Abstract
Lactobacilli are found in a wide variety of habitats. Four species, Lactobacillus crispatus, L. gasseri, L. iners, and L. jensenii, are common and abundant in the human vagina and absent from other habitats. These may be adapted to the vagina and possess characteristics enabling them to thrive in that environment. Furthermore, stable codominance of multiple Lactobacillus species in a single community is infrequently observed. Thus, it is possible that individual vaginal Lactobacillus species possess unique characteristics that confer to them host-specific competitive advantages. We performed comparative functional genomic analyses of representatives of 25 species of Lactobacillus, searching for habitat-specific traits in the genomes of the vaginal lactobacilli. We found that the genomes of the vaginal species were significantly smaller and had significantly lower GC content than those of the nonvaginal species. No protein families were found to be specific to the vaginal species analyzed, but some were either over- or underrepresented relative to nonvaginal species. We also found that within the vaginal species, each genome coded for species-specific protein families. Our results suggest that even though the vaginal species show no general signatures of adaptation to the vaginal environment, each species has specific and perhaps unique ways of interacting with its environment, be it the host or other microbes in the community. These findings will serve as a foundation for further exploring the role of lactobacilli in the ecological dynamics of vaginal microbial communities and their ultimate impact on host health.
INTRODUCTION
Lactobacillus species are commonly found in the vaginas of healthy women. In a study describing the vaginal bacterial communities of 396 women, Ravel et al. (1) identified five community states based on similarity in community composition and structure. Four of these were dominated by one of four Lactobacillus species (L. crispatus, L. gasseri, L. iners, and L. jensenii), and the fifth was composed mainly of a mixture of strict anaerobes such as Prevotella, Megasphaera, and Atopobium. In the four community states characterized by a high abundance of lactobacilli, only one Lactobacillus species typically dominated a community. Furthermore, the relative proportions of community types differed among women of different ethnicities (1). The same four species have been reported to dominate the vaginal communities of healthy women across the world (2–9). Even though the four Lactobacillus species commonly found and abundant in the human vagina have been isolated from other sites, their occurrence is extremely rare (10–14). Furthermore, the vaginal microbial community of nonhuman primates is not dominated by Lactobacillus species (15). For instance, in an analysis of the genital microbiota of the rhesus macaque, Spear et al. (16) found that the communities were somewhat similar to human vaginal communities of women with bacterial vaginosis, which is characterized by a shift from a flora dominated by lactobacilli to a mixed flora (17), and when individuals belonging to the genus Lactobacillus were detected, they were present at a low relative abundance. This suggests that the human vaginal Lactobacillus species possess unique characteristics that enable them to thrive in this environment and exhibit habitat specialization.
The Lactobacillus genus is comprised of over 130 lactic-acid-producing species (18) that inhabit a variety of environments, namely, foodstuffs such as yogurt, wine, and cheese and various human body sites (19). However, some species are found almost exclusively in specific habitats. In recent years, genomic analyses of lactobacilli have demonstrated that the pan-genome of this genus is very large, suggesting a relatively high degree of genetic and functional diversity among species (20). Furthermore, these studies have facilitated a more comprehensive characterization of the genetic and phenotypic characteristics of these organisms (21). Genomic studies of a wide variety of Lactobacillus species have been completed and can be divided into three categories: single-genome characterization (see, e.g., references 22 and 23), comparative genomics of a wide range of species occupying several habitats (see, e.g., references 20, 24, 25, and 26), and comparisons between two species occupying distinct habitats (see, e.g., reference 27). Work on the latter category has been aimed at identifying functional traits that could explain the existence of these organisms in their respective habitats. For example, O'Sullivan et al. (27) compared the genomes of 11 Lactobacillus species and identified sets of genes that were specific to the habitat commonly occupied by the species compared. The genome of Lactobacillus helveticus, a species that inhabits dairy products, coded for six dairy-specific genes that were absent from the genomes of the other species. The genome of Lactobacillus acidophilus, a gut inhabitant, coded for three genes that the authors considered to be specific to the gut environment. Lactobacillus bulgaricus is commonly used in the production of yogurt. While it has complete transport systems for lactose, mannose/glucose, fructose, and glycerol, the transport systems for cellobiose, sucrose, and maltose seem to have been lost after transition of the ancestral of this species from what appears to have been a plant-associated environment (28). Likewise, the genome of L. rhamnosus strain LC705, a common probiotic, also contains genes encoding proteins such as three canonical pilus subunits and a sortase protein involved in pilus assembly, which may facilitate host-cell attachment and thus reflect its adaptation to the intestinal tract environment (25).
While the vaginal lactobacilli have long been considered to be important for the maintenance of women's reproductive health (29, 30), of the four dominant and most abundant species, only L. gasseri and L. iners have been the focus of genomic and genetic characterization. L. gasseri has been isolated from several sites in the human body (mouth, intestines, feces, and vagina [31]). An analysis of the genome of a strain isolated from humans uncovered a high number of putative mucus-binding proteins, the highest among the lactobacilli (31). L. iners is the species with the smallest genome among the lactobacilli, and its size is within the range of the genome sizes of several obligate symbionts and parasites (23). L. iners AB-1 lacks genes necessary to synthesize any amino acids de novo, with the possible exception of serine. On the other hand, 15.6% of its genome is dedicated to various transport mechanisms (23), suggesting that it may acquire many of the components needed for survival from the host or other organisms in the community.
We used comparative functional genomics to determine the potential protein repertoire and explore the ecology of the four Lactobacillus species most commonly found in the human vagina. Two main observations guided this work: L. crispatus, L. gasseri, L. iners, and L. jensenii are the most prominent Lactobacillus species found in the human vagina and do not seem to thrive in other environments; and each of these four species usually dominates the community in which it is found, with stable codominance of lactobacilli not frequently observed. We hypothesized that the functional profiles of all vaginal Lactobacillus species show signatures of adaptation to this environment. Because of the near-exclusive dominance of each of the four most abundant Lactobacillus species in the communities, we also expected to find differential representation of protein families involved in interactions with the environment in each of their profiles.
We found that the genomes of the vaginal Lactobacillus species show some features (reduced genome size and GC content) commonly associated with an obligately symbiotic lifestyle and present a plethora of genes encoding protein families that are overrepresented relative to their occurrence in the genomes of nonvaginal species. Analysis of the protein families coded by the genomes of each of the vaginal species suggests that they may have different mechanisms to interact with their environment, be it the host or other microbes. These findings will aid in making predictions about how each species contributes to the ecological dynamics of vaginal bacterial communities and, in turn, modulates host health.
MATERIALS AND METHODS
We retrieved the coding sequences of the genomes of 67 strains from 25 Lactobacillus species. These were reportedly isolated from three different sites: the gastrointestinal tract, food products, and the human vagina. We then identified homologous proteins and clustered them into protein families, yielding a total of 11,047 protein families. These make up the total functional potential of the strains used in this analysis. From this, we recorded the presence or absence of each protein family in the genomes and used the resulting matrix to estimate distances between pairs of strains. If different species are well adapted to an environment, they may be functionally more similar among themselves than to species that inhabit different environments and consequently may form a cluster in a distance representation. We also assigned functional annotations to each protein family based on information from multiple databases. We then estimated the over- and underrepresentation of the functional categories in the vaginal relative to nonvaginal Lactobacillus species to understand how different protein categories may be functionally more relevant to specifically occupying the vaginal environment. We also used Cramer's V to measure the degree of association between each protein family and specific groups of strains or species.
Sequences.
Complete and draft genome sequences of bacteria in GenBank format (32) were retrieved from the National Center for Biotechnology Information (NCBI) FTP site at ftp://ftp.ncbi.nih.gov/genomes/Bacteria/ and from the PATRIC FTP site at ftp://ftp.patricbrc.org/patric2/ (33). The downloaded material included data for the comparative genomics analyses of 67 Lactobacillus strains (Table 1).
TABLE 1.
Genomic characteristics of the strains analyzed in this studya
| Organism | Source | Size (bp) | %G+C | No. of CDS | No. of MCL |
|---|---|---|---|---|---|
| Lactobacillus acidophilus 30SC* | Gastrointestinal tract | 2,097,766 | 38.1 | 2,059 | 1,601 |
| Lactobacillus acidophilus ATCC 4796 | Gastrointestinal tract | 2,020,500 | 34.2 | 2,020 | 1,498 |
| Lactobacillus acidophilus NCFM* | Gastrointestinal tract | 1,993,560 | 34.7 | 1,862 | 1,476 |
| Lactobacillus amylolyticus DSM_11664 | Gastrointestinal tract | 1,540,806 | 38.3 | 1,684 | 1,330 |
| Lactobacillus amylovorus GRL1118* | Gastrointestinal tract | 1,977,087 | 38.0 | 1,920 | 1,508 |
| Lactobacillus antri DSM_16041 | Gastrointestinal tract | 2,302,896 | 49.8 | 2,224 | 1,729 |
| Lactobacillus brevis ATCC 367* | Food | 2,340,228 | 46.1 | 2,218 | 1,723 |
| Lactobacillus brevis subsp. gravesensis ATCC 27305 | Gastrointestinal tract | 3,144,656 | 39.1 | 3,041 | 2,125 |
| Lactobacillus buchneri ATCC 11577 | Gastrointestinal tract | 2,906,028 | 38.9 | 3,002 | 2,093 |
| Lactobacillus casei BD-II* | Food | 3,127,288 | 46.3 | 3,204 | 2,529 |
| Lactobacillus casei LC2W* | Food | 3,077,434 | 46.3 | 3,164 | 2,466 |
| Lactobacillus casei str. Zhang* | Food | 2,898,335 | 46.4 | 2,848 | 2,338 |
| Lactobacillus coleohominis 101-4-CHN | Vagina | 1,725,829 | 40.8 | 1,652 | 1,410 |
| Lactobacillus crispatus 125-2-CHN | Vagina | 2,305,246 | 33.9 | 2,082 | 1,652 |
| Lactobacillus crispatus 214-1 | Vagina | 2,068,805 | 36.9 | 2,163 | 1,639 |
| Lactobacillus crispatus CTV-05 | Vagina | 2,364,583 | 36.0 | 2,248 | 1,634 |
| Lactobacillus crispatus JV-V01 | Vagina | 2,221,719 | 34.3 | 2,209 | 1,637 |
| Lactobacillus crispatus MV-1A-US | Vagina | 2,311,882 | 34.5 | 2,151 | 1,730 |
| Lactobacillus crispatus MV-3A-US | Vagina | 2,437,083 | 34.4 | 2,330 | 1,787 |
| Lactobacillus crispatus ST1* | Vagina | 2,043,161 | 36.9 | 2,024 | 1,513 |
| Lactobacillus delbrueckii subsp. bulgaricus 2038* | Gastrointestinal tract | 1,872,918 | 49.7 | 1,792 | 1,504 |
| Lactobacillus delbrueckii subsp. bulgaricus ATCC 11842* | Food | 1,864,998 | 49.7 | 1,562 | 1,508 |
| Lactobacillus delbrueckii subsp. bulgaricus ATCC BAA-365* | Food | 1,856,951 | 49.7 | 1,721 | 1,504 |
| Lactobacillus delbrueckii subsp. lactis DSM_20072 | Gastrointestinal tract | 1,875,881 | 49.8 | 2,006 | 1,604 |
| Lactobacillus fermentum ATCC 14931 | Gastrointestinal tract | 1,867,005 | 50.8 | 1,866 | 1,460 |
| Lactobacillus fermentum IFO_3956* | Gastrointestinal tract | 2,098,685 | 51.5 | 1,843 | 1,565 |
| Lactobacillus gasseri 202-4 | Vagina | 1,820,250 | 34.9 | 1,773 | 1,431 |
| Lactobacillus gasseri 224-1 | Vagina | 2,005,522 | 35.0 | 2,252 | 1,584 |
| Lactobacillus gasseri JV-V03 | Vagina | 2,013,030 | 34.5 | 2,001 | 1,586 |
| Lactobacillus gasseri MV-22 | Vagina | 1,929,551 | 34.2 | 1,917 | 1,462 |
| Lactobacillus helveticus DPC_4571* | Gastrointestinal tract | 2,080,931 | 37.1 | 1,610 | 1,489 |
| Lactobacillus helveticus DSM_20075 | Gastrointestinal tract | 2,039,942 | 33.0 | 2,091 | 1,534 |
| Lactobacillus helveticus H10* | Food | 2,172,383 | 36.8 | 1,978 | 1,600 |
| Lactobacillus iners AB-1 | Vagina | 1,287,456 | 32.7 | 1,209 | 1,098 |
| Lactobacillus iners ATCC 55195 | Vagina | 1,238,993 | 32.0 | 1,144 | 1,041 |
| Lactobacillus iners DSM_13335 | Vagina | 1,277,649 | 32.3 | 1,214 | 1,095 |
| Lactobacillus iners LactinV_01V1-a | Vagina | 1,294,173 | 32.7 | 1,527 | 1,103 |
| Lactobacillus iners LactinV_03V1-b | Vagina | 1,303,958 | 32.7 | 1,459 | 1,154 |
| Lactobacillus iners LactinV_09V1-c | Vagina | 1,312,334 | 32.7 | 1,361 | 1,144 |
| Lactobacillus iners LactinV_11V1-d | Vagina | 1,310,250 | 32.6 | 1,338 | 1,152 |
| Lactobacillus iners SPIN_1401G | Vagina | 1,281,027 | 32.5 | 1,238 | 1,119 |
| Lactobacillus iners SPIN_2503V10-D | Vagina | 1,283,897 | 32.6 | 1,273 | 1,096 |
| Lactobacillus iners UPII_143-D | Vagina | 1,257,583 | 32.6 | 1,186 | 1,080 |
| Lactobacillus iners UPII_60-B | Vagina | 1,323,615 | 32.7 | 1,276 | 1,145 |
| Lactobacillus jensenii 115-3-CHN | Vagina | 1,648,660 | 33.4 | 1,470 | 1,275 |
| Lactobacillus jensenii 1153 | Vagina | 1,756,479 | 33.7 | 1,578 | 1,312 |
| Lactobacillus jensenii 269-3 | Vagina | 1,688,275 | 34.4 | 1,575 | 1,331 |
| Lactobacillus jensenii 27-2-CHN | Vagina | 1,658,778 | 33.1 | 1,476 | 1,276 |
| Lactobacillus jensenii JV-V16 | Vagina | 1,590,829 | 33.7 | 1,475 | 1,202 |
| Lactobacillus jensenii SJ-7A-US | Vagina | 1,722,751 | 33.3 | 1,630 | 1,395 |
| Lactobacillus kefiranofaciens ZW3* | Food | 2,354,088 | 37.4 | 2,162 | 1,725 |
| Lactobacillus paracasei subsp. paracasei 8700:2* | Gastrointestinal tract | 3,001,787 | 45.9 | 3,021 | 2,483 |
| Lactobacillus paracasei subsp. paracasei ATCC 25302 | Gastrointestinal tract | 2,991,737 | 44.9 | 3,042 | 2,437 |
| Lactobacillus plantarum JDM1* | Gastrointestinal tract | 3,197,759 | 44.7 | 2,948 | 2,072 |
| Lactobacillus plantarum subsp. plantarum ST-III* | Food | 3,307,936 | 44.5 | 3,038 | 2,127 |
| Lactobacillus reuteri 100-23 | Gastrointestinal tract | 2,315,924 | 38.7 | 2,186 | 1,759 |
| Lactobacillus reuteri CF48-3A | Gastrointestinal tract | 2,107,903 | 37.3 | 2,164 | 1,673 |
| Lactobacillus reuteri DSM_20016* | Gastrointestinal tract | 1,999,618 | 38.9 | 1,900 | 1,601 |
| Lactobacillus reuteri JCM_1112* | Gastrointestinal tract | 2,039,414 | 38.9 | 1,820 | 1,624 |
| Lactobacillus reuteri MM2-3 | Gastrointestinal tract | 2,015,721 | 37.3 | 2,045 | 1,635 |
| Lactobacillus reuteri MM4-1A | Gastrointestinal tract | 1,968,087 | 38.0 | 2,023 | 1,621 |
| Lactobacillus reuteri SD2112* | Gastrointestinal tract | 2,203,235 | 35.9 | 2,235 | 1,700 |
| Lactobacillus rhamnosus LMS2-1 | Gastrointestinal tract | 3,159,402 | 45.7 | 3,155 | 2,341 |
| Lactobacillus ruminis ATCC 25644 | Gastrointestinal tract | 2,159,237 | 42.1 | 2,264 | 1,955 |
| Lactobacillus ruminis SPM0211 | Gastrointestinal tract | 2,172,227 | 43.7 | 2,326 | 1,987 |
| Lactobacillus salivarius CECT_5713* | Gastrointestinal tract | 2,136,138 | 33.0 | 1,552 | 1,517 |
| Lactobacillus vaginalis ATCC 49540 | Vagina | 1,877,332 | 39.1 | 1,870 | 1,521 |
%G+C, G+C content, defined as 100 × (G+C)/(A+T+G+C). CDS, protein-coding sequences. MCL, protein families built by BLAST and Markov clustering. Asterisks (*) denote strains for which the complete genome sequence was available.
Software.
Sequence data were analyzed using the G-language Genome Analysis Environment, version 1.8.13 (34–36), available at http://www.g-language.org. Statistical analyses were implemented using R, version 2.15.0 (37), available at http://www.R-project.org/.
Protein repertoire analysis.
Protein-coding sequences were retrieved from sequence information of chromosomes and plasmids of the bacterial strains (Table 1). Homologous proteins were identified by BLASTP (38) on the criteria of an E value cutoff of 1e-5 and a minimum aligned sequence length coverage of 50% of a query sequence (abbreviated as E value < 1e-5 and >50% coverage). A cluster of homologous proteins (protein family) was built by all-against-all protein sequence comparison using BLASTP followed by Markov clustering (MCL) with an inflation factor of 1.2 (39) using MCLBLASTLINE (available at http://micans.org/mcl/). This method yielded 11,047 protein families containing 136,962 proteins from all the strains. To reduce annotation inconsistencies caused by differences in the gene-finding algorithms used in different genome-sequencing projects, we performed TBLASTN searches of each strain's proteome against all the other strains' genomes (E-value < 1e-5 and >50% coverage). The resulting protein repertoire (binary data for the presence or absence of each protein family) is shown in Table S1 in the supplemental material.
A value corresponding to 1 minus the Jaccard coefficient was used to measure pairwise distances between genomes based on their protein repertoires. To visualize the similarities in protein repertoire among genomes, we performed hierarchical clustering (average linkage clustering, or the unweighted-pair group method using average linkages [UPGMA]) on the distance matrix, and the clustering result was represented by a dendrogram.
We assigned functional annotations to each protein family by merging all the functional annotations of proteins belonging to the same protein family (deleting uninformative annotations such as “hypothetical protein”). We used multiple databases as follows: Clusters of Orthologous Groups (COG [http://www.ncbi.nlm.nih.gov/COG/; 40]), JCVI/CMR [http://cmr.jcvi.org/; 41]), SEED (http://www.theseed.org/ [42]), UniProtKB/Uniref90 (http://www.uniprot.org), Virulence Factors Database (VFDB) (http://www.mgc.ac.cn/VFs/ [43]), Pfam (http://pfam.sanger.ac.uk), and Gene Ontology (GO) (http://www.geneontology.org). We performed a similarity search of the 136,962 Lactobacillus proteins against the Uniref90 and VFDB protein sequence databases using BLASTP (E-value < 1e−5 and >50% coverage) and assigned the functional annotations of the most similar protein sequences. We searched protein sequences against the Pfam library of hidden Markov models (HMMs) using HMMER (http://hmmer.janelia.org/) and converted Pfam accession numbers to GO terms using “pfam2go” mapping (http://www.geneontology.org/external2go/pfam2go).
Statistical analyses.
We performed several statistical analyses to evaluate the protein family presence, absence, and relative abundance among vaginal and nonvaginal Lactobacillus species. The vaginal group consisted of the species L. crispatus, L. iners, L. jensenii, L. gasseri, L. coleohominis, and L. vaginalis. The latter two species have been found in the vagina but typically occur in very low relative abundance (2, 7, 8). To examine over- or underrepresented functional categories among vaginal relative to nonvaginal species, we calculated odds ratios (OR) and tested their significance using Fisher's exact test. A two-by-two contingency table was constructed for each functional category from the COG, JCVI, SEED, VFDB, and GO databases. This table included the following parameters: the number of protein families among the vaginal bacteria in this category (a); the number of protein families among the vaginal bacteria not in this category (b); the number of protein families among the nonvaginal bacteria in this category (c); and the number of protein families among the nonvaginal bacteria not in this category (d). The odds ratio (defined as ad/bc) was used to rank the relative overrepresentation (odds ratio > 1) or underrepresentation (odds ratio < 1) of each functional category. The P value obtained by Fisher's exact test was adjusted for the multiple comparisons by controlling for the false-discovery rate (FDR [44]).
We calculated Cramer's V to screen protein families showing biased distributions between comparative groups: i.e., vaginal and nonvaginal bacteria. Cramer's V is a measure of the degree of correlation in contingency tables. Cramer's V values close to 0 indicate weak associations between variables, while those close to 1 indicate strong associations.
RESULTS
Even though Lactobacillus species are associated with several sites in the human body, only 4 of the over 130 described species are commonly found in high abundance in the human vagina. Furthermore, each of these species typically inhabits distinct communities and the species do not often display patterns of stable codominance (1, 45). We analyzed functional relationships of different species and strains of the Lactobacillus genus to understand whether the vaginal lactobacilli showed signatures of adaptation to their environment. We then analyzed the functional profile of each of the vaginal species to try to understand how they potentially interact with their environment and each other.
General genomic features.
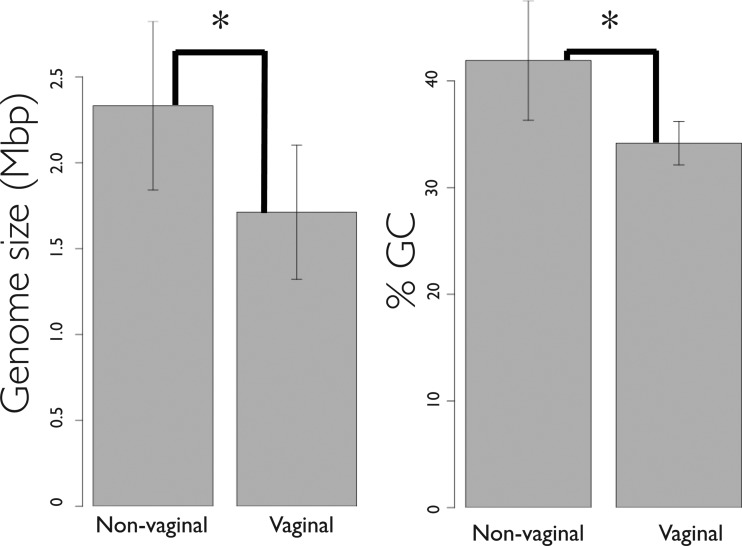
We surveyed the NCBI and PATRIC (33) databases and retrieved coding sequences for all sequences from the genus Lactobacillus marked as having the whole genome sequenced and as having been isolated from food, the gastrointestinal tract, or the human vagina. As of 9 April 2013, there were 75 draft or complete genome sequences. We then analyzed several reported features of the genomes and discarded sequences based on the following parameters: ambiguous isolation site, deviation of genome size or number of coding sequences (CDS) at a level of over 1.5× that of the average genome size or number of CDS for the species, and lack of annotation of rRNA and tRNA gene sequences. The following strains were removed from the analysis: Lactobacillus casei BL23 (rRNAs and tRNAs were not annotated for this strain), L. fermentum 28-3-CHN (rRNAs were not annotated for this strain), L. iners LEAF_2052A-d, L. iners LEAF_2052A-b, L. iners LEAF_2062A-hi, and L. iners LEAF_3008A-a (the number of CDS for these strains was approximately twice the number reported for all the other strains of this species), L. jensenii 208-1 (the genome size reported for this strain was much larger than for the other strains of this species, and the CDS number was much higher), and L. gasseri ATCC 33323 (the isolation site reported for this strain was ambiguous). Table 1 lists the general genomic features of the bacterial strains analyzed. The genome sizes of the strains used in this analysis range from 1.23 Mbp to 3.36 Mbp, and the GC content ranges from 32% to 52%. The values for both the genome size and the GC content of the strains isolated from the vagina are significantly lower than those of the strains isolated from the other sites (Fig. 1) (genome size, 1.71 ± 0.391 Mbp versus 2.33 ± 0.491 Mbp; GC content, 34.17% ± 2.036% versus 41.92% ± 5.609%). Of the 67 genomes tested, 22 were complete and 45 were incomplete (Table 1). The clustering analysis performed did not differentiate the data into two subsets (see Fig. 3).
FIG 1.
Genome size and GC content of Lactobacillus species. Data represent average genome size and % GC content of the Lactobacillus species isolated from the vagina and from other habitats (gastrointestinal tract and food). Error bars denote standard deviations (SD). The Kruskal-Wallis rank sum test was done on vaginal versus nonvaginal Lactobacillus strains to compare the two groups, and significant differences (P < 0.05) are denoted with an asterisk (*).
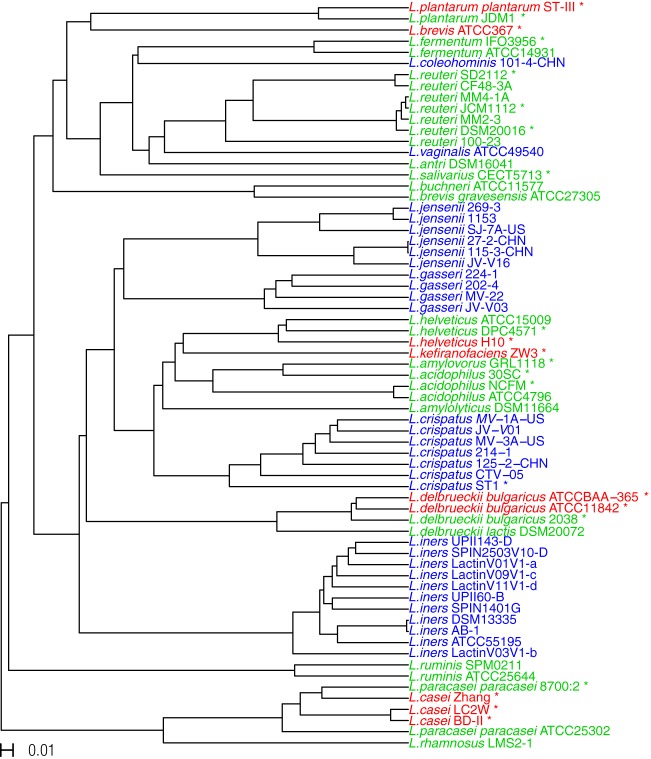
FIG 3.
Hierarchical clustering of Lactobacillus strains based on dissimilarities in gene content. The dissimilarities were measured on binary data using Jaccard distance values, ranging from 0 to 1, to evaluate the presence or absence of each of the protein families. The bar on the left bottom of the figure represents a distance of 0.01. The different colors of the leaves represent the habitats from which the strains were isolated. Red represents strains isolated from food, green represents strains isolated from gastrointestinal tract, and blue represents strains isolated from the vagina.
Protein repertoire of the Lactobacillus species analyzed.
The genus Lactobacillus has a large pan-genome that codes for a wide variety of proteins (46). The 136,962 protein coding sequences from the genomes of 67 strains (25 species) of Lactobacillus were classified into 11,047 homologous gene clusters or protein families (see Materials and Methods for details; see also Table S1 in the supplemental material). Of the 11,047 protein families, 5,091 were present in a single strain (singletons) and 5,956 were present in two or more strains; of those, 311 were conserved across all the strains (core). The core genes that persist over long periods may contain the essential genes required by all the strains within this genus under the conditions they experience.
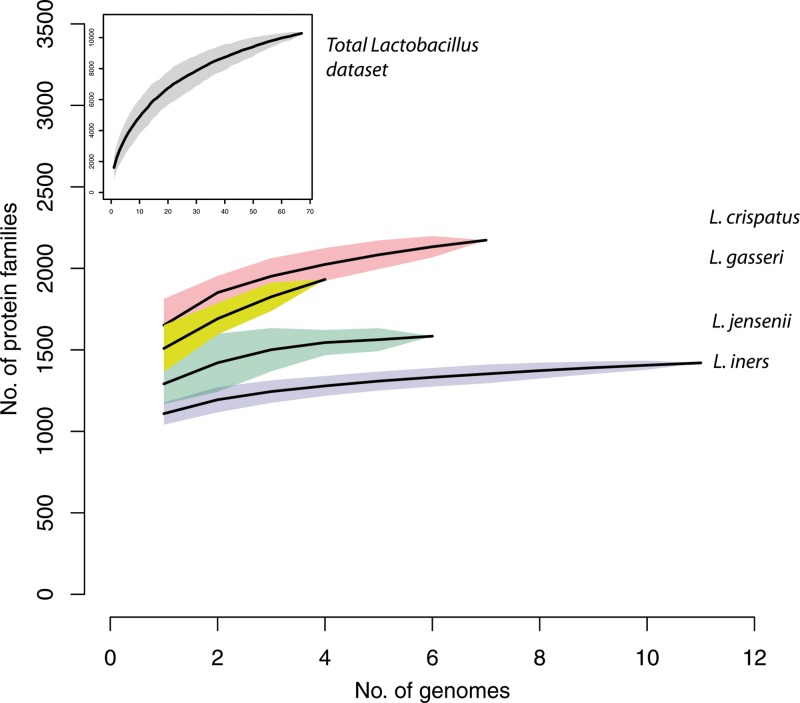
Figure 2 shows a rarefaction curve representing the addition of new protein families when each new genome sequence was added to the data set. Note that a protein family can contain multiple fragmented sequences derived from a single gene in draft genome sequences as well as homologs (e.g., orthologs, paralogs, and xenologs). Collectively, the members of the genus Lactobacillus show the presence of a high number of protein families (>10,000). As a point of reference, the same approach yielded 5,014 protein families containing 74,122 individual proteins from the 28 strains (12 species) of Staphylococcus and Macrococcus caseolyticus JCSCS5402 (47). The high number of protein families is likely related to the wide distribution of lactobacilli in very diverse habitats (19). More specifically, each of the four species that are found more commonly in the vagina, L. iners, L. crispatus, L. jensenii, and L. gasseri (1, 48), has between 1,500 and 2,500 protein families by our measures.
FIG 2.
Gene number in Lactobacillus species. Gene accumulation rarefaction curves across Lactobacillus strains were calculated based on the presence or absence of protein families using the “specaccum” function in the “vegan” package of R (http://www.R-project.org/) and were estimated by bootstrapping 100 permutations of randomized sample orders. The curves for all the genomes analyzed in this study (gray) and for the genomes of L. crispatus (pink), L. gasseri (yellow), L. jensenii (green), and L. iners (blue) are represented.
To understand whether species isolated from the same environment were functionally similar in terms of their potential protein repertoire, we measured pairwise distances between all strains analyzed based on the presence or absence of each of the protein families that make up the total protein family repertoire and visualized their hierarchical clustering in a dendrogram (Fig. 3). We hypothesized that species that were functionally adapted to a particular environment would be more similar to each other and so would form a cluster in the dendrogram. We found no clustering of the Lactobacillus species occupying each of the three sources (i.e., food, gastrointestinal tract, and vagina), suggesting that even if the species are adapted to a specific habitat, their functional potentials may still be very distinct (Fig. 3). L. jensenii and L. gasseri formed a single cluster in the dendrogram, indicating that the protein repertoires of these two species are more similar to each other than to those of any of the other species (Fig. 3).
Relative representation of protein categories and protein families in vaginal Lactobacillus species.
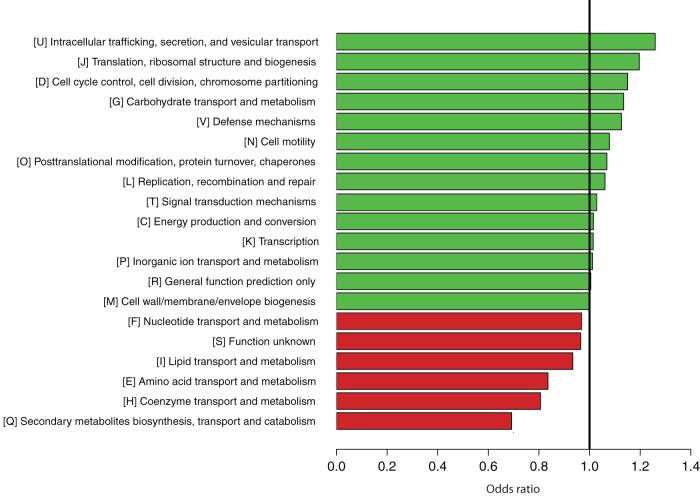
The odds ratio (OR) was used to rank the relative underrepresentation (OR < 1) or overrepresentation (OR > 1) of each functional category in the 30 vaginal strains compared to their representation in the 37 nonvaginal strains isolated from the gastrointestinal tract and food (see Table S2 in the supplemental material for a comprehensive list). Note that a functional category can consist of one or more protein families. For example, 18 of the 11,047 protein families were assigned to COG1846, and the repertoires differed among species (see Table S1). The COG database defines four major functional categories, which are further subdivided into 25 categories, i.e., 5 classified as “information storage and processing,” 10 classified as “cellular processes and signaling,” 8 classified as “metabolism,” and the last 2 classified as “poorly characterized” (ftp://ftp.ncbi.nih.gov/pub/COG/COG/fun.txt). Of the four COG major categories, “Metabolism” (OR = 0.91) was underrepresented, while “Information storage and processing” (OR = 1.12) was overrepresented. Of the 25 COG subcategories, “(Q) Secondary metabolites biosynthesis, transport, and catabolism” (OR = 0.69), “(H) Coenzyme transport and metabolism” (OR = 0.81), and “(E) Amino acid transport and metabolism” (OR = 0.84) were underrepresented, while “(U) Intracellular trafficking, secretion, and vesicular transport” (OR = 1.26), “(J) Translation, ribosomal structure, and biogenesis” (OR = 1.20), and “(G) Carbohydrate transport and metabolism” (OR = 1.14) were overrepresented (Fig. 4).
FIG 4.
Overrepresentation (green) and underrepresentation (red) of the Clusters of Orthologous Genes (COGs) in the genomes of the vaginal Lactobacillus species. The horizontal scale at the bottom represents the odds ratio calculated for each COG.
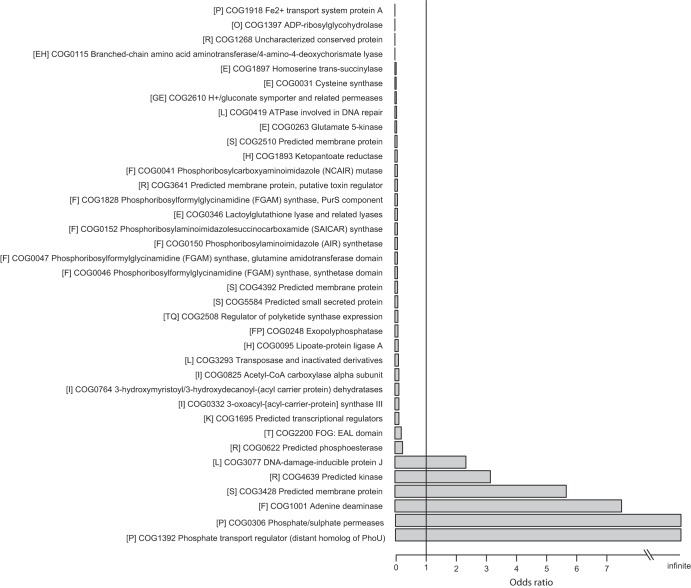
We also found numerous specific COGs that were over- and underrepresented in the vaginal strains relative to the nonvaginal strains. Of the 1,939 COGs analyzed, 123 were significantly differentially represented (FDR < 0.01); 15 were overrepresented and 108 were underrepresented (see Table S2 in the supplemental material). The 108 underrepresented COGs included “COG0031 Cysteine synthase” (OR = 0.043), “COG0620 Methionine synthase II (cobalamin-independent)” (OR = 0.356), and “COG0498 Threonine synthase” (OR = 0.421), which belong to the COG subcategory “(E) Amino acid transport and metabolism.” Two protein families (no. 975 and 1414) were assigned to COG0498. The lack of some of the proteins that are responsible for the synthesis of amino acids supports the idea that the vaginal species had adapted or are experiencing ongoing adaptation to a symbiotic life style (28). Figure 5 shows 37 COGs that were highly significant (FDR < 0.001), including those exclusively present (OR = infinite) or absent (OR = 0) in the vaginal species.
FIG 5.
Cluster of Orthologous Genes (COGs) with over- and underrepresentation in vaginal Lactobacillus species. Data represent Clusters of Orthologous Genes (COGs) that were significantly overrepresented (odds ratio > 1) and underrepresented (odds ratio < 1) in the vaginal relative to nonvaginal Lactobacillus species. CoA, coenzyme A.
We further analyzed the four COGs that were present exclusively in the vaginal Lactobacillus species; two involved in phosphate transport (protein family no. 1481 [COG1392] and 1505 [COG0306]) were found exclusively in L. iners and L. jensenii, and two hydrolases (no. 1966 [COG0499] and 1971 [COG1451]) were found exclusively in L. iners (see Table S2 in the supplemental material). For this, we created a search term for each COG code and the keyword Lactobacillus on the NCBI website. We then ran a BLAST search on the protein sequence results, again restricting our search to the genus Lactobacillus. This allowed us to identify proteins that were similar to our four proteins but that were not considered as such based on the COG annotation. These analyses showed that the protein sequence corresponding to pitA (COG0306) was found exclusively in L. iners and L. jensenii. Furthermore, of all the genetic information relative to the genus Lactobacillus available in GenBank as of August 2013, only the genomes of three other organisms, L. farciminis, L. gigeriorum, and L. pasteurii, carry genes that code for a protein whose maximum alignment score with PitA is higher than 200 (49, 50). PitA is a low-affinity inorganic phosphate transport protein (51, 52). Similarly, the protein corresponding to COG1392 is identified as a phosphate transport regulator that is a distant homolog to PhoU. PhoU was found to be a repressor of phosphate transport (52, 53) in Escherichia coli. The product of the phoU gene is also involved in the persister phenotype that confers temporary resistance of bacterial cultures to multiple antibiotics (54). Interestingly, of all the strains analyzed here, the protein sequence for phoU aligned significantly (E-value < 0.05) only with that of L. iners. Other lactobacilli with which the sequence aligned significantly were again L. farciminis, L. gigeriorum, and L. pasteurii, as well as Lactobacillus sp. 7_1_47FAA. A BLASTN analysis of the 16S rRNA gene sequence of this organism showed a 100% identity match to a partial sequence of the species L. iners. The unique presence of phoU in the genome of L. iners relative to the other genomes analyzed raises the possibility that phosphate acquisition is important to the physiology of this species in the vagina.
The other two protein families exclusively present in the vaginal Lactobacillus species were hydrolases. One was S-adenosylhomocysteine hydrolase (COG0499). Of the genomes used in our analysis, the sequence of this protein aligns significantly only with protein sequences in L. iners. However, a BLAST search that included all available genetic information for the genus Lactobacillus also yielded a significant alignment with protein sequences found in the genomes of L. plantarum 16 and the L. iners strain Lactobacillus sp. 7_1_47FAA. Interestingly, there was also a significant alignment with 31% identity with the d-lactate dehydrogenase protein sequence of L. salivarius. It is possible that there was a co-option of the d-lactate dehydrogenase protein from L. iners to a different function (adenosylhomocysteine hydrolase) since this is absent from the genomes of L. iners analyzed. Also overrepresented in the vaginal species is a metal-dependent hydrolase (COG1451), although this function is characterized as being just a general prediction. Again, this protein is found only in L. iners and the L. iners strain Lactobacillus sp. 7_1_47FAA.
We also used Cramer's V to measure associations between protein families and the vaginal and nonvaginal species. A Cramer's V = 1 indicates a complete association between the presence of a specific protein family and a particular group of species. The fact that we found no complete association between the protein families and the group with vaginal or nonvaginal species (Cramer's V = 1) indicates that there are no protein families that exist exclusively in all the vaginal or all nonvaginal Lactobacillus strains. However, we detected protein families with high Cramer's V values close to 1, indicating a biased distribution between vaginal and nonvaginal strains. We set a threshold Cramer's V value of 0.7 as being biologically significant. This resulted in the 33 protein families showing significant association with either the group of vaginal strains (3 protein families) or the group of nonvaginal strains (30 protein families). Protein family no. 10091 was present in 21 of the 30 vaginal strains and absent in all of the 37 nonvaginal strains. This protein (locus_tag LineA_010100003225) is short (47 amino acids) and is a hypothetical protein without any known functions. Protein family no. 1081 (biotin synthase) and no. 1107 (branched-chain amino acid aminotransferase) were absent in all the 30 vaginal strains and present in 32 and 28 of the 37 nonvaginal strains, respectively (see Table S3 in the supplemental material).
Functional characteristics of each of the vaginal species.
When present in a community, Lactobacillus species tend to dominate numerically, and codominance of Lactobacillus species is observed only occasionally (1). Therefore, we focused our further analyses on the four most abundant Lactobacillus species found in the vagina in an attempt to identify species-specific traits that could explain the community patterns observed. The number of proteins present exclusively in each of the vaginal species was highest in L. crispatus, which has the largest genome on average, and lowest in L. iners, which has the smallest genome on average (Table 1). L. iners shows the highest number of absent protein families that are present in the other three vaginal species analyzed.
Each of the four vaginal species may possess a set of specific genes related to adaptation to vaginal environments that distinguishes it from the other vaginal species and nonvaginal species. Here we highlight some protein families that are present or absent in all strains of each of a given vaginal species in comparison to all strains of the other vaginal species and in comparison to all the other strains of lactobacilli used in our analysis. These are only some examples that we considered biologically interesting given the present knowledge about the biology of the species studied, particularly their interactions with the surrounding environment. A complete list of all protein families with Cramer's V > 0.7 indicating a biased distribution for each species is given in Tables S4 to S11 in the supplemental material.
Lactobacillus iners-specific genes.
Among the species analyzed, L. iners is the one with the smallest genome on average (∼1.3 Mbp). A total of 67 protein families were present in all the 11 L. iners strains but absent in the 17 remaining vaginal strains belonging to L. crispatus, L. gasseri, and L. jensenii (see Table S4 in the supplemental material). All of the L. iners strains lacked several integral membrane proteins that were present in most of the other vaginal strains (protein families no. 106, 385, 844, 329, and 877). On the other hand, several ABC transporter permeases, e.g., COG4587 (protein family no. 1651 and 2566) and COG3694 (protein family no. 1713 and 2565), were exclusively present in the L. iners strains analyzed, but other protein families involved in ABC transport, e.g., protein family no. 715 (COG3221) and 106 (COG4485), were absent from L. iners while present in all other vaginal strains. L. iners strains also lacked several protein families related to the acetyltransferase GNAT family (protein family no. 289, 881, 1019, 1077, 1200, and 1290) that were present in all other vaginal strains.
L. iners strains lacked several protein families that are identified as transcriptional regulators that respond to extracellular environmental signals, e.g., AraC family transcriptional regulators (protein family no. 219, 812, 2019, and 2087), TetR family transcriptional regulators (protein family no. 76, 960, and 1034 [COG1309]), and LysR family transcriptional regulators (protein family no. 13 and 1250 [COG0583). Also of note is the presence of thiol-activated cytolysin (protein family no. 1965) in L. iners and its absence in all other Lactobacillus species analyzed. This likely corresponds to inerolysin (52), a broad-range cholesterol-dependent cytolysin. This toxin forms pores in epithelial cells and activates proinflammatory signaling and so is suggested to be a virulence factor for L. iners (55). In support of this is the fact that this cytolysin is upregulated 6-fold under bacterial vaginosis conditions (56). There are also 17 protein families that are absent from any of the 11 L. iners genomes but present in all of the other 56 Lactobacillus genomes in our analysis (see Table S5 in the supplemental material).
Lactobacillus crispatus-specific genes.
L. crispatus has the largest genome size of all the vaginal lactobacilli. A total of 143 protein families were present in all seven L. crispatus strains but absent in the 21 remaining vaginal strains (see Table S6 in the supplemental material). L. crispatus strains exclusively have a unique DNA polymerase (protein family no. 1490). In addition, the strains of this species have several protein families involved in organismal interactions that are absent from all other vaginal strains. Two of them are related to bacteriocins (protein family no. 1889 and 2918 [PF10439]), and two are involved in toxin-antitoxin systems (protein family no. 1863 and 1506). There are many studies on the possible interaction of vaginal Lactobacillus with phages (57–59) or plasmids (60). Our analysis showed that the L. crispatus-specific genes included those related to mobile genetic elements such as integrase/recombinase (protein family no. 1124), abortive phage resistance protein (no. 1999), phage shock protein C (no. 865), ParE-domain protein (no. 3237), and temperature-sensitive replication protein (no. 1637). Transposases (61), e.g., COG0675 (protein family no. 34 and 1347), COG3385 (protein family no. 207 and 800), and COG3436 (protein family no. 310 and 730), were enriched in L. crispatus. A total of four protein families, uncharacterized conserved secreted or membrane protein (protein family no. 921), bile salt transporter (no. 985), 3-dehydroquinate dehydratase (no. 1092), and acetyltransferases (no. 1379), were absent in all the L. crispatus strains but present in all of the 21 vaginal strains belonging to L. iners, L. gasseri, and L. jensenii (see Table S6). In addition, we found that the genome of L. crispatus codes for 26 protein families that are not found in the other Lactobacillus species analyzed (see Table S7). Among them are two LPXTG-motif cell wall anchor domain proteins (protein family no. 2568 and 2916 [23, 46, 62]). We did not find any protein families that were absent from L. crispatus and present in all other Lactobacillus species (i.e., Cramer's V = 1) or proteins more abundant in L. crispatus than in other Lactobacillus species (i.e., Cramer's V > 0.7). As observed for L. gasseri, this is likely correlated to the large genome size of this species compared to the other vaginal lactobacilli. The number, and thus the presence, of protein families annotated as transposases tended to be lower and the genome size tended to be smaller in vaginal species relative to the other Lactobacillus species, but L. crispatus represents an exception (Table 1). These results suggest that in this species, the genome size may have increased due to transposable element amplification (a proliferation of transposons) (63) and gene gain via horizontal transfer.
Lactobacillus gasseri-specific genes.
A total of 122 protein families were present in all four L. gasseri strains but absent in the 24 strains from the other three vaginal species (see Table S8 in the supplemental material). Among them was a copper resistance/homeostasis protein (protein family no. 1514 [COG3142]). The strains of L. gasseri also have several protein families that are a reflection of organismal interactions. Among them are an addiction module toxin (protein family no. 2863 [COG3668] plasmid stabilization system protein), a toxin-antitoxin addiction module regulator (PF14472), and a protein of the toxin-antitoxin system AbrB family (protein family no. 1688). Of note is the fact that all the strains of this species have a pediocin (Pediococcus bacteriocin) immunity protein (PF08951) that is absent from all the strains of the other vaginal species. Also present is a signal transduction histidine kinase regulating citrate and malate metabolism (protein family no. 2030 [COG3290]), but a citrate lyase regulator is absent from the strains of this species and present in all other vaginal strains (see Table S8). A set of six protein families was absent in all four L. gasseri strains but present in the other three vaginal species (see Table S8). In addition, the genome of L. gasseri contains sequences coding for 57 protein families that are absent from all the other genomes in our analysis (see Table S9). Among them are acetyltransferases (protein family no. 2994), transcriptional regulators (protein family no. 3454 [COG1846]), and an inner membrane permease of ABC transporter (protein family no. 3711).
Lactobacillus jensenii-specific genes.
A total of 83 protein families were present in all six L. jensenii strains but absent in all 22 strains from the other three vaginal species (see Table S10 in the supplemental material). The strains of L. jensenii have several unique versions of protein families functionally equivalent to unique protein families found in the other three vaginal species. They have two families involved in ABC transport (protein family no. 3788 [COG1653] and 3791), four families of transcriptional regulators (no. 1319, 1837, 1987, and 4520 [COG1309]), and a citrate transporter (protein family no. 1783 [COG2851]). Additionally, these strains also have a malate/l-lactate dehydrogenase (protein family no. 1260 [COG2055]), a 4-amino-4-deoxy-l-arabinose transferase-like glycosyltransferase (protein family no. 1546 [COG1807]), an acetoacetate decarboxylase (protein family no. 1673 [COG4689]), a metal-dependent hydrolase of beta-lactamase super family III (protein family no. 1807 [COG1234]), and a competence protein (protein family no. 2706 [COG2165]) that are absent in all other vaginal strains. A total of 14 protein families were absent in all six L. jensenii strains and present in all 22 strains from the other three vaginal species (see Table S10). This species lacks proteins involved in the glucitor/sorbitol phosphotransferase system (protein family no. 804 [COG3731]) that are present in the strains of all the other vaginal species (see Table S10). Finally, L. jensenii has 43 unique protein families that were absent from all other Lactobacillus species in our analysis (see Table S11). Unfortunately, only a small subset of these is annotated. Among them are membrane-associated proteins: a Gram-positive anchor domain protein (protein family no. 2592 [PF00746]), a transcriptional repressor of sporulation and extracellular protein (protein family no. 2604), a LPXTG-motif cell wall anchor domain protein (protein family no. 2979), and a choline binding protein (protein family no. 2987). Three protein families were absent in L. jensenii and present in all of the other 61 Lactobacillus strains analyzed: a carboxylase (protein family no. 705), a orotidin-5′-phosphate decarboxylase (protein family no. 753 [COG0284]), and a pyrimidine operon attenuation protein/uracil phosphoribosyltransferase (protein family no. 244 [COG2065]) (see Table S11). The proteins listed above thus show a lack of particular signatures (exclusive presence or absence) indicative of the potential of this species to interact with the environment.
These examples show that the four most commonly found vaginal Lactobacillus species may potentially have different mechanisms of interacting with the environment, be it the host or other microbes in the community. This in turn may result in the existence of abilities that are unique to each species allowing them to establish a stable population in the vaginal environment and lead to their almost exclusive dominance in the communities they are part of, as each species is more likely to be better adapted to a particular environment.
DISCUSSION
Lactobacillus is a diverse genus with members inhabiting various habitats, including our own bodies as well as various food and dairy products (19). The growing number of sequenced genomes from members of this genus enables comparative genomic studies to address aspects of the biology and natural history of these organisms. For instance, L. bulgaricus (strain ATCC 11842) has undergone significant genome reduction and lost many genes related to carbohydrate metabolism, supporting the view that this species has adapted to its current stable environment (yogurt) following colonization from a plant environment (28). The pan-genome of L. casei also showed that this species has remarkable genetic variation that is associated with its multiniche distribution (14). In this study, we used comparative genomics to understand whether the genomes of the most common and abundant vaginal Lactobacillus species, L. iners, L. jensenii, L. crispatus, and L. gasseri (1), have signatures that suggest that they have specialized to succeed in the human vaginal environment (64).
The genomes of vaginal Lactobacillus species are significantly smaller (Fig. 1) than those of their close relatives residing in other environments, and this suggests that these species show some degree of adaptation to a host-dependent lifestyle, even if the adaptation is facultative (65). Genome size reduction is often observed in species that are dependent on a host for survival, since the host provides essential compounds and a stable environment for its symbionts (64, 66), thus relaxing selection for the maintenance of some genes necessary for survival as a free-living organism. For instance, some of the smallest genomes have been observed in bacteria that are obligate intracellular symbionts, which depend completely on the host for growth (65).
The genomes of host-dependent symbionts also tend to differ significantly from the genomes of free-living bacteria in terms of gene content. Among the differences between bacteria with different lifestyles, Merhej et al. (65) found that the genomes of host-dependent bacteria had an overrepresentation of genes involved in DNA replication, recombination, and repair, RNA processing and modification, translation, posttranslational modification, intracellular trafficking, and secretion. Genes coding for proteins involved in transcription, such as transcriptional regulators, defense mechanisms, transport and metabolism of amino acids, inorganic ions, and secondary metabolites were underrepresented in host-dependent bacteria (65).
We observed that the genomes of the vaginal lactobacilli shared some of these traits with host-dependent bacteria, suggesting a certain degree of association between these species and the host. However, reduced genomes of host-dependent bacteria also tend to maintain genes related to the provision of biological compounds to the host (65, 67), thus establishing a mutualistic relationship with the host where both players benefit from their interaction. This was not observed in the vaginal species, suggesting that, if there is a mutually beneficial relationship between these organisms and the women that harbor them, it is not based solely on the provision of compounds, such as amino acids, that the host is unable to synthesize.
Genes coding for complete biosynthetic pathways were previously found to be absent in vaginal lactobacilli relative to nonvaginal species (23), and this could suggest that the former are somehow utilizing compounds produced by the host. However, the vaginal bacterial communities are composed of hundreds of different species (1, 7) which, in addition to the host, could complement the metabolic needs of the Lactobacillus species. The ability to utilize resources secreted by other members of the community could thus be another factor influencing genome reduction by the loss of function of some genes, as suggested by the Black Queen hypothesis (68). This hypothesis posits that species may lose essential genes if their products are provided by other organisms in their communities (68).
The bacteria that inhabit the human vagina are not uniform across women, and even the same woman can have remarkable changes in the identity and relative abundances of vaginal bacterial species through time (1, 32). Although codominance of Lactobacillus species in a vaginal bacterial community can occur, it is not very common. This may be due to interspecies competition for nutrients or space, which may lead to the establishment of only one Lactobacillus species and exclusion of competitors. For instance, while both L. reuteri and L. johnsonii can inhabit the gut of vertebrates, this is possible only because nutritional adaptations provide niche differentiation that allows cohabitation by the two species through resource partitioning (69).
It is also possible that the relative abundances of the different Lactobacillus species are determined in part by the woman and by the other microbes in the vaginal environment. In this case, the representation of each of the Lactobacillus species depends on how each is able to respond to its particular environment. Our analysis of the genomes of the four most abundant and commonly found vaginal Lactobacillus species revealed they have genes encoding proteins that can facilitate different kinds of interactions between the bacterium and its environment, be it the host or other members in the microbiota. For instance, the genome of L. iners encodes a thiol-activated cytolysin (70). Proteins such as these are intimately involved in pathogenesis, and their main targets seem to be cells involved in host defense against invasion by pathogens (58). Recent work has shown that the gene encoding this protein is upregulated during the occurrence of bacterial vaginosis (56). The genome of L. crispatus codes for several protein families that are involved in organismal interactions, such as resistance to phage infections, bacteriocin-type sequences, toxin-antitoxin systems, and a putative plasmid stabilization system. Interestingly, of the four vaginal Lactobacillus species, lysogeny leading to the release of phage particles is most commonly observed in L. crispatus isolates (57). Furthermore, only the genome of L. gasseri codes for a pediocin immunity protein. This bacteriocin is produced by Pediococcus sp. (26)., individuals of which may also be found in vaginal bacterial communities (7).
In this study, we performed comparative genomics analyses to explore characteristics (e.g., genome size, GC content, and protein family repertoire) of vaginal Lactobacillus species to gain a better insight into their adaptation to the vaginal environment. We found that each of the four vaginal Lactobacillus species uniquely possesses or lacks numerous different kinds of protein families. This suggests that each species has experienced lineage-specific gene gain and loss and thus developed a unique set of protein families. The whole gene repertoire of a genome and consequently its functional potential can reflect various evolutionary events, including both vertical and horizontal transfer of genes and their duplication and loss events (71, 72). Convergent evolution leading to adaptation to a particular environment would be reflected in a similarity of functional profiles. For the Lactobacillus species analyzed here, we found no evidence that the whole-protein profiles of the four vaginal species or the species isolated from the other sites (gastrointestinal tract and food products) are more similar to each other than to those of any of the other species (Fig. 3), and so we cannot conclude that the functional profiles of the Lactobacillus species occupying the same habitat have converged to be similar as a whole. However, the functional profile of the vaginal species can potentially account for the variation found in acid generation, hydrogen peroxide production, bacteriocin production, biofilm formation, plasmid content, and antimicrobial susceptibility of indigenous vaginal lactobacilli from healthy women (73). The increasing number of genome sequences available for strains within the same species will help narrow down species-specific genes that influence their ecological and evolutionary dynamics. Genes gained and lost within each lineage of the four vaginal Lactobacillus species may provide clues to the adaptation of each lineage into vaginal environments, but many of the genes uniquely present and absent in each species are hypothetical or poorly characterized proteins. In the future, studies aimed at functional characterization of these species-specific genes should reveal details of how lactobacilli have adapted to the human vagina.
Supplementary Material
ACKNOWLEDGMENTS
We acknowledge Vandhana Krishnan and Matthew Settles (IBEST Genomics Resources Core, University of Idaho) for conducting preliminary analyses that provided useful insights and prompted further exploration in the present study. We also acknowledge Robert Lyon and Trent Nelson for their technical support with use of the IBEST Computational Resources Core at the University of Idaho.
R.J.H. is supported by a University of Idaho Bioinformatics and Computational Biology Fellowship in partnership with IBEST. This work was supported by grants U19 AI084044 from the National Institute of Allergies and Infectious Diseases and P30 RR033376 from the National Institute of General Medical Sciences of the National Institutes of Health.
Footnotes
Published ahead of print 31 January 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.01439-13.
REFERENCES
- 1.Ravel J, Gajer P, Abdo Z, Schneider GM, Koenig SSK, McCulle SL, Karlebach S, Gorle R, Russel J, Tacket CO, Brotman RM, Davis CC, Ault K, Peralta L, Forney LJ. 2011. Vaginal microbiome of reproductive-age women. Proc. Natl. Acad. Sci. U. S. A. 108:4680–4687. 10.1073/pnas.1002611107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Antonio MAD, Hawes SE, Hillier SL. 1999. The identification of vaginal Lactobacillus species and the demographic and microbiologic characteristics of women colonized by these species. J. Infect. Dis. 180:1950–1956. 10.1086/315109 [DOI] [PubMed] [Google Scholar]
- 3.De Backer E, Verhelst R, Verstraelen H, Alqumber MA, Burton JP, Tagg JR, Temmerman M, Vaneechoutte M. 2007. Quantitative determination by real-time PCR of four vaginal Lactobacillus species, Gardnerella vaginalis and Atopobium vaginae indicates an inverse relationship between L. gasseri and L. iners. BMC Microbiol. 7:115. 10.1186/1471-2180-7-115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pavlova SI, Kilic AO, Kilic SS, So JS, Nader-Macias ME, Simoes JA, Tao L. 2002. Genetic diversity of vaginal lactobacilli from women in different countries based on 16S rRNA gene sequences. J. Appl. Microbiol. 92:451–459. 10.1046/j.1365-2672.2002.01547.x [DOI] [PubMed] [Google Scholar]
- 5.Pendharkar S, Magopane T, Larsson PGR, de Bruyn G, Gray GE, Hammarstrom LH, Marcotte H. 2013. Identification and characterisation of vaginal lactobacilli from South African women. BMC Infect. Dis. 13:43. 10.1186/1471-2334-13-43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vásquez A, Jakobsson T, Ahrné S, Forsum U, Molin G. 2002. Vaginal Lactobacillus flora of healthy Swedish women. J. Clin. Microbiol. 40:2746–2749. 10.1128/JCM.40.8.2746-2749.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Verhelst R, Verstraelen H, Claeys G, Verschraegen G, Van Simaey L, De Ganck C, De Backer E, Temmerman M, Vaneechoutte M. 2005. Comparison between Gram stain and culture for the characterization of vaginal microflora: definition of a distinct grade that resembles grade I microflora and revised categorization of grade I microflora. BMC Microbiol. 5:61. 10.1186/1471-2180-5-61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhou X, Brown CJ, Abdo Z, Davis CC, Hansmann MA, Joyce P, Foster JA, Forney LJ. 2007. Differences in the composition of vaginal microbial communities found in healthy Caucasian and black women. ISME J. 1:121–133. 10.1038/ismej.2007.12 [DOI] [PubMed] [Google Scholar]
- 9.Zhou X, Hansmann MA, Davis CC, Suzuki H, Brown CJ, Schütte U, Pierson JD, Forney LJ. 2010. The vaginal bacterial communities of Japanese women resemble those of women in other racial groups. FEMS Immunol. Med. Microbiol. 58:169–181. 10.1111/j.1574-695X.2009.00618.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Alqumber MA, Burton JP, Devenish C, Tagg JR. 2008. A species-specific PCR for Lactobacillus iners demonstrates a relative specificity of this species for vaginal colonization. Microb. Ecol. Health Dis. 20:135–139. 10.1080/08910600802340967 [DOI] [Google Scholar]
- 11.Faust K, Sathirapongsasuti JF, Izard J, Segata N, Gevers D, Raes J, Huttenhower C. 2012. Microbial co-occurrence relationships in the human microbiome. PLoS Comput. Biol. 8:e1002606. 10.1371/journal.pcbi.1002606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grice EA, Kong HH, Conlan S, Deming CB, Davis J, Young AC, NISC Comparative Sequencing Program. Bouffard GG, Blakesley RW, Murray PR, Green ED, Turner ML, Segre JA. 2009. Topographical and temporal diversity of the human skin microbiome. Science 324:1190–1192. 10.1126/science.1171700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lamont RF, Sobel JD, Akins RA, Hassan SS, Chaiworapongsa T, Kusanovic JP, Romero R. 2011. The vaginal microbiome: new information about genital tract flora using molecular based techniques. BJOG 118:533–549. 10.1111/j.1471-0528.2010.02840.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhou X, Bent SJ, Schneider MG, Davis CC, Islam MR, Forney LJ. 2004. Characterization of vaginal microbial communities in adult healthy women using cultivation-independent methods. Microbiology 150:2565–2573. 10.1099/mic.0.26905-0 [DOI] [PubMed] [Google Scholar]
- 15.Stumpf RM, Wilson BA, Rivera A, Yildirim S, Yeoman CJ, Polk JD, White BA, Leigh SR. 2013. The primate vaginal microbiome: comparative context and implications for human health and disease. Am. J. Phys. Anthropol. 152(Suppl 57):119–134. 10.1002/ajpa.22395 [DOI] [PubMed] [Google Scholar]
- 16.Spear GT, Gilbert D, Sikaroodi M, Doyle L, Green L, Gillevet PM, Landay AL, Veazey RS. 2010. Identification of rhesus macaque genital microbiota by 16S pyrosequencing shows similarities to human bacterial vaginosis: implications for use as an animal model for HIV vaginal infection. AIDS Res. Hum. Retroviruses 26:193–200. 10.1089/aid.2009.0166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nugent RP, Krohn MA, Hillier SL. 1991. Reliability of diagnosing bacterial vaginosis is improved by a standardized method of Gram stain interpretation. J. Clin. Microbiol. 29:297–301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yarza P, Ludwig W, Euzéby J, Amann R, Schleifer K-H, Glöckner FO, Rossello-Mora R. 2010. Update of the All-Species Living Tree Project based on 16S and 23S rRNA sequence analyses. Syst. Appl. Microbiol. 33:291–299. 10.1016/j.syapm.2010.08.001 [DOI] [PubMed] [Google Scholar]
- 19.Kandler O, Weiss N. 1986. Genus Lactobacillus Beijerinck 1901, p 1209–1234 In Bergey's manual of systematic bacteriology, vol 2 Lippincott Williams & Wilkins, Hagerstown, MD [Google Scholar]
- 20.Lukjancenko O, Ussery DW, Wassenaar TM. 2012. Comparative genomics of Bifidobacterium, Lactobacillus, and related probiotic genera. Microb. Ecol. 63:651–673. 10.1007/s00248-011-9948-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Klaenhammer TR, Altermann E, Arigoni F, Bolotin A, Breidt F, Broadbent J, Cano R, Chaillou S, Deutscher J, Gasson M, van de Guchet M, Guzzo J, Hartke A, Hawkins T, Hols P, Hutkins R, Kleerebezem M, Kok J, Kuipers O, Lubbers M, Maguin E, McKay K, Mills D, Nauta A, Overbeekm R, Pel H, Pridmore D, Saier M, van Sinderen D, Sorokin A, Steele J, O'Sullivan D, de Vos W, Wiemer B, Zagorec M, Siexen R. 2002. Discovering lactic acid bacteria by genomics. Antonie Van Leeuwenhoek 82:29–58. 10.1023/A:1020638309912 [DOI] [PubMed] [Google Scholar]
- 22.Broadbent JR, Neeno-Eckwall EC, Stahl B, Tandee K, Cai H, Morovic W, Horvath P, Heidenreich J, Perna NT, Barrangou R, Steele JL. 2012. Analysis of the Lactobacillus casei supragenome and its influence in species evolution and lifestyle adaptation. BMC Genomics 13:533. 10.1186/1471-2164-13-533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Macklaim JM, Gloor GB, Anukam KC, Cribby S, Reid G. 2011. At the crossroads of vaginal health and disease, the genome sequence of Lactobacillus iners AB-1. Proc. Natl. Acad. Sci. U. S. A. 108:4688–4695. 10.1073/pnas.1000086107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Canchaya C, Claesson MJ, Fitzgerald GF, van Sinderen D, O'Toole PW. 2006. Diversity of the genus Lactobacillus revealed by comparative genomics of five species. Microbiology 152:3185–3196. 10.1099/mic.0.29140-0 [DOI] [PubMed] [Google Scholar]
- 25.Kankainen M, Paulin L, Tynkkynen S, von Ossowski I, Reunanen J, Partanen P, Satokari R, Vesterlund S, Hendrickx AP, Lebeer S, de Keersmaecker SCJ, Vaderleyden J, Hamalainen T, Laukkanen S, Salovuori N, Ritan J, Alatalo E, Korpela R, Mattila-Sandholm Lassig A, Hatakka K, Kinnunen KT, Karjalainen H, Saxelin M, Laakso K, Surakka A, Palva A, Salusjarvi T, Auvinen P, de Vos WM. 2009. Comparative genomic analysis of Lactobacillus rhamnosus GG reveals pili containing a human-mucus binding protein. Proc. Natl. Acad. Sci. U. S. A. 106:17193–17198. 10.1073/pnas.0908876106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Makarova KS, Koonin EV. 2007. Evolutionary genomics of lactic acid bacteria. J. Bacteriol. 189:1199–1208. 10.1128/JB.01351-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.O'Sullivan O, O'Callaghan J, Sangrador-Vegas A, McAuliffe O, Slattery L, Kaleta P, Callanan M, Fitzgerald GF, Ross RP, Beresford T. 2009. Comparative genomics of lactic acid bacteria reveals a niche-specific gene set. BMC Microbiol. 9:50. 10.1186/1471-2180-9-50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.van de Guchte M, Penaud S, Grimaldi C, Barbe V, Bryson K, Nicolas P, Robert C, Oztas S, Mangenot S, Couloux A, Loux V, Dervyn R, Bossy R, Bolotin A, Batto J-M, Walunas T, Gibrat J-F, Bessieres P, Weissenbach J, Ehrlich SD, Maguin E. 2006. The complete genome sequence of Lactobacillus bulgaricus reveals extensive and ongoing reductive evolution. Proc. Natl. Acad. Sci. U. S. A. 103:9274–9279. 10.1073/pnas.0603024103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hickey RJ, Zhou X, Pierson JD, Ravel J, Forney LJ. 2012. Understanding vaginal microbiome complexity from an ecological perspective. Transl. Res. 160:267–282. 10.1016/j.trsl.2012.02.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Linhares IM, Summers PR, Larsen B, Giraldo PC, Witkin SS. 2011. Contemporary perspectives on vaginal pH and lactobacilli. Am. J. Obstet. Gynecol. 204:120.e1–120.e5. 10.1016/j.ajog.2010.07.010 [DOI] [PubMed] [Google Scholar]
- 31.Azcarate-Peril MA, Altermann E, Goh YJ, Tallon R, Sanozky-Dawes RB, Pfeiler EA, O'Flaherty S, Buck BL, Dobson A, Duong T, Miller MJ, Barrangou R, Klaenhammer TR. 2008. Analysis of the genome sequence of Lactobacillus gasseri ATCC 33323 reveals the molecular basis of an autochthonous intestinal organism. Appl. Environ. Microbiol. 74:4610–4625. 10.1128/AEM.00054-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Benson DA, Clark K, Karsch-Mizrachi I, Lipman DJ, Ostell J, Sayers EW. 11 November 2013. GenBank. Nucleic Acids Res. 10.1093/nar/gkt1030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gillespie JJ, Wattam AR, Cammer SA, Gabbard JL, Shukla MP, Dalay O, Driscoll T, Hix D, Mane SP, Mao C, Nordberg EK, Scott M, Schulman JR, Snyder EE, Sullivan DE, Wang C, Warren A, William KP, Xue T, Yoo HS, Zhang C, Zhang Y, Will R, Kenyon RW, Sobral BW. 2011. PATRIC: the comprehensive bacterial bioinformatics resource with a focus on human pathogenic species. Infect. Immun. 79:4286–4298. 10.1128/IAI.00207-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Arakawa K, Kido N, Oshita K, Tomita M. 2003. G-language Genome Analysis Environment: a workbench for nucleotide sequence data mining. Bioinformatics 19:305–306. 10.1093/bioinformatics/19.2.305 [DOI] [PubMed] [Google Scholar]
- 35.Arakawa K, Suzuki H, Tomita M. 2008. Computational genome analysis using the G-language system. Genes Genomes Genomics 2:1–13 [Google Scholar]
- 36.Arakawa K, Tomita M. 2006. G-language system as a platform for large-scale analysis of high-throughput omics data. J. Pestic. Sci. 31:282–288. 10.1584/jpestics.31.282 [DOI] [Google Scholar]
- 37.Core Team R. 2012. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria [Google Scholar]
- 38.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389–3402. 10.1093/nar/25.17.3389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.van Dongen S. 2000. Graph clustering by flow simulation. University of Utrecht, Utrecht, Netherlands [Google Scholar]
- 40.Tatusov RL, Natale DA, Garkavtsev IV, Tatusova TA, Shankavaram UT, Rao BS, Kiryutin B, Galperin MY, Fedorova ND, Koonin EV. 2001. The COG database: new developments in phylogenetic classification of proteins from complete genomes. Nucleic Acids Res. 29:22–28. 10.1093/nar/29.1.22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Davidsen T, Beck E, Ganapathy A, Montgomery R, Zafar N, Yang Q, Madupu R, Goetz P, Galinski K, White O, Sutton G. 2010. The comprehensive microbial resource. Nucleic Acids Res. 38:D340–D345. 10.1093/nar/gkp912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Overbeek R, Begley T, Butler RM, Choudhuri JV, Chuang HY, Cohoon M, de Crécy-Lagard V, Diaz N, Disz T, Edwards R, Fonstein M, Frank ED, Gerdes S, Glass EM, Goesmann A, Hanson A, Iwata-Reuyl D, Jensen R, Jamshidi N, Krause L, Kubal M, Larsen N, Linke B, McHardy AC, Meyer F, Neuweger H, Olsen G, Olson R, Osterman A, Portnoy V, Pusch GD, Rodionov DA, Rückert C, Steiner J, Stevens R, Thiele I, Vassieva O, Ye Y, Zagnitko O, Vonstein V. 2005. The subsystems approach to genome annotation and its use in the project to annotate 1000 genomes. Nucleic Acids Res. 33:5691–5702. 10.1093/nar/gki866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen L, Yang J, Yu J, Yao Z, Sun L, Shen Y, Jin Q. 2005. VFDB: a reference database for bacterial virulence factors. Nucleic Acids Res. 33(Suppl 1):D325–D328. 10.1093/nar/gki008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Benjamini Y, Yekutieli D. 2001. The control of the false discovery rate in multiple testing under dependency. Ann. Statist. 29:1165–1188. 10.1214/aos/1013699998 [DOI] [Google Scholar]
- 45.Gajer P, Brotman RM, Bai G, Sakamoto J, Schütte UM, Zhong X, Koenig SS, Fu L, Ma ZS, Zhou X, Abdo Z, Forney LJ, Ravel J. 2012. Temporal dynamics of the human vaginal microbiota. Sci. Transl. Med. 4:132ra52. 10.1126/scitranslmed.3003605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kant R, Blom J, Palva A, Siezen RJ, de Vos WM. 2011. Comparative genomics of Lactobacillus. Microb. Biotechnol. 4:323–332. 10.1111/j.1751-7915.2010.00215.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Suzuki H, Lefébure T, Bitar PP, Stanhope MJ. 2012. Comparative genomic analysis of the genus Staphylococcus including Staphylococcus aureus and its newly described sister species Staphylococcus simiae. BMC Genomics 13:38. 10.1186/1471-2164-13-38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhou X, Brotman RM, Gajer P, Abdo Z, Schüette U, Ma S, Ravel J, Forney LJ. 2010. Recent advances in understanding the microbiology of the female reproductive tract and the causes of premature birth. Infect. Dis. Obstet. Gynecol. 2010:737425. 10.1155/2010/737425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403–410 [DOI] [PubMed] [Google Scholar]
- 50.Stromo GD. 2009. An introduction to sequence similarity (“homology”) searching. Curr. Protoc. Bioinformatics 27:3.1.1–3.1.7. 10.1002/0471250953.bi0301s27 [DOI] [PubMed] [Google Scholar]
- 51.Elvin CM, Dixon NE, Rosenberg H. 1986. Molecular cloning of the phosphate (inorganic) transport (pit) gene of Escherichia coli K-12. Identification of the pit+ gene product and physical mapping of the pit-gor region of the chromosome. Mol. Gen. Genet. 204:477–484 [DOI] [PubMed] [Google Scholar]
- 52.van Veen HW. 1997. Phosphate transport in prokaryotes: molecules, mediators and mechanisms. Antoine van Leeuwenhoek 72:299–315. 10.1023/A:1000530927928 [DOI] [PubMed] [Google Scholar]
- 53.White D, Drummond J, Fuqua C. 2012. The physiology and biochemistry of prokaryotes, 4th ed. Oxford University Press, Oxford, United Kingdom [Google Scholar]
- 54.Li Y, Zhang Y. 2007. PhoU is a persistence switch involved in persister formation and tolerance to multiple antibiotis and stresses in Escherichia coli. Antimicrob. Agents Chemother. 51:2092–2099. 10.1128/AAC.00052-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rampersaud R, Planet PJ, Randis TM, Kulkarni R, Aguilar JL, Lehrer RI, Ratner AJ. 2011. Inerolysin, a cholesterol-dependent cytolysin produced by Lactobacillus iners. J. Bacteriol. 193:1034–1041. 10.1128/JB.00694-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Macklaim JM, Fernandes AD, Di Bella JM, Hammond JA, Reid G, Gloor GB. 2013. Comparative meta-RNA-seq of the vaginal microbiota and differential expression by Lactobacillus iners in health and dysbiosis. Microbiome 1:12. 10.1186/2049-2618-1-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Damelin LH, Paximadis M, Mavri-Damelin D, Birkhead M, Lewis DA, Tiemessen CT. 2011. Identification of predominant culturable vaginal Lactobacillus species and associated bacteriophages from women with and without vaginal discharge syndrome in South Africa. J. Med. Microbiol. 60:180–183. 10.1099/jmm.0.024463-0 [DOI] [PubMed] [Google Scholar]
- 58.Kilic AO, Pavlova SI, Alpay S, Kilic SS, Tao L. 2001. Comparative study of vaginal Lactobacillus phages isolated from women in the United States and Turkey: prevalence, morphology, host range, and DNA homology. Clin. Diagn. Lab. Immunol. 8:31–39. 10.1128/CDLI.8.1.31-39.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pavlova SI, Kilic AO, Mou SM, Tao L. 1997. Phage infection in vaginal lactobacilli: an in vitro study. Infect. Dis. Obstet. Gynecol. 5:36–44. 10.1155/S1064744997000094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Reid G, McGroarty JA, Tomeczek L, Bruce AW. 1996. Identification and plasmid profiles of Lactobacillus species from the vagina of 100 healthy women. FEMS Immunol. Med. Microbiol. 15:23–26. 10.1111/j.1574-695X.1996.tb00354.x [DOI] [PubMed] [Google Scholar]
- 61.Aziz RK, Breitbart M, Edwards RA. 2010. Transposases are the most abundant, most ubiquitous genes in nature. Nucleic Acids Res. 38:4207–4217. 10.1093/nar/gkq140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Schneewind O, Mihaylova-Petkov D, Model P. 1993. Cell wall sorting signals in surface proteins of Gram-positive bacteria. EMBO J. 12:4803–4811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chénais B, Caruso A, Hiard S, Casse N. 2012. The impact of transposable elements on eukaryotic genomes: from genome size increase to genetic adaptation to stressful environments. Gene 509:7–15. 10.1016/j.gene.2012.07.042 [DOI] [PubMed] [Google Scholar]
- 64.McCutcheon JP, Moran NA. 2011. Extreme genome reduction in symbiotic bacteria. Nat. Rev. Microbiol. 10:13–26. 10.1038/nrmicro2670 [DOI] [PubMed] [Google Scholar]
- 65.Merhej V, Royer-Carenzi M, Pontarotti P, Raoult D. 2009. Massive comparative genomic analysis reveals convergent evolution of specialized bacteria. Biol. Direct 4:13. 10.1186/1745-6150-4-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Moran NA. 2002. Microbial minimalism: genome reduction in bacterial pathogens. Cell 108:583–586. 10.1016/S0092-8674(02)00665-7 [DOI] [PubMed] [Google Scholar]
- 67.McCutcheon JP, Moran NA. 2010. Functional convergence in reduced genomes of bacterial symbionts spanning 200 million years of evolution. Genome Biol. Evol. 2:708–718. 10.1093/gbe/evq055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Morris JJ, Lenski RE, Zinser ER. 2012. The Black Queen Hypothesis: evolution of dependencies through adaptive gene loss. mBio 3:e00036–12. 10.1128/mBio.00036-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tannock GW, Wilson CM, Loach D, Cook GM, Eason J, O'Toole PW, Holtrop G, Lawley B. 2012. Resource partitioning in relation to cohabitation of Lactobacillus species in the mouse forestomach. ISME J. 6:927–938. 10.1038/ismej.2011.161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Billington SJ, Jost HB, Songer JG. 2000. Thiol-activated cytolysins: structure, function and role in pathogenesis. FEMS Microbiol. Lett. 182:197–205. 10.1111/j.1574-6968.2000.tb08895.x [DOI] [PubMed] [Google Scholar]
- 71.Galperin MY, Koonin EV. 2000. Who's your neighbor? New computational approaches for functional genomics. Nat. Biotechnol. 18:609–613. 10.1038/ismej.2011.161 [DOI] [PubMed] [Google Scholar]
- 72.Pellegrini M, Marcotte EM, Thompson MJ, Eisenberg D, Yeates TO. 1999. Assigning protein functions by comparative genome analysis: protein phylogenetic profiles. Proc. Natl. Acad. Sci. U. S. A. 96:4285–4288. 10.1073/pnas.96.8.4285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Martín R, Soberón N, Vaneechoutte M, Flórez AB, Vázquez F, Suárez JE. 2008. Characterization of indigenous vaginal lactobacilli from healthy women as probiotic candidates. Int. Microbiol. 11:261–266 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.