Abstract
The peptidoglycan cell wall of bacteria is a complex macromolecule composed of glycan strands that are cross-linked by short peptide bridges. Its biosynthesis involves a conserved group of enzymes, the bifunctional penicillin-binding proteins (bPBPs), which contain both a transglycosylase and a transpeptidase domain, thus being able to elongate the glycan strands and, at the same time, generate the peptide cross-links. The stalked model bacterium Caulobacter crescentus possesses five bPBP paralogs, named Pbp1A, PbpC, PbpX, PbpY, and PbpZ, whose function is still incompletely understood. In this study, we show that any of these proteins except for PbpZ is sufficient for growth and normal morphogenesis when expressed at native or elevated levels, whereas inactivation of all five paralogs is lethal. Growth analyses indicate a central role of PbpX in the resistance of C. crescentus against the noncanonical amino acid d-alanine. Moreover, we show that PbpX and PbpY localize to the cell division site. Their recruitment to the divisome is dependent on the essential cell division protein FtsN and likely involves interactions with FtsL and the putative peptidoglycan hydrolase DipM. The same interaction pattern is observed for Pbp1A and PbpC, although these proteins do not accumulate at midcell. Our findings demonstrate that the bPBPs of C. crescentus are, to a large extent, redundant and have retained the ability to interact with the peptidoglycan biosynthetic machineries responsible for cell elongation, cytokinesis, and stalk growth. Nevertheless, they may preferentially act in specific peptidoglycan biosynthetic complexes, thereby facilitating the independent regulation of distinct growth processes.
INTRODUCTION
Peptidoglycan is a complex macromolecule that constitutes the main part of the bacterial cell wall and is essential for survival in the osmotically challenging environment that most bacteria inhabit. It is composed of glycan strands of alternating N-acetylglucosamine (GlcNAc) and N-acetylmuramic acid (MurNAc) moieties that are cross-linked by short peptides (1). The basic building blocks of peptidoglycan are disaccharide (GlcNAc-MurNAc)-pentapeptide units that are assembled in the cytoplasm and attached to the membrane carrier bactoprenol phosphate, yielding the so-called lipid II precursor. After flipping to the periplasmic side, these units are incorporated into the nascent peptidoglycan structure by a two-step process involving a transglycosylation and a transpeptidation reaction. Transglycosylation results in the formation of a β-1,4-glycosidic bond between the bactoprenol-linked reducing end of a glycan chain and the GlcNAc moiety of lipid II, accompanied by the release of bactoprenol diphosphate (2, 3). Later on, the peptide side chains of MurNAc residues from neighboring glycan strands are cross-linked by a transpeptidation reaction, in which the free amino group of mesodiaminopimelic acid (mDAP) from one side chain attacks the subterminal d-alanine in the adjacent side chain, thereby releasing the terminal d-alanine residue. β-Lactam antibiotics such as penicillin are structurally similar to the terminal d-alanine-d-alanine moiety of the peptide side chain. They can therefore interact with the substrate binding pocket of transpeptidases and covalently attach to the active site residue, thereby irreversibly inhibiting their activity. Based on this effect, transpeptidases are also commonly known as penicillin-binding proteins. Although the two enzymatic activities required for peptidoglycan synthesis can be executed by distinct enzymes, a large group of peptidoglycan synthases contain both a transglycosylase and a transpeptidase domain and are thus bifunctional.
Most Gram-negative bacteria possess several bifunctional penicillin-binding proteins (bPBPs) with overlapping and partially redundant cellular functions. A notable exception is Neisseria gonorrhoeae, as it contains only a single bPBP, which is essential for growth (4, 5). Escherichia coli contains a total of three bPBPs. One of them, PBP1A, is part of the so-called elongasome (6), a protein complex responsible for lateral cell wall elongation that assembles on the bacterial actin homolog MreB (1, 7). Its paralog PBP1B, by contrast, associates with components of the cell division apparatus, including FtsN and the transpeptidase FtsI (PBP3) (8, 9), thus likely contributing to medial cell growth and/or constriction of the cell wall during cytokinesis. In both cases, interaction with the respective biosynthetic complexes stimulates the activity of the transglycosylase domains (6, 9). Each of the two proteins additionally interacts with a cognate outer membrane lipoprotein (LpoA and LpoB, respectively) that is required to trigger its transpeptidase activity, thereby possibly coordinating peptidoglycan synthesis with overall cell growth (10, 11). Linking the outer membrane with the other layers of the cell envelope, PBP1B and LpoB also mediate invagination of the outer membrane during cytokinesis, a function redundant with that of the Tol-Pal complex (10). Although functioning in different contexts, neither PBP1A nor PBP1B is essential for growth, but inactivation of both proteins leads to cell death (12). Notably, E. coli contains a third bPBP, known as PBP1C, which cannot replace its paralogs and is structurally distinct from them (13, 14). It has so far not been possible to detect transpeptidase activity for PBP1C, suggesting that this protein may exclusively function as a transglycosylase. While the function of PBP1C in E. coli has remained unclear, some of its homologs were proposed to have specialized roles, e.g., in nitrogen fixation or the resistance against host defense mechanisms, in other organisms (15, 16). Overall, peptidoglycan synthesis has been studied intensively in E. coli, but little is known about this process in other Gram-negative bacteria. It is interesting that the number of bPBPs is highly variable between species. Moreover, the transpeptidase-activating lipoproteins LpoA and LpoB are conserved only in gammaproteobacteria (11). The mechanisms controlling the activity of bPBPs are therefore likely to vary considerably between different bacterial lineages.
A species known for its unusual morphology is Caulobacter crescentus, an organism that divides asymmetrically into two different offspring: a motile swarmer cell and a sessile stalked cell. Whereas the newborn stalked cell can immediately enter the next round of cell division, the swarmer cell first has to differentiate into a stalked cell before it can continue its life cycle. Notably, swarmer cells elongate by dispersed, MreB-dependent incorporation of peptidoglycan along the sidewalls. Upon transition to the stalked stage, C. crescentus then switches to FtsZ-dependent zonal growth, characterized by localized peptidoglycan synthesis at midcell, which is finally followed by constriction of the cell and formation of the new cell poles (17). Consistent with its complex cell shape, C. crescentus possesses five bPBPs, named Pbp1A, PbpC, PbpX, PbpY, and PbpZ (18–20). While PbpZ shows similarity to E. coli PBP1C, the other proteins are evolutionarily most closely related to E. coli PBP1A (20). So far, the function of bPBPs in C. crescentus has been addressed by only a limited number of studies. PbpC was shown to be recruited to the stalked cell pole through interaction with a patch of the cytoskeletal protein bactofilin, where it contributes to stalk biogenesis (19). Pbp1A, by contrast, was found to condense at midcell in response to an osmotic upshift, thereby potentially contributing to the robustness of cell division under stress conditions (18). Moreover, a recent study has provided first insights into the redundancy of the system and the subcellular localization of the five bPBPs in C. crescentus (20). However, overall, the precise localization dynamics and biological roles of these proteins are still incompletely understood. Moreover, it has remained unclear why this species contains such an unusually high number of bPBP paralogs.
In this study, we investigate the role and localization behavior of the five bPBPs identified in C. crescentus. We show that any of them, except for PbpZ, can sustain growth and normal cell morphology in the absence of its paralogs, whereas inactivation of all bPBPs is lethal. Growth studies further identify a key role of PbpX in the resistance of C. crescentus against the amino acid d-alanine. Analyzing the localization patterns of all bPBP paralogs, we finally demonstrate that PbpX and PbpY are recruited to midcell during the late stages of cell division. Their recruitment to the divisome is dependent on the late cell division protein FtsN and involves interactions with FtsL and the peptidoglycan hydrolase DipM, indicating the formation of a divisome-associated enzyme complex mediating constriction of the cell wall during cytokinesis.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
All C. crescentus strains used in this study were derived from the synchronizable wild-type strain CB15N (NA1000). Cells were grown at 28°C in peptone-yeast extract (PYE) medium (21) or M2-glucose (M2G) minimal medium (22). Induction or repression of the xylX promoter (23) was achieved by supplementing the medium with 0.3% xylose or 0.2% glucose, respectively. To induce the iolC (24) or vanA (25) promoter, media were supplemented with 0.3% myo-inositol and 0.5 mM vanillate, respectively. Plasmids were introduced into C. crescentus by electroporation (22). Cell synchronization for time-lapse microscopy was achieved by Percoll density gradient centrifugation (26). Large-scale preparations of swarmer cells were performed using Ludox AS-40 (Sigma-Aldrich) (22). Cells carrying inducible fluorescent protein fusions were grown to exponential phase in PYE medium and induced for 3 h with 0.3% xylose or 0.3% myo-inositol prior to microscopic analysis. For C. crescentus, antibiotics were used at the concentrations in parentheses (μg ml−1; liquid/solid medium): kanamycin (5/25), gentamicin (0.5/5), spectinomycin (25/50), streptomycin (0/5). E. coli TOP10 (Invitrogen) and E. coli XL1-Blue (Agilent) were used for general cloning purposes. The cells were grown aerobically at 37°C in Luria-Bertani broth (LB) (Carl-Roth, Germany) or Super broth (SB) (27). Antibiotics were used at the concentrations in parentheses: (μg/ml−1; liquid/solid medium): ampicillin (200/200), kanamycin (30/50), gentamicin (15/20), and spectinomycin (50/100).
Construction of plasmids and bacterial strains.
The strains, plasmids and primers used in this study are detailed in Tables S1, S2, S3, and S4 in the supplemental material. All plasmid constructs were verified by DNA sequence analysis. Single homologous recombination was used to integrate nonreplicating plasmids into the C. crescentus chromosome. Correct integration of the plasmids was verified by colony PCR. Chromosomal in-frame deletions and insertions were generated by double-homologous recombination, using a two-step procedure based on the sacB-containing suicide vector pNPTS138 (M. R. K. Alley, unpublished data) (28). To generate conditional mutants, nonreplicating plasmids carrying the gene of interest under the control of the xylX or iolC promoter were integrated at the xylX or the iolC locus, respectively. Subsequently, the corresponding endogenous gene was deleted in the resulting strain in the presence of xylose or myo-inositol, respectively.
Protein depletion and complementation analysis.
To deplete proteins synthesized under the control of the xylose-inducible xylX promoter, cells were grown to exponential phase in PYE medium containing 0.3% xylose and harvested by centrifugation for 2 min at 9,000 × g and 4°C. They were washed three times with 1 volume of M2 salt solution (22) and then resuspended in PYE medium for further cultivation. For complementation analyses, the washed cells were resuspended in PYE medium containing 0.3% myo-inositol to induce expression of the complementing gene that was placed under the control of the iolC promoter. To monitor the depletion of proteins and its phenotypic consequences, samples were taken at regular intervals and analyzed by immunoblotting and differential interference contrast (DIC) microscopy, respectively. Cultures were diluted with fresh medium when necessary to ensure exponential growth throughout the course of the experiments.
Immunoblot analysis.
To generate polyclonal anti-PbpX, anti-PbpY, and anti-PbpC antibodies, rabbits were immunized with mixtures of the PbpX-derived peptides AEAPAGEPPPADQLPY and DYNDSQFDRATTARRQ, the PbpY-derived peptides CAAQPGKPTQKPDPLS and MPDAGELQDYRPPTAT, or the PbpC-derived peptides CVAPAPGQPPPDNLPY and LPPYKFDDGKSPGEPC (Eurogentec). Immunoblot analysis was performed as described previously (28), using anti-CtrA (29) (1:10,000), anti-PbpX (1:2,500), anti-PbpY (1:2,500), anti-PbpC (1:10,000), or anti-CyaA (T25) (30) antiserum or a monoclonal anti-green fluorescent protein (anti-GFP) (Sigma-Aldrich) (1:10,000) or anti-CyaA (T18) (3D1; Santa Cruz Biotechnology) (1:100) antibody. Immunocomplexes were visualized using the Western Lightning chemiluminescence reagent (PerkinElmer) and Hyperfilm ECL (GE Healthcare) autoradiography films.
Microscopy.
For DIC and fluorescence microscopy, exponentially growing cells were spotted onto 1% agarose pads. For time-lapse imaging, isolated swarmer cells were immobilized on pads made of 1% agarose in M2G medium, which was supplemented with 0.3% xylose or 0.3% myo-inositol if necessary. The cover slide was sealed with a 1:1:1 mixture of petrolatum, lanolin, and paraffin. Images were taken with an Axio Observer.Z1 (Zeiss) microscope equipped with a Plan Apochromat 100×/1.45 oil DIC objective and a pco.edge sCMOS camera (PCO). Images were processed with Metamorph 7.7.5 (Universal Imaging Group) and Adobe Illustrator CS5 (Adobe Systems).
Bacterial two-hybrid analysis.
Bacterial two-hybrid analysis was performed as described previously (31). The proteins of interest were fused to either the T25 or the T18 fragment of Bordetella pertussis adenylate cyclase. The adenylate cyclase-deficient strain E. coli BTH101 was then cotransformed with two plasmids carrying the fusion constructs to be analyzed and plated on MacConkey agar (Carl-Roth, Germany) supplemented with ampicillin and kanamycin. For the drop assay, 15 to 20 transformants were combined and grown to exponential phase in LB medium containing ampicillin, kanamycin, and IPTG (isopropyl-β-d-thiogalactopyranoside) (0.5 mM). Subsequently, 4-μl samples of the cultures were dropped onto MacConkey agar plates supplemented with ampicillin and kanamycin and incubated at 28°C for 24 h.
Peptidoglycan analysis.
To determine the composition of the peptidoglycan cell wall, cells were grown for at least 30 h in 250 ml PYE medium supplemented with 0.3% myo-inositol until they reached an optical density at 600 nm (OD600) of 0.7 (CB15N, WS070, WS071, WS072, and WS073) or 0.34 (WS074), respectively. After centrifugation at 4,500 × g, the pelleted cells were resuspended in 1 ml H2O, heat inactivated for 10 min at 95°C, lyophilized, and analyzed essentially as described previously (32) (CeCo Labs, Tübingen, Germany). In brief, peptidoglycan was isolated from 10 mg of lyophilized cells and treated with mutanolysin. After reduction with sodium borohydride, muropeptides were separated on an MZ-Aqua Perfect C18 reversed-phase high-pressure liquid chromatography (HPLC) column (MZ Analysentechnik GmbH, Germany), using a linear gradient from 50 mM NaH2PO4 (buffer A) to 75 mM NaH2PO4 in 30% methanol (buffer B).
Bioinformatic analysis.
The domain composition of proteins was determined by SMART (33) and Pfam (34) analysis. Protein sequences were aligned using Clustal W2 (35) and edited in GeneDoc (v2.6.022; http://www.psc.edu/biomed/genedoc).
RESULTS
Bifunctional penicillin-binding proteins of C. crescentus.
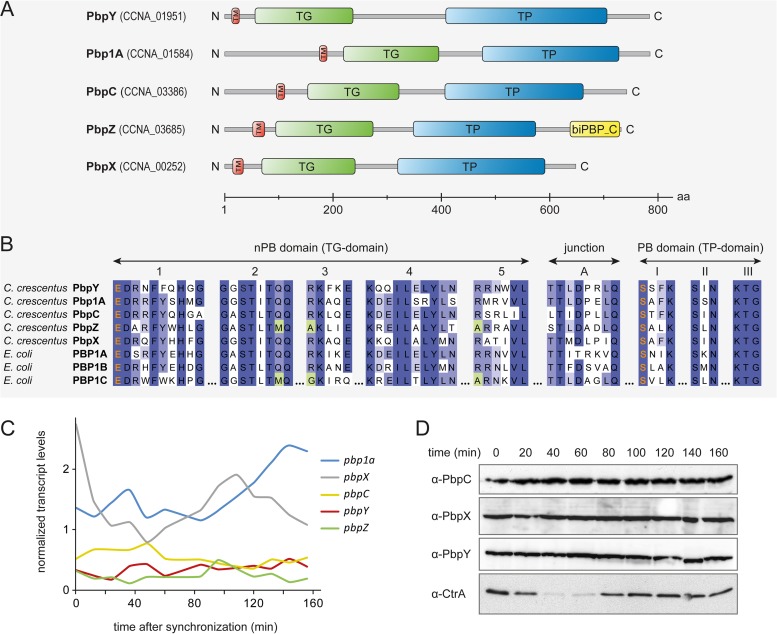
The genome of C. crescentus encodes five bifunctional penicillin-binding proteins. They are composed of a cytoplasmic tail of variable length, a single transmembrane helix, and a large periplasmic region that harbors the catalytic transglycosylase and transpeptidase domains (Fig. 1A). PbpY (CCNA_01951) has the highest molecular weight, followed by Pbp1A (CCNA_01584), PbpC (CCNA_03386), PbpZ (CCNA_03685), and finally PbpX (CCNA_00252). The five proteins share the nine conserved motifs that have been described for the bPBPs of E. coli (36) (Fig. 1B). The active site glutamate of the transglycosylase (non-penicillin-binding [nPB]) domain is found in all C. crescentus bPBPs. The same is true for the active site serine of the transpeptidase (TP) domain. Notably, highly conserved amino acid residues lacking in the nPB motif of E. coli PBP1C are also absent from PbpZ in C. crescentus (Fig. 1B).
FIG 1.
Characteristics of the bifunctional penicillin-binding proteins of C. crescentus. (A) Schematic of the five bifunctional penicillin-binding proteins (bPBPs) of C. crescentus. The proteins are typically composed of an N-terminal cytoplasmic region, a single transmembrane helix (TM), and a large C-terminal periplasmic region containing the catalytic transglycosylase (TG) and transpeptidase (TP) domains. PbpZ additionally features a C-terminal dimerization domain (biPBP_C). (B) Conservation of conserved signature motifs among the bPBPs of C. crescentus and E. coli. Catalytic residues are indicated in orange. Positions in which both C. crescentus PbpZ and E. coli PBP1C deviate from the consensus are highlighted in green. (C) Normalized transcript levels of genes encoding bPBPs over the course of the cell cycle. The graph is based on a microarray-based transcriptome analysis of synchronized C. crescentus wild-type cells (37). (D) Cellular levels of PbpC, PbpX, and PbpY over the course of the cell cycle. Isolated swarmer cells were transferred into M2G medium. Samples were taken at the indicated time points and analyzed by immunoblotting using antisera directed against PbpC, PbpX, PbpY, and CtrA, respectively. The characteristic cell cycle-dependent changes in the abundance of CtrA verify the synchrony of the cells (29).
Using data from a previous microarray analysis (37), we found that out of the five bPBP-encoding genes of C. crescentus, only pbp1a and pbpX displayed significant changes in their expression levels over the course of the cell cycle (Fig. 1C). Specifically, pbp1a is upregulated during the final stage of cell division, whereas pbpX transcript already accumulates early during the cell division process. However, despite these temporal variations in mRNA abundance, the protein levels of PbpX remain constant throughout the cell cycle (Fig. 1D). The same is true for PbpC and PbpY, in line with their relatively constant transcript levels. The cell cycle patterns of the other bPBPs remain unknown, as it has not been possible to obtain functional antibodies so far.
bPBPs are functionally redundant but essential for cell viability.
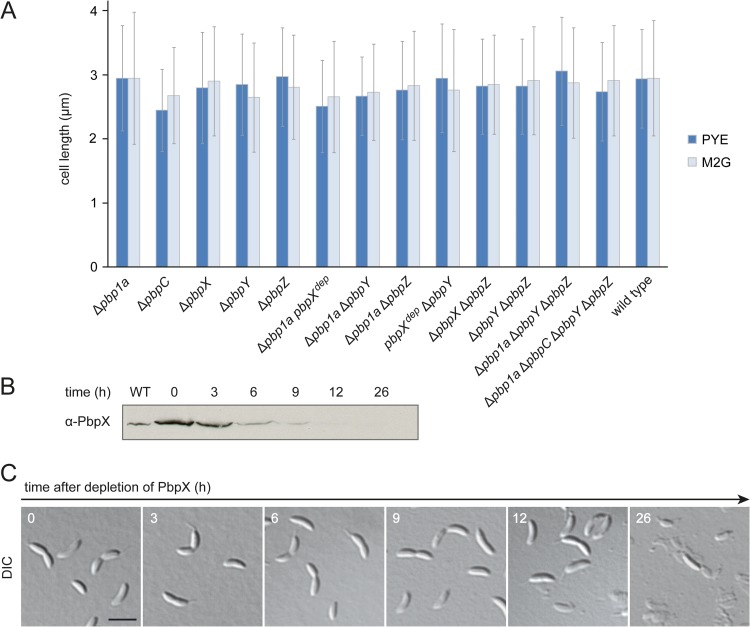
Compared to other bacteria, C. crescentus possesses an unusually large number of bPBPs. To clarify their degree of functional divergence, we generated a series of mutants carrying in-frame deletions in one or several bPBP genes and analyzed them for changes in cell morphology under standard conditions. Visual inspection of the strains did not reveal any obvious cell shape or division defects, even for a strain lacking all bPBPs except for PbpX (data not shown). To reveal potential mild phenotypes, we further determined the lengths of the different strains after growth in complex or minimal medium. However, none of the mutants showed any significant differences from the wild type (Fig. 2A). In order to clarify whether bPBPs were required for cell viability in C. crescentus at all, we constructed a conditional pbpX mutant carrying in-frame deletions in all native bPBP-encoding genes. While the strain grew normally under inducing conditions, depletion of PbpX by cultivation in inducer-free medium led to cell lysis (Fig. 2B and C). Consistent with this result, it was not possible to generate a quintuple mutant lacking all native bPBP-encoding genes unless a complementing copy of pbpX was present in the cells. Collectively, these findings indicate that the bPBPs of C. crescentus may have, at least in part, redundant functions but are required for viability.
FIG 2.
The bifunctional PBPs of C. crescentus are highly redundant but essential. (A) Length of mutant cells lacking one or more bPBPs. Wild-type strain CB15N and derivatives carrying the indicated mutations were grown to exponential phase in PYE and M2G medium, respectively, and the average cell lengths were determined (n > 300 per strain; error bars show standard deviations). The genes labeled with “dep” were not deleted but expressed under the control of the xylX promoter and transcriptionally inactivated by growth of the cells in the absence of inducer for 24 h. (B) Depletion of PbpX in a strain lacking all native bPBPs. Cells of strain WS056 (Δpbp1a ΔpbpC ΔpbpX ΔpbpY ΔpbpZ Pxyl::Pxyl-pbpX) were grown in PYE medium supplemented with 0.3% xylose, washed, and transferred into PYE medium lacking inducer (t = 0). Samples were taken at the indicated time points and subjected to immunoblot analysis using anti-PbpX antiserum. An exponentially growing culture of wild-type strain CB15N was analyzed as a control (WT). (C) Phenotypic consequences of bPBP depletion. Cells from the samples described for panel B were visualized by DIC microscopy. Bar, 3 μm.
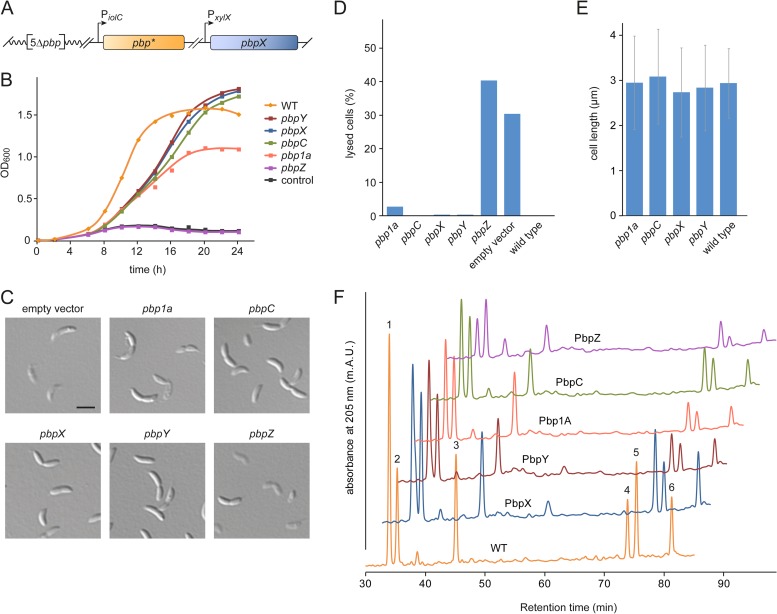
We wondered whether bPBPs other than PbpX were also sufficient to ensure growth and normal cell shape in the absence of their paralogs. To address this question, we used the conditional pbpX mutant lacking all native bPBP-encoding genes and introduced ectopic copies of individual bPBP genes under the control of an inositol-inducible promoter (Fig. 3A). The resulting strains were then cultivated in inositol-containing medium to deplete PbpX and replace it with the bPBP of interest, thereby allowing assessment of the bPBP's ability to complement the loss of PbpX activity. Monitoring growth in batch cultures, we found that cells synthesizing PbpC, PbpX, or PbpY, respectively, as the only bPBP showed very similar growth characteristics, with slightly longer doubling times than the C. crescentus wild type but comparable cell densities in stationary phase (Fig. 3B). Complementation with Pbp1A gave rise to viable cells as well, although the growth yield was significantly lower. In contrast, cells producing only PbpZ were unable to reach high densities, exhibiting the same behavior as control cells lacking an inositol-inducible bPBP gene. To further test the functionality of individual bPBPs, the different strains were analyzed for cell shape defects after 30 h of PbpX depletion in continuous culture (Fig. 3C to E). A control strain lacking an inositol-inducible bPBP gene produced about 30% lysed cells. Upon complementation with PbpC, PbpX, or PbpY, respectively, cells were fully viable and displayed wild-type morphology. Cells producing only Pbp1A also looked normal but showed a tendency to lyse, which may explain the lower cell density reached in batch culture (compare with Fig. 3B). In contrast, expression of pbpZ could not compensate for the loss of PbpX and caused extensive cell lysis. Identical results were obtained upon complementation with fluorescently (Venus-) tagged proteins, confirming the functionality of the fusion constructs (see Fig. S1A, C, and D in the supplemental material). Immunoblot analysis revealed that the Pbp1A fusion accumulated to significantly lower levels than did the PbpC, PbpX, and PbpY fusions (see Fig. S1E), which could explain its impaired ability to support growth in the complementation assay. Moreover, it was clearly possible to detect a signal for Venus-PbpZ, excluding the possibility that the lack of complementation by this construct was due to inefficient gene expression.
FIG 3.
Any bPBP except for PbpZ is sufficient to support cell growth. (A) Assay system for analyzing the functionality of individual bPBPs. In a strain lacking all native bPBP-encoding genes (5Δpbp), a copy of pbpX was placed under the control of the xylose-inducible xylX promoter, while a copy of a second bPBP gene (pbp*) was placed downstream of the inositol-inducible iolC promoter. This setup enabled the depletion of PbpX and concomitant induction of a paralogous bPBP, thereby revealing the ability of individual bPBPs to support cell growth in an otherwise bPBP-free background. (B) Growth behavior of cells synthesizing only a single bPBP. Strains constructed as described for panel A, carrying inositol-inducible copies of pbp1a (WS072), pbpC (WS073), pbpX (WS070), pbpY (WS071), or pbpZ (WS074), were grown in PYE medium containing 0.3% xylose. After washing and transfer of the cells into PYE medium supplemented with 0.3% myo-inositol, the growth of the cultures was followed by monitoring their optical density (OD600). The wild-type strain CB15N (WT) and a mutant (WS076) lacking an inositol-inducible bPBP gene (control) were used as controls. (C) Microscopic analysis of cells expressing only a single bPBP gene. The strains described for panel B were grown in PYE medium containing 0.3% xylose. The cells were washed, transferred into PYE medium supplemented with 0.3% myo-inositol, and analyzed by DIC microscopy after 30 h of growth. The cultures were diluted when necessary to ensure exponential growth throughout the course of the experiment. Bar, 3 μm. (D) Quantification of cell lysis in the cultures described for panel B. At t = 30 h, the fraction of lysed (ghost) cells was determined for each strain (n > 300). Note that the xylX promoter is slightly leaky in the presence of inositol (see Fig. S1B in the supplemental material), explaining the lower fraction of lysed cells than that in Fig. 2C. (E) Average length of the cells in the cultures described for panel B. Measurements were performed at t = 30 h (n = 100 per strain; error bars show standard deviations). (F) Effect of different bPBPs on peptidoglycan composition. Strains synthesizing only a single bPBP were cultivated as described for panel B and harvested after at least 30 h of growth in the absence of xylose. Peptidoglycan was isolated from 10 mg of freeze-dried cell pellet, hydrolyzed by muramidase treatment, and analyzed for the relative ratios of different muropeptides by reversed-phase high-pressure liquid chromatography. Analysis of the peak fractions identified the following muropeptides: 1, GlcNAc-MurNAc-l-Ala-d-Glu-mDAP-d-Ala (Tetra); 2, GlcNAc-MurNAc-l-Ala-d-Glu-mDAP-d-Ala-Gly (PentaGly); 3, GlcNAc-MurNAc-l-Ala-d-Glu-mDAP-d-Ala-d-Ala (Penta); 4, Tetra-PentaGly; 5, Tetra-Tetra; 6, Tetra-Penta.
Our results indicated that each of the bPBPs except for PbpZ was able to support growth in the absence of its paralogs when synthesized under the control of an inducible promoter. However, comparison of the native and induced protein levels showed that PbpY and, in particular, PbpC were significantly overproduced in the complemented strains, although there was little difference for PbpX (see Fig. S2A in the supplemental material). The abundance of the remaining bPBPs could not be tested due to the lack of suitable antibodies. To clarify whether elevated expression levels may be required for complementation, we conducted a more detailed analysis of PbpC. It was not possible to construct a strain that contained only pbpC expressed from the native promoter as the sole source of bPBPs (data not shown), suggesting that PbpC was in fact unable to balance the loss of its paralogs when expressed at wild-type levels. In support of this notion, cells of a mutant (WS090) that lacked all native bPBP genes except for pbpC and carried a conditional allele of pbpX started to lyse after extended growth under nonpermissive conditions, in particular when grown in M2G minimal medium (see Fig. S2B). Thus, while PbpX is sufficient for viability when produced at native levels (Fig. 2A), other bPBPs may require overproduction to support normal growth. Nevertheless, these results suggest that Pbp1A, PbpC, PbpX, and PbpY are, in principle, functionally redundant, whereas PbpZ may have a more specialized biological role.
Despite the overlapping functionality of most bPBPs, it was conceivable that different paralogs might have distinct substrate specificities. For instance, a peculiarity of C. crescentus is the facultative incorporation of glycine instead of d-alanine at the terminal position of the MurNAc pentapeptide side chain (38, 39). To investigate whether the five bPBPs showed preferences for certain types of side chains, we isolated cell walls from strains producing only a single bPBP and analyzed their muropeptide composition (Fig. 3F). For the wild-type control, we obtained the same pattern as described previously (38), with a high proportion of fragments carrying either d-alanine- or glycine-containing pentapeptide side chains (Fig. 3F, peaks 2 and 4). The strain producing only PbpX synthesized a similar amount of peptidoglycan (as defined by the sum of all peak areas) as did the wild-type strain (see Table S5 in the supplemental material), which agrees with the previous finding that this protein represents the most important bPBP in C. crescentus (20). In all other strains, by contrast, peptidoglycan biosynthesis was significantly impaired. Importantly, all mutants showed a reduction in the fraction of tetratetrapeptides (Fig. 3F, peak 5). Since C. crescentus lacks d,d-carboxypeptidase activity (38), this muropeptide species can be generated only through the formation and subsequent cleavage of cross-links. Its lower abundance thus indicates reduced transpeptidase activity, consistent with the observed decrease in overall peptidoglycan content. Apart from that, the percentage of glycine-containing pentapeptides (Fig. 3F, peak 2) was clearly elevated in all mutant strains. However, the ratios of fragments with d-alanine and glycine-containing pentapeptides were largely comparable (except for the PbpZ-producing strain), indicating the lack of major substrate preferences. Of note, PbpX-, PbpY-, and PbpC-producing cells exhibited only slight differences in their muropeptide profiles, which may explain their similar growth behaviors (Fig. 3B and D). The PbpZ-producing strain contained the least peptidoglycan, with the lowest proportion of tetra- and tetratetrapeptides (see Table S5). These results confirm that under the conditions used, PbpZ does not make a major contribution to peptidoglycan biosynthesis, whereas its four paralogs are each able to support growth. However, the full complement of bPBPs appears to be required for normal peptidoglycan content and composition.
PbpX confers resistance to d-alanine.
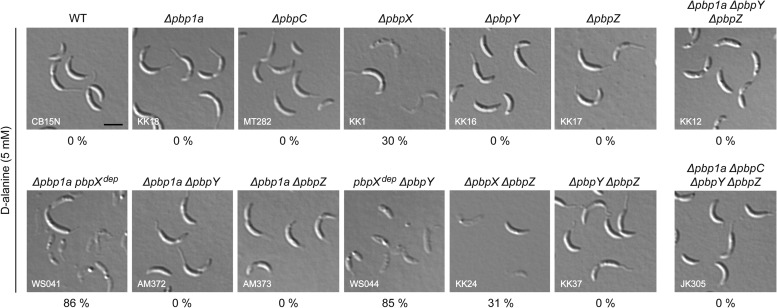
A possible explanation for the existence of multiple redundant bPBP paralogs may be the differential adaptation of individual proteins to distinct environmental conditions, resulting, e.g., in different temperature optima. However, we did not detect any significant differences when comparing the growth of the bPBP-deficient strains at high (37°C) and low (18°C) incubation temperatures (data not shown). Another challenge that cells may frequently face in the environment is d-amino acids, which many bacteria release into the medium in considerable amounts after transition to the stationary growth phase (40). d-Amino acids were shown to significantly affect peptidoglycan biosynthesis, either by replacing the terminal d-alanine residues of the peptide side chains and thus affecting their recognition by peptidoglycan-modifying enzymes or by directly blocking the substrate-binding site of transpeptidases (41, 42). We observed that the C. crescentus wild type was resistant to elevated concentrations of d-alanine. While this was also true for many of the bPBP-deficient strains, all mutants lacking PbpX were prone to cell lysis (Fig. 4). Interestingly, the deleterious effect of d-alanine was more pronounced in the absence of both PbpX and Pbp1A or PbpY. Thus, PbpX appears to be the only bPBP able to cope with elevated d-alanine concentrations, whereas its paralogs are largely inactive under these conditions. Pbp1A and PbpY may retain some residual activity that is not sufficient to support growth on their own but may delay lysis in the absence of PbpX.
FIG 4.
PbpX is required for resistance to d-alanine. Strains carrying the indicated mutations were grown to exponential phase in M2G medium. After addition of 5 mM d-alanine, the cells were cultivated for another 3 h and then visualized by DIC microscopy. The genes labeled with “dep” were not deleted but expressed under the control of the xylX promoter and transcriptionally inactivated by growth of the cells in the absence of inducer for 24 h prior to addition of d-alanine. The percentage of lysed cells in the different cultures is given below the corresponding images (n > 250).
PbpX and PbpY localize to the cell division site.
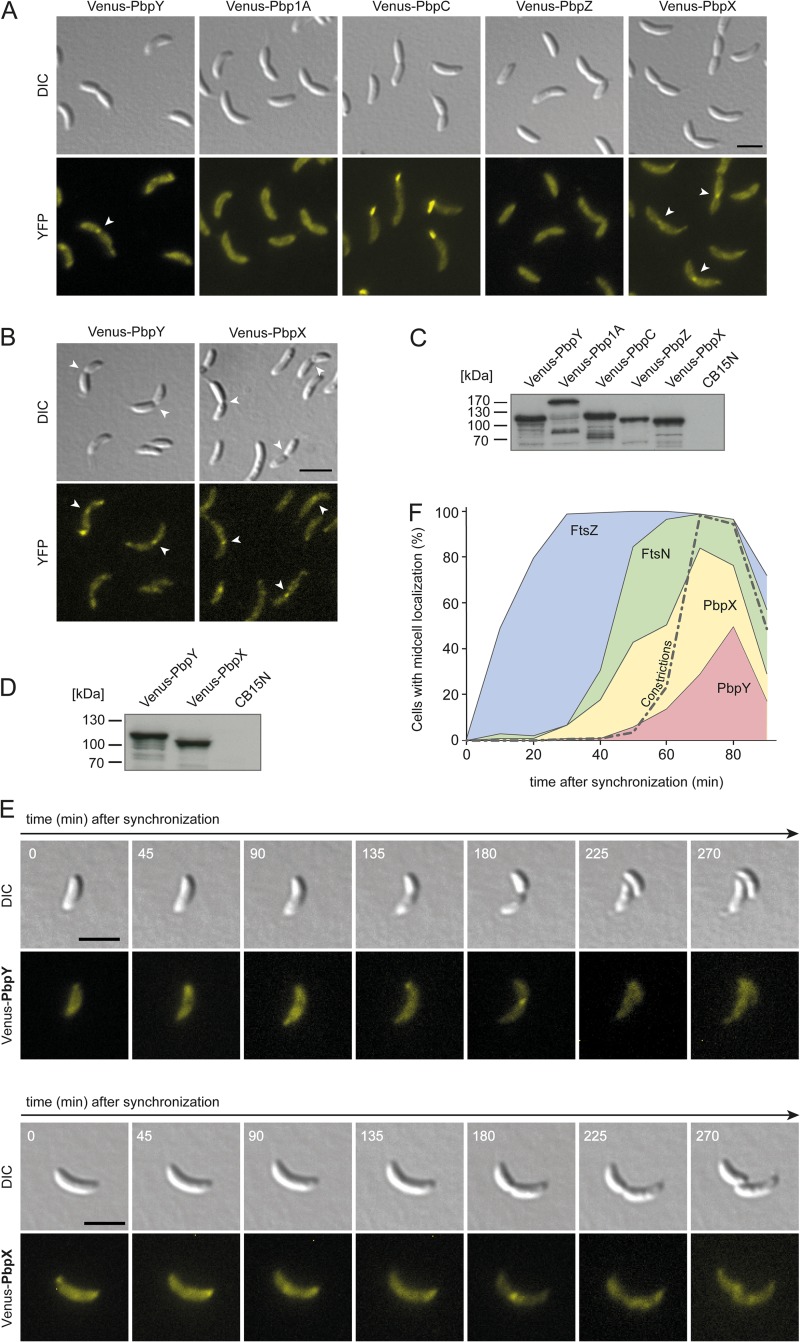
Previous work has shown that bPBPs have distinct localization patterns in C. crescentus (18–20). To analyze their positioning in more detail, we generated derivatives of the C. crescentus wild-type strain producing fluorescent protein (Venus) fusions of individual bPBPs under the control of an inducible promoter. Upon induction, Venus-Pbp1A and Venus-PbpZ were evenly distributed over the cell surface (Fig. 5A), whereas Venus-PbpC accumulated at the stalked cell pole. Interestingly, the PbpX and PbpY fusions were detectable throughout the cell envelope but also formed distinct foci at one or both of the cell poles or at the division site. Similar localization patterns were observed in strains carrying the fusion constructs in place of the respective native genes (Fig. 5B), excluding the possibility that the formation of foci was an artifact resulting from overexpression of the genes. Immunoblot analysis revealed that the fusion proteins were largely stable and accumulated to similar levels (Fig. 5C and D). Interestingly, Venus-Pbp1A displayed an aberrant migration behavior during gel electrophoresis that indicated the formation of stable dimers (Fig. 5C). A similar effect was observed during purification of the proline-rich cytoplasmic tail of Pbp1A (data not shown), suggesting that the long N-terminal extension of Pbp1A mediates self-interaction.
FIG 5.
PbpX and PbpY localize to the division site. (A) Subcellular localization of the five bPBPs of C. crescentus. Derivatives of wild-type strain CB15N expressing venus-pbpY (AM457), venus-pbp1a (KK33), venus-pbpC (MT279), venus-pbpZ (AM458), or venus-pbpX (MT278), respectively, under the control of the xylose-inducible xylX promoter were imaged by DIC and fluorescence microscopy. Bar, 3 μm. YFP, yellow fluorescent protein. (B) Localization of Venus-PbpY and Venus-PbpX in strains carrying the respective hybrid genes in place of native pbpY (WS045) and pbpX (WS055). Bar, 3 μm. (C and D) Immunoblot analysis of the strains shown in panels A and B, respectively, using anti-GFP antiserum. Wild-type strain CB15N served as a negative control. (E) Cell cycle-dependent localization dynamics of Venus-PbpX and Venus-PbpY. Strains carrying venus-pbpX (MT278) or venus-pbpY (AM457) under the control of the xylX promoter were grown to exponential phase in PYE medium and induced for 3 h with 0.3% xylose. After synchronization, swarmer cells were transferred onto M2G agarose pads supplemented with 0.3% xylose and imaged at the indicated time intervals by DIC and fluorescence microscopy. Bar, 3 μm. (F) Timing of the recruitment of PbpX and PbpY to midcell. Swarmer cells of strains producing fluorescent protein fusions of FtsZ and FtsN (AM160), PbpX (MT278), or PbpY (AM457) were transferred into PYE medium and grown for the duration of one cell cycle. Samples were taken at regular intervals and analyzed for the fraction of cells showing a noticeable focus of the indicated protein at midcell (n > 100 for each time point and strain). The quantification of constricted cells is shown for AM160. Similar values were obtained for the other two strains (data not shown).
To determine whether the subcellular localization of PbpX and PbpY varies as a function of the cell cycle, we performed time-lapse analysis on synchronized cells producing an inducible Venus-PbpX or Venus-PbpY fusion, respectively (Fig. 5E). At early time points, each of the two proteins formed patches or foci at one or both of the cell poles. Later on, they additionally formed apparently random accumulations within the cell, before finally condensing at the cell division site. Quantifying these localization dynamics in a large number of cells (Fig. 5F), we found that PbpX starts to accumulate at midcell slightly before or concomitant with the onset of cell constriction. In contrast, PbpY associates with the division site only at the late predivisional stage. These findings suggest that PbpX and PbpY may preferentially be involved in polar morphogenesis and cell division, even though their presence is not essential for these processes (compare with Fig. 3).
The recruitment of PbpX and PbpY to the divisome is dependent on FtsN.
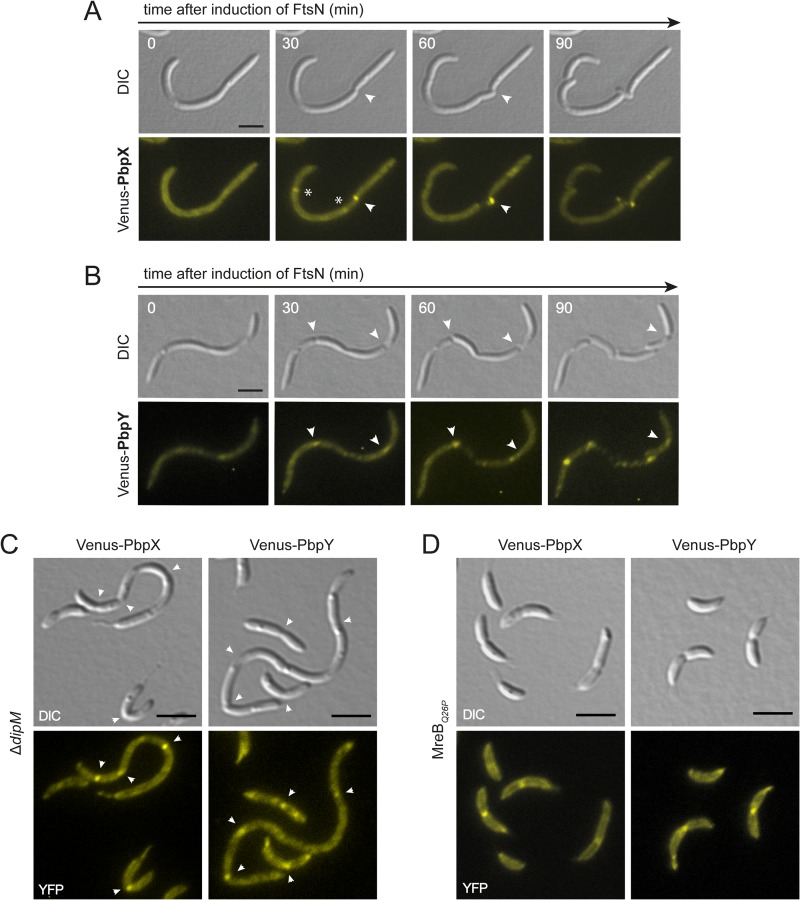
Our localization studies suggest that PbpX and PbpY may interact with the divisome. To gain insight into the underlying mechanism, we analyzed the localization of fluorescently (Venus-) tagged PbpX and PbpY derivatives in a conditional ftsZ mutant. FtsZ polymerizes into a ringlike structure that forms the foundation of the cell division machinery (43, 44). In its absence, none of the other divisome components localizes to midcell (45), leading to the formation of long, smooth filaments (46). We found that Venus-PbpX and Venus-PbpY were evenly distributed in cells depleted of FtsZ. In contrast, when ftsZ expression was reinduced and cell division resumed, fluorescent foci formed at the constriction sites, consistent with a direct role of the divisome in the positioning of the two proteins (see Fig. S3 in the supplemental material). The above time-lapse analyses (Fig. 5F) showed that PbpX and PbpY localized to midcell after FtsN, an essential cell division protein that binds peptidoglycan and has been implicated in the coordination of cell wall remodeling during cytokinesis (9, 47, 48). To test for a role of FtsN in the recruitment of bPBPs, we further analyzed the localization dynamics of the two fusion proteins in a conditional ftsN mutant (Fig. 6A and B). In both cases, the fluorescence signal was evenly distributed in cells depleted of FtsN, whereas foci appeared at the constriction sites after reinduction of ftsN expression. In contrast, Venus-PbpX and Venus-PbpY were still able to localize to the division sites in cells lacking the nonessential cell division protein DipM, a putative peptidoglycan hydrolase that interacts with FtsN and contributes to cell wall invagination (47, 49, 50) (Fig. 6C). Collectively, these results indicate that PbpX and PbpY associate with the division apparatus through direct or indirect interaction with the divisome component FtsN.
FIG 6.
Midcell localization of PbpX and PbpY is dependent on FtsN but independent of MreB. (A) Effect of FtsN depletion on PbpX localization. Cells of strain AM473 (Pvan::Pvan-ftsN ΔftsN Pxyl::Pxyl-venus-pbpX) were cultivated for 12 h in PYE medium to deplete FtsN. Three hours prior to microscopic analysis, expression of venus-pbpX was induced by addition of 0.3% xylose. The cells were then transferred onto an M2G agarose pad supplemented with 0.3% xylose and 0.3% vanillate to reinduce ftsN expression and imaged at the indicated time points. Arrowheads point to constriction sites. Asterisks indicate Venus-PbpX foci at incipient division sites that fade during the course of the experiment, possibly due to photobleaching. Bar, 3 μm. (B) Effect of FtsN depletion on PbpY localization. Cells of strain AM472 (Pvan::Pvan-ftsN ΔftsN Pxyl::Pxyl-venus-pbpY) were treated and analyzed as described for panel A. Bar, 3 μm. (C) DipM-independent localization of PbpX and PbpY to the cell division site. DipM-deficient cells expressing venus-pbpX (WS058) and venus-pbpY (WS057) from the xylose-inducible xylX promoter were imaged by DIC and fluorescence microscopy. Arrowheads point to constriction sites. Bar, 3 μm. (D) MreB-independent localization of PbpX and PbpY to the division site. Cells of an mreBQ26P mutant expressing venus-pbpX (WS084) or venus-pbpY (WS085) under the control of the xylose-inducible xylX promoter were imaged by DIC and fluorescence microscopy. Bar, 3 μm.
Recent work has shown that in E. coli, midcell localization of PBP1B is mediated by the FtsZ-dependent recruitment of MreB to the division apparatus (51). Similarly, the MreB homolog of C. crescentus was found to condense at midcell at the onset of cell division, in a process requiring divisome formation (52, 53). It was therefore conceivable that its relocation could contribute to the cell cycle-regulated localization dynamics of PbpX and PbpY. To test this possibility, we localized fluorescently tagged derivatives of the two proteins in cells synthesizing a mutant form of MreB (MreBQ26P) that no longer accumulates at midcell (17). However, both fusions were still able to form foci at the constriction sites in this background (Fig. 6D), suggesting that their recruitment to midcell occurs independently of the MreB cytoskeleton.
PbpX and PbpY interact with divisome components in vivo.
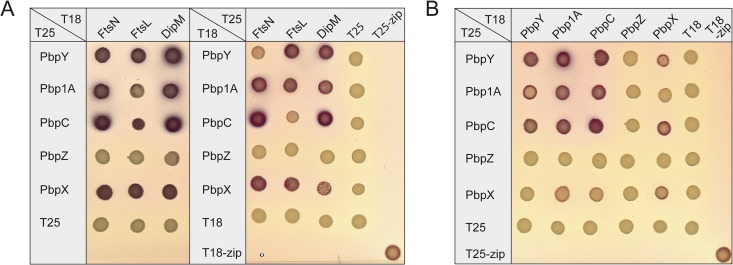
Our results suggest that PbpX and PbpY are targeted to the division site through association with the cell division apparatus. However, given that cells are viable in the absence of these two proteins (compare with Fig. 2A), other bPBPs must be able to functionally replace them during cell division. To identify the divisome components responsible for the recruitment of bPBPs, we tested all five C. crescentus bPBP paralogs for interaction with a variety of cell division proteins using bacterial two-hybrid analysis (31) (data not shown). It turned out that all bPBPs except for PbpZ interacted with FtsN, FtsL, and DipM (Fig. 7A). The precise function of FtsL is still unclear, but its E. coli homolog was shown to interact with FtsN and the divisome-associated monofunctional transpeptidase FtsI (54). These results suggest that bPBPs may be part of a subcomplex within the division machinery that harbors multiple peptidoglycan biosynthetic activities. The lack of interactions with PbpZ may explain the inability of this protein to complement the loss of its paralogs (compare with Fig. 3). Importantly, immunoblot analysis confirmed that the PbpZ hybrids were synthesized and accumulated to approximately the same levels as the corresponding PbpX hybrids, which produced clear interaction signals (see Fig. S4 in the supplemental material). This result excludes the possibility that the lack of interactions for PbpZ was due to inefficient expression or instability of the fusion constructs.
FIG 7.
C. crescentus bPBPs form homo- and heterodimers that interact with the divisome. (A) Bacterial two-hybrid analysis of the interaction between the five C. crescentus bPBPs and selected divisome components. Reporter strain E. coli BTH101 was transformed with plasmids encoding fusions of B. pertussis adenylate cyclase fragments T18 and T25 to the N termini of the indicated bPBPs and divisome components or the yeast GCN4 leucine-zipper region (zip). Transformants were plated on MacConkey agar. Interactions are indicated by the formation of red colonies. In the case of the periplasmic protein DipM, a transmembrane linker (MalG1–77) was inserted between DipM and the T18/T25 fusion partner (47). (B) Bacterial two-hybrid analysis of the interaction between different bPBPs. The T18 and T25 fragments were fused to the N termini of the indicated bPBPs and analyzed as described for panel A.
In E. coli, PBP1A and PBP1B form homodimers but are unable to associate with each other to form heterodimeric complexes (55). To determine the dimerization potential of the C. crescentus homologs, we tested them for mutual interaction in all pairwise combinations using two-hybrid analysis (Fig. 7B). Surprisingly, Pbp1A, PbpC, PbpX, and PbpY were each able to form homodimers as well as heterodimers, consistent with their significant functional overlap. In contrast, PbpZ did not show any dimerization, again suggesting that its cellular role may be distinct from that of its paralogs.
DISCUSSION
Peptidoglycan synthesis is a complex process that requires a delicate balance between synthetic and hydrolytic reactions. To ensure proper cell morphology and cell wall integrity, the enzymatic activities involved need to be tightly regulated in time and space. To this end, cells have evolved a variety of mechanisms, including differential gene expression, the site-specific activation of enzymes, or the targeting of proteins to distinct locations within the cell. Another mechanism to ensure robust cell wall biogenesis is the divergence of peptidoglycan synthases and hydrolases into sets of paralogous proteins that have specialized, yet often overlapping, functions (4, 43). This is also true for bPBPs, which are often found in multiple copies with distinct biological roles. However, in most cases, the precise reason for their multiplicity has so far remained unclear.
Notably, additional bPBP genes can be readily acquired under selective pressure (56). A potential driving force for the diversification of peptidoglycan biosynthetic enzymes may therefore be the need to adapt to specific environmental challenges. For instance, the synthesis of isoforms with distinct catalytic properties may ensure robust peptidoglycan biogenesis over a range of different temperatures, pH values, or salt concentrations. In addition, different paralogs may have acquired resistance against distinct sets of small-molecule inhibitors such as β-lactam antibiotics. Since C. crescentus possesses a potent β-lactamase (57, 58), antibiotic stress may not have been a major driving force for the accumulation of bPBPs, unless the different paralogs can tolerate compounds not hydrolyzed by this enzyme. Moreover, we could not detect any temperature-dependent differences in the growth of bPBP-deficient strains. However, we did observe differential sensitivity to d-alanine, as only PbpX was able to tolerate this metabolite, whereas the other bPBPs were largely inactive in its presence. This finding is consistent with previous results implicating bPBPs in the resistance to d-amino acids that are secreted during stationary phase by various species (40). For instance, a Vibrio cholerae mutant lacking PBP1A was observed to become spherical when exposed to stationary-phase culture supernatants or purified d-amino acids (40). Furthermore, an E. coli pbp1b mutant shows a reduced fitness in stationary phase compared to the wild-type strain (59). C. crescentus does not accumulate d-amino acids in the extracellular milieu. However, it is conceivable that these compounds are produced by surrounding bacteria, especially in the context of stationary-phase biofilms. Interestingly, a previous study demonstrated that among a series of single mutants, only ΔpbpX cells showed a significant reduction in growth rate. This effect was more pronounced for strains with an additional lesion in pbp1a or pbpY (20), reflecting exactly the pattern of d-alanine sensitivity observed in our analyses (Fig. 4). However, the mechanistic basis of this correlation remains unclear.
The different localization patterns of bPBPs in C. crescentus suggest that they have acquired specialized cellular functions. Whereas PbpZ is evenly distributed throughout the cell envelope, Pbp1A shows a patchy pattern that may reflect its involvement in MreB-dependent lateral cell wall elongation. PbpC, in contrast, forms a distinct complex at the stalked pole, while it is evenly distributed throughout the remaining parts of the cell. Although its deletion does not affect growth of the cell body, it significantly reduces stalk length (19), suggesting a specialized function in stalk biogenesis. PbpX and PbpY, on the other hand, show complex cell cycle-dependent localization patterns. In swarmer and early stalked cells, they form polar foci as well as patches with an apparently random distribution along the cell, thus probably contributing to polar morphogenesis and lateral cell wall elongation. At later stages of the cell cycle, both proteins condense at the division site, where they may contribute to formation of the new cell poles during cytokinesis. The foci formed by Venus-PbpX were, in general, more distinct and brighter than those of Venus-PbpY, suggesting that PbpX is the major bPBP involved in cell division. This notion is consistent with the previous finding that inactivation of PbpX, but not PbpY, leads to a strong synthetic growth defect in strains with impaired FtsI activity (20). Despite the apparent functional diversification of bPBPs in C. crescentus, any of the five paralogs except for PbpZ is, in principle, sufficient to support growth, although in some cases only upon overproduction. Thus, each of them must have retained the ability to interact with the elongasome, the cell division apparatus, and the machinery responsible for stalk biogenesis. The different localization patterns may reflect different affinities for the target complexes and/or interactions with specific localization factors. Indeed, midcell localization of Venus-PbpX and Venus-PbpY was much less apparent in the absence of other bPBPs (see Fig. S1 in the supplemental material), suggesting that a lack of competing paralogs leads to redistribution of the proteins to peptidoglycan biosynthetic complexes with low-affinity binding sites. In contrast, PbpC is recruited to the stalked pole through interaction with the bactofilin cytoskeleton (19) and thus retains its polar localization irrespective of the genetic background. Its entrapment in a stable polar complex may normally prevent productive interaction with the cell division or elongation machinery. However, when overproduced, PbpC may be able to saturate the binding capacity of the polar bactofilin cluster, thereby increasing the pool of free protein available for noncognate interactions.
What could be the advantage of having multiple redundant bPBPs that are localized to distinct positions within the cell? It is conceivable that the substrate binding affinities or reaction kinetics of the different paralogs have been fine-tuned for function in specific biosynthetic complexes. In addition, the existence of task-specific bPBPs may allow the cell to regulate distinct growth modes in a mutually independent fashion. For instance, the cell cycle-regulated polar localization of PbpC specifically promotes stalk biogenesis after the swarmer-to-stalked-cell transition (19). Similarly, the observed relocation of Pbp1A to midcell after an osmotic upshift may enhance the robustness of cell division under stress conditions without affecting other growth processes (18). Last, but not least, bPBPs may have evolved additional functions that require them to be positioned at certain cellular locations, as exemplified by the role of PbpC as a localization factor for the stalk-specific protein StpX (60).
Consistent with their functional redundancy, all bPBPs except for PbpZ were found to interact with the divisome components FtsN, FtsL, and DipM by two-hybrid analysis. Moreover, each of them is able to support cell division, although only PbpX and PbpY specifically accumulate at the division site. The condensation of bPBPs at midcell may thus be dispensable for normal cytokinesis, even though it may render the constriction process more robust. Similar observations have previously been made for FtsN (48, 61) and DipM (47, 49, 50), which remain fully active even if their accumulation at midcell is abolished by deletion of their peptidoglycan-binding domains. Our findings underscore the importance of FtsN as a central hub that recruits both synthetic (bPBPs) and hydrolytic (AmiC and DipM) (47) peptidoglycan-modifying enzymes to the division site. However, additional studies are required to verify the interactions observed in this work and exclude artifacts due to nonspecific aggregation or the formation of complexes with divisome components of the E. coli two-hybrid host strain. Overall, the interactions between bPBPs and the divisome in C. crescentus are reminiscent of those observed for E. coli PBP1B. It was shown that PBP1B associates with FtsN (9) as well as FtsI (8), a transpeptidase proposed to be adjacent to FtsL in the divisome complex (54), with both interactions leading to a stimulation of its enzymatic activity. In addition, PBP1B also interacts with a peptidoglycan hydrolase, namely, the lytic transglycosylase MltA, although this protein belongs to a different class of enzymes than does DipM. However, PBP1B requires the outer membrane lipoprotein LpoB for activity, which is not conserved outside the enteric bacteria (10, 11). Therefore, despite similarities in the interplay with core divisome components, the mechanisms controlling the activity of bPBPs during cell division may be different in C. crescentus. The same must be true for the regulation of bPBPs in the elongasome complex, as the PBP1A activator LpoA is also missing in this species. Notably, PbpZ is unable to functionally replace its four paralogs, reminiscent of PBP1C in E. coli (13). However, whereas PBP1C can associate with cell division proteins and other bPBPs (13, 62), no such interactions were detectable for PbpZ, suggesting that this protein has functionally diverged from its paralogs. Therefore, either PbpZ may act in a thus-far-unknown pathway that is induced only under certain environmental conditions, or it may have lost its bifunctionality and exclusively act as a monofunctional transglycosylase or transpeptidase, consistent with the presence of mutations in the respective signature motifs. Collectively, our results suggest that basic features of the paradigmatic peptidoglycan biosynthetic machinery of E. coli are conserved in C. crescentus. However, additional enzymes and new regulatory pathways have been implemented to adjust the system to the specific needs of this species.
Supplementary Material
ACKNOWLEDGMENTS
We thank Nicole Schnass, Juliane Kühn, and Susan Schlimpert for the construction of strains and plasmids; Christine Jacobs-Wagner and Daniel Ladant for providing material; Julia Rosum for expert technical assistance; and members of the Waldor lab for helpful discussions.
This work was supported by the Max Planck Society and the LOEWE Center for Synthetic Biology. W.S. received a fellowship from the International Max Planck Research School (IMPRS) for Environmental, Cellular and Molecular Microbiology.
Footnotes
Published ahead of print 14 February 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.01194-13.
REFERENCES
- 1.Typas A, Banzhaf M, Gross CA, Vollmer W. 2012. From the regulation of peptidoglycan synthesis to bacterial growth and morphology. Nat. Rev. Microbiol. 10:123–136. 10.1038/nrmicro2677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lovering AL, de Castro LH, Lim D, Strynadka NC. 2007. Structural insight into the transglycosylation step of bacterial cell-wall biosynthesis. Science 315:1402–1405. 10.1126/science.1136611 [DOI] [PubMed] [Google Scholar]
- 3.Schwartz B, Markwalder JA, Seitz SP, Wang Y, Stein RL. 2002. A kinetic characterization of the glycosyltransferase activity of Escherichia coli PBP1b and development of a continuous fluorescence assay. Biochemistry 41:12552–12561. 10.1021/bi026205x [DOI] [PubMed] [Google Scholar]
- 4.Sauvage E, Kerff F, Terrak M, Ayala JA, Charlier P. 2008. The penicillin-binding proteins: structure and role in peptidoglycan biosynthesis. FEMS Microbiol. Rev. 32:234–258. 10.1111/j.1574-6976.2008.00105.x [DOI] [PubMed] [Google Scholar]
- 5.Ropp PA, Nicholas RA. 1997. Cloning and characterization of the ponA gene encoding penicillin-binding protein 1 from Neisseria gonorrhoeae and Neisseria meningitidis. J. Bacteriol. 179:2783–2787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Banzhaf M, van den Berg van Saparoea B, Terrak M, Fraipont C, Egan A, Philippe J, Zapun A, Breukink E, Nguyen-Disteche M, den Blaauwen T, Vollmer W. 2012. Cooperativity of peptidoglycan synthases active in bacterial cell elongation. Mol. Microbiol. 85:179–194. 10.1111/j.1365-2958.2012.08103.x [DOI] [PubMed] [Google Scholar]
- 7.den Blaauwen T, de Pedro MA, Nguyen-Disteche M, Ayala JA. 2008. Morphogenesis of rod-shaped sacculi. FEMS Microbiol. Rev. 32:321–344. 10.1111/j.1574-6976.2007.00090.x [DOI] [PubMed] [Google Scholar]
- 8.Bertsche U, Kast T, Wolf B, Fraipont C, Aarsman ME, Kannenberg K, von Rechenberg M, Nguyen-Disteche M, den Blaauwen T, Höltje JV, Vollmer W. 2006. Interaction between two murein (peptidoglycan) synthases, PBP3 and PBP1B, in Escherichia coli. Mol. Microbiol. 61:675–690. 10.1111/j.1365-2958.2006.05280.x [DOI] [PubMed] [Google Scholar]
- 9.Müller P, Ewers C, Bertsche U, Anstett M, Kallis T, Breukink E, Fraipont C, Terrak M, Nguyen-Disteche M, Vollmer W. 2007. The essential cell division protein FtsN interacts with the murein (peptidoglycan) synthase PBP1B in Escherichia coli. J. Biol. Chem. 282:36394–36402. 10.1074/jbc.M706390200 [DOI] [PubMed] [Google Scholar]
- 10.Paradis-Bleau C, Markovski M, Uehara T, Lupoli TJ, Walker S, Kahne DE, Bernhardt TG. 2010. Lipoprotein cofactors located in the outer membrane activate bacterial cell wall polymerases. Cell 143:1110–1120. 10.1016/j.cell.2010.11.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Typas A, Banzhaf M, van den Berg van Saparoea B, Verheul J, Biboy J, Nichols RJ, Zietek M, Beilharz K, Kannenberg K, von Rechenberg M, Breukink E, den Blaauwen T, Gross CA, Vollmer W. 2010. Regulation of peptidoglycan synthesis by outer-membrane proteins. Cell 143:1097–1109. 10.1016/j.cell.2010.11.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yousif SY, Broome-Smith JK, Spratt BG. 1985. Lysis of Escherichia coli by beta-lactam antibiotics: deletion analysis of the role of penicillin-binding proteins 1A and 1B. J. Gen. Microbiol. 131:2839–2845 [DOI] [PubMed] [Google Scholar]
- 13.Schiffer G, Höltje JV. 1999. Cloning and characterization of PBP 1C, a third member of the multimodular class A penicillin-binding proteins of Escherichia coli. J. Biol. Chem. 274:32031–32039. 10.1074/jbc.274.45.32031 [DOI] [PubMed] [Google Scholar]
- 14.Goffin C, Ghuysen JM. 1998. Multimodular penicillin-binding proteins: an enigmatic family of orthologs and paralogs. Microbiol. Mol. Biol. Rev. 62:1079–1093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lazaro S, Fernandez-Pinas F, Fernandez-Valiente E, Blanco-Rivero A, Leganes F. 2001. pbpB, a gene coding for a putative penicillin-binding protein, is required for aerobic nitrogen fixation in the cyanobacterium Anabaena sp. strain PCC7120. J. Bacteriol. 183:628–636. 10.1128/JB.183.2.628-636.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Budd A, Blandin S, Levashina EA, Gibson TJ. 2004. Bacterial alpha2-macroglobulins: colonization factors acquired by horizontal gene transfer from the metazoan genome? Genome Biol. 5:R38. 10.1186/gb-2004-5-6-r38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Aaron M, Charbon G, Lam H, Schwarz H, Vollmer W, Jacobs-Wagner C. 2007. The tubulin homologue FtsZ contributes to cell elongation by guiding cell wall precursor synthesis in Caulobacter crescentus. Mol. Microbiol. 64:938–952. 10.1111/j.1365-2958.2007.05720.x [DOI] [PubMed] [Google Scholar]
- 18.Hocking J, Priyadarshini R, Takacs CN, Costa T, Dye NA, Shapiro L, Vollmer W, Jacobs-Wagner C. 2012. Osmolality-dependent relocation of penicillin-binding protein PBP2 to the division site in Caulobacter crescentus. J. Bacteriol. 194:3116–3127. 10.1128/JB.00260-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kühn J, Briegel A, Mörschel E, Kahnt J, Leser K, Wick S, Jensen GJ, Thanbichler M. 2010. Bactofilins, a ubiquitous class of cytoskeletal proteins mediating polar localization of a cell wall synthase in Caulobacter crescentus. EMBO J. 29:327–339. 10.1038/emboj.2009.358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yakhnina AA, Gitai Z. 2013. Diverse functions for six glycosyltransferases in Caulobacter crescentus cell wall assembly. J. Bacteriol. 195:4527–4535. 10.1128/JB.00600-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Poindexter JS. 1964. Biological properties and classification of the Caulobacter group. Bacteriol. Rev. 28:231–295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ely B. 1991. Genetics of Caulobacter crescentus. Methods Enzymol. 204:372–384. 10.1016/0076-6879(91)04019-K [DOI] [PubMed] [Google Scholar]
- 23.Meisenzahl AC, Shapiro L, Jenal U. 1997. Isolation and characterization of a xylose-dependent promoter from Caulobacter crescentus. J. Bacteriol. 179:592–600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Boutte CC, Srinivasan BS, Flannick JA, Novak AF, Martens AT, Batzoglou S, Viollier PH, Crosson S. 2008. Genetic and computational identification of a conserved bacterial metabolic module. PLoS Genet. 4:e1000310. 10.1371/journal.pgen.1000310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thanbichler M, Iniesta AA, Shapiro L. 2007. A comprehensive set of plasmids for vanillate- and xylose-inducible gene expression in Caulobacter crescentus. Nucleic Acids Res. 35:e137. 10.1093/nar/gkm818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tsai JW, Alley MR. 2001. Proteolysis of the Caulobacter McpA chemoreceptor is cell cycle regulated by a ClpX-dependent pathway. J. Bacteriol. 183:5001–5007. 10.1128/JB.183.17.5001-5007.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Botstein K, Lew KK, Jarvik V, Swanson CA. 1975. Role of antirepressor in the bipartite control of repression and immunity by bacteriophage P22. J. Mol. Biol. 91:439–462. 10.1016/0022-2836(75)90271-5 [DOI] [PubMed] [Google Scholar]
- 28.Thanbichler M, Shapiro L. 2006. MipZ, a spatial regulator coordinating chromosome segregation with cell division in Caulobacter. Cell 126:147–162. 10.1016/j.cell.2006.05.038 [DOI] [PubMed] [Google Scholar]
- 29.Domian IJ, Quon KC, Shapiro L. 1997. Cell type-specific phosphorylation and proteolysis of a transcriptional regulator controls the G1-to-S transition in a bacterial cell cycle. Cell 90:415–424. 10.1016/S0092-8674(00)80502-4 [DOI] [PubMed] [Google Scholar]
- 30.Robichon C, Karimova G, Beckwith J, Ladant D. 2011. Role of leucine zipper motifs in association of the Escherichia coli cell division proteins FtsL and FtsB. J. Bacteriol. 193:4988–4992. 10.1128/JB.00324-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Karimova G, Pidoux J, Ullmann A, Ladant D. 1998. A bacterial two-hybrid system based on a reconstituted signal transduction pathway. Proc. Natl. Acad. Sci. U. S. A. 95:5752–5756. 10.1073/pnas.95.10.5752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.de Jonge BL, Chang YS, Gage D, Tomasz A. 1992. Peptidoglycan composition of a highly methicillin-resistant Staphylococcus aureus strain. The role of penicillin binding protein 2A. J. Biol. Chem. 267:11248–11254 [PubMed] [Google Scholar]
- 33.Schultz J, Milpetz F, Bork P, Ponting CP. 1998. SMART, a simple modular architecture research tool: identification of signaling domains. Proc. Natl. Acad. Sci. U. S. A. 95:5857–5864. 10.1073/pnas.95.11.5857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Punta M, Coggill PC, Eberhardt RY, Mistry J, Tate J, Boursnell C, Pang N, Forslund K, Ceric G, Clements J, Heger A, Holm L, Sonnhammer EL, Eddy SR, Bateman A, Finn RD. 2012. The Pfam protein families database. Nucleic Acids Res. 40:D290–D301. 10.1093/nar/gkr1065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. 2007. Clustal W and Clustal X version 2.0. Bioinformatics 23:2947–2948. 10.1093/bioinformatics/btm404 [DOI] [PubMed] [Google Scholar]
- 36.Vollmer W, Bertsche U. 2008. Murein (peptidoglycan) structure, architecture and biosynthesis in Escherichia coli. Biochim. Biophys. Acta 1778:1714–1734. 10.1016/j.bbamem.2007.06.007 [DOI] [PubMed] [Google Scholar]
- 37.McGrath PT, Lee H, Zhang L, Iniesta AA, Hottes AK, Tan MH, Hillson NJ, Hu P, Shapiro L, McAdams HH. 2007. High-throughput identification of transcription start sites, conserved promoter motifs and predicted regulons. Nat. Biotechnol. 25:584–592. 10.1038/nbt1294 [DOI] [PubMed] [Google Scholar]
- 38.Markiewicz Z, Glauner B, Schwarz U. 1983. Murein structure and lack of DD- and LD-carboxypeptidase activities in Caulobacter crescentus. J. Bacteriol. 156:649–655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Takacs CN, Hocking J, Cabeen MT, Bui NK, Poggio S, Vollmer W, Jacobs-Wagner C. 2013. Growth medium-dependent glycine incorporation into the peptidoglycan of Caulobacter crescentus. PLoS One 8:e57579. 10.1371/journal.pone.0057579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lam H, Oh DC, Cava F, Takacs CN, Clardy J, de Pedro MA, Waldor MK. 2009. D-amino acids govern stationary phase cell wall remodeling in bacteria. Science 325:1552–1555. 10.1126/science.1178123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cava F, de Pedro MA, Lam H, Davis BM, Waldor MK. 2011. Distinct pathways for modification of the bacterial cell wall by non-canonical D-amino acids. EMBO J. 30:3442–3453. 10.1038/emboj.2011.246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cava F, Lam H, de Pedro MA, Waldor MK. 2011. Emerging knowledge of regulatory roles of D-amino acids in bacteria. Cell. Mol. Life Sci. 68:817–831. 10.1007/s00018-010-0571-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Egan AJ, Vollmer W. 2013. The physiology of bacterial cell division. Ann. N. Y. Acad. Sci. 1277:8–28. 10.1111/j.1749-6632.2012.06818.x [DOI] [PubMed] [Google Scholar]
- 44.Lutkenhaus J, Pichoff S, Du S. 2012. Bacterial cytokinesis: from Z ring to divisome. Cytoskeleton (Hoboken) 69:778–790. 10.1002/cm.21054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Goley ED, Yeh YC, Hong SH, Fero MJ, Abeliuk E, McAdams HH, Shapiro L. 2011. Assembly of the Caulobacter cell division machine. Mol. Microbiol. 80:1680–1698. 10.1111/j.1365-2958.2011.07677.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang Y, Jones BD, Brun YV. 2001. A set of ftsZ mutants blocked at different stages of cell division in Caulobacter. Mol. Microbiol. 40:347–360. 10.1046/j.1365-2958.2001.02395.x [DOI] [PubMed] [Google Scholar]
- 47.Möll A, Schlimpert S, Briegel A, Jensen GJ, Thanbichler M. 2010. DipM, a new factor required for peptidoglycan remodelling during cell division in Caulobacter crescentus. Mol. Microbiol. 77:90–107. 10.1111/j.1365-2958.2010.07224.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Möll A, Thanbichler M. 2009. FtsN-like proteins are conserved components of the cell division machinery in proteobacteria. Mol. Microbiol. 72:1037–1053. 10.1111/j.1365-2958.2009.06706.x [DOI] [PubMed] [Google Scholar]
- 49.Goley ED, Comolli LR, Fero KE, Downing KH, Shapiro L. 2010. DipM links peptidoglycan remodelling to outer membrane organization in Caulobacter. Mol. Microbiol. 77:56–73. 10.1111/j.1365-2958.2010.07222.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Poggio S, Takacs CN, Vollmer W, Jacobs-Wagner C. 2010. A protein critical for cell constriction in the Gram-negative bacterium Caulobacter crescentus localizes at the division site through its peptidoglycan-binding LysM domains. Mol. Microbiol. 77:74–89. 10.1111/j.1365-2958.2010.07223.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fenton AK, Gerdes K. 2013. Direct interaction of FtsZ and MreB is required for septum synthesis and cell division in Escherichia coli. EMBO J. 32:1953–1965. 10.1038/emboj.2013.129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Figge RM, Divakaruni AV, Gober JW. 2004. MreB, the cell shape-determining bacterial actin homologue, co-ordinates cell wall morphogenesis in Caulobacter crescentus. Mol. Microbiol. 51:1321–1332. 10.1111/j.1365-2958.2003.03936.x [DOI] [PubMed] [Google Scholar]
- 53.Gitai Z, Dye N, Shapiro L. 2004. An actin-like gene can determine cell polarity in bacteria. Proc. Natl. Acad. Sci. U. S. A. 101:8643–8648. 10.1073/pnas.0402638101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Karimova G, Dautin N, Landant D. 2005. Interaction network among Escherichia coli membrane proteins involved in cell division as revealed by bacterial two-hybrid analysis. J. Bacteriol. 187:2233–2243. 10.1128/JB.187.7.2233-2243.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Charpentier X, Chalut C, Remy MH, Masson JM. 2002. Penicillin-binding proteins 1a and 1b form independent dimers in Escherichia coli. J. Bacteriol. 184:3749–3752. 10.1128/JB.184.13.3749-3752.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zapun A, Contreras-Martel C, Vernet T. 2008. Penicillin-binding proteins and beta-lactam resistance. FEMS Microbiol. Rev. 32:361–385. 10.1111/j.1574-6976.2007.00095.x [DOI] [PubMed] [Google Scholar]
- 57.Simm AM, Higgins CS, Pullan ST, Avison MB, Niumsup P, Erdozain O, Bennett PM, Walsh TR. 2001. A novel metallo-beta-lactamase, Mbl1b, produced by the environmental bacterium Caulobacter crescentus. FEBS Lett. 509:350–354. 10.1016/S0014-5793(01)03152-0 [DOI] [PubMed] [Google Scholar]
- 58.West L, Yang D, Stephens C. 2002. Use of the Caulobacter crescentus genome sequence to develop a method for systematic genetic mapping. J. Bacteriol. 184:2155–2166. 10.1128/JB.184.8.2155-2166.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pepper ED, Farrell MJ, Finkel SE. 2006. Role of penicillin-binding protein 1b in competitive stationary-phase survival of Escherichia coli. FEMS Microbiol. Lett. 263:61–67. 10.1111/j.1574-6968.2006.00418.x [DOI] [PubMed] [Google Scholar]
- 60.Hughes HV, Lisher JP, Hardy GG, Kysela DT, Arnold RJ, Giedroc DP, Brun YV. 2013. Coordinate synthesis and protein localization in a bacterial organelle by the action of a penicillin-binding-protein. Mol. Microbiol. 90:1162–1170. 10.1111/mmi.12422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gerding MA, Liu B, Bendezu FO, Hale CA, Bernhardt TG, de Boer PA. 2009. Self-enhanced accumulation of FtsN at division sites and roles for other proteins with a SPOR domain (DamX, DedD, and RlpA) in Escherichia coli cell constriction. J. Bacteriol. 191:7383–7401. 10.1128/JB.00811-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Vollmer W, von Rechenberg M, Höltje JV. 1999. Demonstration of molecular interactions between the murein polymerase PBP1B, the lytic transglycosylase MltA, and the scaffolding protein MipA of Escherichia coli. J. Biol. Chem. 274:6726–6734. 10.1074/jbc.274.10.6726 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.