Abstract
In torque generation by the bacterial flagellar motor, it has been suggested that electrostatic interactions between charged residues of MotA and FliG at the rotor-stator interface are important. However, the actual role(s) of those charged residues has not yet been clarified. In this study, we systematically made mutants of Vibrio alginolyticus whose charged residues of PomA (MotA homologue) and FliG were replaced by uncharged or charge-reversed residues and characterized the motilities of those mutants. We found that the members of a group of charged residues, 7 in PomA and 6 in FliG, collectively participate in torque generation of the Na+-driven flagellar motor in Vibrio. An additional specific interaction between PomA-E97 and FliG-K284 is critical for proper performance of the Vibrio motor. Our results also reveal that more charged residues are involved in the PomA-FliG interactions in the Vibrio Na+-driven motor than in the MotA-FliG interactions in the H+-driven one. This suggests that a larger number of conserved charged residues at the PomA-FliG interface contributes to the robustness of the Vibrio motor against mutations. The interaction surfaces of the stator and rotor of the Na+-driven motor seem to be more complex than those previously proposed in the H+-driven motor.
INTRODUCTION
The flagellum is an organelle that many bacteria use to move in liquid and viscous surface environments. The flagellum has a long helical structure, works as a screw, and rotates to generate the propulsion force. At the base of each flagellum, there exists a supramolecular complex called the flagellar motor which is embedded in the cytoplasmic membrane and works as an engine to rotate the flagellum. The rotation of the flagellar motor is driven by the flow of ions (H+ or Na+) from outside to inside the cell. The H+-driven flagellar motor of Escherichia coli and Salmonella rotates at about 200 to 300 revolutions per second (1), and the Na+-driven flagellar motor of Vibrio rotates at up to about 1,700 revolutions per second (2). The flagellar motor consists of the rotor and the stator. The rotor is a rotational part composed of several rings, and the most essential components for motor function in the rotor are located at the C-ring, which consists of 3 proteins, FliG, FliM, and FliN. The C-ring is responsible for torque generation and for switching the direction of rotation, and FliG is most directly involved in torque generation (3). The stator is composed of MotA and MotB in the H+-driven flagellar motors of Escherichia coli and Salmonella and is composed of PomA and PomB in the Na+-driven flagellar motors of Vibrio species and Shewanella species (4–7). MotA (PomA) has 4 transmembrane (TM) segments and a long cytoplasmic loop between the second and third TM segments. MotB (PomB) has a single TM segment and a large C-terminal domain at the periplasmic region. Each stator unit is composed of 4 MotA (PomA) and 2 MotB (PomB) subunits and exhibits the ion conducting activity (8, 9). More than 11 stator units can assemble around a rotor (10, 11). It is believed that the ion influx through the stator allows an interaction between the cytoplasmic region of the stator and the rotor that generates torque. The detailed mechanism for rotation of the flagellar motor has not yet been clarified (3, 12, 13).
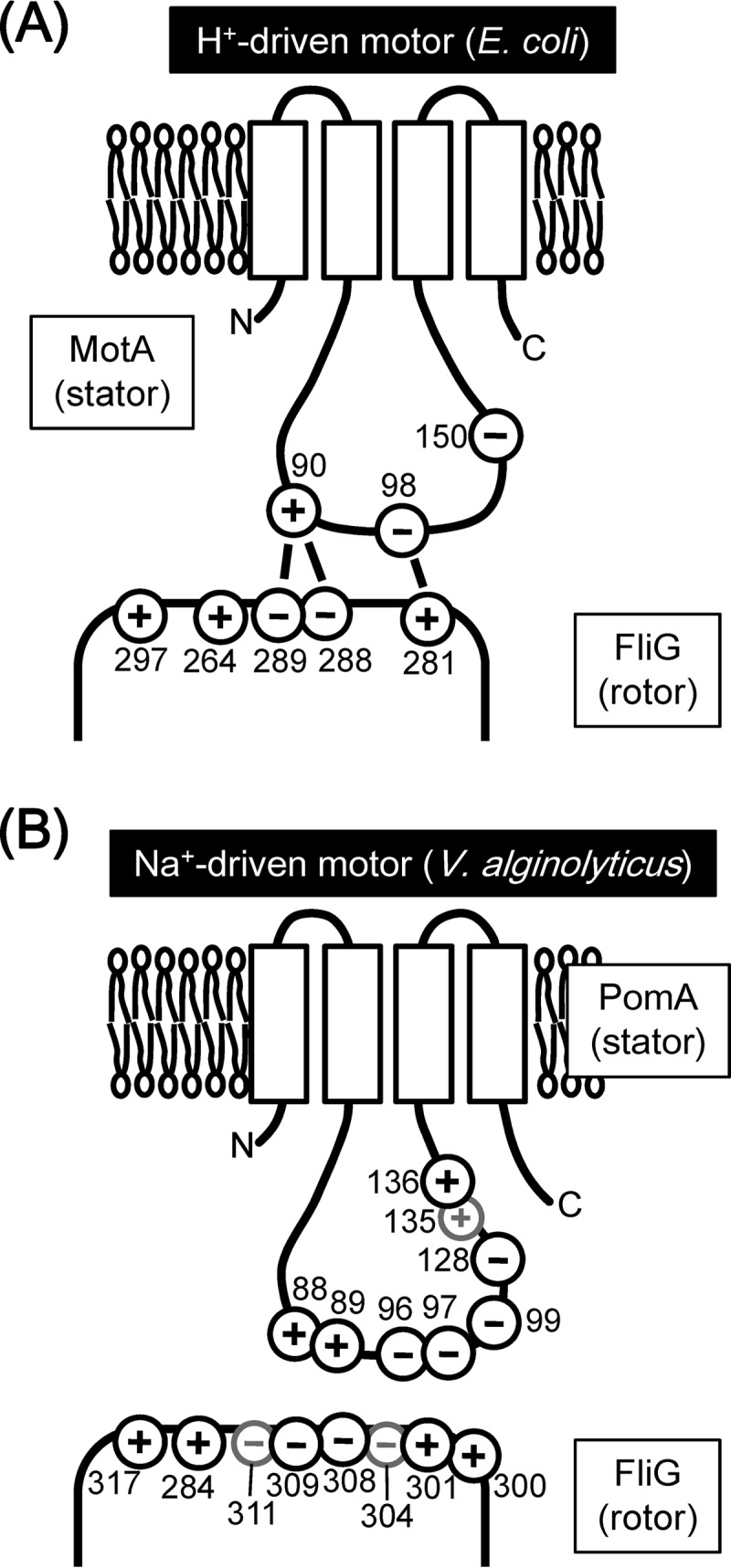
In the H+-driven flagellar motor of E. coli, it is believed that electrostatic interactions between some conserved charged residues of the cytoplasmic loop of MotA (MotAloop) and the C-terminal domain of FliG (FliGC) are important for torque generation (14). The interactions between MotA-R90 and FliG-D289 and between MotA-E98 and FliG-R281 were suggested to be important for the torque generation. Furthermore, in the H+-driven flagellar motor of Salmonella, such charged residues in MotA and FliG are important not only for torque generation but also for efficient stator assembly around the rotor (15, 16). On the other hand, in the Na+-driven flagellar motor of Vibrio alginolyticus, such conserved charged residues of PomA (R88, K89, E96, E97, and E99) and of FliG (K284, R301, D308, D309, and R317) were suggested not to be critical for torque generation, because replacements of those charged residues to uncharged or charge-reversed residues do not strongly affect the motility (17, 18). However, in a hybrid stator composed of PomA and PotB, a chimera composed of the N-terminal region of PomB and the C-terminal region of MotB, these conserved charged residues contribute to proper motor performance in the E. coli flagellar motor (19).
In the Na+-driven flagellar motor of V. alginolyticus, the R88A and E96Q mutations in PomA in combination with mutations in the cytoplasmic domain of PomA, L131F, and T132M caused a temperature-sensitive phenotype with respect to motility (20). Furthermore, mutations in charged residues located in the C-terminal region of PomA (R207E, R215E, and D220K) caused defects in motility (21). PomA-R135Q and H136Y, which are mutations located in the cytoplasmic loop, also caused motility defects (20). This raises the possibility that such mutated regions are involved in the PomA-FliG interaction specific to the Na+-driven flagellar motor. Recently, it has been shown that mutations in the cytoplasmic region of PomA that disrupt the motor function also inhibit the assembly of the stator around the rotor (22). We found that among such mutations, R135Q, R207E, R215E, and D220K considerably inhibit the Na+ conductivity of the PomA/PomB (PomA/B) complex in an unplugged form whereas the H136Y mutation does not inhibit the Na+ conductivity of the PomA/B complex at all, although it results in a loss-of-function phenotype (22, 23). Thus, it was suggested that the cytoplasmic region of PomA is involved not only in the interaction with FliG but also in the Na+ influx through the stator and that the region containing H136 is involved in the stator-rotor interaction, although this effect may be indirect. It has been shown that certain mutations in the core region of the FliG C-terminal domain, which contains conserved charged residues, also disrupt the motor function as well as the stator assembly around the rotor (24). These lines of evidence indicate that interactions between the cytoplasmic region of PomA and the C-terminal domain of FliG are involved in the function of the Na+-driven motor as seen in the H+-driven motor, but the detailed mechanism underlying this remains unclear.
In this study, we systematically replaced the charged residues of PomA and FliG, located on the putative stator-rotor interaction surface, with uncharged or charge-reversed residues and examined the effects of those mutations on motility in the native host V. alginolyticus. The results show strong synergistic effects on motility and weak suppression of the motility defect in certain combinations of PomA and FliG mutations. We also observed the stator assembly around the rotor of some mutants and found that charged residues in the rotor-stator interface could have distinct roles for stator assembly and for torque generation. Based on these results, we discuss the interaction between charged residues at the rotor-stator interface that are required for the function of the Na+-driven flagellar motor of Vibrio.
MATERIALS AND METHODS
Bacterial strains, growth conditions, and media.
Bacterial strains used in this study are listed in Table 1. The pomAB fliG deletion mutants were generated from NMB198 (ΔfliG strain) by homologous recombination with the ΔpomAB gene using a suicide vector, pKY704, with sacB selection, as previously described (25). V. alginolyticus was cultured at 30°C in VC medium (0.5% [wt/vol] polypeptone, 0.5% [wt/vol] yeast extract, 0.4% [wt/vol] K2HPO4, 3% [wt/vol] NaCl, 0.2% [wt/vol] glucose) or in VPG medium (1% [wt/vol] polypeptone, 0.4% [wt/vol] K2HPO4, 3% [wt/vol] NaCl, 0.5% [wt/vol] glycerol). If needed, kanamycin or chloramphenicol was added to reach a final concentration of 2.5 μg ml−1 or 100 μg ml−1, respectively. E. coli was cultured at 37°C in LB medium (1% [wt/vol] bactotryptone, 0.5% [wt/vol] yeast extract, 0.5% [wt/vol] NaCl). If needed, kanamycin or chloramphenicol was added to reach a final concentration of 25 μg ml−1 or 50 μg ml−1, respectively.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Descriptiona | Source or reference |
|---|---|---|
| Strains | ||
| E. coli S17-1 | recA hsdR thi pro ara RP-4 2-Tc::Mu Km::Tn7 (Tpr Smr) | 42 |
| E. coli SM10λpir | thi thr leu tonA lacY supE recA::RP-4 2-Tc::Mu Km λpirRK6 | 43 |
| V. alginolyticus VIO5 | Rifr Pof+ Laf− Pom+ | 28 |
| V. alginolyticus NMB198 | VIO5 ΔfliG (Rifr Pof− Pom−) | 18 |
| V. alginolyticus NMB301 | VIO5 ΔpomAB ΔfliG (Rifr Pof− Pom−) | This study |
| Plasmids | ||
| pKY704 | Suicide vector, Cmr | 44 |
| pKY704-ΔpomAB | pKY704-pomA1–68-pomB126–301, which was constructed by the DraI and HpaI deletion | 29 |
| pKY704-ΔpomAB-sacB | pKY704-ΔpomAB, 2.0-kb XbaI-XbaI fragment (sacB+) from pHFS401 | Hiroyuki Terashima |
| pSU41 | Kmr, Plac | 45 |
| pYA303 | pSU41-pomAB | 46 |
| pBAD33 | Cmr, PBAD | 47 |
| pHFGBA2 | pBAD33-his6-egfp-pomB, pomA | 26 |
| pMMB206 | Cmr, Ptac PlacUV5 | 48 |
| pNT1 | pMMB206-fliG | This study |
| pTY60 | Kmr, PBAD | 24 |
| pNO2 | pTY60-fliG | 24 |
| pNT2 | pTY60-gfp-pomB, pomA | This study |
Tpr, trimethoprim resistant; Smr, streptomycin resistant; Rifr, rifampin resistant; Kmr, kanamycin resistant; Cmr, chloramphenicol resistant; Pof+, normal polar flagellar formation; Laf−, defective in lateral flagellar formation; Pom+, polar flagellar motility; Plac, lac promoter; PBAD, arabinose promoter.
Plasmids and mutagenesis.
Plasmids used in this study are listed in Table 1. The fliG gene on pNT1 was subcloned from pNO2 (24). pMMB206 and pNO2 were digested with HindIII and BamHI and were then separated by agarose gel electrophoresis. The fragments were purified using a gel extraction kit (Qiagen) and were ligated with T4 DNA ligase to generate pNT1. The gfp-pomB and pomA genes on pNT2 were subcloned from pHFGBA2 (26). The gfp-pomB and pomA genes on pHFGBA2 were PCR amplified using an upstream sense primer containing a KpnI site and a downstream antisense primer containing a SalI site. PCR-amplified DNA fragments and pTY60, a plasmid vector, were digested with KpnI and SalI and then were separated using agarose gel electrophoresis. The fragments were purified using a gel extraction kit (Qiagen) and were ligated with T4 DNA ligase to generate pNT2. To introduce mutations into the pomA gene on pYA303 and pNT2 or the fliG gene on pNT1, site-directed mutagenesis was performed using the QuikChange method as described by Stratagene. The sequences of all constructs were checked by DNA sequencing. Transformation of V. alginolyticus by plasmids pYA303 or pNT2 was carried out by the electroporation method as described previously (27). Transformation of V. alginolyticus by plasmid pKY704-ΔpomAB-sacB or pNT1 was carried out by conjugational transfer from E. coli SM10λpir or S17-1, respectively, as described previously (25, 28).
Motility assay in soft-agar plates.
A 2-μl volume of an overnight culture of NMB301 cells harboring the plasmids pYA303 and pNT1 was spotted on VPG soft-agar plates (0.25% [wt/vol] Bacto agar [Difco]) and was incubated at 30°C for the appropriate time as noted in the text.
Detection of proteins.
Overnight cultures grown in VC medium were inoculated at a 100-fold dilution into VPG medium and were cultured at 30°C for 4 h. Cells were harvested by centrifugation and were then suspended in sodium dodecyl sulfate (SDS) loading buffer containing 5% (vol/vol) β-mercaptoethanol to an optical density at 660 nm of 10 and then were boiled at 95°C for 5 min. Samples were separated by SDS-PAGE and transferred to polyvinylidene difluoride (PVDF) membranes, and immunoblotting was performed using anti-PomA (PomA1312) (29) and anti-FliG (FliG B0164) (30) antibodies.
Observation of stator localization with fluorescence microscopy.
Overnight cultures of NMB301 cells harboring plasmids pNT1 and pNT2 were inoculated at a 100-fold dilution into VPG medium containing 0.006% (wt/vol) arabinose, and cells were grown at 30°C for 4 h. Cells were harvested by centrifugation and were then resuspended in TMN500 (50 mM Tris-HCl [pH 7.5], 5 mM MgCl2, 5 mM glucose, 500 mM NaCl) and incubated for several minutes. Cells were fixed to the glass surface using 0.1% (wt/vol) poly-l-lysine and were observed by epifluorescence microscopy, as described previously (31). Images were recorded using a digital charge-coupled-device (CCD) camera (ORCA-AG; Hamamatsu Photonics) and software (Scanalytics, IP lab ver. 3.9.5r2). The rate of polar dot formation was calculated by counting the number of cells that had a fluorescent dot at the cell pole in proportion to all cells in the image.
RESULTS
Charged residues of PomA and FliG at the putative rotor-stator interface.
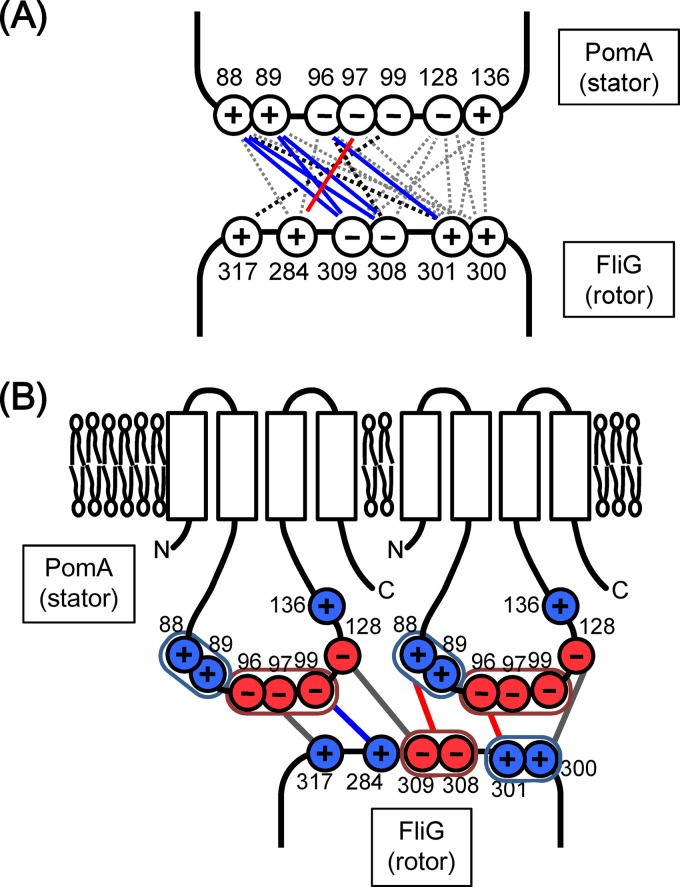
PomA and FliG of the Na+-driven flagellar motor in V. alginolyticus have conserved charged residues that are important for the motor function in the putative PomA-FliG interaction regions, the cytoplasmic loop between TM2 and TM3 of PomA and the C-terminal domain of FliG (Fig. 1; see also Fig. S1 in the supplemental material). PomA and FliG of V. alginolyticus have more charged residues than the E. coli counterparts (see Fig. S1), and these additional charged residues are conserved in Shewanella oneidensis and Bacillus subtilis, whose motors are driven by the Na+ motive force. The crystal structures of FliG have been reported in extreme thermophiles, in Thermotoga maritima for the middle and C-terminal domains of FliG (FliGMC) (32, 33, 34) and in Aquifex aeolicus for full-length FliG (FliGfull) (35). We introduced mutations into the charged residues in PomA and FliG of V. alginolyticus, as shown in Fig. 1 (see also Fig. S1). We then analyzed the effects of those mutations on motility using a pomAB fliG triple-deletion mutant and a two-plasmid system, each component of which carries pomAB or fliG, since stator proteins are more stable when they are produced together from a single mRNA (36). We constructed the ΔpomAB ΔfliG strain (NMB301) and confirmed that it did not form any flagellum, as expected, because FliG is essential for flagellar formation as well as for flagellar motor rotation (37). When strain NMB301 included the two plasmids carrying pomAB and fliG, the motility defect was complemented and the cells were motile on soft-agar plates (see Fig. S2 in the supplemental material). Using this system, we could easily produce mutant PomA and FliG proteins in NMB301 cells in any desired combinations. Our systematic mutational analyses of the rotor-stator interface are described below.
FIG 1.
Charged residues in PomA and FliG characterized in this study. (A) Schematic model for residue interactions at the stator-rotor interface in the H+-driven motor as previously described (14). (B) Charged residues of PomA and FliG in the Na+-driven motor of V. alginolyticus mutated in this study. Black circles indicate residues important for the stator-rotor interaction, and gray circles indicate residues that are not important.
Effect of charge-neutralized pomA fliG combined mutations in highly conserved charged residues on motility.
We introduced mutations into PomA and FliG in which charged residues were replaced by noncharged residues. None of the single mutations affected the expression of PomA and FliG (see Fig. S2B and C in the supplemental material). None of the pomA single mutations strongly affected the motilities of mutant cells in soft-agar plates (Table 2; see also Fig. S3). None of the fliG single mutations affected the motilities of mutant cells in soft-agar plates except for the K284A mutation which caused a weak defect of motility (Table 2; see also Fig. S3). When PomA and FliG proteins with single mutations were coexpressed, some combinations of PomA(R88A)/FliG(R301A), PomA(E96Q)/FliG(D308A), and PomA(E99Q)/FliG(R317A) showed weak synergistic inhibitions of motility whereas other combinations did not (Table 2; see also Fig. S3). These results suggest possible interactions between PomA-R88 and FliG-R301, between PomA-E96 and FliG-D308, and between PomA-E99 and FliG-R317.
TABLE 2.
Relative motilities of charge-neutralized pomA fliG mutants in soft-agar platesa
| fliG strain | Relative diam of pomA strain motility ring |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| WT | R88A | K89A | R88A K89A | E96Q | E97Q | E99Q | E96Q E97Q E99Q | R88A K89A E96Q E97Q E99Q | |
| WT | 1.00 | 1.13 | 0.92 | 0.82 | 1.17 | 0.87 | 0.84 | 0.40 | 0.42 |
| K284A | 0.66 | 0.66 | 0.59 | 0.60 | 0.89 | 0.62 | 0.66 | 0.00 | 0.01 |
| R301A | 1.12 | 0.79 | 1.01 | 0.25 | 0.96 | 0.95 | 0.92 | 0.00 | 0.01 |
| D308A | 0.98 | 1.19 | 1.19 | 0.82 | 0.87 | 1.17 | 0.97 | 0.46 | 0.50 |
| D309A | 1.11 | 1.18 | 1.34 | 0.87 | 1.06 | 1.23 | 0.87 | 0.45 | 0.48 |
| R317A | 0.93 | 1.14 | 1.03 | 0.80 | 0.97 | 1.16 | 0.67 | 0.01 | 0.01 |
The wild-type (WT) control was the pomAB fliG mutant strain expressing wild-type PomA and FliG from the plasmids, and the relative diameters of the motility rings on soft-agar plates compared to wild-type strain are shown for each mutant. Cases of synergistic inhibition of motility are indicated by italics, and cases of suppression are indicated by boldface. Values reported are the means of the results of 3 independent experiments.
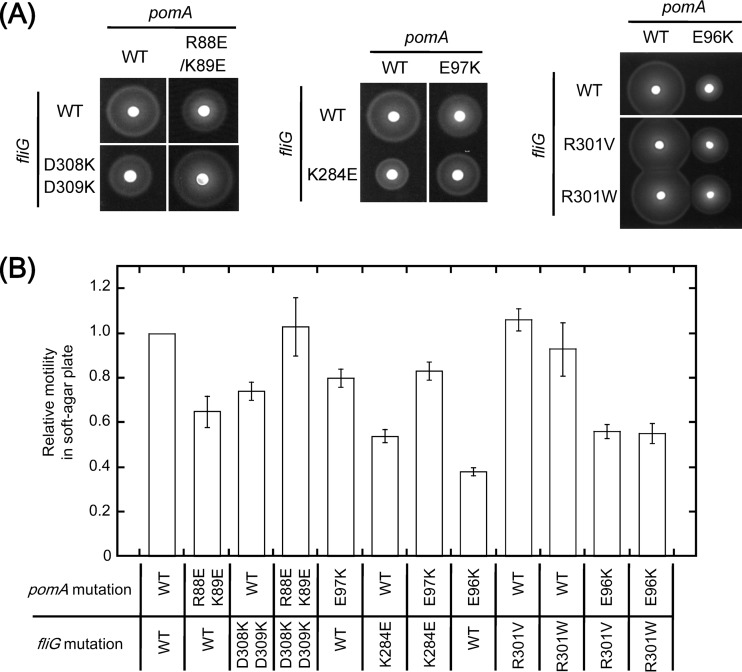
Next, we introduced multiple charge-neutralized mutations into PomA. In the triple mutant (E96Q/E97Q/E99Q) or the quintuple mutant (R88A/K89A/E96Q/E97Q/E99Q) of PomA, the motilities in soft-agar plates were substantially reduced (Table 2 and Fig. 2). In a previous study, it was reported that those mutations did not affect motility at a high Na+ concentration when the mutant PomA was produced from a plasmid carrying the pomA gene in a ΔpomA strain (17). Combined with the fliG mutations, some combinations of the pomA and fliG mutations gave strong synergistic effects (Table 2 and Fig. 2). For example, the motility of PomA(R88A/K89A)/FliG(R301A) was strongly reduced although PomA(R88A)/FliG(R301A) and PomA(K89A)/FliG(R301A) showed only weak effects. The motilities of PomA triple or quintuple mutants combined with some of the fliG mutations (K284A, R301A, or R317A) were completely disrupted (Table 2 and Fig. 2).
FIG 2.
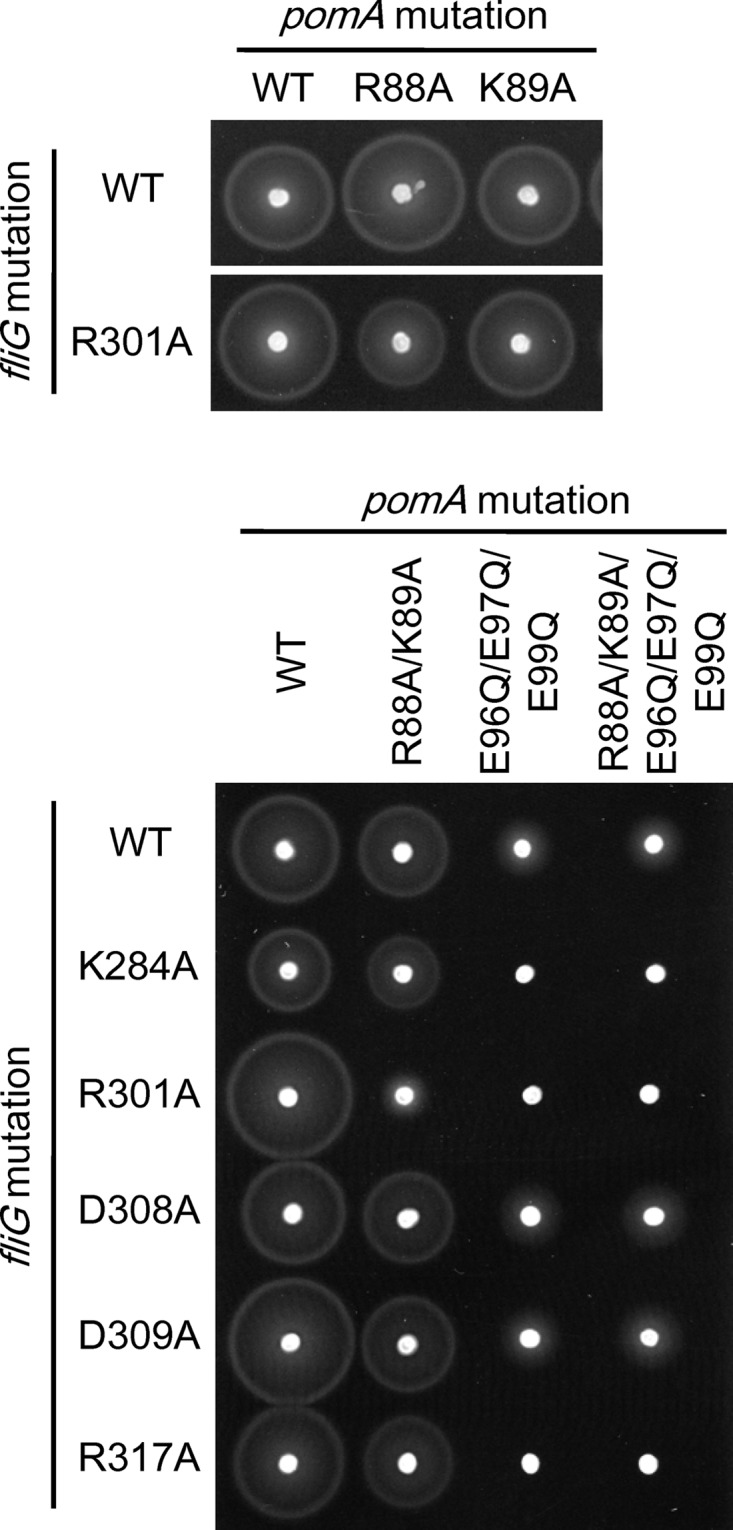
Motilities of pomA fliG combined mutants in soft-agar plates. Data represent swimming abilities in soft-agar plates of pomA fliG combined mutants where highly conserved charged residues were replaced by uncharged residues. NMB301 (ΔpomAB ΔfliG strain) was transformed with two plasmids, pYA303 (carrying the pomAB genes) and pNT1 (carrying the fliG gene), with or without mutations. Cells were inoculated in soft-agar plates and incubated at 30°C for 6 h.
Effect of charge-reversed pomA fliG combined mutations of charged residues on motility.
The charged residues were replaced by charge-reversed residues in PomA and FliG. We confirmed that these single mutations did not affect the expression of PomA and FliG (see Fig. S2B and C in the supplemental material). In the PomA single mutants, the motilities in soft-agar plates were reduced only by the E96K and R135D mutations (Table 3; see also Fig. S4 and S5 in the supplemental material). Among the FliG single mutants, the motility in soft-agar plates was reduced by the K284E mutation and was almost completely inhibited by the R317D mutation (Table 3; see also Fig. S5). The motility inhibition caused by the R317D mutation has been previously reported as a defect in flagellar formation (24). We observed a strong synergistic inhibition of motility in many combinations of mutant PomA and mutant FliG (Table 3; see also Fig. S5). The motility defect caused by the FliG(K284E) mutation was suppressed by the PomA(E97K) mutation (Table 3 and Fig. 3; see also Fig. S4). Combination of the PomA(R88E/K89E) mutation with the FliG(D308K/D309K) mutation suppressed the weak motility defects of each mutant, and the FliG(D309K) single mutation also suppressed the weak motility defects of PomA(R88E/K89E) (Table 3 and Fig. 3; see also Fig. S4). Mutation of PomA(E99K), FliG(E304K), or FliG(E311K) combined with other FliG or PomA mutations did not confer the synergistic effect of either inhibition or suppression (Table 3; see also Fig. S4).
TABLE 3.
Relative motilities of charge-reversed pomA fliG mutants in soft-agar platesa
| fliG strain | Relative diam of pomA strain motility ring |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| WT | R88E | K89E | R88E K89E | E96K | E97K | E99K | D128K | D133K | E134K | R135D | H136D | |
| WT | 1.00 | 0.98 | 1.31 | 0.65 | 0.35 | 0.80 | 1.18 | 1.00 | 1.31 | 1.34 | 0.00 | 1.17 |
| K284E | 0.54 | 0.14 | 0.51 | 0.00 | 0.01 | 0.83 | 0.72 | 0.37 | 0.83 | 0.66 | 0.00 | 0.49 |
| K300E | 1.00 | 0.00 | 1.21 | 0.00 | 0.50 | 0.79 | 1.16 | 0.66 | 1.09 | 1.40 | 0.00 | 0.94 |
| R301D | 0.79 | 0.00 | 0.00 | 0.00 | 0.07 | 0.39 | 0.86 | 0.00 | 0.94 | 1.23 | 0.00 | 0.43 |
| E304K | 0.87 | 0.84 | 0.92 | 0.89 | 0.37 | 0.76 | 0.97 | 0.91 | 0.94 | 1.14 | 0.00 | 1.00 |
| D308K | 1.11 | 1.00 | 1.04 | 0.84 | 0.00 | 0.52 | 1.37 | 0.74 | 1.20 | 1.23 | 0.00 | 1.14 |
| D309K | 0.98 | 0.94 | 0.86 | 1.00 | 0.02 | 0.56 | 1.04 | 0.26 | 1.31 | 1.14 | 0.00 | 0.60 |
| D308K/D309K | 0.74 | 0.89 | 0.74 | 1.03 | 0.00 | 0.50 | 0.74 | 0.00 | 1.00 | 0.91 | 0.00 | 0.69 |
| E311K | 0.87 | 0.84 | 1.21 | 0.76 | 0.47 | 0.74 | 1.21 | 0.89 | 1.17 | 1.06 | 0.00 | 1.09 |
| R317D | 0.16 | 0.03 | 0.20 | 0.00 | 0.03 | 0.16 | 0.14 | 0.14 | 0.17 | 0.20 | 0.00 | 0.14 |
The procedure used to determine the relative diameters of motility rings was the same as that described for Table 2. Cases of synergistic inhibition of motility are indicated by italics, and cases of suppression are indicated by boldface. The data from the strains shown in Fig. 3A and B and in Fig. S5A and B in the supplemental material are underlined. Values reported are the means of the results of 3 independent experiments.
FIG 3.
Motilities of pomA fliG combined mutants in soft-agar plates. Motilities in soft-agar plates of pomA fliG combined mutants which showed suppression of motility were analyzed as described for Fig. 2. Cells were incubated on VPG soft-agar plates at 30°C (A), and data are shown as relative values normalized to wild-type (WT/WT) cells (B).
In the H+-driven motor of E. coli, the defect in motility elicited by the charge-reversed MotA(E98K) mutation was not suppressed by the charge-reversed FliG(R281D) mutation but was suppressed by the replacement of FliG-R281 with V or W (14). We introduced similar mutations into pomA and fliG of V. alginolyticus and examined the effects on motility in soft-agar plates. We observed that the motility reduction caused by the PomA(E96K) mutation was suppressed by the FliG(R301V) and FliG(R301W) mutations in soft-agar plates (Table 4 and Fig. 3). No suppression of the pomA mutation was observed for the charge-reversed mutation of FliG(R301D), as reported for the H+-driven motor in E. coli, but the mutation instead had a synergistic inhibition effect on motility (Table 3; see also Fig. S4 in the supplemental material). PomA-E96 and FliG-R301 interact with each other in the Vibrio motor in a way similar to the interaction between MotA-E98 and FliG-R281 observed in the E. coli motor.
TABLE 4.
Relative motilities of pomA-E96K fliG-R301V or W mutants in soft-agar platesa
| fliG strain | Relative diam of pomA strain motility ring |
|
|---|---|---|
| WT | E96K | |
| WT | 1.00 | 0.38 |
| R301V | 1.06 | 0.56 |
| R301W | 0.93 | 0.50 |
The procedure used to determine the relative diameters of motility rings was same as that described for Table 2. Cases of suppression are indicated by boldface. All strains whose data are reported in this table are shown in Fig. 3A and B and in S5A and B in the supplemental material. Values reported are the means of the results of 3 independent experiments.
In the H+-driven motor of E. coli, it had been suggested that MotA-E150 (corresponding to V. alginolyticus PomA-D128) was a secondarily important residue for the stator-rotor interaction (14). PomA(D128K) did not affect the motility but gave strong synergistic effects with FliG(R301D) and FliG(D308K/D309K) and weak synergistic effects with FliG(K284E), FliG(K300E), and FliG(D309K) (Table 3; see also Fig. S5 in the supplemental material). This suggests that PomA-D128 is a primary important residue for the Na+-driven motor in similarity to the other charged residues.
In the Na+-driven motor of V. alginolyticus, PomA-H136 was thought to be important for the stator-rotor interaction, because the PomA(H136Y) mutation inhibited the motor function without disrupting the Na+ influx through the stators (20, 22, 23). We introduced charge-reversed mutations into adjacent residues in PomA, R135D, and H136D. Those pomA single mutations did not affect the expression of PomA (see Fig. S2B in the supplemental material), and motility was completely impaired by the PomA(R135D) mutation but not by the PomA(H136D) mutation (Table 3; see also Fig. S5). The PomA(R135D) mutation was not suppressed by any fliG mutations examined in this study. PomA(R135D) might cause the defect of ion conductivity previously reported in PomA(R135Q) (22, 23). Regarding the pomA fliG double mutations, PomA(H136D) showed an inhibition of motility that was weakly synergistic with some fliG mutations (K284E, R301D, or D309K) (Table 3; see also Fig. S5).
Effect of pomA fliG mutations in charged residues on the polar localization of the PomA/PomB stator complex.
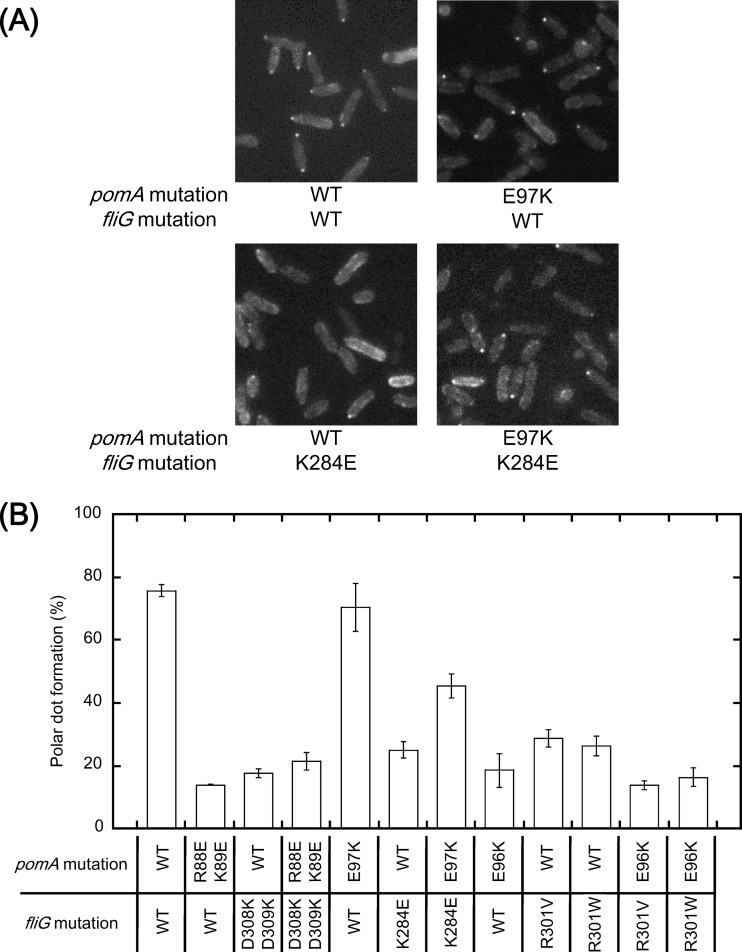
It has been shown that some charge-reversed mutations of conserved charged residues in FliG (K284E and R301D) reduce the efficiency of the stator assembly around the rotor (24). In this study, we observed the polar localization of the stator using a green fluorescent protein (GFP)-fused stator (PomA/GFP-PomB) to examine the effects of double mutations in both pomA and fliG on the stator assembly around the rotor (Fig. 4). Representative examples are shown in Fig. 4A, and the results are summarized in Fig. 4B. We found that the polar localization of the GFP-fused stator was significantly reduced by the pomA mutations corresponding to PomA(R88E/K89E) and PomA(E96K) (Fig. 4B), although those mutations affected motility only slightly (Table 3 and Fig. 3; see also Fig. S4A in the supplemental material). The fliG mutation(s) K284E, R301V, R301W, or D308K/D309K significantly reduced the polar localization of the stator at various levels (Fig. 4), but again this mutation(s) did not cause a significant reduction of motility (Table 3, Table 4, and Fig. 3; see also Fig. S4). In certain combinations of pomA and fliG mutations, stator assemblies were improved. In the PomA(R88E/K89E)/FliG(D308K/D309K) mutant, the polar localization of the stator was slightly increased compared to the PomA(R88E/K89E) mutant (Fig. 4B), though corresponding combinations of motA fliG mutations showed strong suppression of decreased assembly of the stators in the H+-driven motor (16). The PomA(E97K) mutation alone did not affect the polar localization of the stator, but it significantly improved the assembly of stators reduced by the FliG(K284E) mutation (Fig. 4). The combined effects on the polar localization of the stator were not seen in the FliG(R301V) or (R301W) mutation with PomA(E96K) (Fig. 4B). Corresponding mutations in the H+-driven motor, i.e., the combination of MotA(E98K) and FliG(R281V), did not improve the stator assembly either (17).
FIG 4.
Polar localization of the GFP-fused stator in pomA fliG combined mutants. (A) Representative fluorescence microscopic images. NMB301 (ΔpomAB ΔfliG) cells that harbor two plasmids, pNT2 carrying gfp-pomB and pomA with or without mutations and pNT1 carrying fliG with or without mutations, were grown at 30°C for 4 h in VPG medium containing 0.006% arabinose, and the subcellular localization of the GFP-fused stator was observed using fluorescence microscopy. Typical fluorescence images of cells producing wild-type or mutant PomA/FliG are shown. (B) The polar dot formation of the stator complex in cells producing mutant PomA/FliG proteins was evaluated as the percentage of cells with a fluorescent dot at the cell pole.
DISCUSSION
Extra charged residues are involved in motor function at the rotor-stator interface of Vibrio.
In the Na+-driven motor of Vibrio, there are more charged residues at the predicted regions of the interaction surface between PomA and FliG than in the H+-driven motor of E. coli and Salmonella (Fig. 1). For example, K89 is located next to R88, E97 and E99 are neighbors of E96 in PomA, and K300 is next to R301 in FliG. In this report, we show that such extra adjacent charged residues are important for the Na+-driven motor. Combined with the FliG(R301A) mutation, the single pomA mutation PomA(R88A) or PomA(K89A) did not show a synergistic reduction of motility; however, the pomA double mutation (R88A/K89A) showed a strongly synergistic effect on motility reduction. Those results suggest that the adjacent residues R88 and K89 have similar roles in the interaction with FliG and that their roles compensate for each other. However, PomA-R88 might have a more important role for the motor function than K89, because a synergistic inhibition of motility was observed in R88E but not in K89E combined with FliG(K300E). The PomA(E96Q/E97Q/E99Q) mutant showed reduced motility, although single mutants did not, further indicating that these 3 residues can compensate for each other. These results obtained with the charge-neutralized combinations suggest that R301 of FliG interacts with the region composed either of R88 and K89 or of E96, E97, and E99.
A weakly synergistic suppression of motility was observed when the mutation in PomA(R88E/K89E) was combined with FliG(D309K) or FliG(D308K/D309K). This might indicate that there are some electrostatic interactions between PomA-R88/K89 and FliG-D308/D309 and that those contribute to the motor function. This interaction seems to be similar to the electrostatic interaction between MotA-R90 and FliG-D288/D289 described previously for the H+-driven motor of E. coli (14). However, the activity of the Na+-driven motor of Vibrio against the point mutations seems to be more robust such that the single mutations do not cause severe effects on motility, and suppression effects cannot be seen that are as drastic as those in the H+-driven motor of E. coli. The defect of motility caused by the FliG(K284E) mutation was suppressed by the PomA(E97K) mutation, suggesting that there is an interaction between E97 of PomA and K284 of FliG.
In the Na+-driven motor of Vibrio, the PomA(E96K) mutation was suppressed by the FliG(R301V) or (R301W) mutations but not by the charge-reversed FliG(R301D) mutation. These results are similar to the results observed in the H+-driven motor of E. coli (14). Valine and tryptophan are noncharged residues, and the sizes of those residues are considerably different. We do not know why the different putative structures of R301V and R301W mutations, which do not affect the motor function by themselves, confer the suppression for PomA(E96K), but PomA-E96 and FliG-R301 may interact as proposed in the E. coli motor (14).
Taken together, our results suggest that there are at least 3 interaction sites, PomA-R88/K89 and FliG-D308/D309, PomA-E96 and FliG-R301, and PomA-E97 and FliG-K284, in the PomA-FliG interaction surface of the Na+-driven flagellar motor (Fig. 5A). Two of the three interactions correspond to the interactions proposed in E. coli, but one (PomA-E97 plus FliG-K284) is novel and is specific for the Na+-driven motor, which has more charged residues than the H+-driven motor. The motilities of PomA charge-neutralized triple or quintuple mutants were almost completely disrupted by some combinations of the fliG mutations (K284A, R301A, or R317A). The results of the synergistic inhibition of motility suggest that the three charged residues in FliG are primarily important for the force generation. The positive charges of R301 and R317 are located distantly in the FliG structure (32). Thus, as shown in Fig. 5B, we speculate that each charged residue interacts with different PomA molecules of a stator complex which is composed of 4 PomA and 2 PomB molecules (38, 39). Alternatively, it is also possible that the interactions do not occur simultaneously.
FIG 5.
Hypothetical models for residue interactions at the stator-rotor interface of the Na+-driven flagellar motor of V. alginolyticus. (A) Predicted residue interactions between PomA and FliG. Solid lines and dashed lines indicate interactions suggested by the results of suppression from defective motility and of synergistic inhibitions of motility, respectively. Blue lines indicate similar interactions reported in the H+-driven motor, and red lines indicate novel interactions specific in the Na+-driven motor. Black lines and gray lines indicate interactions supported by the results of the effects of charge-neutralized and charge-reversed combined mutations, respectively. (B) Model for the Na+-driven motor based on the present study and previous studies (17, 18). Possible interactions important for the torque generation and the stator assembly are indicated by the red lines and the blue lines, respectively. Possible interactions that contribute to either the torque generation or the stator assembly are shown as gray lines. A group of residues that may work redundantly is circled.
Stator alignment at the rotor-stator interface.
It has been shown that interactions between some charged residues in PomA (MotA) and FliG are necessary for proper assembly of the stators around the rotor (15, 16, 22, 24). Here we observed the stator localization in some PomA/FliG single or double mutants. In some PomA or FliG mutants, the polar localization of the stator was remarkably reduced although their motility was not so different from that of the wild-type strain, probably because a small number of stators, which could not be detected as fluorescent dots by microscopy, is sufficient for the motor rotation. It has been shown that the swimming speed of the motor is independent of the number of stators under a low viscous load (40, 41). This report shows that charged residues in PomA (R88, K89, and E96) and FliG (K284, R301, D308, and D309) might be important for efficient stator assembly around the rotor. In the H+-driven motor of Salmonella, it was suggested that the interaction between MotA-E98 and FliG-R281, which correspond to PomA-E96 and FliG-R301 in the Na+-driven motor of Vibrio, is more important for the torque generation than the interaction between MotA-R90 and FliG-D289, which correspond to PomA-R88 and FliG-D309 in the Na+-driven motor of Vibrio, since the FliG(D289K) mutation recovered the localization of the stators around the rotor of the MotA(R90E) mutant whereas the FliG(R281V) mutation did not recover the stator assembly of the MotA(E98K) mutant (16).
Here we show that in the Na+-driven motor, the FliG(D308K/D309K) mutation improves only slightly the localization of the stators around the rotor of the PomA(R88E/K89E) mutant. The PomA(E97K) mutation improves the localization of the stators in the FliG(K284E) rotor. The FliG(R301V) or (R301W) mutation does not restore but rather inhibits the stator assembly of the PomA(E96K) mutant. These results distinguish the property of the interaction between PomA-R88/K89 and FliG-D308/D309 from that of the interaction between MotA-R90 and FliG R289 in the H+-driven motor. The interaction between PomA-E96 and FliG-R301 is also important for torque generation and is secondarily important for stator assembly, similarly to the H+-driven motor. The interaction between PomA-E97 and FliG-K284 seems to be critical for the proper assembly of the stators around the rotor. However, it is very difficult to distinguish which step (torque generation or stator assembly) is impaired by disrupting the E97-K284 interaction. To clarify the role of that interaction, we need to isolate mutants which disrupt the motor function but retain the stator assembly around the rotor.
E96 and E97 are adjacent to each other in PomA; however, K284 and R301 are distant from each other in the FliG crystal structure. This suggests that interactions between PomA-E96 and FliG-R301 and between PomA-E97 and FliG-K284 do not occur simultaneously in a single PomA molecule. This raises the possibility that more than two PomA molecules interact with one FliG molecule at the same time (Fig. 5B) (38) and/or that interactions occur sequentially through the rotation of the FliG ring. Alternatively, there are two modes, the stator assembly mode and the torque generation mode, for the PomA-FliG interaction. The interaction between PomA-E97 and FliG-K284 may occur in the stator assembly mode, and the interactions between PomA-R88/K89 and FliG-D308/D309 and between PomA-E96 and FliG-R301 may occur in the torque generation mode. In the latter case, these two structural modes are continuously alternately coupled to ion influx through the stator, and such an alternation of the two modes may play a direct role in the torque generation of the motor.
This report supports the possibility that more than 13 charged residues (Fig. 5A) are involved in the PomA-FliG interaction that is required for motor function of the Na+-driven flagellar motor. Compared to the H+-driven motor, a larger number of charged residues at the rotor-stator interface are involved in stator-rotor interactions in the Vibrio motor, thereby conferring robustness against mutations. The distinct roles of charged residues in the Na+-driven motor and in the H+-driven motor seem to be different. These properties may be responsible for the different motor characteristics of the Na+-driven motor and the H+-driven motor and may confer the ability of its high-speed rotation. Abundant numbers of charged residues and more interactions at the rotor-stator interface have been shown in this study, suggesting that the interaction between the stator and the rotor is not a simple interaction between a few residues in the individual molecules but is a more complex interaction between those residues and the surface structures of multiple molecules. Such more complex interactions might be important for the torque generation of the bacterial flagellar motor in various species.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the Japan Society for the Promotion of Science (JSPS) of KAKENHI grants 24117004 and 23247024 to M.H., KAKENHI grant 24657087 to S.K., or a Grant-in-Aid for Encouragement of Scientists (to N.T.). N.T. was partly supported by the Integrative Graduate Education and Research program of Nagoya University.
Footnotes
Published ahead of print 24 January 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.01392-13.
REFERENCES
- 1.Kudo S, Magariyama Y, Aizawa S. 1990. Abrupt changes in flagellar rotation observed by laser dark-field microscopy. Nature 346:677–680. 10.1038/346677a0 [DOI] [PubMed] [Google Scholar]
- 2.Magariyama Y, Sugiyama S, Muramoto K, Kawagishi I, Imae Y, Kudo S. 1995. Simultaneous measurement of bacterial flagellar rotation rate and swimming speed. Biophys. J. 69:2154–2162. 10.1016/S0006-3495(95)80089-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kojima S, Blair DF. 2004. The bacterial flagellar motor: structure and function of a complex molecular machine. Int. Rev. Cytol. 233:93–134. 10.1016/S0074-7696(04)33003-2 [DOI] [PubMed] [Google Scholar]
- 4.Dean GE, Macnab RM, Stader J, Matsumura P, Burks C. 1984. Gene sequence and predicted amino acid sequence of the motA protein, a membrane-associated protein required for flagellar rotation in Escherichia coli. J. Bacteriol. 159:991–999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stader J, Matsumura P, Vacante D, Dean GE, Macnab RM. 1986. Nucleotide sequence of the Escherichia coli motB gene and site-limited incorporation of its product into the cytoplasmic membrane. J. Bacteriol. 166:244–252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Asai Y, Kojima S, Kato H, Nishioka N, Kawagishi I, Homma M. 1997. Putative channel components for the fast-rotating sodium-driven flagellar motor of a marine bacterium. J. Bacteriol. 179:5104–5110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Paulick A, Koerdt A, Lassak J, Huntley S, Wilms I, Narberhaus F, Thormann KM. 2009. Two different stator systems drive a single polar flagellum in Shewanella oneidensis MR-1. Mol. Microbiol. 71:836–850. 10.1111/j.1365-2958.2008.06570.x [DOI] [PubMed] [Google Scholar]
- 8.Braun TF, Al-Mawsawi LQ, Kojima S, Blair DF. 2004. Arrangement of core membrane segments in the MotA/MotB proton-channel complex of Escherichia coli. Biochemistry 43:35–45. 10.1021/bi035406d [DOI] [PubMed] [Google Scholar]
- 9.Sato K, Homma M. 2000. Functional reconstitution of the Na+-driven polar flagellar motor component of Vibrio alginolyticus. J. Biol. Chem. 275:5718–5722. 10.1074/jbc.275.8.5718 [DOI] [PubMed] [Google Scholar]
- 10.Leake MC, Chandler JH, Wadhams GH, Bai F, Berry RM, Armitage JP. 2006. Stoichiometry and turnover in single, functioning membrane protein complexes. Nature 443:355–358. 10.1038/nature05135 [DOI] [PubMed] [Google Scholar]
- 11.Reid SW, Leake MC, Chandler JH, Lo CJ, Armitage JP, Berry RM. 2006. The maximum number of torque-generating units in the flagellar motor of Escherichia coli is at least 11. Proc. Natl. Acad. Sci. U. S. A. 103:8066–8071. 10.1073/pnas.0509932103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Terashima H, Kojima S, Homma M. 2008. Flagellar motility in bacteria structure and function of flagellar motor. Int. Rev. Cell Mol. Biol. 270:39–85. 10.1016/S1937-6448(08)01402-0 [DOI] [PubMed] [Google Scholar]
- 13.Li N, Kojima S, Homma M. 2011. Sodium-driven motor of the polar flagellum in marine bacteria Vibrio. Genes Cells 16:985–999. 10.1111/j.1365-2443.2011.01545.x [DOI] [PubMed] [Google Scholar]
- 14.Zhou J, Lloyd SA, Blair DF. 1998. Electrostatic interactions between rotor and stator in the bacterial flagellar motor. Proc. Natl. Acad. Sci. U. S. A. 95:6436–6441. 10.1073/pnas.95.11.6436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Morimoto YV, Nakamura S, Kami-ike N, Namba K, Minamino T. 2010. Charged residues in the cytoplasmic loop of MotA are required for stator assembly into the bacterial flagellar motor. Mol. Microbiol. 78:1117–1129. 10.1111/j.1365-2958.2010.07391.x [DOI] [PubMed] [Google Scholar]
- 16.Morimoto YV, Nakamura S, Hiraoka KD, Namba K, Minamino T. 2013. Distinct roles of highly conserved charged residues at the MotA-FliG interface in bacterial flagellar motor rotation. J. Bacteriol. 195:474–481. 10.1128/JB.01971-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yorimitsu T, Sowa Y, Ishijima A, Yakushi T, Homma M. 2002. The systematic substitutions around the conserved charged residues of the cytoplasmic loop of Na+-driven flagellar motor component PomA. J. Mol. Biol. 320:403–413. 10.1016/S0022-2836(02)00426-6 [DOI] [PubMed] [Google Scholar]
- 18.Yorimitsu T, Mimaki A, Yakushi T, Homma M. 2003. The conserved charged residues of the C-terminal region of FliG, a rotor component of the Na+-driven flagellar motor. J. Mol. Biol. 334:567–583. 10.1016/j.jmb.2003.09.052 [DOI] [PubMed] [Google Scholar]
- 19.Yakushi T, Yang J, Fukuoka H, Homma M, Blair DF. 2006. Roles of charged residues of rotor and stator in flagellar rotation: comparative study using H+-driven and Na+-driven motors in Escherichia coli. J. Bacteriol. 188:1466–1472. 10.1128/JB.188.4.1466-1472.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fukuoka H, Yakushi T, Homma M. 2004. Concerted effects of amino acid substitutions in conserved charged residues and other residues in the cytoplasmic domain of PomA, a stator component of Na+-driven flagella. J. Bacteriol. 186:6749–6758. 10.1128/JB.186.20.6749-6758.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Obara M, Yakushi T, Kojima S, Homma M. 2008. Roles of charged residues in the C-terminal region of PomA, a stator component of the Na+-driven flagellar motor. J. Bacteriol. 190:3565–3571. 10.1128/JB.00849-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Takekawa N, Li N, Kojima S, Homma M. 2012. Characterization of PomA mutants defective in the functional assembly of the Na+-driven flagellar motor in Vibrio alginolyticus. J. Bacteriol. 194:1934–1939. 10.1128/JB.06552-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Takekawa N, Terauchi T, Morimoto YV, Minamino T, Lo CJ, Kojima S, Homma M. 2013. Na+ conductivity of the Na+-driven flagellar motor complex composed of unplugged wild-type or mutant PomB with PomA. J. Biochem. 153:441–451. 10.1093/jb/mvt011 [DOI] [PubMed] [Google Scholar]
- 24.Kojima S, Nonoyama N, Takekawa N, Fukuoka H, Homma M. 2011. Mutations targeting the C-terminal domain of FliG can disrupt motor assembly in the Na+-driven flagella of Vibrio alginolyticus. J. Mol. Biol. 414:62–74. 10.1016/j.jmb.2011.09.019 [DOI] [PubMed] [Google Scholar]
- 25.Terashima H, Fukuoka H, Yakushi T, Kojima S, Homma M. 2006. The Vibrio motor proteins, MotX and MotY, are associated with the basal body of Na+-driven flagella and required for stator formation. Mol. Microbiol. 62:1170–1180. 10.1111/j.1365-2958.2006.05435.x [DOI] [PubMed] [Google Scholar]
- 26.Fukuoka H, Yakushi T, Kusumoto A, Homma M. 2005. Assembly of motor proteins, PomA and PomB, in the Na+-driven stator of the flagellar motor. J. Mol. Biol. 351:707–717. 10.1016/j.jmb.2005.06.037 [DOI] [PubMed] [Google Scholar]
- 27.Kawagishi I, Okunishi I, Homma M, Imae Y. 1994. Removal of the periplasmic DNAse before electroporation enhances efficiency of transformation in the marine bacterium Vibrio alginolyticus. Microbiology 140:2355–2361. 10.1099/13500872-140-9-2355 [DOI] [Google Scholar]
- 28.Okunishi I, Kawagishi I, Homma M. 1996. Cloning and characterization of motY, a gene coding for a component of the sodium-driven flagellar motor in Vibrio alginolyticus. J. Bacteriol. 178:2409–2415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yorimitsu T, Sato K, Asai Y, Kawagishi I, Homma M. 1999. Functional interaction between PomA and PomB, the Na+-driven flagellar motor components of Vibrio alginolyticus. J. Bacteriol. 181:5103–5106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Koike M, Terashima H, Kojima S, Homma M. 2010. Isolation of basal bodies with C-ring components from the Na+-driven flagellar motor of Vibrio alginolyticus. J. Bacteriol. 192:375–378. 10.1128/JB.01121-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fukuoka H, Wada T, Kojima S, Ishijima A, Homma M. 2009. Sodium-dependent dynamic assembly of membrane complexes in sodium-driven flagellar motors. Mol. Microbiol. 71:825–835. 10.1111/j.1365-2958.2008.06569.x [DOI] [PubMed] [Google Scholar]
- 32.Lloyd SA, Whitby FG, Blair DF, Hill CP. 1999. Structure of the C-terminal domain of FliG, a component of the rotor in the bacterial flagellar motor. Nature 400:472–475. 10.1038/22794 [DOI] [PubMed] [Google Scholar]
- 33.Brown PN, Hill CP, Blair DF. 2002. Crystal structure of the middle and C-terminal domains of the flagellar rotor protein FliG. EMBO J. 21:3225–3234. 10.1093/emboj/cdf332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Minamino T, Imada K, Kinoshita M, Nakamura S, Morimoto YV, Namba K. 2011. Structural insight into the rotational switching mechanism of the bacterial flagellar motor. PLoS Biol. 9:e1000616. 10.1371/journal.pbio.1000616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee LK, Ginsburg MA, Crovace C, Donohoe M, Stock D. 2010. Structure of the torque ring of the flagellar motor and the molecular basis for rotational switching. Nature 466:996–1000. 10.1038/nature09300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Van Way SM, Hosking ER, Braun TF, Manson MD. 2000. Mot protein assembly into the bacterial flagellum: a model based on mutational analysis of the motB gene. J. Mol. Biol. 297:7–24. 10.1006/jmbi.2000.3548 [DOI] [PubMed] [Google Scholar]
- 37.Irikura VM, Kihara M, Yamaguchi S, Sockett H, Macnab RM. 1993. Salmonella typhimurium fliG and fliN mutations causing defects in assembly, rotation, and switching of the flagellar motor. J. Bacteriol. 175:802–810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sato K, Homma M. 2000. Multimeric structure of PomA, the Na+-driven polar flagellar motor component of Vibrio alginolyticus. J. Biol. Chem. 275:20223–20228. 10.1074/jbc.M002236200 [DOI] [PubMed] [Google Scholar]
- 39.Yonekura K, Maki-Yonekura S, Homma M. 2011. The structure of the flagellar motor protein complex PomAB: implications for the torque-generating conformation. J. Bacteriol. 193:3863–3870. 10.1128/JB.05021-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ryu WS, Berry RM, Berg HC. 2000. Torque-generating units of the flagellar motor of Escherichia coli have a high duty ratio. Nature 403:444–447. 10.1038/35000233 [DOI] [PubMed] [Google Scholar]
- 41.Yuan J, Berg HC. 2008. Resurrection of the flagellar rotary motor near zero load. Proc. Natl. Acad. Sci. U. S. A. 105:1182–1185. 10.1073/pnas.0711539105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Simon R, Priefer U, Pühler A. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in Gram negative bacteria. Nat. Biotechnol. 1:784–791. 10.1038/nbt1183-784 [DOI] [Google Scholar]
- 43.Miller VL, Mekalanos JJ. 1988. A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. J. Bacteriol. 170:2575–2583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xu M, Yamamoto K, Honda T, Ming X. 1994. Construction and characterization of an isogenic mutant of Vibrio parahaemolyticus having a deletion in the thermostable direct hemolysin-related hemolysin gene (trh). J. Bacteriol. 176:4757–4760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bartolomé B, Jubete Y, Martínez E, de la Cruz F. 1991. Construction and properties of a family of pACYC184-derived cloning vectors compatible with pBR322 and its derivatives. Gene 102:75–78. 10.1016/0378-1119(91)90541-I [DOI] [PubMed] [Google Scholar]
- 46.Kojima S, Asai Y, Atsumi T, Kawagishi I, Homma M. 1999. Na+-driven flagellar motor resistant to phenamil, an amiloride analog, caused by mutations in putative channel components. J. Mol. Biol. 285:1537–1547. 10.1006/jmbi.1998.2377 [DOI] [PubMed] [Google Scholar]
- 47.Guzman LM, Belin D, Carson MJ, Beckwith J. 1995. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 177:4121–4130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Morales VM, Backman A, Bagdasarian M. 1991. A series of wide-host-range low-copy-number vectors that allow direct screening for recombinants. Gene 97:39–47. 10.1016/0378-1119(91)90007-X [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.