Abstract
LapA and LapF are large extracellular proteins that play a relevant role in biofilm formation by Pseudomonas putida. Current evidence favors a sequential model in which LapA is first required for the initial adhesion of individual bacteria to a surface, while LapF participates in later stages of biofilm development. In agreement with this model, lapF transcription was previously shown to take place at late times of growth and to respond to the stationary-phase sigma factor RpoS. We have now analyzed the transcription pattern of lapA and other regulatory elements that influence expression of both genes. The lapA promoter shows a transient peak of activation early during growth, with a second increase in stationary phase that is independent of RpoS. The same pattern is observed in biofilms although expression is not uniform in the population. Both lapA and lapF are under the control of the two-component regulatory system GacS/GacA, and their transcription also responds to the intracellular levels of the second messenger cyclic diguanylate (c-di-GMP), although in surprisingly reverse ways. Whereas expression from the lapA promoter increases with high levels of c-di-GMP, the opposite is true for lapF. The transcriptional regulator FleQ is required for the modulation of lapA expression by c-di-GMP but has a minor influence on lapF. This work represents a further step in our understanding of the regulatory interactions controlling biofilm formation in P. putida.
INTRODUCTION
One of the strategies that allow microorganisms to thrive in different habitats is the formation of surface-associated multicellular communities called biofilms (1). Intensive research on diverse bacterial species has provided an inventory of molecular determinants that participate in the process of bacterial colonization of surfaces and biofilm development. However, the regulatory networks that connect environmental and cellular signals with the transition from planktonic to sessile growth and then modulate the sequence of events in the different stages of biofilm formation are less well defined and can vary from one species to another (2).
In a variety of bacteria, changes in the intracellular levels of the secondary messenger cyclic dimeric guanosine phosphate (c-di-GMP) participate in the control of that switch between lifestyles (3, 4). Increasing the intracellular concentration of c-di-GMP by the overexpression of diguanylate cyclase (DGC) enzymes leads to increased attachment and reduced biofilm dispersal, as well as to variations in the global expression pattern (5, 6). In Pseudomonas fluorescens, intracellular levels of the secondary messenger c-di-GMP contribute to the molecular mechanism that controls the release from the cell surface of the large adhesin LapA (7), an essential element for biofilm development by P. fluorescens and Pseudomonas putida (8, 9).
Several two-component signal transduction systems consisting of a sensor histidine kinase and a transcriptional regulator have also been related to biofilm regulation in different ways and in diverse bacteria: ArsIRS in Staphylococcus epidermidis (10); CpxRA in Escherichia coli (11); and PprBA, PilRS, FleRS, RcsBC, and GacSA in Pseudomonas aeruginosa (12, 13). The GacS/GacA two-component system is known to regulate processes such as the expression of virulence factors and the synthesis of secondary metabolites with antimicrobial activity, as well as social behaviors (14, 15). In response to an as-yet-unidentified environmental signal, GacS activates by phosphotransfer the transcriptional regulator GacA, which subsequently triggers expression of genes corresponding to small RNAs. This results in a regulatory cascade that involves derepression of the translation of target genes. In Azotobacter vinelandii this system regulates the synthesis of alginate (16), an exopolysaccharide that is part of the extracellular matrix of biofilms in different bacteria. In P. fluorescens, it participates in the regulatory network controlling flagellar motility (17).
The Gram-negative bacterium Pseudomonas putida KT2440 is an efficient colonizer of the root system of different plants that activates induced systemic resistance in the plant and can be used in rhizoremediation (18, 19, 20). P. putida KT2440 is also able to develop biofilms on biotic and abiotic surfaces, and molecular determinants participating in these processes have been described. They include surface proteins, exopolysaccharides (EPS), and elements involved in the synthesis, regulation, and functionality of the flagellar system (8, 9, 21–25). Two very large secreted proteins, LapA and LapF, have been recognized as significant players with different roles (8, 22). We have proposed a sequential model for P. putida biofilm formation in which the surface protein LapA is mainly involved in the first stages of biofilm formation, facilitating the irreversible cell-surface interaction; later on, LapF mediates cell-cell interactions, providing support for microcolony formation and maturation of the biofilm (22). Activity of the lapF promoter is consistent with this role of LapF since it is activated at the beginning of the stationary phase of growth and at advanced stages of biofilm development and is dependent on the alternative, stationary-phase sigma factor RpoS (22).
In this work, we have analyzed the transcription pattern of lapA and the influence of different global regulators on expression of both adhesins. The results significantly increase our understanding of biofilm formation in P. putida KT2440 and add essential details to the current model of the regulatory connections that drive this process.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Strains and plasmids used in this work are listed in Table 1. Pseudomonas putida KT2440 is a plasmid-free derivative of P. putida mt-2, originally isolated from a vegetable orchard in Japan (26). Strains mus-20 (lapF mutant), mus-42 (lapA mutant), and mus-69 (fleQ mutant), which were obtained by random transposon mutagenesis with mini-Tn5 Km1 and identified as defective in attachment to corn seeds, have been previously described (9, 21). C1R1 is an rpoS null derivative of KT2440 (27). A gacS mutant of KT2440 was obtained by random transposon mutagenesis with mini-Tn5 (′luxCDABE Km2) (28) and identified after selection of mutants deficient in biofilm formation on abiotic surfaces. Escherichia coli DH5α was used as a host for cloning.
TABLE 1.
Strains and plasmids used in this work
| Strain or plasmid | Description | Source or reference |
|---|---|---|
| P. putida strains | ||
| KT2440 | Wild type | Lab stock |
| mus-20 | lapF mutant (mini-Tn5 Km1) | 21 |
| mus-42 | lapA mutant (mini-Tn5 Km1) | 9 |
| mus-69 | fleQ mutant (mini-Tn5 Km1) | 9 |
| MM2 | gacS mutant (mini-Tn5 luxCDABE Km) | This work |
| C1R1 | rpoS null mutant | 27 |
| E. coli strains | ||
| DH5α | λ− ϕ80dlacZΔM15 Δ(lacZYA-argF)U196 recA1 endA1 hsdR17(rK− mK−) supE44 thi-1 gyrA relA1 | Lab stock |
| MKW909 | S17-1 λpir (pUT mini-Tn5 ′luxCDABE Km2) | 28 |
| Plasmids | ||
| pMMG1 | lapF::lacZ transcriptional fusion, Tcr | 22 |
| pMMGA | lapA::lacZ transcriptional fusion, Tcr | 32 |
| pMMG2 | lapF::gfp transcriptional fusion, Gmr | 22 |
| pMMG5 | lapA::gfp transcriptional fusion, Gmr | This work |
| pMMG6 | lapA::mCherry transcriptional fusion, Tcr | This work |
| pMAMV1 | Diguanylate cyclase gene rup4959 in pBBR1MCS5, Gmr | 31 |
| pMAMV21 | rpoS::lacZ translational fusion in pMP220, Tcr | 31 |
| pMP220 | Promoter probe vector, promoterless lacZ, Tcr | 30 |
| pRU1097 | Promoter probe vector, promoterless gfp, Gmr | 29 |
Plasmids pRU1097 (29) and pMP220 (30) were used to construct pMMG5 (PlapA::gfp) and pMMG6 (PlapA::mCherry) as detailed below. Plasmids pMMG1 (PlapF::lacZ), pMMG2 (PlapF::gfp), pMMGA (PlapA::lacZ), pMAMV21 harboring an rpoS::lacZ translational fusion, and pMAMV1 carrying the diguanylate cyclase-encoding gene rup4959 have been described elsewhere (22, 31, 32). E. coli was grown at 37°C in LB medium (33). P. putida was grown at 30°C in either LB or M9 minimal medium with MgSO4, Fe citrate, and trace metals (9) with glucose (20 mM) as a carbon source. When appropriate, antibiotics were used at the following concentrations: kanamycin (Km), 25 μg/ml; tetracycline (Tc), 10 μg/ml; gentamicin (Gm), 10 μg/ml (for E. coli) or 100 μg/ml (for P. putida).
Plasmid construction.
To generate plasmid pMMG5 (PlapA::gfp), a 403-bp PCR fragment containing the lapA promoter region was obtained with primers LapAFwPstI (TACGGCTGCAGAGGTGTATG) and LapARevKpnI (CAGGCGGGTACCTTCGATA). The PCR product was digested with PstI and KpnI and cloned in vector pRU1097, which harbors a promoterless gfp derivative. Plasmid pMMG6 was obtained by overlapping PCR amplification of the lapA promoter and the coding sequence of mCherry from Bacillus subtilis DR-40 (34) with primers FwLapAmch (TTTTTTTGAATTCTGTCGAGTAAGTCGGTCGCG), RvLapAmch (ATCCTCGCCCTTGCTCACCATCGGACCGGTGAGCACTTCCTC), mcherryFWD (ATGGTGAGCAAGGGCGAGGAT), and mcherryREV (TTTTTTGGTACCTTACTTGTACAGCTCGTC), followed by digestion with EcoRI and Acc65I and cloning in the same sites of pMP220. This plasmid was used to allow for dual incorporation of plasmids harboring green fluorescent protein (GFP; derived from pRU1097) and mCherry fusions in the same cell. Absence of mutations was confirmed in all of the constructs by sequencing.
Biofilm formation analysis.
Biofilm formation was examined during growth in polystyrene microtiter plates (Sterilin) or in borosilicate glass tubes, without medium replacement, as described previously (9). Biomass attached to the surface was visually inspected by crystal violet (0.4%) staining and quantified by solubilizing the dye with 70% ethanol and measuring the absorbance at 580 nm (35). Microscopy analysis of biofilm formation under static conditions was done by obliquely placing 40- by 20-mm glass coverslips into the wells of a six-well plate where cultures were allowed to grow in 1:10 LB medium at 30°C. Visualization was done on a Zeiss Axioscope fluorescence microscope with appropriate filter sets. A Nikon C-1 microscope was used for confocal laser scanning microscopy (CLSM) at the Microscopy Service (Estación Experimental del Zaidín).
Assays for β-galactosidase activity.
β-Galactosidase activity was measured as described before (36). Overnight cultures were inoculated (1:100 dilution) in fresh LB medium and grown for 1.5 h; cultures were diluted 1:1 three times (every half hour) before the start of sample collection. These steps were done to ensure proper dilution of β-galactosidase accumulated after overnight growth. No detectable differences were observed in planktonic growth of the strains. Cultures harboring pMAMV1 were vortexed in the presence of glass beads (diameter, 0.5 mm) to disrupt aggregates that appeared as a consequence of high levels of c-di-GMP (31). Experiments were repeated at least three times with two technical repeats per sample, and data are given in Miller units.
RNA extraction and preparation of cDNA.
Total RNA from exponentially growing cells in LB medium was extracted with TRI reagent (Ambion) as recommended by the manufacturer, except that TriPure Isolation Reagent was preheated at 70°C. RNA was pretreated with RNase-free DNase I (Roche) plus RNaseOUT (Invitrogen), followed by purification with RNeasy columns (Qiagen) and a second DNase I treatment to remove DNA traces with a Turbo DNA-free kit (Ambion). Reverse transcription reactions to generate the corresponding cDNA were performed with 1 μg of RNA using SuperScript II Reverse Transcriptase (Invitrogen) with random hexamers as primers, according to the manufacturer's protocol.
qRT-PCR.
Primers used for quantitative real-time PCR (qRT-PCR) analyses were as follows (5′-3′ sequences): LAPAQF (ATGAGCAGCGTTGTAGCC) and LAPAQR (TGCTACTGTCAGGCGCAT) for analysis of lapA and LAPFQF (CCATGGACAACATCGTCG) and LAPFQR (CCACGGCGAAGAAGTTAC) for analysis of lapF. The PCR products were 220 and 204 bp, respectively. 16S rRNA was used as an internal control for normalization, using previously described primers (31). Real-time PCR amplification was carried out on a MyiQ2 system (Bio-Rad) associated with iQ5 Optical System Software (version 2.1.97.1001). Each 25-μl reaction mixture contained 12.5 μl of iQ SYBR green Supermix (100 mM KCl, 40 mM Tris-HCl, pH 8.4, 0.4 mM each deoxynucleoside triphosphate [dNTP], 50 U ml−1 iTaq DNA polymerase, 6 mM MgCl2, SYBR green I, 20 nM fluorescein, and stabilizers) (Bio-Rad) and 2 μl of template cDNA (diluted 1- or 1,000-fold). Thermal cycling conditions were as follows: one cycle at 95°C for 5 min and then 40 cycles at 95°C for 30 s, 65°C for 30 s, and 72°C for 20 s, with a single fluorescence measurement per cycle, according to the manufacturer's recommendations. A final extension cycle (at 72°C for 1 min) was performed. Melting curve analysis was performed by gradually heating the PCR mixture from 55 to 95°C at a rate of 0.5°C per 10 s for 80 cycles. The results were analyzed by means of the comparative threshold cycle (ΔΔCT) method (37) to determine the relative expression of each gene in the mutants with respect to the wild type.
Flow cytometry.
Cells of P. putida KT2440 were grown in LB medium at 30°C in a six-well plate with a 40- by 20-mm glass coverslip placed obliquely into the well. At different time points, cells attached to the coverslip (biofilm cells) were recovered as follows: the coverslip was washed twice with sterile phosphate-buffered saline (PBS) buffer and then introduced in a 50-ml tube with 25 ml of PBS buffer; cells were detached with three repetitive passes of 30 s of mild sonication (20% power) with an UltraSonics sonicator equipped with a small-tip probe. For the analysis of planktonic cells, 2 ml of culture from the well was taken. Cells obtained from either the wells or the coverslips were centrifuged at 13,000 rpm for 15 min, and the pellets were fixed in 4% paraformaldehyde for 7 min. After fixation, cells were washed with PBS, resuspended in GTE buffer (50 mM glucose, 20 mM Tris-HCl at pH 8, 10 mM EDTA at pH 8), and stored at 4°C. Prior to flow cytometric analysis, cells were subjected to mild sonication (two rounds of 10 1-s pulses at 20% power), conditions that remove cells from any extracellular matrix but do not cause cell lysis at detectable levels (38). This procedure gave a preparation of single cells. For flow cytometric analysis, cells were diluted in PBS and directly measured on a BD LSR II flow cytometer (BD Biosciences) with solid-state lasers at 405/488/594 nm. For each sample, at least 30,000 events were analyzed. Data from the fluorescent signals were collected using 505-nm long-pass (LP) and 530/30-nm band-pass (BP) filters, with the photomultiplier voltage set between 300 and 500 V. Data were captured using FACS Diva software (BD Biosciences) and further analyzed using FlowJo, version 8.5.2, software (Ashland, OR).
RESULTS
The lapA promoter is active at early and late stages of growth.
In order to test if gene expression data support the sequential model of LapA and LapF activity proposed previously (22), the expression pattern of lapA was studied in P. putida KT2440 using plasmid pMMGA, which carries an lapA::lacZ transcriptional fusion. Expression from the lapA promoter was followed during growth in liquid LB medium. As shown in Fig. 1, there was a peak of β-galactosidase activity at early time points, followed by a decrease until the culture entered the stationary phase, when activity increased again. The early transient peak of activity was consistently observed when sampling was done with sufficient frequency during the first hours but was easily missed when periods between samples were longer. We have previously reported that the lapF promoter is dependent on RpoS, which is active in stationary phase and at late times during biofilm formation (22). Data for the β-galactosidase activity of an lapF::lacZ fusion are included in Fig. 1 for comparison. We considered the possibility that RpoS influenced expression of the lapA promoter in stationary phase, as is the case for lapF. However, when plasmid pMMGA was introduced in strain C1R1, an rpoS mutant derivative of KT2440 (27), no differences in expression levels were observed with respect to the wild type (see Fig. S1 in the supplemental material).
FIG 1.
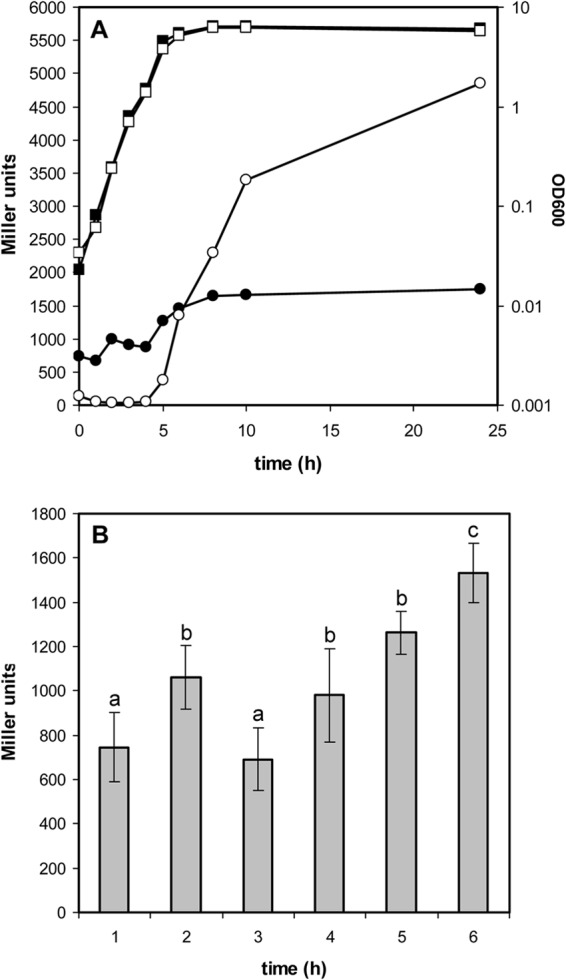
Expression profile of the lapA promoter. (A) β-Galactosidase activity (filled circles) of an lapA::lacZ fusion during growth (filled squares) of KT2440(pMMGA) in liquid LB medium, showing a transient peak during early exponential phase and a later rise in stationary phase. Expression (open circles) of an lapF::lacZ fusion during growth (open squares) of KT2440(pMMG1) is shown for comparison. (B) Detailed analysis of expression of the lapA::lacZ fusion in the first hours of growth. Data are averages and standard deviations of four experiments with duplicate samples. Statistically significant differences between time points are indicated by different letters on top of the bars, based on an analysis of variance test with Tukey's posttest. OD600, optical density at 600 nm.
Expression of lapA was then examined in biofilms grown under static conditions on glass coverslips. For that purpose, an lapA::gfp transcriptional fusion was constructed in plasmid pRU1097, yielding pMMG5, which was then introduced in P. putida KT2440. Expression was followed during biofilm formation by fluorescence microscopy (Fig. 2). In agreement with the β-galactosidase activity results, expression of the lapA::gfp fusion was evident at very early times (30 min after inoculation), with every cell attached to the coverslip showing green fluorescence even though the intensity was not uniform. During the following stages of biofilm development, a decrease in fluorescence intensity was observed, followed by a significant increase that remained after 24 h although at this time not all the cells on the coverslip showed fluorescence.
FIG 2.
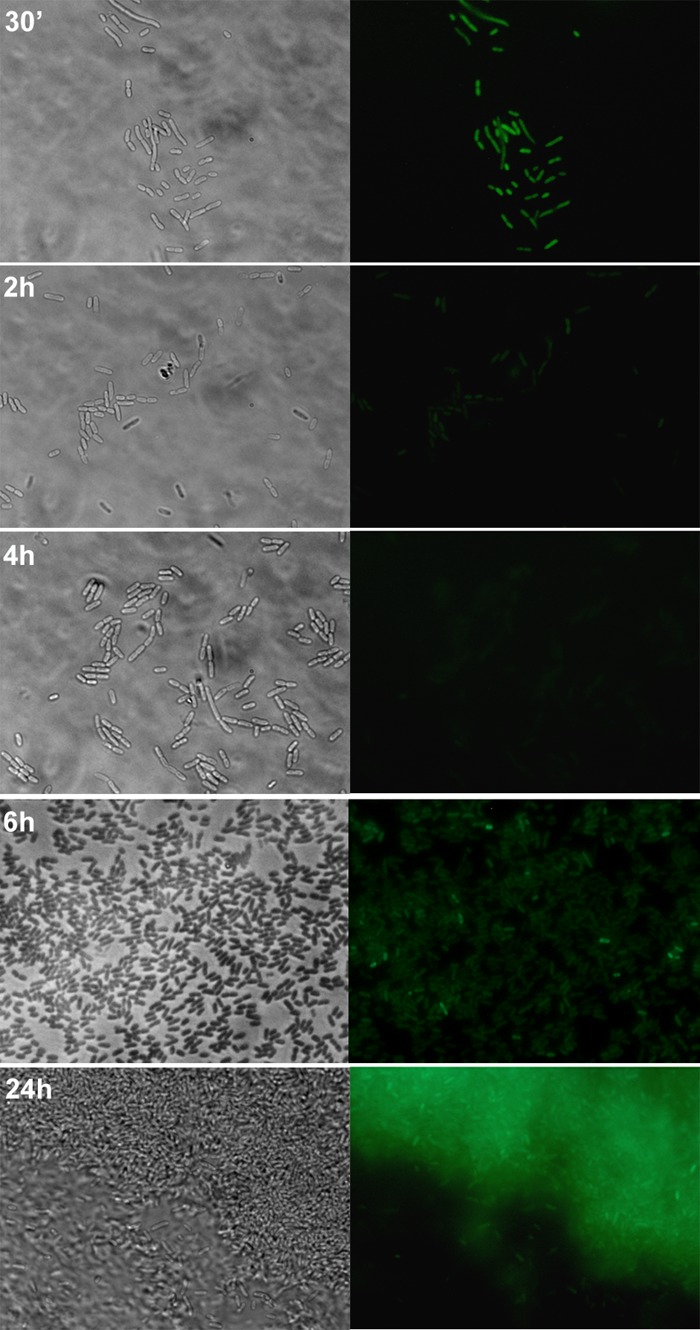
Expression of lapA::gfp in biofilms of KT2440 harboring pMMG5. Cells were grown on glass coverslips under static conditions as detailed in Materials and Methods. The same field on the coverslip was examined by phase-contrast (left panels) and fluorescence (right panels) microscopy. Magnification, ×1,000 (30 min, 2 h, 4 h, and 6 h) or ×400 (24 h).
Heterogeneous populations of cells expressing lapA or lapF within a biofilm.
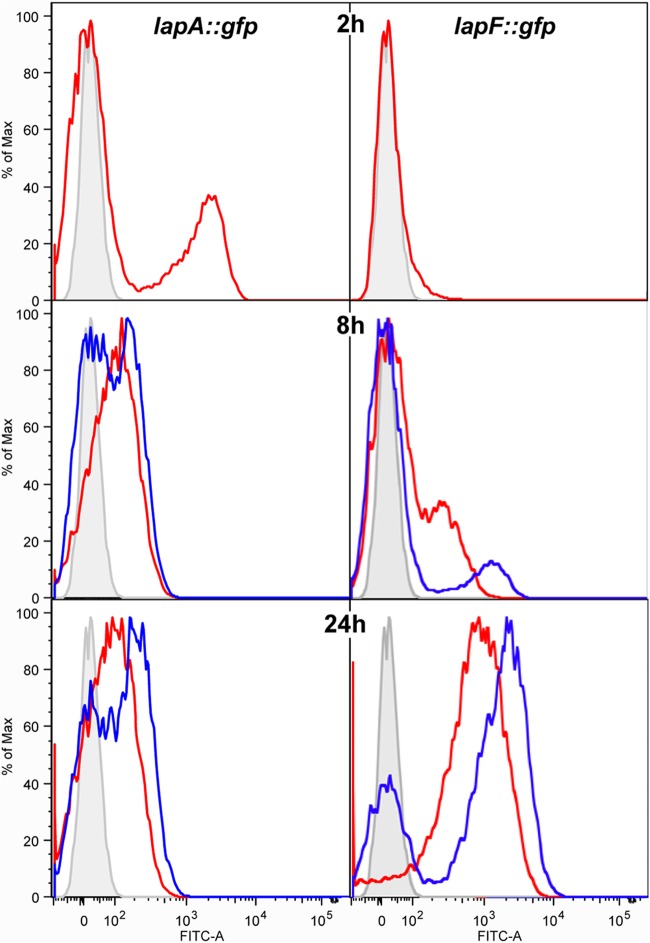
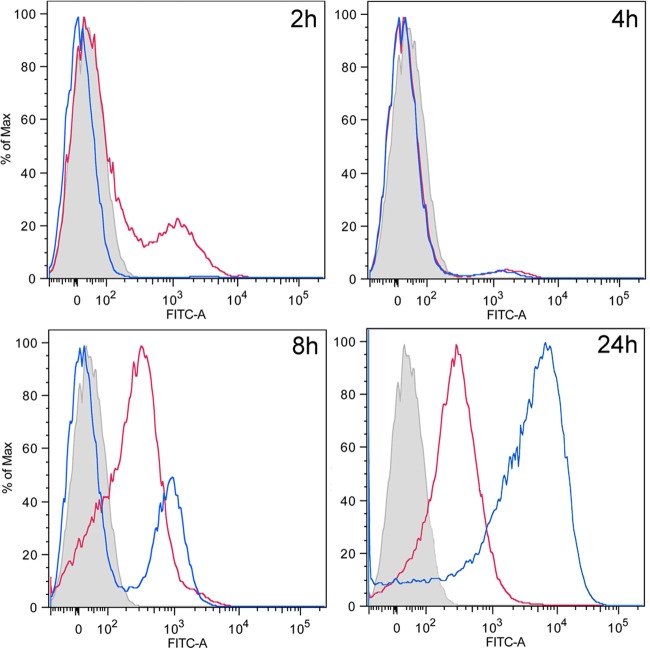
The results of fluorescence microscopy revealed heterogeneity in the expression of lapA within a biofilm, especially at 24 h. Thus, we decided to follow the population dynamics of cells of P. putida KT2440 expressing lapA::gfp (pMMG5) and lapF::gfp (pMMG2) using flow cytometry. Cells were grown under static conditions in six-well plates with an immersed coverslip. At different time points cells were harvested from the wells (planktonic cells) and the coverslips (biofilm cells) as described in Materials and Methods. Results show that after 2 h of incubation, a small subpopulation of planktonic cells were expressing the lapA fusion, while expression of lapF was undetectable (Fig. 3). At this time, the number of cells attached to the coverslip was insufficient for analysis by flow cytometry. Small subpopulations of cells expressing lapF were detected at 4 and 6 h in both coverslips and wells (data not shown). These subpopulations increased at 8 h (Fig. 3). At this time the overall intensity of the signal of planktonic cells expressing lapA was slightly higher than that of the negative control, suggesting that the majority of the population showed some basal expression of lapA. However, on the coverslip two subpopulations could be discerned, one expressing lapA and the other matching the negative control, thus indicating lack of expression (Fig. 3). This situation remained invariable over 24 h, with little change in the expression of lapA (Fig. 3). At 24 h, the expression of lapF increased considerably, not only in the number of cells wherein fluorescence was observed but also in the intensity of the fluorescence signal. Although two subpopulations could be differentiated in the coverslip at this time, one expressing lapF and the other showing no expression, similar to the negative control, the entire planktonic population was expressing lapF, even if the intensity was lower than that of the subpopulation on the coverslip. It is noteworthy that the intensity of lapF expression showed an increase over time, while the lapA promoter presented its highest activity at early time points.
FIG 3.
Flow cytometry analysis of lapA::gfp and lapF::gfp expression in planktonic and biofilm populations. The y axis represents relative cell counts. The x axis shows arbitrary fluorescence units in a logarithmic scale. Red lines correspond to planktonic cells recovered from the well, and blue lines correspond to cells recovered from the surface of the coverslip. The gray peak corresponds to KT2440 cells with no gfp reporter and is shown as a nonfluorescent negative control to identify populations of gfp-harboring cells with no above-background fluorescence. Shown are results from samples taken after 2, 8, and 24 h. FITC, fluorescein isothiocyanate; Max, maximum.
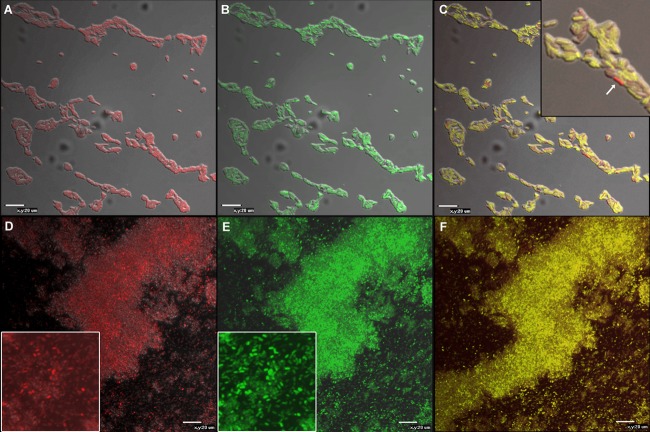
These data were further explored by confocal laser scanning microscopy in biofilms of KT2440 harboring plasmids pMMG1 (lapF::gfp) and pMMG6 (containing an lapA::mCherry transcriptional fusion) (see Materials and Methods for details), grown as described in the previous section, to determine if different subpopulations were “specialized” in expression of one or the other fusion construct. As shown in Fig. 4, most cells in the attached population expressed both fusions at 6 and 24 h although the lapF::gfp fusion shows an overall higher intensity. Some cells expressing one of the fusions with significantly more intensity than the other could be observed, but in general no clear specialization was evident under these conditions. Heterogeneous fluorescence intensity was observed for both fusions, but it was more manifest for the lapA::mCherry fusion.
FIG 4.
Confocal microscopy analysis of biofilms of KT2440 harboring both an lapA::mCherry and an lapF::gfp fusion. Biofilms were observed after 6 h (A, B, and C) or 24 h (D, E, and F) of growth. Fluorescence and pseudo-Nomarski images were combined to detect cells expressing lapA (A and D), lapF (B and E), or both (C and F). Scale bar, 20 μm. In panel F, output from the red channel was increased to correct for the higher fluorescence of the lapF fusion at this time. Insets are close-ups to show heterogeneity at the individual cell level more clearly. The arrow in the panel C inset points to an example of high lapA expression at 6 h.
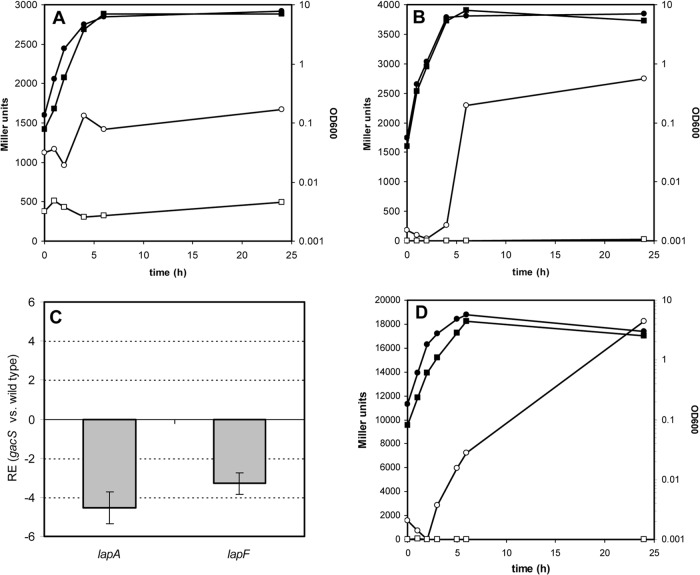
A mutant in the sensor kinase GacS is affected in biofilm formation and shows reduced expression of lapA and lapF.
The involvement of the two-component system GacS/GacA in biofilm formation by P. putida has been recently reported (25). Independently of these results, a P. putida mutant affected in gacS was obtained after random transposon mutagenesis and selection of mutants showing reduced biofilm formation in an effort to unveil regulators of this process. As shown in Fig. 5, the gacS mutant showed reduced attachment during growth in glass tubes or on microtiter plates. In the latter, the wild type showed the characteristic dynamic of attachment during growth, followed by detachment due to nutrient/oxygen depletion (9, 22), whereas the mutant showed very little attached biomass throughout the experiment (Fig. 5B). No differences in planktonic growth levels were observed between the two strains (data not shown).
FIG 5.
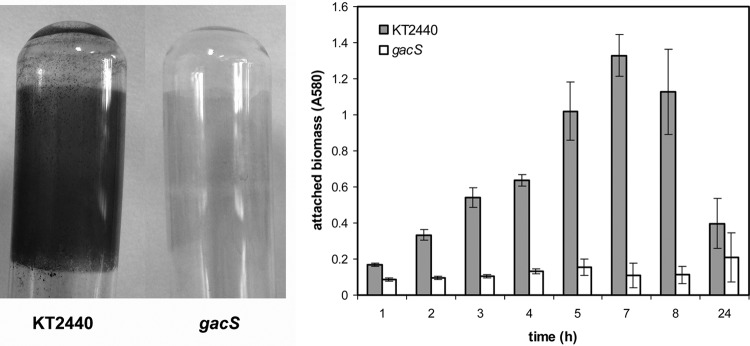
A gacS mutant is impaired in biofilm formation. The left panel shows attachment of the wild-type KT2440 and the gacS mutant strains to borosilicate glass after 4 h of growth in LB medium under orbital shaking, assessed by staining with crystal violet. The right panel shows data for biofilm formation during static growth in LB medium in microtiter plates. Data correspond to averages and standard deviations from three experiments with duplicate samples. Differences between the wild type and the mutant are statistically significant at all time points except at 24 h (Student's t test, P ≤ 0.05).
Since LapA and LapF are the main structural components involved in P. putida biofilm formation, we decided to test if this defect might be due to altered expression of lapA and/or lapF. For that purpose, pMMGA and pMMG1 were introduced in the gacS mutant, and β-galactosidase assays were performed. As shown in Fig. 6, expression of both promoters is affected in similar but not identical ways in the mutant. A clear reduction in the level of activity of the lapA::lacZ fusion was observed, and no increase could be detected in stationary phase (Fig. 6A). Expression of lapF::lacZ, on the other hand, was nearly abolished in the gacS mutant (Fig. 6B). Similar results were obtained when the gfp reporter fusions were introduced in a gacA mutant, where no fluorescence was detected in either case (data not shown), indicating that the effect of GacS on lapA and lapF is via its cognate response regulator GacA.
FIG 6.
Expression of lapA and lapF is regulated by the Gac system. Growth (filled symbols) and β-galactosidase activity (open symbols) of KT2440 (circles) and the gacS mutant (squares) harboring the lapA::lacZ fusion in pMMGA (A) and the lapF::lacZ fusion in pMMG1 (B). Experiments were repeated twice with duplicate samples, and results of a representative experiment are shown. (C) Analysis of the relative expression (RE) of lapA and lapF in the gacS mutant versus KT2440 by qRT-PCR. (D) Growth (filled symbols) and β-galactosidase activity (open symbols) of KT2440 (circles) and the gacS mutant (squares) harboring an rpoS::lacZ fusion in pMAMV21.
These results were further validated by quantitative real-time PCR (qRT-PCR). As shown in Fig. 6C, the relative amounts of mRNAs of lapA and lapF were significantly reduced in the gacS mutant with respect to the wild type. The apparent divergences between β-galactosidase activity and the qRT-PCR results in the case of lapF reflect the stage at which RNA samples were taken (exponential phase, when lapF expression is not maximal in the wild type).
The alternative sigma factor σs (RpoS) is positively regulated by the two-component system GacS/GacA in P. fluorescens Pf-5 (39). Given that the expression of lapF is under the control of this sigma factor (22), we checked rpoS expression in the gacS mutant using plasmid pMAMV21, which harbors an rpoS::lacZ translational fusion (31). Expression of rpoS was abolished in the mutant (Fig. 6D), indicating that GacS/GacA regulate rpoS in P. putida and suggesting that the influence of the two-component system on lapF is indirect, via RpoS.
High intracellular levels of c-di-GMP influence transcription of lapA and lapF through different pathways.
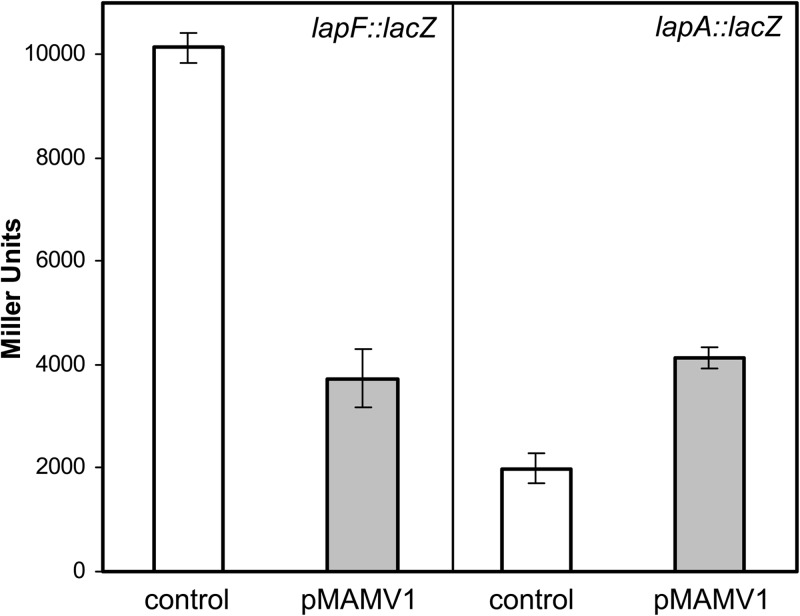
It has been recently shown that the response regulator Rup4959 of P. putida KT2440 contains GGDEF/EAL domains and that when the gene is present in multicopy, it confers high levels of intracellular c-di-GMP due to the diguanylate cyclase activity of the protein (31). As a consequence, a variety of phenotypic effects are apparent, including enhanced biofilm formation, wrinkly colonies, pellicle formation at the air-liquid interphase, EPS overproduction, and cellular clumping. Given that the levels of c-di-GMP influence gene expression, in particular, of genes involved in EPS production and biofilm formation (5, 40), we next studied if a high intracellular level of this secondary messenger could have an influence on the transcriptional regulation of lapA or lapF. For this purpose pMMG1 or pMMGA was introduced in KT2440 harboring pMAMV1 (rup4959 in multicopy), and β-galactosidase activity was studied. We observed variations in the expression intensities of both promoters (Fig. 7). Surprisingly, each was affected in different ways. In the presence of high levels of c-di-GMP, the expression of lapA was around 2-fold higher than physiological concentrations of the secondary messenger. In contrast, the expression of lapF was negatively affected by high levels of c-di-GMP.
FIG 7.
Influence of high levels of c-di-GMP on expression of lapA and lapF. Plasmid pMAMV1, carrying the diguanylate cyclase gene rup4959, was introduced in KT2440 harboring pMMGA (lapA::lacZ) or pMMG1 (lapF::lacZ), and β-galactosidase activity was measured after overnight growth in LB medium. Data correspond to averages and standard deviations of three experiments with duplicate samples, and differences between each strain harboring pMAMV1 and their respective control are statistically significant (Student's t test, P ≤ 0.05).
In a recent work, the flagellar regulator FleQ has been reported to modulate expression of EPS genes in P. aeruginosa through its interaction with c-di-GMP (41). A fleQ mutant of P. putida KT2440 shows reduced attachment to plant seeds and biofilm formation on abiotic surfaces (9). Analysis of expression of the lapA::lacZ and lapF::lacZ fusions in this mutant (Fig. 8) revealed that FleQ functions as a positive regulator of lapA, while it has little effect on lapF (only a detectable but minor increase in β-galactosidase activity was observed in the mutant at 24 h). Similar results were obtained by qRT-PCR, showing a clear reduction in the relative expression of lapA in the fleQ mutant and a slight increase (less than 2-fold) in the relative expression of lapF (Fig. 8C). When plasmid pMAMV1 was introduced in the fleQ mutant, the increase in activity observed for the lapA::lacZ fusion in the wild-type strain harboring this plasmid was lost, indicating that FleQ is required for c-di-GMP-dependent activation of the lapA promoter (Fig. 8). In contrast, in the case of lapF, there was a reduction in β-galactosidase activity in the mutant with high c-di-GMP levels similar to that previously observed in the wild type, suggesting that the negative effect of the second messenger is not through FleQ. Analysis of the sequence of the promoter region of lapA revealed the existence of a potential recognition site for FleQ (see Fig. S2 in the supplemental material), but no similar site was detected in the lapF promoter (not shown).
FIG 8.
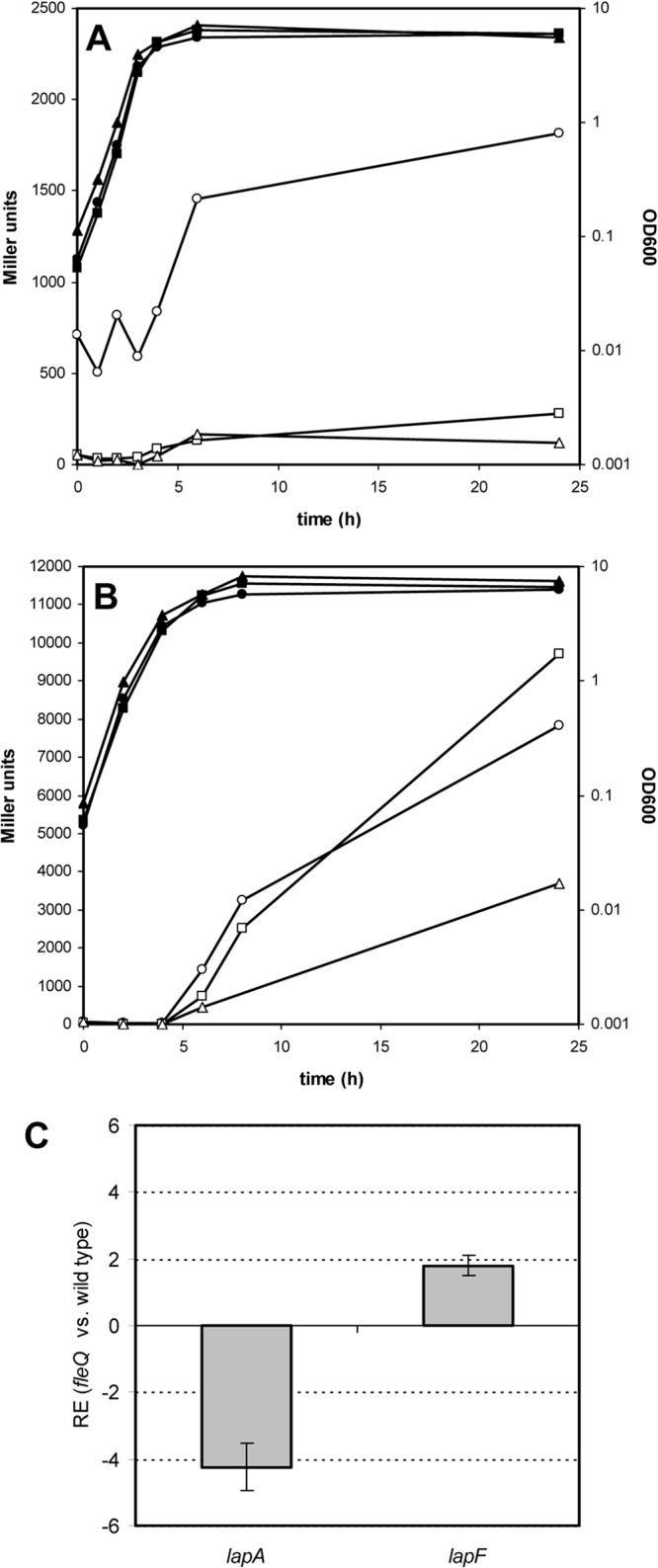
Growth (filled symbols) and β-galactosidase activity (open symbols) of KT2440 (circles), the fleQ mutant mus-69 (squares), and mus-69 with pMAMV1 (triangles) harboring the lapA::lacZ fusion in pMMGA (A) and the lapF::lacZ fusion in pMMG1 (B). Experiments were repeated three times with duplicate samples, and results of a representative experiment are shown. (C) Analysis of the relative expression (RE) of lapA and lapF in the fleQ mutant versus KT2440 by qRT-PCR.
Lack of LapA induces changes in lapF expression.
We have recently reported that mutations in lapA or lapF resulted in increased expression and production of EPS (32). To determine if such an effect could also exist between the two adhesins, expression of their promoters in either mutant was analyzed with plasmids pMMGA and pMMG1. A significant reduction in β-galactosidase activity was detected for the lapF::lacZ fusion in the lapA mutant after 24 h (Fig. 9). This effect seems to be independent of the alterations observed in high levels of c-di-GMP since the increase in lapA expression and the decrease in lapF expression in the presence of pMAMV1 were similar in all of the strains (see Fig. S3 in the supplemental material).
FIG 9.
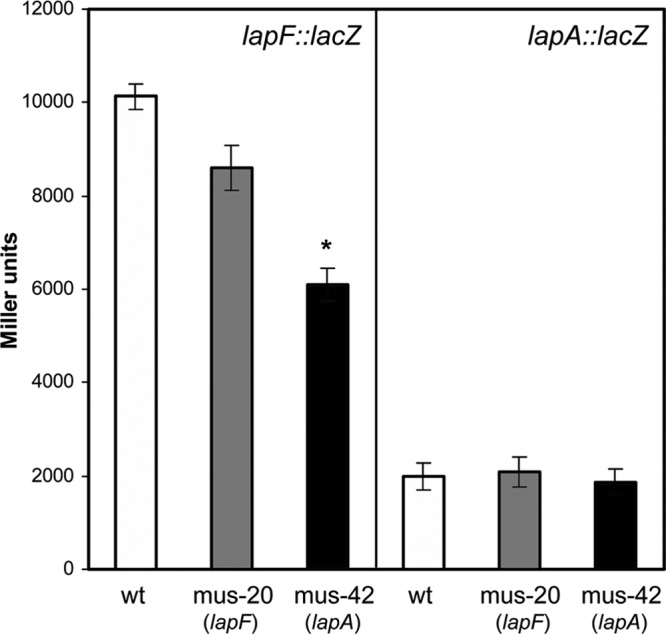
Expression of lapF::lacZ is reduced in an lapA mutant. Plasmid pMMGA or pMMG1 was introduced in KT2440 (white bars), mus-20 (lapF mutant; gray bars), and mus-42 (lapA mutant; black bars), and β-galactosidase activity was measured after overnight growth in LB medium. Results are averages and standard deviations from three experiments with duplicate samples. The asterisk indicates statistically significant differences relative to the wild type (wt) (Student's t test, P ≤ 0.05).
These results were further explored using flow cytometry. Plasmids pMMG5 (lapA::gfp) and pMMG2 (lapF::gfp) were introduced in the mus-42 and mus-20 mutants, and cells were grown on six-well plates under static conditions with a coverslip in each well. At different time points cells were harvested from the culture in the well or from the coverslips and treated for analysis as described in Materials and Methods. It was not possible to recover sufficient cells from the coverslip in the lapA mutant (mus-42), given that this mutant is impaired in biofilm formation. For this mutant, we compared the expression levels only in cells recovered from the culture in the well. Expression of lapF in mus-20 (lapF mutant) and expression of lapA in either mutant did not vary compared to the wild-type levels (data not shown). On the other hand, as shown in Fig. 10, lapF expression in the lapA mutant showed a clearly altered pattern. A new subpopulation of cells expressing lapF, absent in the wild type, appeared at 2 h. At 4 h the two strains behaved identically, with only a very small subpopulation showing expression of lapF. At 8 h, we observed a shift of the entire mus-42 population, indicating lapF expression, whereas in the wild type only a subpopulation of cells expressed the lapF promoter. Similar to the results obtained in β-galactosidase assays, expression of lapF was lower in the lapA mutant than in the wild type after 24 h.
FIG 10.
Flow cytometry analysis of lapF::gfp expression in planktonic populations of KT2440 and the lapA mutant mus-42 grown in LB medium under static conditions. The y axis represents relative cell counts. The x axis shows arbitrary fluorescence units in a logarithmic scale. Blue and red lines correspond to planktonic wild-type and lapA mutant cells, respectively, recovered from the well. The gray peak corresponds to control cells with no gfp reporter and is shown as a nonfluorescent negative control. Shown are results from samples taken after 2, 4, 8, and 24 h.
DISCUSSION
Bacterial communities living on surfaces are commonly surrounded by an extracellular matrix composed mainly of polysaccharides and surface proteins, which confer adhesiveness, cohesiveness, and stability. In P. putida KT2440, biofilm formation is essentially dependent on the surface adhesion proteins LapA and LapF, which have been shown to play different roles (8, 22). The transcription pattern deduced for lapA correlates with its participation in the initial, irreversible attachment of bacteria to the surface (8) and also suggests a later role as part of the matrix of mature biofilms. Such a role had been previously proposed (24, 42) along with the idea that LapA may interact directly with EPS. In fact, in P. fluorescens and P. putida, LapA is essential for the maintenance of the biofilm structure, given that the activity of the protease LapG causes disassembly of the biofilm due to the release of LapA from the cell surface (7, 43).
Results from flow cytometry experiments indicate that planktonic cells and cells recovered from the biofilm behave differently with respect to lapA and lapF expression, which is dynamic during biofilm development. It is worth noting that at the early stages of surface colonization, when only a few cells are attached, all of them seem to express lapA (from microscopy observations with the lapA::gfp fusion), whereas in the planktonic population only a subset of cells show lapA expression. Further analysis would be required to establish whether this could represent a bistability mechanism, whereby LapA is produced by a subpopulation of free-living cells, and those are the ones that attach and initiate the build-up of a biofilm. The subsequent reduction in lapA expression might indicate that it is repressed in response to surface attachment although a similar reduction is observed in liquid cultures harboring an lapA::lacZ fusion. One possibility is that in liquid cultures a mixture of planktonic cells and cells detached from the surface is present. At later stages, subpopulations of cells expressing lapA or lapF appear within the biofilm, which is consistent with the notion that the different parts of a biofilm are heterogeneous with respect to physiology, metabolic state, and gene expression (1). This is exemplified in the micrographs shown in Fig. 4, where most cells are expressing lapF::gfp and lapA::mCherry but in both cases at various intensities.
Our current view of the regulatory network controlling biofilm formation in P. putida at the transcriptional level is summarized in Fig. 11. The two-component system GacS/GacA appears as a master regulator of the process, influencing expression of both adhesins. In the case of LapF, the influence is via RpoS, while it remains to be determined what the exact pathway is for LapA. This is the first direct evidence in P. putida connecting that two-component regulatory system with genes involved in biofilm formation. We cannot exclude the possibility that other structural components of the biofilm are also under the control of GacS/GacA. In P. aeruginosa, disruption of gacS results in accelerated attachment followed by early detachment under flow conditions (44) when biofilms are analyzed for prolonged periods (72 to 144 h). However, under static conditions and shorter incubation times, similar to those used here, a gacA mutant was shown to be defective in biofilm maturation (45). In the plant root-colonizing strain P. fluorescens F113, mutations in gacS or gacA cause a hypermotile phenotype (46, 47) as well as the loss of biocontrol activity (48). Although the production of certain secondary metabolites is regulated by this system, biocontrol activity also requires efficient colonization of the rhizosphere and of root surfaces. Based on our results, it seems possible that reduced biofilm formation may contribute to the decrease in biocontrol activity observed in gac mutants (49). It is also worth noting that GacA and RpoS have a significant influence on the global transcription profile of Pseudomonas protegens Pf-5 (formerly P. fluorescens Pf-5) colonizing the surface of plant seeds. Intriguingly, expression of lapA is reported to be upregulated on seed surfaces in an rpoS mutant of Pf-5 (50). In this bacterium there is no LapF homolog, so there may be differences with P. putida in the regulation and function of LapA.
FIG 11.
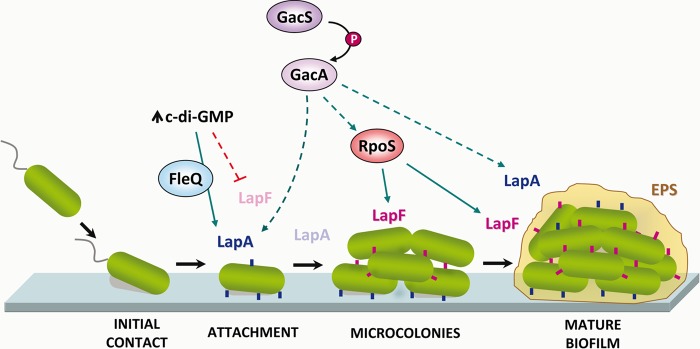
Current model of the regulatory network controlling transcription of lapA and lapF in P. putida. Positive and negative effects are shown with green and red lines, respectively. Broken lines indicate pathways that are likely indirect, while solid lines indicate those putatively direct. Dimmed labels for LapA and LapF indicate times when expression is turned off. EPS, exopolysaccharide; P, phosphotransfer.
The intracellular levels of the secondary messenger c-di-GMP affect the expression of lapA and lapF in reverse ways. The role of c-di-GMP in the transition between motile and sessile lifestyles has been widely demonstrated although many studies have focused on aspects related to flagellar motility. Recent results have demonstrated that high intracellular concentrations of c-di-GMP contribute to maintain LapA on the cell surface (7, 42, 43). Our results show increased expression of the lapA promoter in response to high c-di-GMP, suggesting that the levels of this secondary messenger regulate biofilm formation at both the transcriptional and posttranslational levels. Interestingly, the influence of c-di-GMP on lapA expression requires the regulator FleQ, a result that correlates in P. aeruginosa with its role in expression of biofilm-related elements, namely, exopolysaccharides (41). In P. fluorescens F113, a connection between GacS/GacA and FleQ has been described in the regulation of swimming motility, which is negatively controlled by GacS/GacA through FleQ, while a high intracellular concentration of c-di-GMP represses motility through WspR (17, 47).
The fact that lapF expression is reduced when c-di-GMP levels are increased could be indicative of a mechanism for controlling the timing of events during biofilm development. Furthermore, the lack of LapA alters the expression pattern of lapF so that cells expressing lapF at early time points arise in LapA-defective populations although the level of expression is lower at late time points. These results, along with our previous observation that the lack of either adhesin results in increased exopolysaccharide expression (32), suggest that there is an internal mechanism for sensing the balance of structural components of the biofilm. This balance could be viewed from an energy investment standpoint, whereby during rapid growth the cell ensures that energy is devoted to other processes and that only LapA is made early on since it is essential for attachment. It could also reflect that the need for this adhesin in the establishment of a sessile population might be circumvented under certain environmental conditions yet to be defined. How such interplay between biofilm matrix components is sensed and how the “biofilm timetable” is controlled are the next challenges to face in the analysis of bacterial multicellularity.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by grants BFU2010-17946 from Plan Nacional de I+D and P08-3869-CVI from Junta de Andalucía and by EDFR funds. M.M.-G. was supported by an FPI fellowship and an EMBO Short-Term Fellowship.
We thank M. I. Soriano and M. L. Travieso for excellent technical assistance and M. Matilla for providing plasmids pMAMV1 and pMAMV21. We thank P. Rogers from the FAS Center for Systems Biology at Harvard University for advice on the use of flow cytometry, A. Rodríguez from the Microscopy Service of the Estación Experimental del Zaidín for CLSM, and R. Kolter for helpful discussions.
Footnotes
Published ahead of print 31 January 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.01287-13.
REFERENCES
- 1.Nadell CD, Xavier JB, Foster KR. 2009. The sociobiology of biofilms. FEMS Microbiol. Rev. 33:206–224. 10.1111/j.1574-6976.2008.00150.x [DOI] [PubMed] [Google Scholar]
- 2.Petrova OE, Sauer K. 2012. Sticky situations: key components that control bacterial surface attachment. J. Bacteriol. 194:2413–2425. 10.1128/JB.00003-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boyd CD, O'Toole GA. 2012. Second messenger regulation of biofilm formation: breakthroughs in understanding c-di-GMP effector systems. Annu. Rev. Cell Dev. Biol. 28:439–462. 10.1146/annurev-cellbio-101011-155705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Römling U, Galperin MY, Gomelsky M. 2013. Cyclic di-GMP: the first 25 years of a universal bacterial second messenger. Microbiol. Mol. Biol. Rev. 77:1–52. 10.1128/MMBR.00043-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beyhan S, Tischler AD, Camilli A, Yildiz FH. 2006. Transcriptome and phenotypic responses of Vibrio cholerae to increased cyclic di-GMP level. J. Bacteriol. 188:3600–3613. 10.1128/JB.188.10.3600-3613.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Méndez-Ortiz MM, Hyodo M, Hayakawa Y, Membrillo-Hernández J. 2006. Genome-wide transcriptional profile of Escherichia coli in response to high levels of the second messenger 3′,5′-cyclic diguanylic acid. J. Biol. Chem. 281:8090–8099. 10.1074/jbc.M510701200 [DOI] [PubMed] [Google Scholar]
- 7.Newell PD, Boyd CD, Sondermann H, O'Toole GA. 2011. A c-di-GMP effector system controls cell adhesion by inside-out signaling and surface protein cleavage. PLoS Biol. 9:e1000587. 10.1371/journal.pbio.1000587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hinsa SM, Espinosa-Urgel M, Ramos JL, O'Toole GA. 2003. Transition from reversible to irreversible attachment during biofilm formation by Pseudomonas fluorescens WCS365 requires an ABC transporter and a large secreted protein. Mol. Microbiol. 49:905–918. 10.1046/j.1365-2958.2003.03615.x [DOI] [PubMed] [Google Scholar]
- 9.Yousef-Coronado F, Travieso ML, Espinosa-Urgel M. 2008. Different, overlapping mechanisms for colonization of abiotic and plant surfaces by Pseudomonas putida. FEMS Microbiol. Lett. 288:118–124. 10.1111/j.1574-6968.2008.01339.x [DOI] [PubMed] [Google Scholar]
- 10.Wu Y, Wang J, Xu T, Liu J, Yu W, Lou Zhu QT, He N, Ben H, Hu J, Götz F, Qu D. 2012. The two-component signal transduction system ArlRS regulates Staphylococcus epidermidis biofilm formation in an ica-dependent manner. PLoS One 7:e40041. 10.1371/journal.pone.0040041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ma Q, Wood TK. 2009. OmpA influences Escherichia coli biofilm formation by repressing cellulose production through the CpxRA two-component system. Environ. Microbiol. 11:2735–2746. 10.1111/j.1462-2920.2009.02000.x [DOI] [PubMed] [Google Scholar]
- 12.Davies JA, Harrison JJ, Marques LLR, Foglia GR, Stremick CA, Storey DG, Turner RJ, Olson ME, Ceri H. 2007. The GacS sensor kinase controls phenotypic reversion of small colony variants isolated from biofilms of Pseudomonas aeruginosa PA14. FEMS Microbiol. Ecol. 59:32–46. 10.1111/j.1574-6941.2006.00196.x [DOI] [PubMed] [Google Scholar]
- 13.Mikkelsen H, Sivaneson M, Filloux A. 2011. Key two-component regulatory systems that control biofilm formation in Pseudomonas aeruginosa. Environ. Microbiol. 13:1666–1681. 10.1111/j.1462-2920.2011.02495.x [DOI] [PubMed] [Google Scholar]
- 14.Heeb S, Haas D. 2001. Regulatory roles of the GacS/GacA two-component system in plant-associated and other Gram-negative bacteria. Mol. Plant Microbe Interact. 14:1351–1363. 10.1094/MPMI.2001.14.12.1351 [DOI] [PubMed] [Google Scholar]
- 15.Lapouge K, Schubert M, Allain FH, Haas D. 2008. Gac/Rsm signal transduction pathway of gamma-proteobacteria: from RNA recognition to regulation of social behaviour. Mol. Microbiol. 67:241–253. 10.1111/j.1365-2958.2007.06042.x [DOI] [PubMed] [Google Scholar]
- 16.Manzo J, Cocotl-Yañez M, Tzontecomani T, Martínez VM, Bustillos R, Velásquez C, Goiz Y, Solís Y, López L, Fuentes LE, Nuñez C, Segura D, Espín G, Castañeda M. 2011. Post-transcriptional regulation of the alginate biosynthetic gene algD by the Gac/Rsm system in Azotobacter vinelandii. J. Mol. Microbiol. Biotechnol. 21:147–159. 10.1159/000334244 [DOI] [PubMed] [Google Scholar]
- 17.Martínez-Granero F, Navazo A, Barahona E, Redondo-Nieto M, Rivilla R, Martín M. 2012. The Gac-Rsm and SadB signal transduction pathways converge on AlgU to downregulate motility in Pseudomonas fluorescens. PLoS One 7:e31765. 10.1371/journal.pone.0031765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Molina L, Ramos C, Duque E, Ronchel MC, García JM, Wyke L, Ramos JL. 2000. Survival of Pseudomonas putida KT2440 in soil and in the rhizosphere of plants under greenhouse and environmental conditions. Soil Biol. Biochem. 32:315–321. 10.1016/S0038-0717(99)00156-X [DOI] [Google Scholar]
- 19.Matilla MA, Ramos JL, Bakker PAHM, Doornbos R, Badri DV, Vivanco JM, Ramos-González MI. 2010. Pseudomonas putida KT2440 causes induced systemic resistance and changes in Arabidopsis root exudation. Environ. Microbiol. Rep. 2:381–388. 10.1111/j.1758-2229.2009.00091.x [DOI] [PubMed] [Google Scholar]
- 20.Fernández M, Niqui-Arroyo JL, Conde S, Ramos JL, Duque E. 2012. Enhanced tolerance to naphthalene and enhanced rhizoremediation performance for Pseudomonas putida KT2440 via the NAH7 catabolic plasmid. Appl. Environ. Microbiol. 78:5104–5110. 10.1128/AEM.00619-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Espinosa-Urgel M, Salido A, Ramos JL. 2000. Genetic analysis of functions involved in adhesion of Pseudomonas putida to seeds. J. Bacteriol. 182:2363–2369. 10.1128/JB.182.9.2363-2369.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Martínez-Gil M, Yousef-Coronado F, Espinosa-Urgel M. 2010. LapF, the second largest Pseudomonas putida protein, contributes to plant root colonization and determines biofilm architecture. Mol. Microbiol. 77:549–561. 10.1111/j.1365-2958.2010.07249.x [DOI] [PubMed] [Google Scholar]
- 23.Nielsen L, Li X, Halverson LJ. 2011. Cell–cell and cell–surface interactions mediated by cellulose and a novel exopolysaccharide contribute to Pseudomonas putida biofilm formation and fitness under water-limiting conditions. Environ. Microbiol. 13:1342–1356. 10.1111/j.1462-2920.2011.02432.x [DOI] [PubMed] [Google Scholar]
- 24.Nilsson M, Chiang W-C, Fazli M, Gjermansen M, Givskov M, Tolker-Nielsen T. 2011. Influence of putative exopolysaccharide genes on Pseudomonas putida KT2440 biofilm stability. Environ. Microbiol. 13:1357–1369. 10.1111/j.1462-2920.2011.02447.x [DOI] [PubMed] [Google Scholar]
- 25.Duque E, de la Torre J, Bernal P, Molina-Henares MA, Alaminos M, Espinosa-Urgel Roca MA, Fernández M, de Bentzmann S, Ramos JL. 2013. Identification of reciprocal adhesion genes in pathogenic and non-pathogenic Pseudomonas. Environ. Microbiol. 15:36–48. 10.1111/j.1462-2920.2012.02732.x [DOI] [PubMed] [Google Scholar]
- 26.Nakazawa T. 2002. Travels of a Pseudomonas, from Japan around the world. Environ. Microbiol. 4:782–786. 10.1046/j.1462-2920.2002.00310.x [DOI] [PubMed] [Google Scholar]
- 27.Ramos-González MI, Molin S. 1998. Cloning, sequencing, and phenotypic characterization of the rpoS gene from Pseudomonas putida KT2440. J. Bacteriol. 180:3421–3431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Winson MK, Swift S, Hill PJ, Sims CM, Griesmayr G, Bycroft BW, Williams P, Stewart GS. 1998. Engineering the luxCDABE genes from Photorhabdus luminescens to provide a bioluminescent reporter for constitutive and promoter probe plasmids and mini-Tn5 constructs. FEMS Microbiol. Lett. 163:193–202. 10.1111/j.1574-6968.1998.tb13045.x [DOI] [PubMed] [Google Scholar]
- 29.Karunakaran R, Mauchline TH, Hosie HF, Poole PS. 2005. A family of promoter probe vectors incorporating autofluorescent and chromogenic reporter proteins for studying gene expression in Gram-negative bacteria. Microbiology 151:3249–3256. 10.1099/mic.0.28311-0 [DOI] [PubMed] [Google Scholar]
- 30.Spaink HP, Okker RJ, Wijffelman CA, Pees E, Lugtenberg BJ. 1987. Promoters in the nodulation region of the Rhizobium leguminosarum Sym plasmid pRL1JI. Plant Mol. Biol. 9:27–39. 10.1007/BF00017984 [DOI] [PubMed] [Google Scholar]
- 31.Matilla MA, Travieso ML, Ramos JL, Ramos-González MI. 2011. Cyclic diguanylate turnover mediated by the sole GGDEF/EAL response regulator in Pseudomonas putida: its role in the rhizosphere and an analysis of its target processes. Environ. Microbiol. 13:1745–1766. 10.1111/j.1462-2920.2011.02499.x [DOI] [PubMed] [Google Scholar]
- 32.Martínez-Gil M, Quesada JM, Ramos-González MI, Soriano MI, de Cristóbal RE, Espinosa-Urgel M. 2013. Interplay between extracellular matrix components of Pseudomonas putida biofilms. Res. Microbiol. 164:382–389. 10.1016/j.resmic.2013.03.021 [DOI] [PubMed] [Google Scholar]
- 33.Lennox ES. 1955. Transduction of linked genetic characters of the host by bacteriophage P1. Virology 1:190–206. 10.1016/0042-6822(55)90016-7 [DOI] [PubMed] [Google Scholar]
- 34.Kolodkin-Gal I, Romero D, Cao S, Clardy J, Kolter R, Losick R. 2010. D-amino acids trigger biofilm disassembly. Science 328:627–629. 10.1126/science.1188628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.O'Toole GA, Kolter R. 1998. Initiation of biofilm formation in Pseudomonas fluorescens WCS365 proceeds via multiple, convergent signalling pathways: a genetic analysis. Mol. Microbiol. 28:449–461. 10.1046/j.1365-2958.1998.00797.x [DOI] [PubMed] [Google Scholar]
- 36.Miller JH. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 37.Pfaffl MW. 2001. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29:e45. 10.1093/nar/29.9.e45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Branda SS, Chu F, Kearns DB, Losick R, Kolter R. 2006. A major protein component of the Bacillus subtilis biofilm matrix. Mol. Microbiol. 59:1229–1238. 10.1111/j.1365-2958.2005.05020.x [DOI] [PubMed] [Google Scholar]
- 39.Whistler CA, Corbell NA, Sarniguet A, Ream W, Loper JE. 1998. The two-component regulators GacS and GacA influence accumulation of the stationary-phase sigma factor σS and the stress response in Pseudomonas fluorescens Pf-5. J. Bacteriol. 180:6635–6641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lee VT, Matewish JM, Kessler JL, Hyodo M, Hayakawa Y, Lory S. 2007. A cyclic-di-GMP receptor required for bacterial exopolysaccharide production. Mol. Microbiol. 65:1474–1484. 10.1111/j.1365-2958.2007.05879.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Baraquet C, Murakami K, Parsek MR, Harwood CS. 2012. The FleQ protein from Pseudomonas aeruginosa functions as both a repressor and an activator to control gene expression from the pel operon promoter in response to c-di-GMP. Nucleic Acids Res. 40:7207–7218. 10.1093/nar/gks384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gjermansen M, Nilsson M, Yang L, Tolker-Nielsen T. 2010. Characterization of starvation-induced dispersion in Pseudomonas putida biofilms: genetic elements and molecular mechanisms. Mol. Microbiol. 75:815–826. 10.1111/j.1365-2958.2009.06793.x [DOI] [PubMed] [Google Scholar]
- 43.Navarro M, Newell PD, Krasteva PV, Chatterjee D, Madden DR, O'Toole GA, Sondermann H. 2011. Structural basis for c-di-GMP-mediated inside-out signaling controlling periplasmic proteolysis. PLoS Biol. 9:e1000588. 10.1371/journal.pbio.1000588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Petrova OE, Sauer K. 2009. A novel signaling network essential for regulating Pseudomonas aeruginosa biofilm development. PLoS Pathog. 5:e1000668. 10.1371/journal.ppat.1000668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Parkins MD, Ceri H, Storey DG. 2001. Pseudomonas aeruginosa GacA, a factor in multihost virulence, is also essential for biofilm formation. Mol. Microbiol. 40:1215–1226. 10.1046/j.1365-2958.2001.02469.x [DOI] [PubMed] [Google Scholar]
- 46.Martínez-Granero F, Rivilla R, Martín M. 2006. Rhizosphere selection of highly motile phenotypic variants of Pseudomonas fluorescens with enhanced competitive colonization ability. Appl. Environ. Microbiol. 72:3429–3434. 10.1128/AEM.72.5.3429-3434.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Navazo A, Barahona E, Redondo-Nieto M, Martínez-Granero F, Rivilla R, Martín M. 2009. Three independent signalling pathways repress motility in Pseudomonas fluorescens F113. Microb. Biotechnol. 2:489–498. 10.1111/j.1751-7915.2009.00103.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Barahona E, Navazo A, Martínez-Granero F, Zea-Bonilla T, Pérez-Jiménez RM, Martín M, Rivilla R. 2011. Pseudomonas fluorescens F113 mutant with enhanced competitive colonization ability and improved biocontrol activity against fungal root pathogens. Appl. Environ. Microbiol. 77:5412–5419. 10.1128/AEM.00320-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Laville J, Voisard C, Keel C, Maurhofer M, Défago G, Haas D. 1992. Global control in Pseudomonas fluorescens mediating antibiotic synthesis and suppression of black root rot of tobacco. Proc. Natl. Acad. Sci. U. S. A. 89:1562–1566. 10.1073/pnas.89.5.1562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kidarsa TA, Shaffer BT, Goebel NC, Roberts DP, Buyer JS, Johnson A, Kobayashi DY, Zabriskie TM, Paulsen I, Loper JE. 2013. Genes expressed by the biological control bacterium Pseudomonas protegens Pf-5 on seed surfaces under the control of the global regulators GacA and RpoS. Environ. Microbiol. 15:716–735. 10.1111/1462-2920.12066 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.