Abstract
The AMP-forming acyl coenzyme A (acyl-CoA) synthetases are a large class of enzymes found in both anabolic and catabolic pathways that activate fatty acids to acyl-CoA molecules. The protein acetyltransferase (Pat) from Rhodopseudomonas palustris (RpPat) inactivates AMP-forming acyl-CoA synthetases by acetylating the ε-amino group of a conserved, catalytic lysine residue. In all of the previously described RpPat substrates, this lysine residue is located within a PX4GK motif that has been proposed to be a recognition motif for RpPat. Here, we report five new substrates for RpPat, all of which are also AMP-forming acyl-CoA synthetases. This finding supports the idea that Pat enzymes may have evolved to control the activity of this family of enzymes. Notably, RpPat did not acetylate the wild-type long-chain acyl-CoA synthetase B (RpLcsB; formerly Rpa2714) enzyme of this bacterium. However, a single amino acid change two residues upstream of the acetylation site was sufficient to convert RpLcsB into an RpPat substrate. The results of mutational and functional analyses of RpLcsB and RpPimA variants led us to propose PK/RTXS/T/V/NGKX2K/R as a substrate recognition motif. The underlined positions within this motif are particularly important for acetylation by RpPat. The first residue, threonine, is located 4 amino acids upstream of the acetylation site. The second residue can be S/T/V/N and is located two positions upstream of the acetylation site. Analysis of published crystal structures suggests that the side chains of these two residues are very close to the acetylated lysine residue, indicating that they may directly interact with RpPat.
INTRODUCTION
N-acetylation of lysine side chains is a posttranslational modification found in all domains of life (1, 2). Although most studies of protein acetylation originally focused on modification of eukaryotic histone tails (3), it is now clear that a wide variety of proteins, including metabolic enzymes, are modified by acetylation (4). Several recent high-throughput proteomics studies reported hundreds of putatively acetylated proteins in Escherichia coli and Salmonella enterica (5–8), suggesting that lysine acetylation may be a common regulatory mechanism in bacteria. Despite this evidence of widespread acetylation, the identity of the acetyltransferases responsible for the observed modifications remains largely unknown. In addition, it appears that much, although not all, of this acetylation likely occurs at low levels and might be chemically mediated by acetyl donors such as acetyl phosphate (9).
The best-characterized bacterial lysine acetyltransferase is the S. enterica Pat enzyme (hereafter SePat) (10), which is also known as Pka and PatZ in E. coli (11, 12). In both bacteria, Pat has a large N-terminal domain of unknown function homologous to NDP-forming acyl coenzyme A (acyl-CoA) synthetase enzymes and a smaller C-terminal acetyltransferase domain homologous to the GCN5-related N-acetyltransferases (GNATs). SePat was originally identified as a regulator of the AMP-forming acetyl-CoA synthetase (Acs) enzyme, which activates acetate to acetyl-CoA (10). SePat acetylates SeAcs once at a conserved catalytic lysine within what is known as the acetylation motif, and acetylation inactivates the enzyme (13, 14). When the gene encoding the S. enterica Sir2-type lysine deacetylase CobB is deleted, this bacterium fails to grow on low (10 mM) concentrations of acetate. Deletion of pat in a CobB-deficient S. enterica strain restores growth, demonstrating that lysine acetylation control of Acs in vivo is reversible (10). SePat and SeCobB also regulate propionyl-CoA synthetase (PrpE) activity, in this case not by acetylation but by propionylation (15).
Subsequent studies proposed that SePat or EcPka could acetylate the response regulator RcsB (8), RNase R (11, 16), and many other proteins (7, 17). Later work indicated that Pat is not responsible for acetylation of the RNA polymerase alpha subunit (18) or of GapA, AceA, or AceK (19). These contradictory results have brought into question the substrate specificity of SePat, and of its homologues in other organisms, and raised the question of whether Pat is a global regulator of metabolism or an enzyme that specifically evolved to acylate AMP-forming acyl-CoA synthetase enzymes.
We previously reported studies of the specificity of the Pat homologue in the purple photosynthetic alphaproteobacterium Rhodopseudomonas palustris, hereafter referred to as RpPat (20). RpPat is 38% identical over 886 amino acids to SePat and can acetylate RpAcs as well as the related enzymes RpBadA, RpHbaA, and RpAliA, which are involved in aromatic and alicyclic acid degradation (20).
Proteomics analysis identified six new substrates for RpPat, all of which were acyl-CoA synthetases (19). We subsequently showed that RpPat also acetylates pimeloyl-CoA synthetase (PimA) (21), bringing the total number of RpPat substrates to 11 acyl-CoA (AMP-forming) synthetases, all of which are acetylated at the catalytic lysine residue (Lys534 in PimA). Examination of these substrates, as well as SeAcs and SePrpE, showed that they all contain a conserved PX4GK motif that includes the site of acetylation (19). We and others have speculated that the presence of the PX4GK motif is required for acetylation of the catalytic lysine residue in AMP-forming acyl-CoA synthetases (13, 19, 22, 23), yet to our knowledge, this has not been experimentally tested. At least one example of an acyl-CoA synthetase that contains the PX4GK motif and is not acetylated by RpPat has been reported (21). In that case, residues needed for recognition of the protein by RpPat were located outside the PX4GK motif.
In this study, we determined which residues surrounding the catalytic lysine residue in acyl-CoA synthetases are important for acetylation by RpPat. It appears that several residues within the acetylation motif were necessary for acetylation. Specifically, residues 2 and 4 amino acids upstream of the target lysine play crucial roles in acetylation and recognition. In addition, we characterized six new acyl-CoA synthetase enzymes from R. palustris and found five of them to be acetylated by RpPat. Interestingly, RpPat did not acetylate one of the newly characterized acyl-CoA synthetases, LcsB. However, a single amino acid variation of LcsB rendered it a substrate for RpPat.
MATERIALS AND METHODS
Strains, culture media, and growth conditions.
Bacterial strains and plasmids used in this work are listed in Table 1. E. coli strains were grown at 37°C on lysogeny broth (LB) medium (24, 25). Radiolabeled [1-14C]acetyl-CoA (54 mCi/mmol) was purchased from Moravek, and all other chemicals were from Sigma. Monocarboxylic acids are abbreviated as CX and dicarboxylic acids as DCX, where X is carbon chain length. Acids tested in this work were acetate (C2), propionate (C3), butyrate (C4), valerate (C5), hexanoate (C6), heptanoate (C7), octanoate (C8), nonanoate (C9), decanoate (C10), undecanoate (C11), laurate/dodecanoate (C12), tridecanoate (C13), myristate (C14), malonate (DC3), glutarate (DC5), adipate (DC6), pimelate (DC7), suberate (DC8), azelate (DC9), sebacate (DC10), dodecanedioate (DC12), tetradecanedioate (DC14), and benzoate (Ben). Acids longer than 10 carbon atoms were solubilized with Triton X-100 (1% [vol/vol] in a 2 mM carboxylic acid stock solution).
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Genotype | Reference, source, or method |
|---|---|---|
| Strains | ||
| Rhodopseudomonas palustris CGA009 | Wild-type genome | 45 |
| E. coli derivatives | ||
| JE6090 | C41(λDE3) | Laboratory collection |
| JE9314 | C41(λDE3) pka::kan+ | Laboratory collection |
| JE4242 | BL21(λDE3)/pLysS | Laboratory collection |
| Plasmids | ||
| pTEV5 | Purification vector; N-terminal His6 tag | 26 |
| pRpPAT13 | pat+ in pTEV5; bla+ | 21 |
| pRpPIMA1 | pimA+ in pTEV5; bla+ | 21 |
| pRpPIMA16 | pimA1 encodes PimAK534A in pTEV5; bla+ | 21 |
| pRpBTYA1 | btyA+ in pTEV5; bla+ | Restriction enzyme cloning |
| pRpBTYA2 | btyA1 encodes BtyAK531A in pTEV5; bla+ | QuikChange |
| pRpVCSA1 | vcsA+ in pTEV5; bla+ | Restriction enzyme cloning |
| pRpVCSA2 | vcsA1 encodes VcsAK499A in pTEV5; bla+ | QuikChange |
| pRpDCAA1 | dcaA+ in pTEV5; bla+ | Restriction enzyme cloning |
| pRpDCAA2 | dcaA1 encodes DcaAK490A in pTEV5; bla+ | QuikChange |
| pRpLCSB1 | lcsB+ in pTEV5; bla+ | Restriction enzyme cloning |
| pRpLCSB3 | lcsB 492-500 pimA in pTEV5; bla+ | QuikChange |
| pRpLCSB4 | lcsB1 encodes LcsBV497R in pTEV5; bla+ | QuikChange |
| pRpLCSB5 | lcsB2 encodes LcsBL500V in pTEV5; bla+ | QuikChange |
| pRpDCAB1 | dcaB+ in pTEV5, bla+ | PIPE cloning |
| pRpDCAB2 | dcaB1 encodes DcaBK506A in pTEV5; bla+ | QuikChange |
| pRpDCAC1 | dcaC+ in pTEV5; bla+ | PIPE cloning |
| pRpDCAC2 | dcaC1 encodes DcaCK514A in pTEV5; bla+ | QuikChange |
| pRpPIMA5 | pimA2 encodes PimAG533A in pTEV5; bla+ | QuikChange |
| pRpPIMA10 | pimA3 encodes PimAV532A in pTEV5; bla+ | QuikChange |
| pRpPIMA11 | pimA4 encodes PimAP528A in pTEV5; bla+ | QuikChange |
| pRpPIMA12 | pimA5 encodes PimAR529A in pTEV5; bla+ | QuikChange |
| pRpPIMA13 | pimA6 encodes PimAT530A in pTEV5; bla+ | QuikChange |
| pRpPIMA14 | pimA7 encodes PimAT531A in pTEV5; bla+ | QuikChange |
| pRpPIMA15 | pimA8 encodes PimAL535A in pTEV5; bla+ | QuikChange |
| pRpPIMA17 | pimA9 encodes PimAL527A in pTEV5; bla+ | QuikChange |
| pRpPIMA18 | pimA10 encodes PimAL540A in pTEV5; bla+ | QuikChange |
| pRpPIMA19 | pimA11 encodes PimAR541A in pTEV5; bla+ | QuikChange |
| pRpPIMA20 | pimA12 encodes PimAR537A in pTEV5; bla+ | QuikChange |
| pRpPIMA21 | pimA13 encodes PimAH538A in pTEV5; bla+ | QuikChange |
| pRpPIMA22 | pimA14 encodes PimAE539A in pTEV5; bla+ | QuikChange |
| pRpPIMA23 | pimA15 encodes PimAS536A in pTEV5; bla+ | QuikChange |
Genetic and recombinant DNA techniques.
To construct overexpression vectors, most acyl-CoA synthetase genes were amplified from R. palustris genomic DNA, cut with the appropriate restriction enzymes (Fermentas), and ligated into plasmid pTEV5 (26). Expression vectors for rpa1763 and rpa2142 were generated using polymerase incomplete primer extension (PIPE) cloning (27). All overexpression plasmids directed the synthesis of the AMP-forming acyl-CoA synthetase with an N-terminal His6 tag that was removed using recombinant tobacco etch virus (rTEV) protease (28). Site-directed mutants were constructed using the QuikChange protocol (Stratagene). Gene sequences were confirmed using BigDye (ABI PRISM) protocols, and sequencing reactions were resolved and analyzed at the University of Wisconsin-Madison Biotechnology Center.
Protein expression and purification.
Plasmids encoding wild-type and variant AMP-forming acyl-CoA synthetases were transformed into strain JE9314, a Pka (formerly YfiQ)-deficient derivative of E. coli C41 (λDE3) (29). All AMP-forming acyl-CoA synthetases were subsequently overexpressed and purified as described previously for RpPimA (21). RpPat and rTEV protease were purified as described previously (21, 28).
In vitro protein acetylation assay.
Proteins were acetylated using radiolabeled acetyl-CoA as described previously (10). Reaction mixtures contained HEPES buffer (50 mM; pH 7.0), tris(2-carboxyethyl)phosphine (TCEP) (1 mM), [1-14C]acetyl-CoA (19 μM; 26 nCi), protein substrate (3 μM), and RpPat (0.06 μM). Reaction mixtures (25-μl total volume) were incubated for 60 min at 30°C. Samples (10 μl each) were resolved using SDS-PAGE (30) and visualized by staining with Coomassie blue R250 (31). Gels were dried and exposed overnight to a multipurpose phosphor screen (Packard). Radioactivity associated with protein was quantified using a Typhoon FLA9000 biomolecular imager (GE) and OptiQuant software version 4.0 (Packard).
Acyl-CoA synthetase activity assays.
Acyl-CoA synthetases (1.5 μM each) were individually incubated with RpPat (0.5 μM) in the presence or absence of acetyl-CoA (50 μM) for 1 h at 30°C using the same buffer system as described above for 14C acetylation assays. Specific activity was quantified using an NADH consumption assay (32). Reaction mixtures contained HEPES buffer (50 mM, pH 7.5), TCEP (1 mM), ATP (2.5 mM), free coenzyme A (CoASH; 0.5 mM), MgCl2 (5 mM), phosphoenolpyruvate (3 mM), NADH (0.1 mM), pyruvate kinase (1 U), myokinase (5 U), lactate dehydrogenase (1.5 U), and organic acid substrate (0.2 mM). All reactions were started by the addition of acyl-CoA synthetase (3 pmol per 100 μl of reaction mixture), and the change in the absorbance at 340 nm was monitored for 8 min in a 96-well plate format using a Spectramax Plus UV-visible spectrophotometer (Molecular Devices). Each point was measured in triplicate, and the entire experiment was repeated on a separate day. Values represent averages and standard deviations of all six data points.
RESULTS
Residues important for activity and acetylation of PimA.
AMP-forming acyl-CoA synthetases are composed of two domains connected by a flexible linker, and the catalytic mechanism includes a large rotation between two different stable conformations (33). The catalytic lysine residue is located within the smaller C-terminal domain and is either buried in the active site at the interface between the N- and C-terminal domains or surface exposed, depending on the conformation of the enzyme. It has been proposed that the conserved PX4GK motif surrounding the acetylation site in RpPat substrates is important for acetylation by RpPat (19). This motif is shown in Fig. 1, in an alignment of 11 RpPat substrates, all of which are AMP-forming acyl-CoA synthetases (19–21).
FIG 1.

Sequence conservation around acetylated lysine residues of RpPat substrates. Shown is an alignment of the regions surrounding the acetylation sites of 11 previously identified RpPat substrates from R. palustris. The acetylated lysine residue is indicated with a star, and the PX4GK motif is underlined. Numbering refers to the PimA sequence. GenBank accession numbers for the sequences are as follows: PimA, NP_949053; BadA, NP_946014; Acs, NP_945564; PrpE, NP_949838; HbaA, NP_946022; AliA, NP_946004; IbuA, NP_947647; HcsA, NP_946354; FadD, NP_949603; LcsA, NP_949757; and FcsA, NP_947048. The alignment was generated using ClustalW (41) and formatted using ESPript (42).
We tested which residues in the vicinity of the acetylation site were important for acetylation by performing an alanine scan of PimA. Each residue within seven residues of the acetylation site, Lys534, was individually changed to alanine, the specific activity of the resulting variants was determined using octanoate as the substrate, and their susceptibility to acetylation by RpPat was also investigated.
Very few of the residues surrounding the catalytic lysine residue were important for activity of PimA (Fig. 2, black bars). As expected, Lys534 (the catalytic residue) was crucial for activity; substitution by alanine decreased activity by 99%. The only other variant with substantially lower activity than that of the wild-type enzyme was PimAT530A, which retained only 11% of wild-type activity. In contrast, more than half of the 15 residues surrounding Lys534 were important for acetylation by RpPat (Fig. 2, white bars), suggesting that conservation in this region of acyl-CoA synthetases is maintained primarily for regulation by acetylation rather than for catalytic activity. Within the PX4GK motif, alanine substitutions at residues Pro528, Arg529, Thr530, Val532, and Gly533 resulted in variants that were acetylated less than 50% relative to the wild-type protein. C terminal to the acetylation site, alanine substitutions at residues Arg537, Leu540, and Arg541 yielded variants that also showed <50% of wild-type acetylation. Among these alanine variants, PimAT530A (−4 position) and PimAV532A (−2 position) were the most severely affected, with just 5% and 9% of wild-type acetylation levels, respectively.
FIG 2.
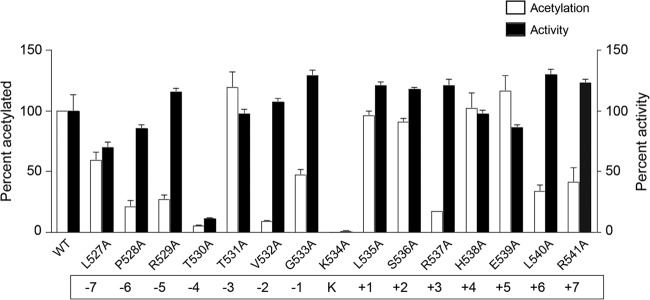
Alanine scan mutagenesis of residues located within the acetylation site of RpPimA to assess the importance of each residue for enzyme activity and recognition by RpPat. The activity of each RpPimA variant was measured using octanoate as the substrate, and each value was normalized to that of wild-type RpPimA (black bars). To assess acetylation (thus recognition), each variant was incubated with RpPat and [1-14C]acetyl-CoA for 1 h, and the extent of acetylation was quantified and normalized to that of wild-type RpPimA (white bars). Values represent averages and standard deviations of three (acetylation) or six (activity) replicates. Numbers below variant names indicate the position of each residue relative to the acetylation site.
Characterization of six putative acyl-CoA synthetases from R. palustris.
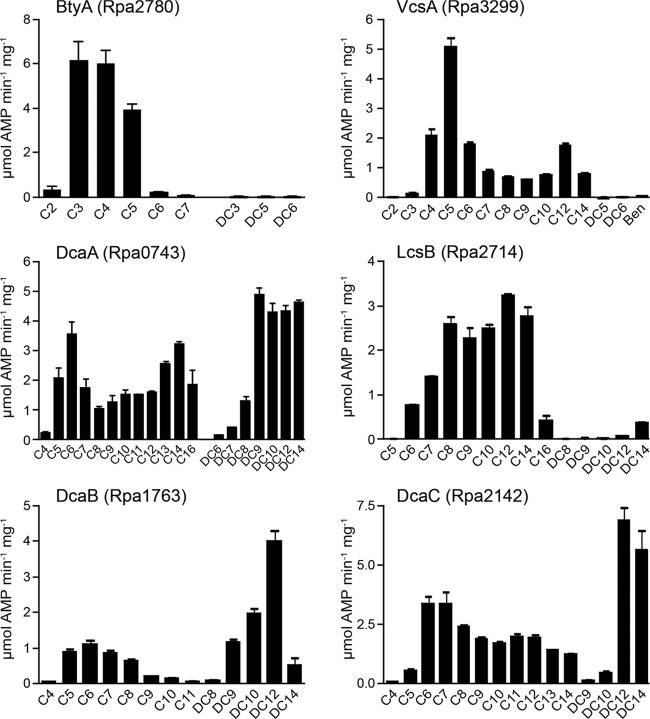
We sought to test whether natural variations in the residues around the catalytic lysine residue of acyl-CoA synthetases would affect their susceptibility to acetylation. To do this, we characterized six putative acyl-CoA synthetases (Rpa0743, Rpa1763, Rpa2142, Rpa2714, Rpa2780, and Rpa3299) that were not previously identified as potentially acetylated in a high-throughput proteomics study of acetylated proteins in R. palustris (19). These six putative acyl-CoA synthetases were all soluble proteins that showed activity with a variety of fatty acid substrates in vitro (Fig. 3).
FIG 3.
Enzymatic activities of newly identified AMP-forming acyl-CoA synthetases. The activities of six putative AMP-forming acyl-CoA synthetases were tested in vitro with a variety of organic acids. Mono- and dicarboxylic acids are indicated as CX and DCX, respectively, where X specifies the carbon chain length. Compound names are listed in Materials and Methods.
Protein Rpa3299 had the highest activity with the C5 fatty acid valerate, although it used C4 to C14 monocarboxylic acids as well (Fig. 3). Based on its strong activity with valerate, we propose naming Rpa3299 VcsA for valeryl-CoA synthetase A. Protein Rpa2780 also used short- to medium-chain fatty acids and had the highest activity with the C3, C4, and C5 fatty acids propionate, butyrate, and valerate (Fig. 3). Based on its strong activity using butyrate, we propose naming Rpa2780 BtyA for butyryl-CoA synthetase A. BtyA is distinct from the previously characterized R. palustris propionyl-CoA synthetase PrpE (19), and at the amino acid sequence level BtyA is more similar to E. coli medium-chain acyl-CoA synthetase FadK (30% identical) than to E. coli PrpE (25% identical).
Proteins Rpa0743 and Rpa2714 both used a broad range of monocarboxylic acids (Fig. 3), ranging in length from C5 to C16 (Rpa0743) and C6 to C16 (Rpa2714). We propose naming Rpa2714 LcsB for long-chain acyl-CoA synthetase B. Interestingly, protein Rpa0743 used medium- to long-chain dicarboxylic acids (C9 to C14) even more efficiently than monocarboxylic acids, and we propose naming it DcaA for dicarboxylic acid acyl-CoA synthetase A. Proteins Rpa1763 and Rpa2142 also used dicarboxylic acids, and both exhibited the highest activity with the C12 dicarboxylic acid dodecanedioic acid (Fig. 3). In light of their preference for dicarboxylic acids, we suggest naming Rpa1763 and Rpa2142 DcaB and DcaC, respectively.
Susceptibility of newly characterized acyl-CoA synthetases to acetylation.
We tested whether these six new acyl-CoA synthetases could be acetylated in vitro by incubating them individually with or without RpPat (1:50 ratio of RpPat to acyl-CoA synthetase) in the presence of [1-14C]acetyl-CoA (Fig. 4). As a positive control for acetylation we used PimA, which is specifically acetylated by Pat at Lys534 (21). At this relatively low ratio of RpPat to substrate, RpPat acetylated five of the newly identified acyl-CoA synthetases, namely, BtyA, VcsA, DcaA, DcaB, and DcaC. Changing the conserved catalytic lysine residue to alanine abolished acetylation (Fig. 4), demonstrating that acetylation occurs at the expected lysine. Notably, LcsB was not a substrate for RpPat under these conditions (Fig. 4). All of the acyl-CoA synthetases that were acetylated by RpPat lost a substantial amount of activity upon acetylation (Table 2). VcsA activity decreased by 86% after incubation with RpPat, and all of the other enzymes lost >98% of their activity after acetylation by RpPat. In contrast, under the conditions of this activity assay (1:3 molar ratio of RpPat to LcsB incubated for 1 h), LcsB lost only 13% of its activity, demonstrating that it is a poor substrate for RpPat.
FIG 4.
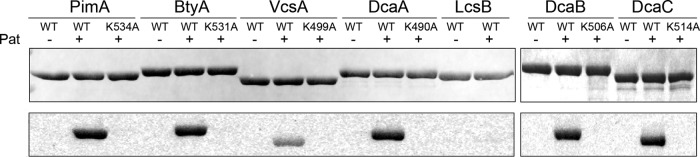
RpPat acetylates all of the newly identified AMP-forming acyl-CoA synthetases except LcsB. Each putative acyl-CoA synthetase was incubated with [1-14C]acetyl-CoA with or without RpPat (1:50 ratio of RpPat to acyl-CoA synthetase). For acyl-CoA synthetases that were acetylated by RpPat, the putative acetylation site was changed to an alanine residue and acetylation by RpPat was tested. The top image shows an SDS-PAGE gel of the reactions, and the bottom image is a phosphor image of the same gel.
TABLE 2.
Effect of acetylation on acyl-CoA synthetase activity
| Locus tag | Gene name | Substrate | Sp act (μmol of AMP min−1 mg−1)a |
Decrease (%) | |
|---|---|---|---|---|---|
| Unmodified | +RpPat | ||||
| Rpa3716 | pimA | Octanoate | 6.9 ± 0.2 | 0.04 ± 0.02 | 99.5 |
| Rpa0743 | dcaA | Azelate (C9, dioic) | 4.4 ± 0.3 | 0.05 ± 0.00 | 98.8 |
| Rpa1763 | dcaB | Dodecanedioate (C12, dioic) | 3.4 ± 0.1 | 0.06 ± 0.02 | 98.1 |
| Rpa2142 | dcaC | Heptanoate (C7) | 3.4 ± 0.3 | 0.03 ± 0.00 | 99.2 |
| Rpa2714 | lcsB | Laurate (C12) | 3.6 ± 0.04 | 3.2 ± 0.1 | 12.6 |
| Rpa2780 | btyA | Butyrate (C4) | 7.5 ± 0.1 | 0.07 ± 0.01 | 99.1 |
| Rpa3299 | vcsA | Valerate (C5) | 5.3 ± 0.5 | 0.7 ± 0.1 | 86.3 |
Specific activity (means ± standard deviations) was measured after incubating acyl-CoA synthetase with acetyl-CoA with or without RpPat (1:3 ratio of RpPat to acyl-CoA synthetase) for 1 h.
A single amino acid substitution makes LcsB a substrate for RpPat.
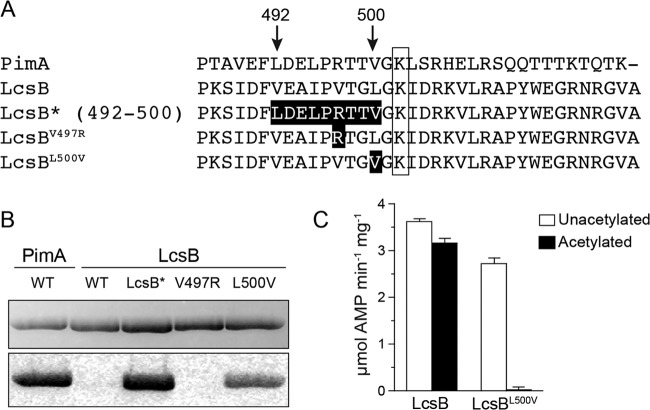
PimA and LcsB have similar activities with octanoate (C8) and decanoate (C10) (Fig. 3) (34), and at the protein sequence level they are 33% identical. Based on this similarity, we compared the PimA and LcsB sequences surrounding the catalytic lysine residue (Fig. 5A) and tried to identify changes that would render LcsB a substrate for RpPat. We began by replacing a stretch of 9 amino acids N terminal to the catalytic lysine residue between Phe491 and Gly501 of LcsB with the corresponding sequence from PimA. The resulting variant was designated LcsB492–500(PimA), or LcsB* for simplicity (Fig. 5A). LcsB* was readily acetylated by RpPat (Fig. 5B), demonstrating that this 9-amino-acid region N terminal to the acetylation site was important for recognition by RpPat.
FIG 5.
Conversion of LcsB into a substrate of RpPat by a single amino acid change. (A) Alignment of the C-terminal ends of RpPimA, RpLcsB, and three RpLcsB variants. Changes in the RpLcsB variants are indicated by black boxes, and residue numbers are noted with arrows above the sequence. The acetylation site is shown in the open box. (B) Acetylation of RpLcsB variants compared to wild-type RpPimA and RpLcsB. Each protein was incubated with [1-14C]acetyl-CoA and RpPat (1:50 ratio of RpPat to acyl-CoA synthetase). The top image shows the SDS-PAGE gel, and the bottom image is a phosphor image of the same gel. (C) Activity of wild-type RpLcsB and RpLcsBL500V using laurate as the substrate. Wild-type RpLcsB lost ∼13% of its activity after incubation with RpPat (1:3 ratio of RpPat to acyl-CoA synthetase) and acetyl-CoA, whereas RpLcsBL500V lost ∼99% of its activity.
Our results from the alanine scan of PimA suggested that the −2 and −4 positions relative to the acetylated lysine residue might be particularly important for recognition by RpPat (Fig. 2). Like PimA, LcsB also had a threonine in the −4 position, but LcsB differed from PimA in the −2 position, with leucine (Leu500) present instead of valine (Fig. 5A). LcsB also has a valine (Val497) instead of arginine in the −5 position, which we thought could be important for recognition by RpPat. We made single amino acid changes at the −5 position (LcsBV497R) and at the −2 position (LcsBL500V) to determine whether RpPat could acetylate LcsBV497R or LcsBL500V. While RpPat did not acetylate LcsBV497R, it did acetylate LcsBL500V (Fig. 5B). LcsBL500V was not as good a substrate for RpPat as LcsB*, based on the weaker acetylation observed in the radioactivity assay with low RpPat/substrate ratios (Fig. 5B). However, when the relative amount of RpPat was increased (1:3 ratio of RpPat to substrate) and LcsB activity was measured, it was evident that RpPat substantially inactivated LcsBL500V (Fig. 5C). The LcsBL500V variant retained 75% of wild-type LcsB activity using sodium laurate as the substrate, demonstrating that Leu500 was not critical for catalytic activity of LcsB. In contrast, the LcsBL500V variant lost 99% of its activity upon acetylation by RpPat, compared with only 13% loss of activity of wild-type LcsB upon acetylation (Fig. 5C). These results demonstrated that the −2 position of LcsB was important for recognition by RpPat and that RpPat would effectively recognize LcsB after the removal of a methylene group in the side chain of Leu500.
DISCUSSION
A refined acetylation motif for RpPat.
We have previously shown that the protein acetyltransferase RpPat specifically acetylates AMP-forming acyl-CoA synthetases (19), a large class of enzymes that activate fatty acids to acyl-CoA thioesters. In this study, we identified five new acyl-CoA synthetases from R. palustris that are substrates of RpPat (Fig. 4), bringing the total number of acyl-CoA synthetase substrates for RpPat to 16. These substrates all share the conserved PX4GK motif surrounding the acetylated lysine residue, and this motif has been suggested to be required for acetylation. Yet this assertion had not been experimentally tested.
An acetylation logo generated from all 16 bona fide substrates is shown in Fig. 6A. Based on the results of an alanine scan of RpPimA, we conclude that the PX4GK motif is important for acetylation (Fig. 2) but that there are additional residues within this region that are also critical for recognition by RpPat. We suggest that a more accurate motif would be PK/RTXS/V/T/NGKX2K/R, where the acetylation site is in italic type. We demonstrated that the underlined residues two and four positions upstream of the acetylated lysine are particularly important. Analysis of the crystal structure of benzoyl-CoA synthetase from Burkholderia xenovorans (PDB code 2V7B [35]) indicates that the −2 and −4 positions are surface exposed and located within a loop in close proximity to the acetylation site (Fig. 6B, loop 1), suggesting that they may directly interact with RpPat during acetylation.
FIG 6.
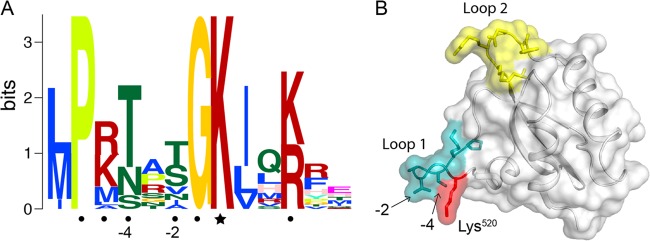
Regions of AMP-forming acyl-CoA synthetases important for RpPat recognition. (A) An acetylation site motif made from all 16 known R. palustris acyl-CoA synthetase substrates of RpPat was generated using MEME (43). The acetylated lysine residue is indicated with a star, and surrounding residues that were found to be important for recognition by RpPat are marked with circles. The −2 and −4 positions, relative to the acetylation site, were particularly important for recognition by RpPat. (B) Surface representation of BxBclM (PDB code 2V7B [35]). The C-terminal domain was generated using Pymol (44). The acetylated lysine residue (Lys520) is shown in red, the rest of the PX4GK motif is shown in teal and labeled loop 1, and the other loop shown to be important for acetylation by RpPat (21) is shown in yellow and labeled loop 2.
Conversion of LcsB into a substrate for RpPat by the removal of a methylene group highlights how small changes can block lysine acetylation.
LcsB (formerly Rpa2714), which activates medium- to long-chain monocarboxylic acids, is not a substrate for RpPat, largely due to the presence of a bulky leucine residue at the −2 position (Fig. 5). Changing this leucine to a valine renders LcsB subject to RpPat control while maintaining nearly wild-type activity with the C12 fatty acid laurate. The fact that wild-type LcsB is not acetylated demonstrates that the region around this lysine residue is critical for recognition by RpPat and that even minor changes (e.g., the presence of a methylene group) can prevent regulation by acetylation. It is somewhat surprising that the −2 position is so important for recognition by RpPat, given the variability at this position (Fig. 6A). This suggests that sequence variability in this region may impact the overall structure of the loop (loop 1 in Fig. 6B), which, in turn, could affect interactions with RpPat.
RpPat recognition determinants are located inside and outside the acetylation motif of AMP-forming acyl-CoA synthetases.
LcsB is the second example of an AMP-forming acyl-CoA synthetase from R. palustris that is not a substrate for RpPat. We previously found that the methylmalonyl-CoA synthetase (RpMatB) of this bacterium is not acetylated by RpPat either, largely due to changes in a loop region relatively far from the acetylation site, labeled loop 2 in Fig. 6B (21). Replacement of loop 2 of RpMatB with the corresponding sequence from RpPimA resulted in a chimeric protein that could be acetylated by RpPat, demonstrating that both loop 1 and loop 2 of acyl-CoA synthetases are important for recognition by RpPat (21). These results further support our claim that RpPat makes relatively extensive contact with its substrates and that it is specific for a subset of AMP-forming acyl-CoA synthetases. Recent work by others has shown that only 8 out of 22 Mycobacterium tuberculosis AMP-forming acyl-CoA synthetases that could be purified were acetylated by the M. tuberculosis Pat homologue (22), suggesting that it may be that only a subset of acyl-CoA synthetases in any given organism are regulated by acetylation. This is in contrast to previous reports claiming that the S. enterica and E. coli Pat homologues have broad substrate ranges and little substrate specificity (7).
Role of reversible lysine acetylation in fatty acid metabolism.
Previous work demonstrated that growth on aromatic compounds such as benzoate is controlled by acetylation in R. palustris (20) and that many different AMP-forming acyl-CoA synthetases involved in mono- and dicarboxylic acid activation are acetylated in vivo (19). While a deacetylase-deficient strain struggles to grow on benzoate, due to acetylation and concomitant inactivation of benzoyl-CoA synthetase, we have not seen a similar phenotype on noncyclic fatty acids (data not shown). One explanation could be that RpPat is expressed only during growth on aromatic acids. Alternatively, since not all AMP-forming acyl-CoA synthetases are regulated by acetylation, the cell may rely on these nonacetylatable acyl-CoA synthetases during growth on linear mono- and dicarboxylic acids.
Many Gram-negative bacteria encode either one or two RpPat homologues, including other organisms that can degrade aromatic compounds, such as members of the Burkholderia, Azoarcus, and Thauera genera. For the most part their activities and substrate specificities have yet to be explored. In addition, R. palustris encodes a second lysine acetyltransferase, RpKatA, which contains only an acetyltransferase domain yet is also involved in regulating acyl-CoA synthetases (19). Unlike RpPat and its S. enterica and E. coli homologues, which have a distinctive ∼700-amino-acid N-terminal domain of unknown function, RpKatA homologues are difficult to distinguish from other members of the large GNAT family of acetyltransferases. It is still unclear why growth on aromatic acids is regulated by acetylation in R. palustris and if this regulation extends to other species. We speculate that acetylation serves as a feedback mechanism to turn off the aromatic acid degradation pathway when the acetyl-CoA concentration is high, since acetyl-CoA and CO2 are the ultimate breakdown products of benzoate.
Acetylation of AMP-forming acyl-CoA synthetases by Pat homologues is unlikely to be regulated strictly by the intracellular acetyl-CoA concentration, though, as the substrate concentration at half the maximal velocity (K0.5) of SePat is ∼10 μM (36), which is well below measured cellular acetyl-CoA concentrations (37, 38). However, there is evidence that SePat may be cooperatively regulated and that its large N-terminal domain is involved in this regulation (36). Similarly, the structurally unrelated M. tuberculosis protein acetyltransferase that acetylates acyl-CoA synthetases also contains two domains, one of which binds cyclic AMP (cAMP) to allosterically regulate the acetyltransferase activity of the other (39, 40).
In addition to allosteric control, the timing and extent of acetylation may also be determined by the relative expression of lysine acetyltransferases and deacetylases in the cell. Little is known about the expression of Pat or the two deacetylases SrtN and LdaA in R. palustris. It has been shown that an R. palustris mutant strain lacking both SrtN and LdaA accumulates acetylated acyl-CoA synthetases and consequentially is unable to grow using benzoate as a carbon and energy source (20), demonstrating that the acetylation/deacetylation system is active during photoheterotrophic growth on benzoate. In E. coli, the Pat homologue Pka is regulated at the transcriptional level by cAMP, resulting in upregulation during stationary phase, whereas the deacetylase CobB is constitutively expressed (12). Taken together, these results suggest that acyl-CoA synthetase regulation occurs at the level of acetylation, rather than deacetylation, and that acetyltransferase activity is likely controlled transcriptionally and/or allosterically. Additional work is needed to determine when these acetylation/deacetylation systems are active in R. palustris and other species.
ACKNOWLEDGMENTS
This work was supported by grant R01 GM062203 to J.C.E.-S. H.A.C. was supported in part by USPHS Biotechnology Training Grant T32-GM08349.
Footnotes
Published ahead of print 31 January 2014
REFERENCES
- 1.Thao S, Escalante-Semerena JC. 2011. Control of protein function by reversible N(epsilon)-lysine acetylation in bacteria. Curr. Opin. Microbiol. 14:200–204. 10.1016/j.mib.2010.12.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kim GW, Yang XJ. 2011. Comprehensive lysine acetylomes emerging from bacteria to humans. Trends Biochem. Sci. 36:211–220. 10.1016/j.tibs.2010.10.001 [DOI] [PubMed] [Google Scholar]
- 3.Kouzarides T. 2007. Chromatin modifications and their function. Cell 128:693–705. 10.1016/j.cell.2007.02.005 [DOI] [PubMed] [Google Scholar]
- 4.Xiong Y, Guan KL. 2012. Mechanistic insights into the regulation of metabolic enzymes by acetylation. J. Cell Biol. 198:155–164. 10.1083/jcb.201202056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yu BJ, Kim JA, Moon JH, Ryu SE, Pan JG. 2008. The diversity of lysine-acetylated proteins in Escherichia coli. J. Microbiol. Biotechnol. 18:1529–1536 [PubMed] [Google Scholar]
- 6.Zhang J, Sprung R, Pei J, Tan X, Kim S, Zhu H, Liu CF, Grishin NV, Zhao Y. 2009. Lysine acetylation is a highly abundant and evolutionarily conserved modification in Escherichia coli. Mol. Cell. Proteomics 8:215–225. 10.1074/mcp.M800187-MCP200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang Q, Zhang Y, Yang C, Xiong H, Lin Y, Yao J, Li H, Xie L, Zhao W, Yao Y, Ning ZB, Zeng R, Xiong Y, Guan KL, Zhao S, Zhao GP. 2010. Acetylation of metabolic enzymes coordinates carbon source utilization and metabolic flux. Science 327:1004–1007. 10.1126/science.1179687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Thao S, Chen CS, Zhu H, Escalante-Semerena JC. 2010. N(epsilon)-lysine acetylation of a bacterial transcription factor inhibits its DNA-binding activity. PLoS One 5:e15123. 10.1371/journal.pone.0015123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Weinert BT, Iesmantavicius V, Wagner SA, Scholz C, Gummesson B, Beli P, Nystrom T, Choudhary C. 2013. Acetyl-phosphate is a critical determinant of lysine acetylation in E. coli. Mol. Cell 51:265–272. 10.1016/j.molcel.2013.06.003 [DOI] [PubMed] [Google Scholar]
- 10.Starai VJ, Escalante-Semerena JC. 2004. Identification of the protein acetyltransferase (Pat) enzyme that acetylates acetyl-CoA synthetase in Salmonella enterica. J. Mol. Biol. 340:1005–1012. 10.1016/j.jmb.2004.05.010 [DOI] [PubMed] [Google Scholar]
- 11.Liang W, Deutscher MP. 2012. Post-translational modification of RNase R is regulated by stress-dependent reduction in the acetylating enzyme Pka (YfiQ). RNA 18:37–41. 10.1261/rna.030213.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Castaño-Cerezo S, Bernal V, Blanco-Catala J, Iborra JL, Canovas M. 2011. cAMP-CRP co-ordinates the expression of the protein acetylation pathway with central metabolism in Escherichia coli. Mol. Microbiol. 82:1110–1128. 10.1111/j.1365-2958.2011.07873.x [DOI] [PubMed] [Google Scholar]
- 13.Starai VJ, Celic I, Cole RN, Boeke JD, Escalante-Semerena JC. 2002. Sir2-dependent activation of acetyl-CoA synthetase by deacetylation of active lysine. Science 298:2390–2392. 10.1126/science.1077650 [DOI] [PubMed] [Google Scholar]
- 14.Starai VJ, Takahashi H, Boeke JD, Escalante-Semerena JC. 2003. Short-chain fatty acid activation by acyl-coenzyme A synthetases requires SIR2 protein function in Salmonella enterica and Saccharomyces cerevisiae. Genetics 163:545–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Garrity J, Gardner JG, Hawse W, Wolberger C, Escalante-Semerena JC. 2007. N-lysine propionylation controls the activity of propionyl-CoA synthetase. J. Biol. Chem. 282:30239–30245. 10.1074/jbc.M704409200 [DOI] [PubMed] [Google Scholar]
- 16.Liang W, Malhotra A, Deutscher MP. 2011. Acetylation regulates the stability of a bacterial protein: growth stage-dependent modification of RNase R. Mol. Cell 44:160–166. 10.1016/j.molcel.2011.06.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lima BP, Antelmann H, Gronau K, Chi BK, Becher D, Brinsmade SR, Wolfe AJ. 2011. Involvement of protein acetylation in glucose-induced transcription of a stress-responsive promoter. Mol. Microbiol. 81:1190–1204. 10.1111/j.1365-2958.2011.07742.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lima BP, Thanh Huyen TT, Basell K, Becher D, Antelmann H, Wolfe AJ. 2012. Inhibition of acetyl phosphate-dependent transcription by an acetylatable lysine on RNA polymerase. J. Biol. Chem. 287:32147–32160. 10.1074/jbc.M112.365502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Crosby HA, Pelletier DA, Hurst GB, Escalante-Semerena JC. 2012. System-wide studies of N-lysine acetylation in Rhodopseudomonas palustris reveal substrate specificity of protein acetyltransferases. J. Biol. Chem. 287:15590–15601. 10.1074/jbc.M112.352104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Crosby HA, Heiniger EK, Harwood CS, Escalante-Semerena JC. 2010. Reversible N(epsilon)-lysine acetylation regulates the activity of acyl-CoA synthetases involved in anaerobic benzoate catabolism in Rhodopseudomonas palustris. Mol. Microbiol. 76:874–888. 10.1111/j.1365-2958.2010.07127.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Crosby HA, Rank KC, Rayment I, Escalante-Semerena JC. 2012. Structural insights into the substrate specificity of the protein acetyltransferase RpPat: identification of a loop critical for recognition by RpPat. J. Biol. Chem. 287:41392–41404. 10.1074/jbc.M112.417360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nambi S, Gupta K, Bhattacharya M, Ramakrishnan P, Ravikumar V, Siddiqui N, Thomas AT, Visweswariah SS. 2013. Cyclic AMP-dependent protein lysine acylation in mycobacteria regulates fatty acid and propionate metabolism. J. Biol. Chem. 288:14114–14124. 10.1074/jbc.M113.463992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vergnolle O, Xu H, Blanchard JS. 2013. Mechanism and regulation of mycobactin fatty acyl-AMP ligase FadD33. J. Biol. Chem. 288:28116–28125. 10.1074/jbc.M113.495549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bertani G. 1951. Studies on lysogenesis. I. The mode of phage liberation by lysogenic Escherichia coli. J. Bacteriol. 62:293–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bertani G. 2004. Lysogeny at mid-twentieth century: P1, P2, and other experimental systems. J. Bacteriol. 186:595–600. 10.1128/JB.186.3.595-600.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rocco CJ, Dennison KL, Klenchin VA, Rayment I, Escalante-Semerena JC. 2008. Construction and use of new cloning vectors for the rapid isolation of recombinant proteins from Escherichia coli. Plasmid 59:231–237. 10.1016/j.plasmid.2008.01.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Klock HE, Koesema EJ, Knuth MW, Lesley SA. 2008. Combining the polymerase incomplete primer extension method for cloning and mutagenesis with microscreening to accelerate structural genomics efforts. Proteins 71:982–994. 10.1002/prot.21786 [DOI] [PubMed] [Google Scholar]
- 28.Blommel PG, Fox BG. 2007. A combined approach to improving large-scale production of tobacco etch virus protease. Protein Expr. Purif. 55:53–68. 10.1016/j.pep.2007.04.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miroux B, Walker JE. 1996. Over-production of proteins in Escherichia coli: mutant hosts that allow synthesis of some membrane proteins and globular proteins at high levels. J. Mol. Biol. 260:289–298. 10.1006/jmbi.1996.0399 [DOI] [PubMed] [Google Scholar]
- 30.Laemmli UK. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680–685. 10.1038/227680a0 [DOI] [PubMed] [Google Scholar]
- 31.Sasse J. 1991. Detection of proteins. Curr. Protoc. Mol. Biol. 1:10.16.11–10.16.18 [Google Scholar]
- 32.Reger AS, Carney JM, Gulick AM. 2007. Biochemical and crystallographic analysis of substrate binding and conformational changes in acetyl-CoA synthetase. Biochemistry 46:6536–6546. 10.1021/bi6026506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wu R, Reger AS, Lu X, Gulick AM, Dunaway-Mariano D. 2009. The mechanism of domain alternation in the acyl-adenylate forming ligase superfamily member 4-chlorobenzoate: coenzyme A ligase. Biochemistry 48:4115–4125. 10.1021/bi9002327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Harrison FH, Harwood CS. 2005. The pimFABCDE operon from Rhodopseudomonas palustris mediates dicarboxylic acid degradation and participates in anaerobic benzoate degradation. Microbiology 151:727–736. 10.1099/mic.0.27731-0 [DOI] [PubMed] [Google Scholar]
- 35.Bains J, Boulanger MJ. 2007. Biochemical and structural characterization of the paralogous benzoate CoA ligases from Burkholderia xenovorans LB400: defining the entry point into the novel benzoate oxidation (box) pathway. J. Mol. Biol. 373:965–977. 10.1016/j.jmb.2007.08.008 [DOI] [PubMed] [Google Scholar]
- 36.Thao S, Escalante-Semerena JC. 2011. Biochemical and thermodynamic analyses of Salmonella enterica Pat, a multidomain, multimeric N(epsilon)-lysine acetyltransferase involved in carbon and energy metabolism. mBio 2:e00216–11. 10.1128/mBio.00216-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bennett BD, Kimball EH, Gao M, Osterhout R, Van Dien SJ, Rabinowitz JD. 2009. Absolute metabolite concentrations and implied enzyme active site occupancy in Escherichia coli. Nat. Chem. Biol. 5:593–599. 10.1038/nchembio.186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chohnan S, Izawa H, Nishihara H, Takamura Y. 1998. Changes in size of intracellular pools of coenzyme A and its thioesters in Escherichia coli K-12 cells to various carbon sources and stresses. Biosci. Biotechnol. Biochem. 62:1122–1128. 10.1271/bbb.62.1122 [DOI] [PubMed] [Google Scholar]
- 39.Lee HJ, Lang PT, Fortune SM, Sassetti CM, Alber T. 2012. Cyclic AMP regulation of protein lysine acetylation in Mycobacterium tuberculosis. Nat. Struct. Mol. Biol. 19:811–818. 10.1038/nsmb.2318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nambi S, Basu N, Visweswariah SS. 2010. Cyclic AMP-regulated protein lysine acetylases in mycobacteria. J. Biol. Chem. 285:24313–24323. 10.1074/jbc.M110.118398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. 2007. Clustal W and Clustal X version 2.0. Bioinformatics 23:2947–2948. 10.1093/bioinformatics/btm404 [DOI] [PubMed] [Google Scholar]
- 42.Gouet P, Courcelle E, Stuart DI, Metoz F. 1999. ESPript: multiple sequence alignments in PostScript. Bioinformatics 15:305–308. 10.1093/bioinformatics/15.4.305 [DOI] [PubMed] [Google Scholar]
- 43.Bailey TL, Elkan C. 1994. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc. Int. Conf. Intell. Syst. Mol. Biol. 2:28–36 [PubMed] [Google Scholar]
- 44.Delano WL. 2002. The Pymol molecular graphics system. DeLano Scientific, San Carlos, CA: http://www.pymol.org [Google Scholar]
- 45.Larimer FW, Chain P, Hauser L, Lamerdin J, Malfatti S, Do L, Land ML, Pelletier DA, Beatty JT, Lang AS, Tabita FR, Gibson JL, Hanson TE, Bobst C, Torres JL, Peres C, Harrison FH, Gibson J, Harwood CS. 2004. Complete genome sequence of the metabolically versatile photosynthetic bacterium Rhodopseudomonas palustris. Nat. Biotechnol. 22:55–61. 10.1038/nbt923 [DOI] [PubMed] [Google Scholar]