Abstract
Spores of Bacillus species can remain in their dormant and resistant states for years, but exposure to agents such as specific nutrients can cause spores' return to life within minutes in the process of germination. This process requires a number of spore-specific proteins, most of which are in or associated with the inner spore membrane (IM). These proteins include the (i) germinant receptors (GRs) that respond to nutrient germinants, (ii) GerD protein, which is essential for GR-dependent germination, (iii) SpoVA proteins that form a channel in spores' IM through which the spore core's huge depot of dipicolinic acid is released during germination, and (iv) cortex-lytic enzymes (CLEs) that degrade the large peptidoglycan cortex layer, allowing the spore core to take up much water and swell, thus completing spore germination. While much has been learned about nutrient germination, major questions remain unanswered, including the following. (i) How do nutrient germinants penetrate through spores' outer layers to access GRs in the IM? (ii) What happens during the highly variable and often long lag period between the exposure of spores to nutrient germinants and the commitment of spores to germinate? (iii) What do GRs and GerD do, and how do these proteins interact? (iv) What is the structure of the SpoVA channel in spores' IM, and how is this channel gated? (v) What is the precise state of the spore IM, which has a number of novel properties even though its lipid composition is very similar to that of growing cells? (vi) How is CLE activity regulated such that these enzymes act only when germination has been initiated? (vii) And finally, how does the germination of spores of clostridia compare with that of spores of bacilli?
INTRODUCTION
The germination of the dormant and highly resistant spores formed by members of the Firmicutes phylum, in particular bacilli and clostridia, has long been of significant research interest for four major reasons, as follows: (i) fascinating regulatory systems allow such spores to remain in their dormant, resistant state for years and yet return to active growth in minutes; (ii) while spores of most Firmicutes do not cause disease, spores of some bacilli and clostridia cause food spoilage and food-borne disease, as well as human diseases like gas gangrene, tetanus, botulism, anthrax, and pseudomembranous colitis; (iii) spores of Bacillus anthracis are a major bioterrorism threat; and (iv) spores of Clostridium difficile are an emerging public health threat (1–3). Invariably, it is the germination of spores of these organisms that leads to disease or food spoilage, and yet, when spores germinate and lose their dormancy, they lose their extreme resistance properties and become relatively easy to kill. Germination is thus both an essential part of disease pathogenesis or food spoilage and a major weak spot in these organisms' life cycle. Consequently, there has long been applied interest in spore germination, with researchers seeking to understand this process better in order to either prevent spore germination or accelerate it and then kill the newly sensitive germinated spores.
This review will concentrate on the germination of spores of bacilli, primarily because of the large amount of detailed knowledge of the germination of spores of these species compared to that of spores of clostridia. However, some of the differences and similarities between the germination of spores of these related genera will also be presented. Most discussion will focus on the germination of the model sporeformer, Bacillus subtilis, although the mechanisms of germination of B. subtilis spores appear to be similar for spores of other bacilli. The properties of the various proteins that are specifically involved in the germination process will not be discussed in great detail, since these have recently been reviewed comprehensively (4). Rather, this review will focus on major unanswered questions about the mechanisms of spore germination, focusing primarily on germination of spores by nutrient germinants. Detailed information on other aspects of the germination of spores of bacilli and clostridia, as well as spore outgrowth that follows germination, can be found in other reviews on these topics (4–8).
OVERVIEW OF SPORE FORMATION AND STRUCTURE
Many members of the Bacillales and Clostridiales orders of bacteria can form spores when the environment is not conducive to growth. These spores are formed within the mother cell compartment of a sporulating cell, are released into the environment when the mother cell lyses, and are survival forms that are extremely resistant to most environmental stress factors. Spores of these species also have little or no metabolic activity and are thus considered dormant, although just after their formation, there may be a brief period when spores exhibit some metabolic activity (9). However, after this period, spore metabolic activity appears to be minimal and possibly nonexistent.
Spore resistance and dormancy are due to both unique spore components and spores' unique structure (10) (Fig. 1). Thus, spores have several layers not found in growing cells, including an outermost exosporium in spores of some species, a coat layer that plays major roles in spore resistance to chemicals and predation, and a layer between the exosporium and the coat layers termed the interspace (11, 12). The outer membrane (OM) is under the coat layer, and the OM could be the permeability barrier observed in dormant spores' outer layers (13, 14) but also may be only a vestigial structure (12). Under the OM is the peptidoglycan (PG) cortex and then the PG germ cell wall. The structures of PG in the germ cell wall and growing cell wall appear identical, and the cortex PG structure is similar. However, cortex PG has several unique features, at least one of which, muramic acid-δ-lactam (MAL), is recognized by cortex-lytic enzymes (CLEs) that hydrolyze cortex PG but not germ cell wall PG during spore germination. Under the germ cell wall is the inner spore membrane (IM) that has a number of novel features discussed below, and most of the major proteins involved in spore germination are present in or adjacent to the spores' IM. Finally, there is the central core where DNA, ribosomes, and most spore enzymes are located. The core has a low water content (25 to 50% of wet weight) and a huge amount (∼10% of total spore dry weight) of the spore-specific molecule pyridine-2,6-dicarboxylic acid (dipicolinic acid [DPA]) in a 1:1 chelate with divalent cations, predominantly Ca2+ (CaDPA) (15). Spores' low core water content is likely the major reason for spores' minimal metabolic activity, and proteins appear to be immobile in the core (16). However, the great majority of water in the spore core appears to be freely mobile (17).
FIG 1.
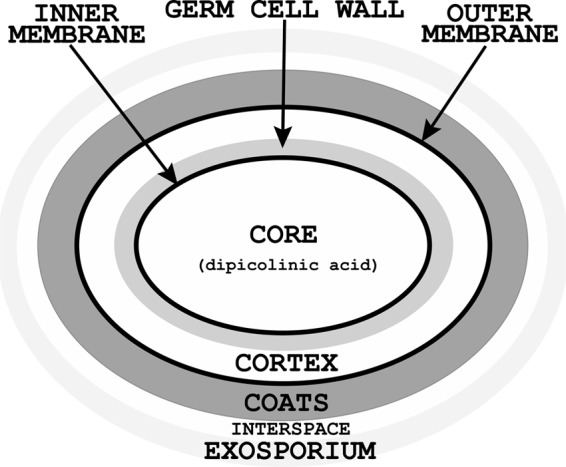
Outline of the structure of a Bacillus spore. The sizes of the various spore layers are not drawn to scale, spores of some species lack an exosporium, and at least for B. subtilis spores, there are several coat layers, as well as a layer outside the coats termed the “crust” (11). All spore dipicolinic acid is in the spore core. Note that a defining feature of spores that have an exosporium is the layer between the coat and exosporium designated the interspace (11), although the precise composition of this interspace layer is not known.
OVERVIEW OF SPORE GERMINATION
Spores can survive for years in their dormant state, but if given the proper stimulus, termed a germinant, spores can rapidly lose their dormancy and resistance properties in germination (Fig. 2). The germination process is followed by outgrowth that converts the germinated spore into a growing cell. There are a number of different types of agents that trigger spore germination (Table 1), and some of these, such as CaDPA and cationic surfactants like dodecylamine, probably are important only in the laboratory. One of these germination agents, high pressure (HP) of the order of many thousands of atmospheres, has attracted increasing interest in the food industry, in particular when HP is combined with moderately high temperatures (19). However, in nature, it is likely that the presence of specific nutrients is what triggers spore germination. The available evidence is consistent with nutrient germinants binding in a stereospecific manner to spore-specific protein complexes, termed germinant receptors (GRs), in the IM. For example, with B. subtilis spores, l-alanine, l-valine, and l-asparagine trigger germination, while the d-amino acids are inert (20, 21). In addition, specific amino acid changes in GR subunits can alter either the specificity or concentration dependence of a GR's response to a nutrient germinant (22, 23). However, there are no studies showing that purified GRs bind specific germinants in vitro, which would be definitive proof that these proteins are indeed deserving of being called GRs. Spores of the great majority of bacilli and clostridia have multiple GRs with various germinant specificities, and GR subunits exhibit obvious sequence homology throughout the spore-forming Firmicutes (4, 6, 8). However, spores of some clostridia, notably Clostridium difficile, do not contain GRs homologous to those in bacilli and many other clostridia (see below).
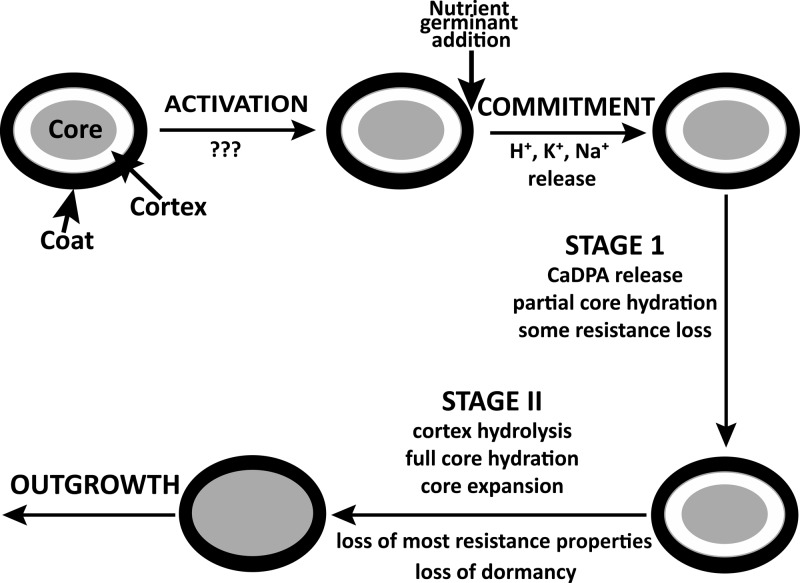
FIG 2.
Schematic outline of nutrient germination of spores of Bacillus species. The precise events in the activation step are not known and are therefore denoted as question marks. The first step seen following the addition of a nutrient germinant to an activated spore is commitment, and the release of monovalent cations is associated with commitment. The germ cell wall is not shown in the figure, but this expands somehow as the cortex is hydrolyzed in stage II of germination.
TABLE 1.
Mechanisms of Bacillus subtilis spore germination by various agentsa
| Germinant | Germination component activated |
|---|---|
| Nutrients | GRs |
| CaDPA | CwlJ (GR independent) |
| Dodecylamine | SpoVA channel (GR independent) |
| Peptidoglycan fragments | Protein kinase (GR independent) |
| HP of 100 to 350 megaPascals | GRs |
| HP of 500 to 1,000 megaPascals | SpoVA channel (GR independent) |
After mixing a nutrient germinant with spores, there is a lag period that varies in length from a few minutes to >24 h for individual spores in populations, and we know essentially nothing about what is happening during this lag period (4). The small fraction of spore populations that exhibit extremely long lag periods in nutrient germination are often termed superdormant (SD), and a major reason for spore superdormancy is very low levels of GRs in the IM (4, 24). Ultimately, GR-nutrient germinant binding results in commitment of a spore to continue through germination some minutes later even if the nutrient germinant is removed (25). Precisely what happens in commitment is not known, although it is associated with a major change in IM permeability and perhaps IM structure, such that monovalent cations, including H+, K+, and Na+ are released, followed by the release of all CaDPA (4, 26, 27). The release of most CaDPA takes only a few minutes for individual spores and is most likely via channels composed of the multiple spore-specific SpoVA proteins (seven in B. subtilis spores) (4). These proteins are encoded in one or more operons in all spore-forming bacilli (one heptacistronic operon in B. subtilis) and clostridia, and at least three of these proteins (SpoVAC, SpoVAD, and SpoVAEb) are present in all of these organisms (6, 8; Perez-Valdespino A, Korza G, Hao B, Setlow P, unpublished results). CaDPA release completes stage I of germination and also triggers entry into stage II, when CLEs degrade the PG cortex. Spores of Bacillus species generally contain two major CLEs, CwlJ and SleB, either of which alone is sufficient to allow the completion of spore germination. CwlJ and SleB are likely to be lytic transglycosylases, although this has only been shown directly for SleB (4). Completion of cortex degradation allows the germ cell wall to expand and the core to expand and take up water. As a result of the latter changes, upon completion of stage II of germination, the core water content has risen to ∼80% of wet weight, equal to that in growing cells. This increased core water content allows metabolism in the core to begin, followed by macromolecular synthesis, ultimately converting the germinated spore into a growing cell in the process of outgrowth, although there are likely several distinct periods during outgrowth (7, 28).
Most of the proteins involved in spore germination are present in or adjacent to the IM, including GRs and the GerD protein, which is essential for normal GR function. In addition, recent work has shown that GRs in B. subtilis spores are in a single cluster in the IM, termed the germinosome, and GerD is essential for germinosome assembly (29). Other proteins present in or adjacent to the IM include the SpoVA proteins, which appear to be uniformly distributed in the IM (29). Much of the spores' SleB is also localized in the IM, probably by its partner protein YpeB (4). Some of these proteins are integral IM proteins, including the A and B subunits of GRs, probably the GR's likely D subunits, and all but two of the seven SpoVA proteins in B. subtilis spores, while SleB, YpeB, GerD, and the GRs' C subunits are peripheral IM proteins. Two SpoVA proteins, SpoVAD and SpoVAEa, are soluble proteins with no obvious membrane-anchoring segments, although these proteins are associated with the IM (4; Perez-Valdespino et al., unpublished results).
The lipid composition of the IM is not notably different from that of the plasma membrane of growing cells or even fully germinated spores. However, the IM appears to be in a gel or semisolid state, as indicated by its passive low permeability even to water, its high viscosity, and the immobility of lipid probes in this membrane (17, 26, 30–32). The fact that so many of the germination proteins act in the IM, a membrane that has rather novel properties, makes the understanding of the structure of and protein action in this membrane important. Some germination proteins are not in the IM, including the CLE CwlJ, which is at the cortex-coat boundary, perhaps associated with its partner protein GerQ (33–35); some SleB is also likely in this region of the spore (4, 35). In addition, the multiple small GerP proteins that appear to facilitate the access of a variety of low-molecular-weight germinants to the IM (36–38) are likely in the spore coat (see below).
MAJOR UNANSWERED QUESTIONS ABOUT SPORE GERMINATION BY NUTRIENTS
While much has been learned about spore germination in recent years, a number of fundamental questions about nutrient germination remain unanswered. Answers to these questions will be crucial to a detailed understanding of spore germination. These major questions are discussed below.
What are the precise effects of activation treatments that potentiate spore germination?
It is well known that the rates and extents of spore germination can be greatly increased by treatments termed activation, with sublethal heat being the most widely used and studied activation treatment (4, 39). The effects of heat activation on spore germination can be dramatic, as for some species, heat activation markedly increases the apparent viability of spore preparations. The effects of heat activation are generally seen only on GR-dependent germination, suggesting that heat activation makes GRs more accessible or receptive to nutrient germinants, although how this is accomplished is not clear. Since the activation is reversible, it may be due to a reversible conformational change in a protein, as such changes accompany heat activation (40). However, specific proteins affected by heat activation have not been identified.
How do germinants gain access to GRs or the SpoVA channel in the IM?
The spore coats restrict the access of molecules of 2 to 8 kDa to inner spore layers (41), and therefore, lysozyme does not hydrolyze the PG cortex in intact dormant spores. However, with spores treated to remove the OM and spore coat protein, lysozyme can hydrolyze the cortex and trigger germination, but lysozyme also degrades the germ cell wall, which can lead to osmotic rupture of germinated spores. There are reports that there may be a significant permeability barrier to small molecules in the coat/cortex region of intact spores (13, 14), although whether this is the OM or some other structure is not clear. Since intact spores respond well to low-molecular-weight germinants, such as nutrients and dodecylamine, both of which act on IM proteins, these germinants must find their way to the IM from the medium. However, the access of at least dodecylamine is slow in intact spores, as spores lacking most coat proteins and their OM germinate much faster with dodecylamine than do intact spores (42). In at least three Bacillus species, mutants lacking one or all of the GerP proteins germinate more slowly than intact spores with exposure to nutrients (36–38). The gerP phenotype is suppressed either by spore coat/OM removal chemically or in appropriate coat assembly mutants or by using nutrient germinant concentrations far above what are saturating for intact spores (36–38). These observations suggest that the GerP proteins facilitate nutrient germinant access to GRs in the IM. While GerP proteins are most likely coat proteins (36), their specific location in the coat is not known. Unfortunately, the multiple GerP proteins exhibit no sequence homology to known proteins, and structures of these proteins have not been determined. Clearly, the mechanism whereby low-molecular-weight germinants access the IM is not yet clear.
What is happening in the lag period between nutrient germinant addition and commitment?
Following mixing of nutrient germinants and spores of bacilli, there is a lag period prior to commitment and rapid DPA release, and the length of this lag period is extremely variable between individual spores in populations (4). This lag period is also seen in germination triggered by HP of ∼150 megaPascals that activates GRs (43), as well as in germination with CaDPA and dodecylamine that trigger germination in a GR-independent fashion (4). Germination of spores of Clostridium perfringens with various germinants also exhibits this variable lag period (44). The length of the lag period in nutrient germination of spores of bacilli or C. perfringens is determined by multiple factors and is decreased by (i) increasing germinant concentration up to saturation, (ii) heat activation, and (iii) increased GR levels (4). However, exactly what is happening during the lag period, other than slow CaDPA release in some cases, is not clear.
What is involved in commitment?
Commitment is defined as the point at which dissociation of a nutrient germinant by a strong competitive inhibitor (d-alanine for some l-alanine-dependent germinations) or acidification to a pH of ∼ 4.5 no longer block completion of germination. Commitment is generally one of the earliest events in nutrient germination and precedes CaDPA release by a few minutes (25). The binding of nutrient germinants to GRs is essential for commitment (25), but the event that causes commitment is not known. Other events taking place prior to CaDPA release include loss of Hg2+ resistance, a large change in the elastic light-scattering intensity from individual spores, the release of H+, Na+, and K+, and the loss of most spore wet heat resistance (26, 27, 45). The latter event is surprising, since spore core water content, the major factor in spore resistance to wet heat, does not change until CaDPA is released. However, recent work has shown that spore wet heat resistance appears to be lost at about the time of commitment because committed spores readily lose their CaDPA during the incubation at elevated temperatures (70 to 80°C) that is used to measure spore wet heat resistance (Luu S, Setlow P, unpublished results). The resulting DPA-less spores that are generated are now heat sensitive due to their elevated core water content (10; Luu S, Setlow P, unpublished results). Overall, it appears likely that commitment involves a change in the IM's permeability and/or strength such that monovalent cations are readily released and CaDPA is not retained during a heat treatment that has no effect on dormant spores. However, the precise change in spores' IM associated with commitment is not known. Perhaps analysis of the kinetics of changes in the IM permeability of individual spores coupled with analysis of these spores' commitment will lead to further information on the precise mechanism of commitment.
What do GRs do and how do they do it?
The GRs are clearly of crucial importance to nutrient germination, as spores of several Bacillus species that have been engineered to lack functional GRs exhibit extremely low levels of germination—as low as 0.1% per day on nutrient media (46, 47). The precise mechanism of this spontaneous GR-independent germination is not clear, but it requires at least one CLE, since Bacillus spores that lack CwlJ and SleB exhibit <0.001% of the colony-forming ability of wild-type spores (48). In spores of bacilli, as well as in spores of clostridia that have GRs (see below), GRs are composed of A, B, and C subunits, and at least in bacilli, all three subunits are required for normal GR function (4). However, there are recent data suggesting that a GR C subunit alone can facilitate the germination of Clostridium perfringens spores (49). There is also recent evidence suggesting that at least some GRs have an additional D subunit (50). Genes encoding a GR's subunits are often cotranscribed, although there are many examples of orphan GR subunits, some of which can interact with other GR subunits. Spores can have multiple GRs, each with a different nutrient germinant specificity, and in some cases, multiple GRs interact in various ways to determine spores' responses to nutrient mixtures, either by increasing or decreasing germination rates (4, 51–53). Unfortunately, how these GR interactions are mediated is not known. The amino acid sequences of the various GR subunits show obvious conservation across the Bacillales and Clostridiales, and a B. subtilis A, B, or C subunit sequence will readily identify a C. perfringens A, B, or C subunit by BLAST analyses. However, despite the significant similarity (both identity and homology) in GR subunits' amino acid sequences, these sequences match extremely poorly with those of other proteins in the available databases. The only exception is a weak homology between some GR B proteins and members of a family of amino acid transporters. However, this homology is rather weak and could be somewhat biased, because both types of proteins contain almost exclusively transmembrane domains (TMDs). The fact that at least two GR subunits are almost certainly integral membrane proteins has hindered the purification of these proteins and, thus, the study of their biochemical properties.
The GR's C subunits are largely on the IM's outer surface, where they are held by a diacylglycerol anchor and, presumably, interaction with their cognate A and B proteins (4, 54, 55). The structure of the C protein of the B. subtilis GerB GR has been determined to 2.3-Å resolution (56). Unfortunately, this novel structure gives no hints as to this protein's function, although predictive programs suggest that all GR C proteins have a similar structure. The GR B proteins are comprised primarily of TMDs with 10 to 12 helices spanning the IM, while the A proteins have 6 to 8 TMDs but also a small C-terminal globular domain and a large, likely N-terminal soluble domain that is probably on the IM's outer surface (54, 55). The amino acid sequences of the putative GR D subunits suggest that these proteins are composed primarily of 2 TMDs, but these proteins' sequences are minimally conserved both within and across species.
What, then, do GRs do? It seems certain that it is the GRs that bind nutrient germinants, since a number of specific mutations in GR subunits affect the apparent affinity of the mutant GR for specific germinants (4, 22, 23, 57). Similarly, spores with elevated levels of specific GRs germinate more effectively with low germinant concentrations (4). While GRs thus are almost certainly the sensors of nutrient germinants, what does germinant-GR binding do? One possibility is that the GRs transport something, with germinant binding gating an IM channel made up of GRs. This is possible, but CaDPA is most likely not transported by GRs, since there is much evidence that a SpoVA protein channel does this (4; Perez-Valdespino et al., unpublished results). However, a GR activated by germinant binding could form a channel for monovalent cations. Another possibility is that GRs signal to another germination protein, either the SpoVA channel or perhaps the GerD protein that is essential for normal GR function. Clearly, more work is needed to resolve exactly what GRs do. A stumbling block to a thorough understanding of what GRs do is that there is currently no system in which to study GRs in vitro. Perhaps reconstituted membrane vesicles containing GRs would be helpful in determining what these proteins do, even if the properties of a membrane in vitro would likely not duplicate those of the IM.
What does the GerD protein do?
While the GRs are essential for nutrient germination, the normal function of GRs requires the GerD protein (4, 58). This small protein is also on the outer surface of the IM and is held there, at least in part, by a lipid anchor (54, 59). The sequence of GerD is well conserved among bacilli, but GerD is not present in spores of clostridia (6, 8; Li Y, Jin K, Ghosh S, Devarakonda P, Carlson K, Davis A, Stewart K-AV, Cammett E, Pelczar-Rossi P, Setlow B, Lu M, Setlow P, Hao B, unpublished data). Spores of Bacillus mutants lacking GerD exhibit much slower germination with all GR-dependent germinants but not with GR-independent germinants (58; Gupta S, Christie G, personal communication). The lack of GerD also results in the disruption of the GR cluster in spores' IM termed the germinosome (see below), such that GRs are dispersed throughout the IM (29). The structure of the C-terminal two-thirds of GerD has been determined at 2.7-Å resolution; the protein is made up of 8 short α-helices that are wrapped around each other in a trimer in the structure, and the protein is also a trimer in solution (Li et al., unpublished data). Unfortunately the GerD structure is novel and has yet to give definitive insight into GerD function, although this protein may function as a scaffold in the germinosome.
What is the structure of the germinosome, and how does this structure accelerate GR-dependent germination?
The formation of the cluster of GRs in the IM termed the germinosome requires the GerD protein, which also is found almost exclusively in this structure. GerD forms a single IM cluster even in the absence of GRs, but in spores lacking GerD, the GRs are dispersed throughout the IM (29). While the slow nutrient germination of gerD spores suggests that germinosome formation is essential for rapid GR-dependent germination, this has not been proven. Other unanswered questions about this novel structure include the following. (i) Are there other proteins in the germinosome besides GRs and GerD? (ii) What is the germinosome structure, and might this promote GR-GR interactions? (iii) How and when is the germinosome assembled during sporulation? (iv) Finally, what is the fate of the germinosome during spore germination and outgrowth? The latter question is of particular interest, since GerD is reported to disappear soon after germination is initiated (59).
What is the structure and function of the SpoVA protein channel and how is it gated?
The SpoVA proteins are involved in DPA movement into the developing spore in sporulation, and genes for many SpoVA proteins are conserved in Bacillus and Clostridium species (4, 6, 8). SpoVA proteins have also been strongly implicated in DPA release in germination, as (i) a temperature-sensitive spoVA mutant is defective in CaDPA release in spore germination, (ii) overexpression of the spoVA operon in spores markedly increases the rates of CaDPA release in germination, and (iii) loss of the SpoVAEa and SpoVAF proteins from B. subtilis spores has no effect on DPA uptake in sporulation but decreases the rate of DPA release in spore germination ∼3-fold (4; Perez-Valdespino et al., unpublished results). Five of the B. subtilis SpoVA proteins are likely integral membrane proteins that are almost certainly in the IM, and one of these proteins, SpoVAC, appears to have mechanosensitive properties, while the hydrophilic SpoVAEa and SpoVAD proteins are also associated with the IM, presumably interacting with other SpoVA proteins (4, 60; Perez-Valdespino et al., unpublished results). Together, these data are consistent with SpoVA proteins forming an IM channel for CaDPA. However, there is minimal information on the structure of this channel, although most if not all of the SpoVAD and SpoVAEa proteins are likely on the IM's outer surface and SpoVAD is a DPA binding protein (54, 61; Perez-Valdespino et al., unpublished results).
While it appears that the SpoVA channel is a conduit for CaDPA, this same channel could also allow passage of other compounds across the IM, including the monovalent cations released before CaDPA in germination. However, this channel has some selectivity, as compounds such as AMP, inorganic phosphate, and 3-phosphoglycerate are not released in the first minutes of germination (62). It is unclear how this channel is gated, and determination of this mechanism will be a major advance in our understanding of the detailed mechanisms of spore germination.
How is the gel-like state of the IM generated and maintained?
As noted above, the IM has some quite different properties than the plasma membrane of growing cells or fully germinated spores, in that the IM appears to be in a gel or semisolid state (17, 26, 30–32). These novel IM properties are lost upon completion of spore germination, when the volume of the IM-encompassed spore core increases 1.5- to 2-fold in the absence of ATP synthesis (30). Two major unanswered questions about the IM are (i) what causes the novel IM structure and (ii) how this IM structure affects the function of germination components, such as GRs, GerD, and SpoVA proteins, in this membrane. The answers to the second question are unknown, although it appears most likely that the dormant spores' low core water and high DPA content are not important in the IM structure, since lipid probe mobility in stage I germinated spores that have lost DPA and have gained some core water is still very low and only increases when the cortex is degraded and the core expands (30). Thus, it is presumably some action of the cortex that constrains the IM and gives it its semisolid characteristics. However, the precise mechanism of this effect and the exact structure of the IM are unclear.
How is CLE activity triggered by stage I germination events?
To complete spore germination after CaDPA is released in stage I of germination, CLEs must degrade the PG cortex to allow core expansion and full hydration (4). Spores of bacilli and clostridia have spore-specific CLEs, although these proteins are rather different in spores of these two groups of organisms (see below). In Bacillus species, the CLE CwlJ's molecular weight does not change upon germination, so the enzyme is not activated by proteolysis. Rather, CwlJ is activated during germination by CaDPA, with the CaDPA released from a spore activating that same spore's CwlJ. Exogenous CaDPA can also activate CwlJ, although only at relatively high concentrations, and this is how exogenous CaDPA triggers spore germination. While this simple signal transduction system ensures that stage I germination events trigger stage II of germination, several questions about this activation mechanism remain, including whether CaDPA activates CwlJ directly or indirectly and, since CwlJ is present in spores when DPA is taken up, how CwlJ activity is suppressed during spore maturation when DPA is taken up by the developing forespore.
While we have a reasonable idea of how CwlJ is activated to allow progression to stage II of germination, how the other redundant CLE in Bacillus species, SleB, is activated is not clear, although it is not directly by CaDPA. Like CwlJ, SleB is also present in spores in a form that is potentially active, and SleB is also not modified proteolytically early in germination. Since DPA-less spores of cwlJ mutants normally germinate spontaneously very rapidly, the presence of DPA in the dormant spore core somehow prevents SleB activation, but how is not clear. There is some evidence that SleB's partner protein, YpeB, can inhibit SleB's catalytic activity (63), but it is not clear how this inhibition is achieved, since no direct interactions have been seen in vitro between YpeB and SleB. The structure of SleB from several Bacillus species has been solved by X-ray crystallography, and these structures are consistent with SleB being a lytic transglycosylase (64, 65). However, the structures alone provide no insight into how SleB activity is regulated.
What is the mechanism of germination of spores of Clostridium species?
Spores of clostridia exhibit some similarities in their germination to that of spores of bacilli but also some notable differences (Table 2) (4, 6, 8). There are also some significant differences in the germination of spores of different clostridia, presumably because this is such an extremely diverse group of organisms (66). The similarities between nutrient germination of spores of clostridia and bacilli include the following: (i) spores of clostridia contain multiple SpoVA proteins that are essential at least for CaDPA uptake in sporulation, (ii) spores of clostridia most often contain GRs that are composed of subunits quite similar to those in spores of bacilli (6), and (iii) germination of individual C. perfringens spores in populations with nutrients, CaDPA, and dodecylamine exhibit the same varying lag periods seen during germination of spores of bacilli (44). However, there are also four major differences. First, while GR function in germination of spores of bacilli requires all three GR subunits, it appears that a GR C subunit alone can facilitate C. perfringens spore germination (49). Second, spores of some clostridia, most notably C. difficile, lack GRs with any similarity to those in spores of bacilli (6, 67, 68), likely a reflection of the great diversity in the clostridia. Indeed, recent work suggests that C. difficile should be moved to the Peptococcaceae family (66), and it will be interesting to track the presence and absence of genes encoding GRs as the taxonomy of the class clostridia improves due to genome sequencing. However, despite lacking GRs, C. difficile spores do germinate well with specific bile salts and also respond to various amino acids as cogerminants for bile salts (6, 8, 68, 69). How, then, do these GR-less spores germinate in response to specific low-molecular-weight compounds? Third, even spores of clostridia that contain GRs lack GerD, but they can still germinate with specific low-molecular-weight germinants. Are GRs in these latter species present in the IM in a germinosomelike structure, and if so, is there a protein that replaces GerD in germinosome assembly and GR function?
TABLE 2.
Comparison of the germination of spores of bacilli and clostridiaa
| Germination aspect | Bacilli | Clostridia |
|---|---|---|
| Presence of similar GRs | Yesb | Mostb but not allc |
| SpoVA proteins | Yesd | Yesd |
| CLEs | CwlJe and SleB | SleCf |
| Regulation of CLE activity | CwlJ activated by CaDPA | Proteolytic activation by Cspg |
| SleB regulation unclearg | ||
| CaDPA germination mechanism | Activates CwlJ | Not clear—may activate GRs |
| Ddah germination mechanism | Activates SpoVA channel | Likely activates SpoVA channel |
Information for the conclusions in this table is described in the text and in references 4, 6, 44, 49, 70, 74, and 79.
All three GR subunits are required for GR function, but perhaps not in C. perfringens.
An example of a Clostridium species with no GRs is C. difficile.
The precise number of SpoVA proteins varies between species.
Spores of some bacilli have multiple CwlJs.
SleC is present in dormant spores as an inactive zymogen, proSleC.
SleB is activated in DPA-less spores of Bacilli, while ProSleC, and CspB are not activated in DPA-less C. perfringens spores.
Dda, dodecylamine.
The fourth major difference is that spores of clostridia appear to regulate CLE activity differently than spores of bacilli (4, 6, 8). Thus, at least C. perfringens and C. difficile spores have only a single CLE, SleC, that is essential for completion of spore germination (4, 70, 71). Like its Bacillus spore counterparts, SleC has lytic transglycosylase activity and is also in the spores' outer layers (4, 70, 72, 73). However, SleC is present in spores as an inactive zymogen, proSleC, which is activated by protease cleavage in the first minutes of germination (4, 74–77). One or multiple subtilisin-like serine proteases, termed Csp proteases, cleave and activate proSleC in the germination of spores of clostridia (74–77). Like subtilisins, Csp proteases are synthesized as zymogens that autoactivate by cleavage of a small prodomain. The structure of CspB from C. difficile spores has been solved by X-ray crystallography, and while the structure is similar to that of subtilisins, there are some major differences (75). These include the following: (i) at least in vitro, the prodomain of CspB remains bound to and inhibits enzyme activity even after the cleavage between the prodomain and the protease domain, and (ii) there is a large insertion in the protease domain that is only rarely seen in subtilisins, and this insertion may stabilize the protein in spores' outer layers (75).
To date, the mechanism(s) that regulate the action of Csp proteases on proSleC in spore germination are not known. One clue to this regulation is the recent identification of missense mutations in the C. difficile cspC gene that essentially eliminate these spores' germination in response to the bile salt taurocholate in vitro (78). Notably, another cspC mutation alters the spectrum of bile salts that trigger C. difficile spore germination (78). Together, these findings suggest that CspC is the protein that interacts with bile salts to trigger C. difficile spore germination. However, CspC most likely has no protease activity, since it lacks several amino acids that comprise subtilisin's catalytic triad, including the catalytic serine (75). Indeed, the only functional Csp protease that can act on proSleC in C. difficile spores is almost certainly CspB, and how the binding of specific bile salts to CspC activates CspB is not known. In addition, it is most likely that additional signals need to be integrated in C. difficile spore germination, since glycine is an essential cogerminant, at least in vitro (69).
In addition to the differences in nutrient germination between spores of bacilli and clostridia, there are notable differences in germination with other agents, as well as at least one similarity. Thus, the surfactant dodecylamine appears to trigger germination of spores of bacilli by opening the SpoVA protein channel and causing CaDPA release, and available data are consistent with this also being the case with spores of C. perfringens (49, 70). The two notable differences are as follows. First, while DPA-less spores of bacilli are extremely unstable and germinate spontaneously, most likely via activation of SleB (4, 6), DPA-less C. perfringens spores are stable (79). This suggests that DPA release is not part of the signal transduction pathway in the germination of spores of clostridia. The second difference is in the mechanism of spore germination by exogenous CaDPA, which with spores of bacilli is via activation of the CLE CwlJ (4). C. perfringens spores lacking CspB or SleC do germinate extremely poorly with CaDPA (44, 74). However, GR subunits also appear to play an important role in CaDPA germination of spores of clostridia, since C. perfringens spores lacking their only GR C subunit, GerKC, germinate very poorly with CaDPA (49). Thus, the mechanism of CaDPA germination of C. perfringens spores is not as simple as activation of a CLE.
Can spore germination be manipulated efficiently in applied settings?
Since spores must germinate to exert their deleterious effects, the food industry has expended significant effort to identify compounds that effectively inhibit spore germination yet are compatible with food safety, generally with no great success. There has also been interest in stimulating spore germination to allow spores to be more readily killed. While the inhibition or stimulation of spore germination in applied settings is somewhat off the focus of this review, these topics have attracted renewed interest in recent years because of the need for effective decontamination regimens for spores of organisms such as C. difficile and B. anthracis but without the concerns unique to the food industry. Thus, there are some recent reports of promising results in using a germination step prior to spore decontamination for promoting inactivation of B. anthracis and C. difficile spores (80–82), as well as enzymatic spore coat removal by lytic enzymes such as lysozyme to allow spore killing (83). Several compounds have also been identified that may be effective in inhibiting the germination or outgrowth of spores of organisms such as B. anthracis and C. difficile (84–88), and perhaps compounds analogous to these could be useful in applied settings. However, at least in the case of C. difficile, it is not clear whether germination inhibitors identified in laboratory studies with one strain will have similar potency against all clinical isolates (89).
SUMMARY
Work in the past 5 years has provided a huge amount of new information about the germination of spores of Bacillus and Clostridium species. Major advances in this area have come from the development of sophisticated methodology for analyzing the germination of individual spores in populations and from the vast amount of bioinformatics data contained in the large number of spore-forming Bacillales and Clostridiales genomes that have been sequenced. However, despite these advances, major, fundamental questions about the function, structure, and regulation of proteins that play central roles in spore germination remain. Clearly, spores do not give up their dormancy simply!
ACKNOWLEDGMENTS
Work in my laboratory described in this communication has been supported in the past by grants from the National Institutes of Health, the U.S. Department of Agriculture, and the Army Research Office. Work over the past 4 years has been supported by a U.S. Department of Defense Multi-disciplinary University Research Initiative through the U.S. Army Research Office under contract number W911NF-09-1-0286.
I am extremely grateful to all laboratory members and collaborators who have contributed so much to our understanding of spore germination.
Footnotes
Published ahead of print 31 January 2014
REFERENCES
- 1.Setlow P, Johnson EA. 2012. Spores and their significance, p 45–79 In Doyle MP, Buchanan R. (ed), Food microbiology, fundamentals and frontiers, 4th ed. ASM Press, Washington, DC [Google Scholar]
- 2.Logan NA. 2012. Bacillus and relatives in foodborne illness. J. Appl. Microbiol. 112:417–429. 10.1111/j.1365-2672.2011.05204.x [DOI] [PubMed] [Google Scholar]
- 3.Mallozzi M, Viswanathan VK, Vedantam G. 2010. Spore-forming bacilli and clostridia in human disease. Future Microbiol. 5:1109–1123. 10.2217/fmb.10.60 [DOI] [PubMed] [Google Scholar]
- 4.Setlow P. 2013. When the sleepers wake: the germination of spores of Bacillus species. J. Appl. Microbiol. 115:1251–1268. 10.1111/jam.12343 [DOI] [PubMed] [Google Scholar]
- 5.Setlow P. 2003. Spore germination. Curr. Opin. Microbiol. 6:550–556. 10.1016/j.mib.2003.10.001 [DOI] [PubMed] [Google Scholar]
- 6.Paredes-Sabja D, Setlow P, Sarker MR. 2011. Germination of spores of Bacillales and Clostridiales species: mechanisms and proteins involved. Trends Microbiol. 19:85–94. 10.1016/j.tim.2010.10.004 [DOI] [PubMed] [Google Scholar]
- 7.Paidhungat M, Setlow P. 2002. Spore germination and outgrowth, p 537–548 In Sonenshein AL, Hoch JA, Losick R. (ed), Bacillus subtilis and its relatives: from genes to cells. American Society for Microbiology, Washington, DC [Google Scholar]
- 8.Xiao Y, Francke C, Abee T, Wells-Bennik MH. 2011. Clostridial spore germination versus bacilli: genome mining and current insights. Food Microbiol. 28:266–274. 10.1016/j.fm.2010.03.016 [DOI] [PubMed] [Google Scholar]
- 9.Segev E, Smith Y, Ben-Yehuda S. 2012. RNA dynamics in aging bacterial spores. Cell 148:139–149. 10.1016/j.cell.2011.11.059 [DOI] [PubMed] [Google Scholar]
- 10.Setlow P. 2006. Spores of Bacillus subtilis: their resistance to radiation, heat and chemicals. J. Appl. Microbiol. 101:514–525. 10.1111/j.1365-2672.2005.02736.x [DOI] [PubMed] [Google Scholar]
- 11.McKenney PT, Driks A, Eichenberger P. 2013. The Bacillus subtilis endospore: assembly and functions of the multilayered coat. Nat. Rev. Microbiol. 11:33–44. 10.1038/nrmicro2921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Henriques AO, Moran CP., Jr 2007. Structure, assembly, and function of the spore surface layers. Annu. Rev. Microbiol. 61:555–588. 10.1146/annurev.micro.61.080706.093224 [DOI] [PubMed] [Google Scholar]
- 13.Rode LJ, Lewis CW, Jr, Foster JW. 1962. Electron microscopy of spores of Bacillus megaterium with special reference to the effects of fixation and thin sectioning. J. Cell Biol. 13:423–435. 10.1083/jcb.13.3.423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gerhardt P, Black SH. 1961. Permeability of bacterial spores. II. Molecular variables affecting solute permeation. J. Bacteriol. 82:750–760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gerhardt P, Marquis RE. 1989. Spore thermoresistance mechanisms, p 43–63 In Smith I, Slepecky RA, Setlow P. (ed), Regulation of procaryotic development: structural and functional analysis of bacterial sporulation and germination. American Society for Microbiology, Washington, DC [Google Scholar]
- 16.Cowan AE, Koppel DE, Setlow B, Setlow P. 2003. A soluble protein is immobile in dormant spores of Bacillus subtilis but is mobile in germinated spores: implications for spore dormancy. Proc. Natl. Acad. Sci. U. S. A. 100:4209–4214. 10.1073/pnas.0636762100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kaieda S, Setlow B, Setlow P, Halle B. 2013. Mobility of core water in Bacillus subtilis spores by 2H NMR. Biophys. J. 105:2016–2123. 10.1016/j.bpj.2013.09.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shah IM, Laaberki MH, Popham DL, Dworkin J. 2008. A eukaryotic Ser/Thr kinase signals bacteria to exit dormancy in response to peptidoglycan fragments. Cell 135:486–496. 10.1016/j.cell.2008.08.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Reineke K, Mathys A, Heinz V, Knorr D. 2013. Mechanisms of endospore inactivation under high pressure. Trends Microbiol. 21:296–304. 10.1016/j.tim.2013.03.001 [DOI] [PubMed] [Google Scholar]
- 20.Gould GW. 1969. Germination, p 397–444 In Gould GW, Hurst A. (ed), The bacterial spore. Academic Press, London, United Kingdom [Google Scholar]
- 21.Atluri S, Ragkousi K, Cortezzo DE, Setlow P. 2006. Cooperativity between different nutrient receptors in germination of spores of Bacillus subtilis and reduction of this cooperativity by alterations in the GerB receptor. J. Bacteriol. 188:28–36. 10.1128/JB.188.1.28-36.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Christie G, Gotzke H, Lowe CR. 2010. Identification of a receptor subunit and putative ligand-binding residues involved in the Bacillus megaterium QM B1551 spore germination response to glucose. J. Bacteriol. 192:4317–4326. 10.1128/JB.00335-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mongkolthanaruk W, Cooper GR, Mawer JSP, Allan RN, Moir A. 2011. Effect of amino acid substitutions in the GerAA protein on the function of the alanine-responsive germinant receptor of Bacillus subtilis spores. J. Bacteriol. 193:2268–2275. 10.1128/JB.01398-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Setlow P, Liu J, Faeder JR. 2012. Heterogeneity in bacterial spore populations, p 201–216 In Abel-Santos E. (ed), Bacterial spores: current research and applications. Horizon Scientific Press, Norwich, United Kingdom [Google Scholar]
- 25.Yi X, Setlow P. 2010. Studies of the commitment step in the germination of spores of Bacillus species. J. Bacteriol. 192:3424–3433. 10.1128/JB.00326-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Swerdlow BM, Setlow B, Setlow P. 1981. Levels of H+ and other monovalent cations in dormant and germinated spores of Bacillus megaterium. J. Bacteriol. 148:20–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Peng L, Chen D, Setlow P, Li Y-q. 2009. Elastic and inelastic light scattering from single bacterial spores in an optical trap allows the monitoring of spore germination dynamics. Anal. Chem. 81:4035–4042. 10.1021/ac900250x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Segev E, Rosenberg A, Mamou G, Sinai L, Ben-Yehuda S. 2013. Molecular kinetics of reviving bacterial spores. J. Bacteriol. 195:1875–1882. 10.1128/JB.00093-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Griffiths KK, Zhang J, Cowan AE, Yu J, Setlow P. 2011. Germination proteins in the inner membrane of dormant Bacillus subtilis spores colocalize in a discrete cluster. Mol. Microbiol. 81:1061–1077. 10.1111/j.1365-2958.2011.07753.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cowan AE, Olivastro EM, Koppel DE, Loshon CA, Setlow B, Setlow P. 2004. Lipids in the inner membrane of dormant spores of Bacillus species are immobile. Proc. Natl. Acad. Sci. U. S. A. 101:7733–7738. 10.1073/pnas.0306859101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Loison P, Hosny NA, Gervais P, Champion D, Kulmova MK, Perrier-Cornet JM. 2013. Direct investigation of viscosity of an atypical inner membrane of Bacillus spores: a molecular rotor/FLIM study. Biochim. Biophys. Acta 1828:2436–2443. 10.1016/j.bbamem.2013.06.028 [DOI] [PubMed] [Google Scholar]
- 32.Sunde EP, Setlow P, Hederstedt L, Halle B. 2009. The physical state of water in bacterial spores. Proc. Natl. Acad. Sci. U. S. A. 106:19334–19339. 10.1073/pnas.0908712106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bagyan I, Setlow P. 2002. Localization of the cortex lytic enzyme CwlJ in spores of Bacillus subtilis. J. Bacteriol. 184:1219–1224. 10.1128/jb.184.4.1219-1224.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ragkousi K, Eichenberger P, Van Ooij C, Setlow P. 2003. Identification of a new gene essential for germination of Bacillus subtilis spores with Ca2+-dipicolinate. J. Bacteriol. 185:2315–2329. 10.1128/JB.185.7.2315-2329.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chirakkal H, O'Rourke M, Atrih A, Foster SJ, Moir A. 2002. Analysis of spore cortex lytic enzymes and related proteins in Bacillus subtilis endospore germination. Microbiology 148:2383–2392 [DOI] [PubMed] [Google Scholar]
- 36.Behravan J, Chirakkal H, Masson A, Moir A. 2000. Mutations in the gerP locus of Bacillus subtilis and Bacillus cereus affect access of germinants to their targets in spores. J. Bacteriol. 182:1987–1994. 10.1128/JB.182.7.1987-1994.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Carr KA, James BK, Hanna PC. 2010. Role of the gerP operon in germination and outgrowth of Bacillus anthracis spores. PLoS One 5:e9128. 10.1371/journal.pone.0009128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Butzin XY, Troiano AJ, Coleman WH, Griffiths KK, Doona CJ, Feeherry FE, Wang G, Li Y-q, Setlow P. 2012. Analysis of the effects of a gerP mutation on the germination of spores of Bacillus subtilis. J. Bacteriol. 194:5749–5758. 10.1128/JB.01276-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Keynan A, Evenchick Z. 1969. Activation, p 359–396 In Gould GW, Hurst A. (ed), The bacterial spore. Academic Press, London, United Kingdom [Google Scholar]
- 40.Zhang P, Setlow P, Li Yq. 2009. Characterization of single heat-activated Bacillus spores using laser tweezers Raman spectroscopy. Optics Express 17:16480–16491. 10.1364/OE.17.016480 [DOI] [PubMed] [Google Scholar]
- 41.Driks A. 1999. Bacillus subtilis spore coat. Microbiol. Mol. Biol. Rev. 63:1–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Setlow B, Cowan AE, Setlow P. 2003. Germination of spores of Bacillus subtilis with dodecylamine. J. Appl. Microbiol. 95:637–648. 10.1046/j.1365-2672.2003.02015.x [DOI] [PubMed] [Google Scholar]
- 43.Kong L, Doona CJ, Setlow P, Li Y-q. 2014. Monitoring rates and heterogeneity of high pressure germination of Bacillus spores using phase contrast microscopy of individual spores. Appl. Environ. Microbiol. 80:345–353. 10.1128/AEM.03043-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang G, Zhang P, Paredes-Sabja D, Green C, Setlow P, Sarker MR, Li Y-Q. 2011. Analysis of the germination of individual Clostridium perfringens spores and its heterogeneity. J. Appl. Microbiol. 111:1212–1223. 10.1111/j.1365-2672.2011.05135.x [DOI] [PubMed] [Google Scholar]
- 45.Levinson HS, Hyatt MT. 1966. Sequence of events during Bacillus megaterium spore germination. J. Bacteriol. 91:1811–1817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Paidhungat M, Setlow P. 2000. Role of Ger proteins in nutrient and non-nutrient triggering of spore germination in Bacillus subtilis. J. Bacteriol. 182:2513–2519. 10.1128/JB.182.9.2513-2519.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gupta S, Üstok FI, Johnson CL, Bailey DMD, Lowe CR, Christie G. 2013. Investigating the functional hierarchy of Bacillus megaterium PV361 spore germinant receptors. J. Bacteriol. 195:3045–3053. 10.1128/JB.00325-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Paidhungat M, Ragkousi K, Setlow P. 2001. Genetic requirements for induction of germination of spores of Bacillus subtilis by Ca2+-dipicolinate. J. Bacteriol. 183:4886–4893. 10.1128/JB.183.16.4886-4893.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Banawas S, Paredes-Sabja D, Korza G, Li Y, Hao B, Setlow P, Sarker MR. 2013. The Clostridium perfringens germinant receptor protein GerKC is located in the spore inner membrane and is crucial for spore germination. J. Bacteriol. 195:5084–5091. 10.1128/JB.00901-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ramirez-Peralta A, Gupta S, Butzin XY, Setlow B, Korza G, Leyva-Vazquez M-A, Christie G, Setlow P. 2013. Identification of new proteins that modulate the germination of spores of Bacillus species. J. Bacteriol. 195:3009–3021. 10.1128/JB.00257-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Luu H, Akoachere M, Patra M, Abel-Santos E. 2011. Cooperativity and interference of germination pathways in Bacillus anthracis spores. J. Bacteriol. 193:4192–4198. 10.1128/JB.05126-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yi X, Liu J, Faeder JR, Setlow P. 2011. Synergism between different germinant receptors in the germination of Bacillus subtilis spores. J. Bacteriol. 193:4664–4671. 10.1128/JB.05343-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Stewart K-AV, Yi X, Ghosh S, Setlow P. 2012. Germination protein levels and rates of germination of spores of Bacillus subtilis with overexpressed or deleted genes encoding germination proteins. J. Bacteriol. 194:3156–3164. 10.1128/JB.00405-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Korza G, Setlow P. 2013. Topology and accessibility of germination proteins in the Bacillus subtilis spore inner membrane. J. Bacteriol. 195:1484–1491. 10.1128/JB.02262-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wilson MJ, Carlson PE, Janes BK, Hanna PC. 2012. Membrane topology of the Bacillus anthracis GerH germinant receptor proteins. J. Bacteriol. 194:1369–1377. 10.1128/JB.06538-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Li Y, Setlow B, Setlow P, Hao B. 2010. Crystal structure of the GerBC component of a Bacillus subtilis spore germinant receptor. J. Mol. Biol. 402:8–16. 10.1016/j.jmb.2010.07.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cooper GR, Moir A. 2011. Amino acid residues in the GerAB protein important in the function and assembly of the alanine spore germination receptor of Bacillus subtilis 168. J. Bacteriol. 193:2261–2267. 10.1128/JB.01397-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pelczar PL, Igarashi T, Setlow B, Setlow P. 2007. Role of GerD in Bacillus subtilis spores. J. Bacteriol. 189:1090–1098. 10.1128/JB.01606-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pelczar PL, Setlow P. 2008. Localization of the germination protein GerD to the inner membrane in Bacillus subtilis spores. J. Bacteriol. 190:5635–5641. 10.1128/JB.00670-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Velasquez-Gusman JC, Kocer A, Abee T, Poolman B. 2012. SpoVA, putative channels involved in spore germination of Bacillus spores, abstr. 101. 5th Eur. Spores Conf., Egham, United Kingdom, 16 to 19 April 2012 [Google Scholar]
- 61.Li Y, Korza G, Zhang P, Li Y-q, Setlow B, Setlow P, Hao B. 2012. Role of a SpoVA protein in dipicolinic acid uptake into developing spores of Bacillus subtilis. J. Bacteriol. 194:1875–1884. 10.1128/JB.00062-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Setlow B, Wahome PG, Setlow P. 2008. Release of small molecules during germination of spores of Bacillus species. J. Bacteriol. 190:4759–4763. 10.1128/JB.00399-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Li Y, Butzin XY, Davis A, Setlow B, Korza G, Ustok F, Christie G, Setlow P, Hao B. 2013. Activity and regulation of various forms of CwlJ, SleB, and YpeB proteins in degrading cortex peptidoglycan of spores of Bacillus species in vitro and during spore germination. J. Bacteriol. 195:2530–2540. 10.1128/JB.00259-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Li Y, Jin K, Setlow B, Setlow P, Hao B. 2012. Crystal structure of the catalytic domain of the Bacillus cereus SleB protein important in cortex degradation during spore germination. J. Bacteriol. 194:4537–4545. 10.1128/JB.00877-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Jing X, Robinson HR, Heffron JD, Popham DL, Schubot FD. 2012. The catalytic domain of the germination-specific lytic transglycosylase SleB from Bacillus anthracis displays a unique active site topology. Proteins 80:2469–2475. 10.1002/prot.24140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yutin N, Galperin MY. 2013. A genomic update on clostridial phylogeny: Gram-negative spore formers and other misplaced clostridia. Environ. Microbiol. 15:2631–2641. 10.1111/1462-2920.12173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sebaihia M, Wren BW, Mullany P, Fairweather NF, Minton N, Stabler R, Thomson NR, Roberts AP, Cerdeño-Tárraga AM, Wang H, Holden MTG, Wright A, Churcher C, Quail MA, Baker S, Bason N, Brooks K, Chillingworth T, Cronin A, Davis P, Dowd L, Fraser A, Feltwell T, Hance Z, Holroyd S, Jagels K, Moule S, Mungall K, Price C, Rabbinowitsch E, Sharp S, Simmonds M, Stevens K, Unwin L, Whithead S, Dupuy B, Dougan G, Barrell B, Parkhill J. 2006. The multidrug-resistant human pathogen Clostridium difficile has a highly mobile, mosaic genome. Nat. Genet. 38:779–786. 10.1038/ng1830 [DOI] [PubMed] [Google Scholar]
- 68.Burns DA, Heap JT, Minton NP. 2010. Clostridium difficile spore germination: an update. Res. Microbiol. 161:730–734. 10.1016/j.resmic.2010.09.007 [DOI] [PubMed] [Google Scholar]
- 69.Sorg JA, Sonenshein AL. 2008. Bile salts and glycine as cogerminants for Clostridium difficile spores. J. Bacteriol. 190:2505–2512. 10.1128/JB.01765-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Paredes-Sabja D, Setlow P, Sarker MR. 2009. SleC is essential for cortex peptidoglycan hydrolysis during germination of spores of the pathogenic bacterium Clostridium perfringens. J. Bacteriol. 191:2711–2720. 10.1128/JB.01832-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Burns DA, Heap JT, Minton NP. 2010. SleC is essential for germination of Clostridium difficile spores in nutrient-rich medium supplemented with the bile salt taurocholate. J. Bacteriol. 192:657–664. 10.1128/JB.01209-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Gutelius D, Hokeness K, Logan SM, Reid CW. 2014. Functional analysis of SleC from Clostridium difficile: an essential lytic transglycosylase involved in spore germination. Microbiology 160:209–216. 10.1099/mic.0.072454-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kumazawa T, Masayama A, Fukuoka S, Makino S, Yoshimura T, Moriyama R. 2007. Mode of action of a germination-specific cortex-lytic enzyme, SleC, of Clostridium perfringens S40. Biosci. Biotechnol. Biochem. 71:884–892. 10.1271/bbb.60511 [DOI] [PubMed] [Google Scholar]
- 74.Paredes-Sabja D, Setlow P, Sarker MR. 2009. The protease CspB is essential for initiation of cortex hydrolysis and DPA release during germination of spores of Clostridium perfringens. Microbiology 155:3464–3472. 10.1099/mic.0.030965-0 [DOI] [PubMed] [Google Scholar]
- 75.Adams CM, Eckenroth BE, Putnam EE, Doublié S, Shen A. 2013. Structural and functional analysis of the CspB protease required for Clostridium spore germination. PLoS Pathol. 9:e1003165. 10.1371/journal.ppat.1003165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Cartman ST, Minton NP. 2010. A mariner-based transposon system for in vivo random mutagenesis of Clostridium difficile. Appl. Environ. Microbiol. 76:1103–1109. 10.1128/AEM.02525-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Shimamoto S, Moriyama R, Sugimoto K, Miyata S, Makino S. 2001. Partial characterization of an enzyme fraction with protease activity which converts the spore peptidoglycan hydrolase (SleC) precursor to an active enzyme during germination of Clostridium perfringens S40 spores and analysis of a gene cluster involved in the activity. J. Bacteriol. 183:3742–3751. 10.1128/JB.183.12.3742-3751.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Francis MB, Allen CA, Shrestha R, Sorg JA. 2013. Bile acid recognition by the Clostridium difficile germinant receptor, CspC, is important for establishing infection. PLoS Pathog. 9:e1003356. 10.1371/journal.ppat.1003356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Paredes-Sabja D, Setlow B, Setlow P, Sarker MR. 2008. Characterization of Clostridium perfringens spores that lack SpoVA proteins and dipicolinic acid. J. Bacteriol. 190:4648–4659. 10.1128/JB.00325-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Nerandzic MM, Donskey CJ. 2013. Activate to eradicate: inhibition of Clostridium difficile spore outgrowth by the synergistic effects of osmotic activation and nisin. PLoS One 8:e54740. 10.1371/journal.pone.0054740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Nerandzic MM, Donskey CJ. 2010. Triggering germination represents a novel strategy to enhance killing of Clostridium difficile spores. PLoS One 5:e12285. 10.1371/journal.pone.0012285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Omotade TO, Heffron JD, Klimko CP, Marchand CL, Miller LL, Halasahoris SA, Bozue JA, Welkos SL, Cote CK. 2013. d-Cycloserine or similar physiochemical compounds may be uniquely suited for use in Bacillus anthracis spore decontamination strategies. J. Appl. Microbiol. 115:1343–1356. 10.1111/jam.12322 [DOI] [PubMed] [Google Scholar]
- 83.Mundra RV, Mehta KK, Paskaleva EE, Kane RS, Dordick JS. Enzyme-driven Bacillus spore coat degradation leading to spore killing. Biotechnol. Bioeng. In press. 10.1002/bit.25132 [DOI] [PubMed] [Google Scholar]
- 84.Akoachere M, Squires RC, Nour AM, Angelov L, Brojatsch J, Abel-Santos E. 2007. Identification of an in vivo inhibitor of Bacillus anthracis spore germination. J. Biol. Chem. 282:12112–12118. 10.1074/jbc.M611432200 [DOI] [PubMed] [Google Scholar]
- 85.Giel JL, Sorg JA, Sonenshein AL, Zhu J. 2010. Metabolism of bile salts in mice influences spore germination in Clostridium difficile. PLoS One 15:e8740. 10.1371/journal.pone.0008740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.van Melis CC, Almeida CB, Kort R, Groot MN, Abee T. 2012. Germination inhibition of Bacillus cereus spores: impact of the lipophilic character of inhibiting compounds. Int. J. Food Microbiol. 160:124–130. 10.1016/j.ijfoodmicro.2012.10.006 [DOI] [PubMed] [Google Scholar]
- 87.Sorg JA, Sonenshein AL. 2010. Inhibiting the initiation of Clostridium difficile spore germination using analogs of chenodeoxycholic acid, a bile acid. J. Bacteriol. 192:4983–4990. 10.1128/JB.00610-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.McKevitt MT, Bryant KM, Shakir SM, Larabee JL, Blanke SR, Lovchik J, Lyons CR, Ballard JD. 2007. Effects of endogenous d-alanine synthesis and autoinhibition of Bacillus anthracis germination on in vitro and in vivo infections. Infect. Immun. 75:5726–5734. 10.1128/IAI.00727-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Heeg D, Burns DA, Cartman ST, Minton NP. 2012. Spores of Clostridium difficile clinical isolates display a diverse germination response to bile salts. PLoS One 7:e32381. 10.1371/journal.pone.0032381 [DOI] [PMC free article] [PubMed] [Google Scholar]