human population (1, 2). However, through the use of its numerous virulence determinants, this organism has the ability to breach the host defensive barriers to cause invasive diseases such as endocarditis, toxic shock syndrome, osteomyelitis, and sepsis (3, 4). One of these virulence factors is the iron-regulated surface determinant (Isd) system which enables S. aureus to acquire and utilize host iron stores, the bulk of which are coordinated to heme molecules that are bound by hemoglobin and sequestered within erythrocytes (5).
In addition to utilizing heme as a nutrient iron source, S. aureus can employ both endogenously synthesized and exogenously acquired heme as a respiratory cofactor (6). During respiration, electrons are extracted from catabolized carbon sources and passed through a series of electron carriers to the heme molecules housed within cytochromes. The redox potential of heme enables it to accept electrons from membrane-associated quinones and donate them to a terminal electron acceptor, which is oxygen in the case of aerobic respiration, in order to generate a proton motive force (7, 8). The proton motive force is harnessed by respiring bacteria in order to efficiently synthesize the energy transfer molecule ATP.
Although heme is a useful nutrient and cofactor, high levels of this molecule become toxic to most organisms (9). While this toxicity is likely multifaceted, it can be at least in part attributed to the nonspecific redox cycling that occurs between heme and oxygen when excess heme accumulates within the cell membrane. Upon exposure to atmospheric oxygen, membrane-associated heme molecules can auto-oxidize to generate damaging superoxide radicals. This reaction is potentiated by the presence of menaquinone, which reduces heme and allows multiple rounds of auto-oxidation to occur (10).
Because heme toxicity is widespread among numerous organisms, various types of heme detoxification systems are utilized by a diverse array of bacteria. Mechanisms to counteract the effects of heme exposure include degradation, export, and sequestration of heme (9). One of the best-characterized examples of the export mechanism of heme detoxification is the heme-regulated transporter HrtAB. HrtA was first identified in S. aureus as the gene most starkly upregulated in response to heme exposure (11). Further characterization revealed that this transporter is regulated by the heme sensor system HssRS. Upon exposure to toxic levels of heme, the HssRS two-component system activates expression of HrtAB (12, 13). Subsequent work in Lactococcus lactis demonstrated that HrtAB detoxifies heme by exporting excess heme from the cell (14, 15). Deletion of either HssRS or HrtAB results in extreme sensitivity to heme exposure (12, 13, 16). The importance of HssRS/HrtAB for bacterial survival within heme-rich environments such as the vertebrate host is evidenced by the presence of this system in numerous Gram-positive pathogens such as Bacillus anthracis, Enterococcus faecalis, Corynebacterium diphtheriae, Listeria monocytogenes, and Streptococcus agalactiae (13, 17, 18). HrtAB has been shown to be upregulated in a B. anthracis mouse model of infection, further highlighting the role of this system as a virulence determinant (16).
Heme is composed of a tetrapyrrole ring known as protoporphyrin IX (PPIX) bound to an iron atom. However, PPIX can be artificially or naturally complexed to other metals. These molecules are collectively known as noniron metalloporphyrins (MPs). A subset of these heme analogues were identified by previous studies as potential antimicrobial agents capable of inducing bacterial death at low micromolar concentrations (6, 19). MPs are thought to enter the bacterial cell by exploiting endogenous heme acquisition systems (19). Once they are taken up by the cell, these molecules are trafficked to the membrane and exert their toxicity in part by displacing cytochrome-associated heme and inhibiting respiration (6).
In this study, we sought to determine the role that the endogenous heme detoxification system of S. aureus plays in protecting against these heme analogues. Our findings indicate that toxic MPs are maximally sensed by S. aureus HssRS; however, HrtAB is ineffective at detoxifying most of these molecules. The inability of HrtAB to detoxify noniron MPs provides an explanation for their increased antimicrobial activity relative to heme. Additionally, these studies defined the specificity of HssRS/HrtAB, providing functional insight that could assist in the elucidation of the biochemical mechanisms of these important systems.
MATERIALS AND METHODS
Chemicals.
Metalloporphyrins (MPs) were purchased from Frontier Biosciences. All other chemicals used in this study were purchased from Sigma-Aldrich unless otherwise specified.
Bacterial strains and growth conditions.
All experiments in this study were performed in S. aureus clinical isolate Newman (20). Generation of the isogenic mutants lacking hrtB, hssRS, hemA, or menB and the strain expressing the phrtB::xylE reporter fusion was previously described (6, 10, 12, 13, 21). S. aureus cultures were grown on tryptic soy broth (TSB) solidified with 1.5% agar at 37°C or in TSB at 37°C with shaking at 180 rpm. Growth curve analyses were performed in 96-well round-bottomed plates containing 100 μl of culture per well, and all other experiments were performed with 5 ml of bacterial culture grown in 15-ml conical tubes. All cultures were grown in the dark to avoid the generation of singlet oxygen by MPs in the presence of ambient light.
Analysis of oxidative protein damage.
Oxidative protein damage was monitored by immunoblotting as previously described (10). Briefly, cells were cultured overnight in untreated TSB and diluted to an optical density at 600 nm (OD600) of 0.2 in the morning. Five milliliters of bacterial cultures was grown for 6 h in TSB in the presence or absence of a 5 μM concentration of each MP tested. These cultures were pelleted, washed, and suspended in 500 μl TSM (100 mM Tris [pH 7]; 500 mM sucrose; 10 mM MgCl2). Cell walls were removed by addition of 20 μg lysostaphin and incubation for 30 min at 37°C. The resulting protoplasts were pelleted, suspended in lysis buffer (50 mM Tris HCl [pH 7]; 60 mM KCl; 10 mM MgCl2; 2% β-mercaptoethanol), and lysed by sonication. Proteins affected by oxidative damage were detected using an OxyBlot protein oxidation detection kit (Millipore) following the manufacturer's instructions. Briefly, the oxidatively damaged proteins were derivatized with DNP (2,4-dinitrophenylhydrazine). DNP-derivatized proteins were detected via standard immunoblotting methods using a primary antibody specific to the DNP moiety on the damaged proteins (Millipore) and a goat anti-rabbit IgG (Alexa Fluor 680-conjugated) secondary antibody (Invitrogen). Immunoblots were visualized using an Odyssey Imager (Li-COR Biosciences). Staining intensity of individual sample lanes was quantified using the Odyssey software.
ICP-MS ion measurements.
Bacterial cells were cultured overnight in untreated TSB and diluted to an OD600 of 0.1 in the morning. Five milliliters of bacterial cultures was grown for 5 h in TSB in the presence or absence of a 5 μM concentration of each MP tested. These cultures were pelleted, washed, and suspended in 500 μl phosphate-buffered saline (PBS). Cell walls were removed by addition of 20 μg of lysostaphin and incubation for 30 min at 37°C. The resulting protoplasts were pelleted, suspended in 500 μl Chelex-treated PBS, and lysed by sonication. Cell membranes were pelleted at 100,000 × g for 30 min. Membranes were washed with 500 μl Chelex-treated PBS and suspended in 200 μl Chelex-treated PBS. Samples were normalized to a concentration of 1.33 μg/μl of total protein, and 100 μl of each sample was digested by adding 1 ml 50% HNO3 (Optima grade; Fisher) followed by overnight incubation at 50°C in a metal-free 15-ml conical tube. After digestion, samples were diluted to a 10-ml final volume in Milli-Q water and submitted for inductively coupled plasma mass spectrometry (ICP-MS) analysis at the Vanderbilt Mass Spectrometry Research Center. Levels of cobalt, chromium, copper, iron, gallium, manganese, nickel, tin, and zinc were measured as a proxy for CoPPIX, CrPPIX, CuPPIX, FePPIX, GaPPIX, MnPPIX, NiPPIX, SnPPIX, and ZnPPIX accumulation, respectively. Ion levels were normalized to the endogenous ion levels of untreated samples.
Spectrophotometric measurement of MP accumulation.
MP accumulation was measured similarly to previously described methods (10). Briefly, cells were cultured overnight in untreated TSB and diluted to an OD600 of 0.2 in the morning. Five milliliters of the diluted cultures was grown for 6 h in the presence or absence of a 5 μM concentration of each MP tested. These cultures were pelleted, washed, suspended in 500 μl TSM, and treated with 20 μg of lysostaphin to remove cell walls. The resulting protoplasts were pelleted, suspended in TKM (50 mM Tris HCl [pH 7]; 60 mM KCl; 10 mM MgCl2), and lysed by sonication. Samples were matched to equivalent protein concentrations (500 μg total protein) and diluted into a final volume of 550 μl in TKM containing 300 mM NaCl and 24% dimethyl sulfoxide (DMSO). Samples were then acidified through the addition of 0.2 ml of 50 mM glycine-HCl (pH 1), and the MP was subsequently extracted by adding 0.2 ml of chloroform and vortex mixing several times. Spectral absorbance readings were taken of the chloroform phase at wavelengths ranging from 300 to 500 nm. CuPPIX, FePPIX (heme), GaPPIX, MnPPIX, NiPPIX, SnPPIX, and ZnPPIX were monitored by absorbance levels at 408 nm, 388 nm, 411 nm, 372 nm, 404 nm, 415 nm, and 418 nm, respectively. Prior to the analysis, the peak absorbance values for individual MPs had been determined using solutions of MP suspended in acidified chloroform. Absorbance values for untreated samples were subtracted from the absorbance values for treated samples.
XylE assays.
XylE reporter assays were performed similarly to previously described methods (13). Cultures of S. aureus Newman harboring a hrtAB promoter-xylE fusion plasmid were grown for 18 h with shaking at 37°C in 15-ml conical tubes containing 5 ml of TSB and 10 μg/ml chloramphenicol. Bacteria were subcultured 1:100 into 1.5-ml snap-cap tubes preloaded with 0.5 ml TSB supplemented with 10 μg/ml chloramphenicol and metalloporphyrin in triplicate. Subcultures were shaken for 2 h at 37°C and centrifuged at 16,000 × g for 5 min. Culture supernatants were aspirated, and pellets were resuspended in 0.5 ml of wash buffer (20 mM potassium phosphate [pH 7.6]). Washed bacterial suspensions were centrifuged at 16,000 × g for 5 min and resuspended in 150 μl lysis solution (100 mM potassium phosphate buffer [pH 8]; 10% acetone; 25 μg/ml lysostaphin). Bacterial suspensions were incubated for 20 min at 37°C, transferred to ice for 5 min, and then centrifuged at 20,000 × g for 30 min at 4°C. Twenty microliters of supernatant was added to a 96-well plate, and 200 μl of substrate buffer (100 mM potassium phosphate [pH 8.0]; 0.2 mM pyrocatechol) was added to each well. Formation of 2-hydroxymuconic semialdehyde was tracked by measuring the absorbance at 375 nm every minute for 30 min on a Varian MP 50 microplate reader. Protein concentrations in samples were determined by a bicinchoninic acid (BCA) assay (Pierce). One unit of XylE-specific activity is defined as the formation of 1 nmol of 2-hydroxymuconic semialdehyde per min per mg of cellular protein at 30°C (22).
qRT-PCR.
Bacterial cells were cultured overnight in untreated TSB and diluted to an OD600 of 0.1 in the morning. Five milliliters of the diluted cultures was grown for 5 h in the presence or absence of a 5 μM concentration of each MP tested. These cultures were pelleted and suspended in 750 μl of LETS buffer (0.1 M LiCl; 10 mM EDTA; 10 mM Tris-HCl [pH 7.4]; 1% sodium dodecyl sulfate). Cell walls were removed by bead beating the samples in Lysing Matrix B tubes (MP Biomedicals) followed by incubation at 55°C for 5 min. The supernatant was removed from the lysis beads and mixed with 1 ml of TRI reagent (Sigma). RNA purification was accomplished following the manufacturer's instructions. DNA contamination was removed by incubating the samples in the presence of RQ1 DNase (Promega) for 2 h at 37°C followed by phenol-chloroform extraction and ethanol precipitation. cDNA was generated by mixing 1.2 μg RNA, 2 μl random primers (Promega), and 1.5 μl Moloney murine leukemia virus (M-MLV) reverse transcriptase (RT) (Promega) in a 25-μl final volume containing a 1× final concentration of M-MLV buffer and incubating for 1 h at 37°C. RNasin (Promega) was included during DNase treatment and cDNA synthesis to minimize RNA degradation. Quantitative RT-PCRs (qRT-PCRs) were set up using diluted cDNA and iQ SYBR green Supermix (Bio-Rad) in a 12.5-μl final volume following the manufacturer's instructions. The primer set used to monitor hrtB gene expression was TAGCTCAAGGGCTTGGTAGG and TCTCAATTTGCGGCTCTTTC. The primer set used to monitor 16S expression was GCTGCAGCTAACGCATTAAGCACT and TTAAACCACATGCTCCACCGCTTG. The hrtB expression of each sample was normalized to the 16S expression prior to determining the fold increase relative to untreated samples.
RESULTS AND DISCUSSION
Certain MPs are toxic to S. aureus.
Previous studies have identified certain noniron metalloporphyrins (MPs) as potent, broad-range antimicrobial compounds (6, 19, 23, 24). At least a portion of MP toxicity has been attributed to the structural homology shared between these molecules and the iron-containing heme cofactor utilized by various organisms to perform numerous cellular functions ranging from catalase activity to respiration (6, 19). Additionally, some of these porphyrin compounds act as photosensitizers, reacting with light to generate damaging singlet oxygen (25). While the broad-spectrum antimicrobial activity of many MPs is widely recognized, MP potencies can differ between studies using different bacterial strains and growth conditions. Therefore, we first sought to establish the subset of MPs that were toxic to S. aureus strain Newman under our laboratory growth conditions. To eliminate the compounding issue of photosensitization, all cultures were grown in the absence of light.
In order to determine the effects of a wide range of MP heme analogues on S. aureus growth, we employed PPIX molecules that were complexed to cobalt, chromium, copper, iron, gallium, manganese, nickel, tin, and zinc atoms. These compounds are commonly denoted CoPPIX, CrPPIX, CuPPIX, FePPIX (heme), GaPPIX, MnPPIX, NiPPIX, SnPPIX, and ZnPPIX, respectively. These compounds were supplemented at a final concentration of 10 μM into S. aureus cultures grown in TSB, and growth was monitored over a 24-h time course (Fig. 1). Under these conditions, the only MPs that exhibit antimicrobial activity against S. aureus are FePPIX (heme), GaPPIX, MnPPIX, and ZnPPIX.
FIG 1.
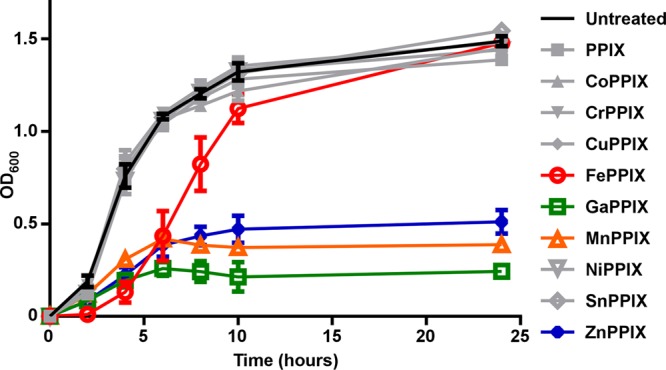
Specific MPs have antimicrobial activity against S. aureus. S. aureus was cultured in TSB in the presence or absence of 10 μM nonmetalated PPIX or various MPs. Growth was monitored by measurement of optical density at 600 nm (OD600). Toxic MPs, which reduce growth compared to untreated culture, are highlighted in color. FePPIX (heme) is shown in red, GaPPIX in green, MnPPIX in orange, and ZnPPIX in blue. Growth assays were performed at least three times on separate days. Additionally, technical triplicates were performed on each day. Error bars represent standard errors of the means (SEM) of the results of the biological replicates.
Most toxic and nontoxic MPs accumulate to similar levels in the cell membrane.
Because previous studies have shown that the toxic MPs FePPIX (heme) and GaPPIX are preferentially trafficked to the cell membrane (5, 6, 10), we sought to determine whether or not a lack of membrane accumulation could provide an explanation for the decreased toxicity of other MPs. The membranes of cells that had been treated with various MPs were isolated and analyzed using inductively coupled plasma mass spectrometry (ICP-MS) to detect levels of the ions unique to each MP. Most toxic and nontoxic MPs were found in similar levels of abundance within the cell membrane, indicating that the inability to traffic to the membrane was not responsible for the lack of toxicity exhibited by certain MPs (Fig. 2). Interestingly, we observed that CrPPIX did not enter the cell membrane as efficiently as other MPs, which might be a factor contributing to the decreased toxicity of this molecule. The reason behind the lower levels of CrPPIX accumulation in the membrane is unknown, but they might be due to the limited target range of a heme uptake system. However, a staphylococcal heme uptake system that is expressed under the iron-replete growth conditions used in these experiments has not yet been characterized.
FIG 2.
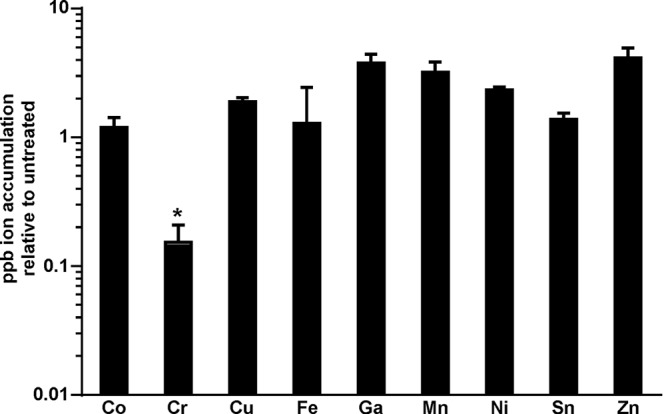
Most MPs accumulate to approximately equivalent levels within the staphylococcal membrane. Cell membranes were isolated from bacteria grown in the presence of 5 μM concentrations of the various MPs highlighted in this study. Samples were normalized based on protein content and analyzed for the accumulation of the metal ions unique to each MP using ICP-MS. Error bars represent standard deviations of the results from triplicate samples. The asterisk (*) denotes samples with decreased ion accumulation (P < 0.05) as determined by a two-tailed Student's t test.
Toxic MP exposure results in maximal upregulation of heme detoxification machinery.
Previous studies have demonstrated that toxic MPs can exploit endogenous heme uptake systems in order to gain entry into the cell (19). In S. aureus, various components of the Isd heme acquisition system have been biochemically shown to interact with MPs. For example, a heme receptor site on the IsdH surface protein has high affinity for MnPPIX and GaPPIX in addition to its cognate ligand heme but for not CuPPIX, MgPPIX, or ZnPPIX (26). This characterization of the ligand specificity of IsdH led to the discovery that NEAT-domain-containing heme receptors recognize the oxidation state of target MPs. Additional studies have found that IsdG and IsdI, the terminal enzymes of the S. aureus Isd system, can recognize CoPPIX, GaPPIX, MnPPIX, and ZnPPIX in addition to heme (27). These enzymes function as heme oxygenases that extract iron from heme for use as a nutrient source under iron-depleted growth conditions. Interestingly, despite the ability of IsdG and IsdI to associate to noniron MPs, the heme oxygenases are incapable of degrading these molecules. These studies enabled high-resolution crystallographic visualization of the ligand-bound precleavage state of a catalytically active IsdI bound to CoPPIX (27).
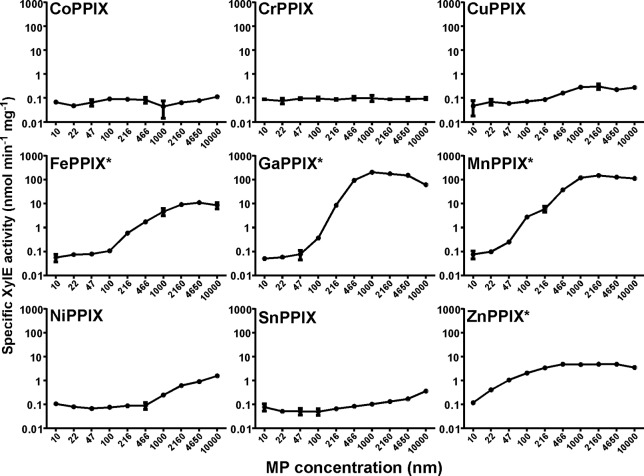
Because MP studies have yielded invaluable insight into the biochemical function of various components of the S. aureus heme acquisition system, we sought to determine the impact of MP treatment on the S. aureus heme detoxification system in order to better understand the specificity of HssRS/HrtAB. We first assessed whether or not exposure to MPs could be sensed by the heme-sensing two-component system HssRS. In order to monitor HssRS activation, we employed a xylE reporter gene fused to the hrtAB promoter, the only characterized target for HssRS activation (12, 13). S. aureus strains expressing this reporter construct were exposed to various concentrations of each MP, and XylE enzymatic activity was monitored as a readout for HssRS activation (Fig. 3). The compounds that most dramatically activated the heme-sensing system were the natural stimulator FePPIX (heme) and the toxic MPs MnPPIX and GaPPIX. These compounds activate HssRS when present at submicromolar levels. Exposure to the toxic compound ZnPPIX also elicited a more modest HssRS activation at submicromolar levels. Certain nontoxic MPs such as NiPPIX were capable of slight activation of the heme detoxification system only when present at higher concentrations, while others compounds did not exhibit any HssRS activation under the conditions tested.
FIG 3.
Toxic MPs activate the S. aureus heme detoxification machinery. Activation of the HssRS/HrtAB system was monitored using a xylE reporter gene under the control of the hrtAB promoter as previously described (13). Growth curve analyses were performed in triplicate. Error bars correspond to 1 standard deviation of the mean. The asterisks (*) highlight compounds that were found to be toxic to S. aureus by growth curve analysis.
The ability of these molecules to activate HssRS was further assessed using a more sensitive qRT-PCR analysis to monitor hrtB expression. In this set of experiments, we sought to determine whether or not the slight activation elicited by micromolar concentrations of certain MPs was statistically significant. qRT-PCR analyses revealed that, at low micromolar concentrations, the nontoxic MPs CoPPIX, NiPPIX, and SnPPIX were capable of moderate HssRS activation, although not to the same extent as the toxic MPs (Fig. 4). However, due to a high level of variability in the ability of CoPPIX to elicit hrtB expression, this data set did not reach statistical significance. The qRT-PCR data are consistent with the slight XylE activation that was observed in some of the nontoxic MP treatments when these MPs were provided at increasing micromolar concentrations (Fig. 3). However, because nontoxic MPs cannot induce hrtB expression at submicromolar concentrations and elicit much lower levels of hrtB expression than toxic MPs when present at low micromolar concentrations, we conclude that only toxic MPs are capable of maximal activation of the heme detoxification system. In all cases, activation of hrtB expression is dependent upon the presence of HssRS because no increase is observed in an ΔhssRS mutant (Fig. 4).
FIG 4.
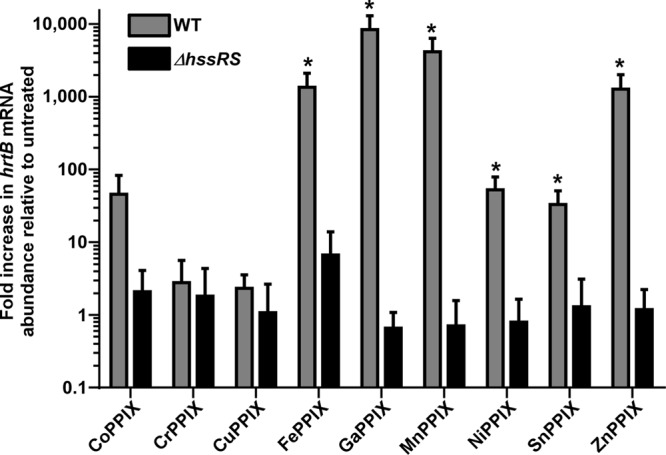
MP induction of hrtB gene expression is HssRS dependent. RNA was isolated from S. aureus grown in the presence or absence of 5 μM MP. Error bars correspond to standard deviations of the results from triplicate samples. The asterisks (*) denotes statistically significant changes (P < 0.05) relative to untreated samples as determined by a two-tailed Student's t test.
Interestingly, other structurally unrelated heme sensing systems such as the L. lactis HrtR cytoplasmic regulator have MP specificities that result in target gene activation (15). HrtR is robustly activated by heme and GaPPIX, modestly activated by MnPPIX, and perhaps weakly activated by ZnPPIX, while remaining insensitive to other MPs (15). The findings showing that toxic MPs are more readily sensed by multiple heme-sensing systems than nontoxic MPs could have multiple implications. One explanation for this trend could be that the characteristics that enable these molecules to be sensed by HssRS and HrtR are the same characteristics that induce toxicity. Alternatively, these heme-sensing systems might be capable of sensing a component of the associated toxicity such as oxidative stress in addition to the presence of the MP in order to fully activate gene expression.
Oxidative damage is a component of some MP toxicity.
Because oxidative damage is the only component of heme toxicity that has been characterized (10), we sought to determine whether or not oxidative stress contributes to the toxic effects of other MPs. In order to monitor the presence of MP-induced oxidative stress, we used an OxyBlot kit (Millipore), which detects oxidative protein damage through standard immunoblotting techniques that can be visualized as high levels of antibody staining throughout the oxidatively damaged samples (Fig. 5A and B). Because heme-induced oxidative stress is primarily evident in S. aureus strains lacking functional heme detoxification systems, oxidative damage was assessed in both wild-type (WT) S. aureus and a ΔhrtB mutant. These strains were grown for 6 h in the presence or absence of 5 μM concentrations of various MPs prior to analysis. As expected, WT S. aureus displayed no detectable protein damage in the presence of FePPIX (heme) whereas ΔhrtB S. aureus samples accrued high levels of heme-induced oxidative damage due to the absence of a functional heme detoxification system. Interestingly, high levels of oxidative damage were observed upon exposure to the toxic GaPPIX and ZnPPIX MPs in both WT and ΔhrtB S. aureus strains, indicating that these MPs share a toxicity with heme that cannot be fully overcome by the presence of a detoxification system. Surprisingly, a more modest but reproducible level of oxidative damage was observed upon treatment with nontoxic SnPPIX, revealing that a certain level of oxidative protein damage can be tolerated by S. aureus with no apparent growth detriment. Intriguingly, the toxic MnPPIX MP did not induce a noticeable increase in oxidative damage, hinting at additional previously uncharacterized aspects of MP toxicity that remain to be discovered. In total, these data indicate that oxidative stress is not the component of MP toxicity that is recognized by heme-sensing systems.
FIG 5.
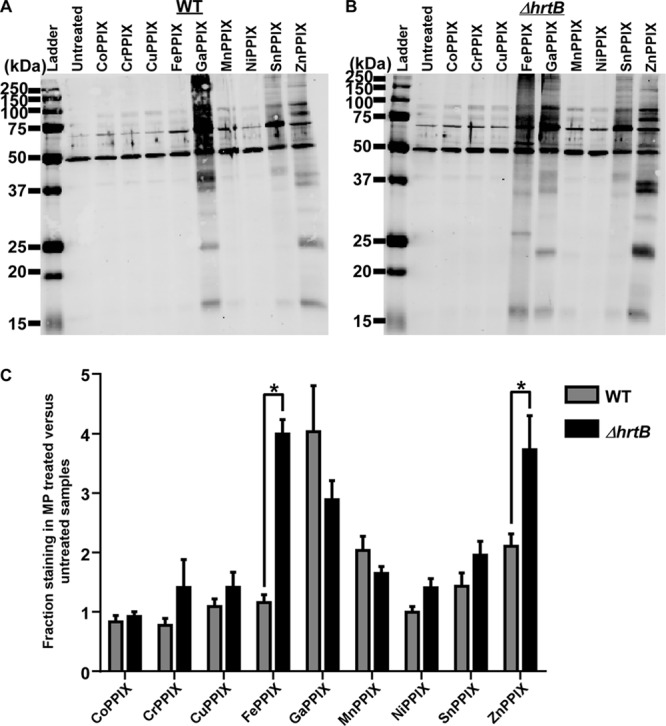
A subset of MPs induces oxidative protein damage in S. aureus. Total protein was extracted from the protoplasts of WT S. aureus (A) or the ΔhrtB S. aureus mutant strain that was grown in the presence or absence of 5 μM MP (B). Representative immunoblots are shown containing protein samples that had been derivatized with DNP (2,4-dinitrophenylhydrazine) to modify sites of oxidative damage. Blots were probed with antibodies specific to the DNP moiety on the proteins. The ladder shown in the image is the Precision Plus Protein All Blue Standard (Bio-Rad). (C) Quantification of immunoblot replicates depicting the fraction of oxidative protein damage in MP-treated samples compared with untreated samples as determined by densitometry. Graphs represent averages of the results determined with at least three independent immunoblots from samples processed on separate days. Error bars represent SEM. The asterisk (*) denotes P < 0.05 as determined by a two-tailed Student's t test.
The quantification of the WT and ΔhrtB S. aureus data sets indicated that the presence of a functional detoxification system differentially influenced the oxidative damage induced by individual MPs (Fig. 5C). ZnPPIX-induced oxidative damage was reduced in the HrtAB-expressing cells in a manner similar to that seen with heme-induced stress, albeit to a lesser extent. However, the HrtAB system was incapable of reducing GaPPIX-associated protein damage. In fact, the presence of the HrtAB detoxification pump appeared to exacerbate GaPPIX-induced oxidative damage to levels verging on statistical significance. These observations indicated that the specificity of the HrtAB pump might differ from the specificity of the HssRS sensing system.
Toxic noniron MPs inhibit bacterial respiration.
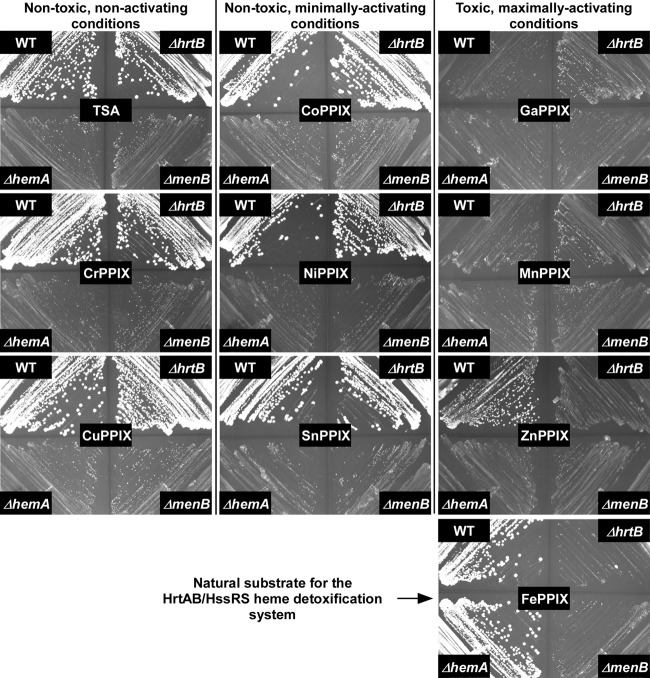
Because oxidative stress does not occur in the presence of all toxic MPs, we sought to identify another source of toxicity shared by these heme analogues. GaPPIX and ZnPPIX have previously been shown to inhibit aerobic respiration, presumably by displacing the critical heme cofactors housed within cytochromes (6). This mechanism of toxicity is akin to that of the gallium toxicity previously described for Pseudomonas aeruginosa in which gallium poisons iron-based processes due to chemical similarities between the two elements (28). Nonrespiring staphylococcal cells display a slow-growth phenotype known as a small-colony variant (SCV). SCVs can be created genetically through mutation of heme and menaquinone biosynthetic pathways. WT S. aureus and ΔhrtB S. aureus were streaked onto TSA plates in the presence or absence of various MPs adjacent to the genetically defined SCVs, the ΔhemA and ΔmenB S. aureus strains, which are deficient in heme and menaquinone biosynthesis, respectively (Fig. 6). Neither nontoxic noninducers nor nontoxic moderate inducers of the HssRS system impacted the ability of WT or ΔhrtB S. aureus cells to respire as evidenced by the large-colony morphology of these strains relative to the genetically defined SCV strains. In contrast, cells grown on toxic, noniron MPs reverted to the SCV phenotype. In the presence of GaPPIX and MnPPIX, WT and ΔhrtB strains were phenotypically indistinguishable from the genetic SCVs, ΔhemA and ΔmenB S. aureus. However, on plates impregnated with ZnPPIX, WT cells displayed an intermediate-growth phenotype relative to the definitive SCV morphology of ΔhrtB S. aureus, indicating that the HrtAB heme detoxification pump can afford some level of protection from the respiratory inhibition induced by this heme analogue. However, the HrtAB-mediated detoxification of ZnPPIX is far less dramatic than the impact of this system in the presence of heme, the natural substrate of the detoxification pump. On plates infused with FePPIX (heme), ΔhemA S. aureus displays a normal colony morphology that is indistinguishable from that of WT cells because the exogenously acquired heme can substitute for the biosynthetic deficiency of this mutant (Fig. 6). In contrast, the ΔhrtB mutant is incapable of growth on this substrate. This inhibition of growth is completely reversed in WT cells through the action of the HrtAB heme detoxification system.
FIG 6.
Toxic noniron MPs induce the small-colony variant (SCV) phenotype. WT and ΔhrtB strains were streaked onto TSA plates adjacent to the characterized SCV controls, the ΔhemA and ΔmenB mutants. These plates were either left untreated or impregnated with 5 μM MP. Images were taken after 20 h of growth. The induction of the SCV phenotype is indicative of respiration deficiency.
These findings distinguish the ability of some toxic MPs to induce oxidative stress from the ability of all toxic noniron MPs to inhibit respiration, highlighting at least two separate antimicrobial activities of these heme analogues. While the inhibition of bacterial respiration is common among all toxic noniron MPs, we do not believe that this feature plays any role in the activation of HssRS due to the fact that this system is designed to sense heme, a compound required for respiration. This belief is further supported by the fact that the HssRS system is not constitutively activated in genetically defined SCVs lacking heme or menaquinone biosynthesis (data not shown). Therefore, if HssRS is sensing a component of MP-associated toxicity, these data indicate that additional aspects of MP toxicity remain to be discovered.
HrtAB does not export noniron MPs as efficiently as FePPIX (heme).
In order to further assess the specificity of the HrtAB component of the S. aureus heme detoxification system, we grew WT and ΔhrtB strains in the presence or absence of 5 μM concentrations of various MPs and monitored the accumulation of these compounds after 6 h of exposure (Fig. 7). The method of MP detection employed for these experiments utilized spectrophotometric measurements to detect only intact MPs whereas ICP-MS analyses, while more sensitive, cannot distinguish between intact or degraded MPs. As expected, WT cells accumulated significantly smaller amounts of FePPIX (heme) compared to the ΔhrtB strain lacking a functional heme export pump. ZnPPIX was also present at lower levels in WT S. aureus cells compared to the ΔhrtB strain, indicating that this MP can be exported by HrtAB. However, ZnPPIX export by HrtAB does not appear to be nearly as efficient as the HrtAB-mediated efflux of heme. The ZnPPIX accumulation data corroborate the oxidative protein damage analyses and colony morphology on MP-infused plates, which revealed modest protection from ZnPPIX-induced damage and respiration deficiency through the action of the HrtAB detoxification system (Fig. 5C and 6). HrtAB was not capable of exporting the toxic MPs GaPPIX and MnPPIX or the nontoxic MPs CuPPIX, NiPPIX, and SnPPIX as evidenced by the fact that no significant decrease in MP accumulation was detected in WT versus ΔhrtB cells (Fig. 7). While CoPPIX and CrPPIX measurements were also attempted, no detectable levels of these compounds were observed in either strain (data not shown). The lack of CrPPIX detection is likely due to the fact that this compound accumulates to significantly lower levels in the cells relative to other MPs (Fig. 2) whereas CoPPIX simply does not have as strong of a spectral absorbance profile as other compounds. Interestingly, MnPPIX levels were actually increased in the WT strain compared to the ΔhrtB mutant (Fig. 7). This surprising result suggests that the presence of high levels of HrtAB within the cell membrane assists in MnPPIX uptake for unknown reasons. In total, the measurements of MP accumulation indicate that the system specificity of HrtAB is narrower than that of HssRS, with heme being most efficiently transported and ZnPPIX being exported to a lesser extent.
FIG 7.
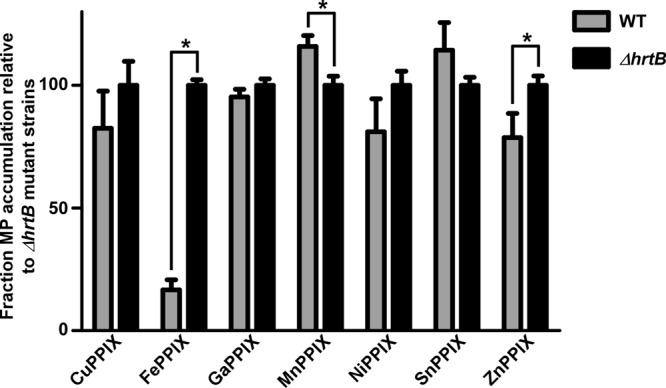
The HrtAB pump does not efficiently export most MPs. MP accumulation was measured in WT S. aureus and in a ΔhrtB mutant grown in the presence or absence of 5 μM MP. Error bars represent standard deviations of the results from triplicate samples. The asterisk (*) indicates P < 0.05 as determined by a two-tailed Student's t test.
The HrtAB detoxification pump does not protect against most toxic MPs and can actually exacerbate certain instances of MP toxicity.
Due to the narrow target specificity of HrtAB, we sought to assess the role of the S. aureus heme detoxification system in protection against toxic MPs through a direct comparison of WT and ΔhrtB S. aureus strains. Therefore, these strains were cultured in the presence or absence of 10 μM MP, and growth was monitored after 24 h of incubation (Fig. 8). As expected, the ΔhrtB strain was much more susceptible to FePPIX (heme) exposure than WT S. aureus. Consistent with the oxidative damage, respiratory inhibition, and MP accumulation data, strains incapable of producing HrtAB were modestly more susceptible to ZnPPIX treatment. Interestingly, the ΔhrtB strain grew slightly better in the presence of both GaPPIX and MnPPIX than WT S. aureus. These surprising data indicate that expression of the S. aureus heme detoxification system actually becomes detrimental upon exposure to certain toxic MPs, increasing the bactericidal activities of these already potently antimicrobial compounds. The source of this increased sensitivity is unknown but could potentially be attributed to a number of factors. One explanation could be ascribed to the relatively high levels of HssRS activation observed upon GaPPIX or MnPPIX treatment. XylE and qRT-PCR data indicate that hrtAB expression is approximately 10-fold higher upon MnPPIX or GaPPIX treatment than upon heme treatment (Fig. 3 and 4). This increased expression might be caused by the fact that both GaPPIX and MnPPIX are capable of activating HssRS but incapable of being effluxed by HrtAB. Alternatively, GaPPIX and MnPPIX may simply be better biochemical activators of HssRS. In either case, because of these high levels of hrtAB expression, GaPPIX- and MnPPIX-treated cells were likely expending needless amounts of energy on the massive upregulation of HrtAB, which could potentially impact growth rates. Additionally, the overexpression of a heme export pump may act to deplete beneficial cellular heme stores, inactivating heme-dependent enzymes such as cytochromes and catalase.
FIG 8.
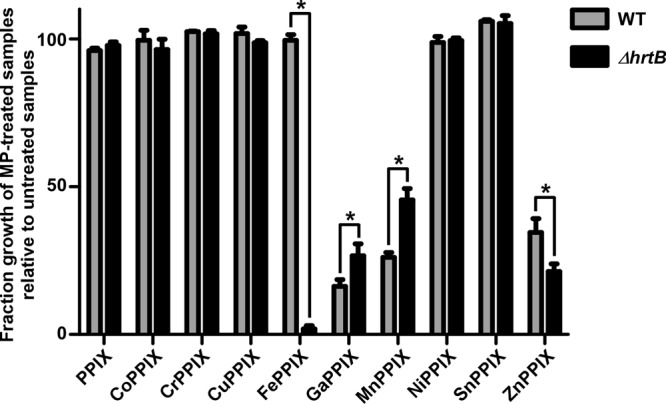
The ΔhrtB S. aureus strain is variably sensitive to exposure to toxic MPs. WT and ΔhrtB S. aureus strains were cultured in TSB in the presence or absence of 10 μM nonmetalated PPIX or various MPs. Growth was monitored at 24 h postinoculation through measurement of OD600. Growth assays were performed at least three times on separate days. Additionally, technical triplicates were performed on each day. Error bars represent SEM of the biological replicates. The asterisk (*) indicates P < 0.05 as determined by a two-tailed Student's t test.
Conclusions.
The data presented herein define the MP specificity of the S. aureus HssRS/HrtAB heme detoxification system, demonstrating that HssRS is broadly responsive to toxic MPs and minimally activated by certain nontoxic MPs whereas HrtAB is capable of detoxifying only a subset of these compounds. Additionally, attempts to determine the source of MP stress revealed that only certain toxic MPs induce oxidative protein damage whereas all toxic noniron MPs inhibit bacterial respiration. Finally, these studies demonstrated that activation of the S. aureus heme detoxification system can actually exacerbate the toxic effects of GaPPIX and MnPPIX. Therefore, these potently antimicrobial heme analogues not only hijack cellular heme uptake systems in order to exert their bactericidal effects but can also exploit the S. aureus heme detoxification machinery in order to efficiently kill this problematic pathogen.
ACKNOWLEDGMENTS
We thank members of the Skaar laboratory for critical reading of the manuscript.
Work in the Skaar laboratory is supported by grants AI069233 and AI073843 from the National Institutes of Health. C.A.W. was supported by grant F32-AI100535 from the National Institute of Allergy and Infectious Diseases.
The content of this article is solely our responsibility and does not necessarily represent the official views of the National Institute of Allergy and Infectious Diseases or the National Institutes of Health.
Footnotes
Published ahead of print 17 January 2014
REFERENCES
- 1.Kuehnert MJ, Kruszon-Moran D, Hill HA, McQuillan G, McAllister SK, Fosheim G, McDougal LK, Chaitram J, Jensen B, Fridkin SK, Killgore G, Tenover FC. 2006. Prevalence of Staphylococcus aureus nasal colonization in the United States, 2001–2002. J. Infect. Dis. 193:172–179. 10.1086/499632 [DOI] [PubMed] [Google Scholar]
- 2.DeLeo FR, Otto M, Kreiswirth BN, Chambers HF. 2010. Community-associated meticillin-resistant Staphylococcus aureus. Lancet 375:1557–1568. 10.1016/S0140-6736(09)61999-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gordon RJ, Lowy FD. 2008. Pathogenesis of methicillin-resistant Staphylococcus aureus infection. Clin. Infect. Dis. 46(Suppl 5):S350–S359. 10.1086/533591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Klevens RM, Morrison MA, Nadle J, Petit S, Gershman K, Ray S, Harrison LH, Lynfield R, Dumyati G, Townes JM, Craig AS, Zell ER, Fosheim GE, McDougal LK, Carey RB, Fridkin SK. 2007. Invasive methicillin-resistant Staphylococcus aureus infections in the United States. JAMA 298:1763–1771. 10.1001/jama.298.15.1763 [DOI] [PubMed] [Google Scholar]
- 5.Skaar EP, Humayun M, Bae T, DeBord KL, Schneewind O. 2004. Iron-source preference of Staphylococcus aureus infections. Science 305:1626–1628. 10.1126/science.1099930 [DOI] [PubMed] [Google Scholar]
- 6.Hammer ND, Reniere ML, Cassat JE, Zhang Y, Hirsch AO, Hood MI, Skaar EP. 2013. Two heme-dependent terminal oxidases power Staphylococcus aureus organ-specific colonization of the vertebrate host. mBio 4:e00241–13. 10.1128/mBio.00241-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Simon J, van Spanning RJ, Richardson DJ. 2008. The organisation of proton motive and non-proton motive redox loops in prokaryotic respiratory systems. Biochim. Biophys. Acta 1777:1480–1490. 10.1016/j.bbabio.2008.09.008 [DOI] [PubMed] [Google Scholar]
- 8.Borisov VB, Gennis RB, Hemp J, Verkhovsky MI. 2011. The cytochrome bd respiratory oxygen reductases. Biochim. Biophys. Acta 1807:1398–1413. 10.1016/j.bbabio.2011.06.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Anzaldi LL, Skaar EP. 2010. Overcoming the heme paradox: heme toxicity and tolerance in bacterial pathogens. Infect. Immun. 78:4977–4989. 10.1128/IAI.00613-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wakeman CA, Hammer ND, Stauff DL, Attia AS, Anzaldi LL, Dikalov SI, Calcutt MW, Skaar EP. 2012. Menaquinone biosynthesis potentiates haem toxicity in Staphylococcus aureus. Mol. Microbiol. 86:1376–1392. 10.1111/mmi.12063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Friedman DB, Stauff DL, Pishchany G, Whitwell CW, Torres VJ, Skaar EP. 2006. Staphylococcus aureus redirects central metabolism to increase iron availability. PLoS Pathog. 2:e87. 10.1371/journal.ppat.0020087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stauff DL, Torres VJ, Skaar EP. 2007. Signaling and DNA-binding activities of the Staphylococcus aureus HssR-HssS two-component system required for heme sensing. J. Biol. Chem. 282:26111–26121. 10.1074/jbc.M703797200 [DOI] [PubMed] [Google Scholar]
- 13.Torres VJ, Stauff DL, Pishchany G, Bezbradica JS, Gordy LE, Iturregui J, Anderson KL, Dunman PM, Joyce S, Skaar EP. 2007. A Staphylococcus aureus regulatory system that responds to host heme and modulates virulence. Cell Host Microbe 1:109–119. 10.1016/j.chom.2007.03.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pedersen MB, Garrigues C, Tuphile K, Brun C, Vido K, Bennedsen M, Mollgaard H, Gaudu P, Gruss A. 2008. Impact of aeration and heme-activated respiration on Lactococcus lactis gene expression: identification of a heme-responsive operon. J. Bacteriol. 190:4903–4911. 10.1128/JB.00447-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lechardeur D, Cesselin B, Liebl U, Vos MH, Fernandez A, Brun C, Gruss A, Gaudu P. 2012. Discovery of intracellular heme-binding protein HrtR, which controls heme efflux by the conserved HrtB-HrtA transporter in Lactococcus lactis. J. Biol. Chem. 287:4752–4758. 10.1074/jbc.M111.297531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stauff DL, Skaar EP. 2009. Bacillus anthracis HssRS signalling to HrtAB regulates haem resistance during infection. Mol. Microbiol. 72:763–778. 10.1111/j.1365-2958.2009.06684.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bibb LA, Schmitt MP. 2010. The ABC transporter HrtAB confers resistance to hemin toxicity and is regulated in a hemin-dependent manner by the ChrAS two-component system in Corynebacterium diphtheriae. J. Bacteriol. 192:4606–4617. 10.1128/JB.00525-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fernandez A, Lechardeur D, Derre-Bobillot A, Couve E, Gaudu P, Gruss A. 2010. Two coregulated efflux transporters modulate intracellular heme and protoporphyrin IX availability in Streptococcus agalactiae. PLoS Pathog. 6:e1000860. 10.1371/journal.ppat.1000860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stojiljkovic I, Kumar V, Srinivasan N. 1999. Non-iron metalloporphyrins: potent antibacterial compounds that exploit haem/Hb uptake systems of pathogenic bacteria. Mol. Microbiol. 31:429–442. 10.1046/j.1365-2958.1999.01175.x [DOI] [PubMed] [Google Scholar]
- 20.Duthie ES, Lorenz LL. 1952. Staphylococcal coagulase; mode of action and antigenicity. J. Gen. Microbiol. 6:95–107. 10.1099/00221287-6-1-2-95 [DOI] [PubMed] [Google Scholar]
- 21.Attia AS, Benson MA, Stauff DL, Torres VJ, Skaar EP. 2010. Membrane damage elicits an immunomodulatory program in Staphylococcus aureus. PLoS Pathog. 6:e1000802. 10.1371/journal.ppat.1000802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chien Y, Manna AC, Projan SJ, Cheung AL. 1999. SarA, a global regulator of virulence determinants in Staphylococcus aureus, binds to a conserved motif essential for sar-dependent gene regulation. J. Biol. Chem. 274:37169–37176. 10.1074/jbc.274.52.37169 [DOI] [PubMed] [Google Scholar]
- 23.Olczak T, Maszczak-Seneczko D, Smalley JW, Olczak M. 2012. Gallium(III), cobalt(III) and copper(II) protoporphyrin IX exhibit antimicrobial activity against Porphyromonas gingivalis by reducing planktonic and biofilm growth and invasion of host epithelial cells. Arch. Microbiol. 194:719–724. 10.1007/s00203-012-0804-3 [DOI] [PubMed] [Google Scholar]
- 24.Bozja J, Yi K, Shafer WM, Stojiljkovic I. 2004. Porphyrin-based compounds exert antibacterial action against the sexually transmitted pathogens Neisseria gonorrhoeae and Haemophilus ducreyi. Int. J. Antimicrob. Agents 24:578–584. 10.1016/j.ijantimicag.2004.06.008 [DOI] [PubMed] [Google Scholar]
- 25.Glaeser J, Nuss AM, Berghoff BA, Klug G. 2011. Singlet oxygen stress in microorganisms. Adv. Microb. Physiol. 58:141–173. 10.1016/B978-0-12-381043-4.00004-0 [DOI] [PubMed] [Google Scholar]
- 26.Moriwaki Y, Caaveiro JM, Tanaka Y, Tsutsumi H, Hamachi I, Tsumoto K. 2011. Molecular basis of recognition of antibacterial porphyrins by heme-transporter IsdH-NEAT3 of Staphylococcus aureus. Biochemistry 50:7311–7320. 10.1021/bi200493h [DOI] [PubMed] [Google Scholar]
- 27.Lee WC, Reniere ML, Skaar EP, Murphy ME. 2008. Ruffling of metalloporphyrins bound to IsdG and IsdI, two heme-degrading enzymes in Staphylococcus aureus. J. Biol. Chem. 283:30957–30963. 10.1074/jbc.M709486200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kaneko Y, Thoendel M, Olakanmi O, Britigan BE, Singh PK. 2007. The transition metal gallium disrupts Pseudomonas aeruginosa iron metabolism and has antimicrobial and antibiofilm activity. J. Clin. Invest. 117:877–888. 10.1172/JCI30783 [DOI] [PMC free article] [PubMed] [Google Scholar]