Abstract
The mobile genetic element ICEBs1 is an integrative and conjugative element (ICE) found in Bacillus subtilis. One of the ICEBs1 genes, cwlT, encodes a cell wall hydrolase with two catalytic domains, a muramidase and a peptidase. We found that cwlT is required for ICEBs1 conjugation. We examined the role of each of the two catalytic domains and found that the muramidase is essential, whereas the peptidase is partially dispensable for transfer of ICEBs1. We also found that the putative signal peptide in CwlT is required for CwlT to function in conjugation, consistent with the notion that CwlT is normally secreted from the cytoplasm. We found that alteration of the putative lipid attachment site on CwlT had no effect on its role in conjugation, indicating that if CwlT is a lipoprotein, the lipid attachment is not required for conjugation. Finally, we found conditions supporting efficient transfer of ICEBs1 into and out of Bacillus anthracis and that cwlT was needed for ICEBs1 to function in B. anthracis. The mature cell wall of B. anthracis is resistant to digestion by CwlT, indicating that CwlT might act during cell wall synthesis, before modifications of the peptidoglycan are complete.
INTRODUCTION
Integrative and conjugative elements (ICEs) are mobile genetic elements that are found stably integrated into a bacterial chromosome. Under certain conditions, an ICE can excise from the chromosome, circularize, and transfer to a recipient cell via the ICE-encoded conjugation machinery (reviewed in references 1 and 2). ICEs are found in a wide variety of bacterial species, both Gram positive and Gram negative (3), and they often bestow physiologically and clinically relevant traits, including nitrogen fixation, biofilm formation, virulence, and antibiotic resistance.
ICEBs1 is a mobile genetic element found in many isolates of Bacillus subtilis (4–6). It is approximately 21 kb in length with 24 open reading frames (Fig. 1A). ICEBs1 is found integrated in trnS-leu2, the gene for a leucine-tRNA, and it remains stably integrated as long as its major operon is repressed. Derepression of ICEBs1 gene expression and subsequent excision occur in response to DNA damage, or when the cell-cell signaling regulator RapI is produced and becomes active, usually when cells are crowded by potential recipients that do not have ICEBs1 (4, 7, 8).
FIG 1.
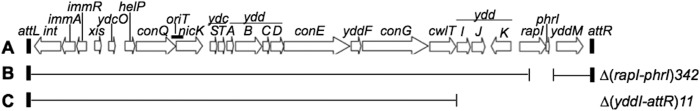
Map of ICEBs1 and derivatives. (A) Linear genetic map of ICEBs1 integrated in the chromosome. Open arrows indicate the open reading frames and direction of transcription. Gene names are indicated above the arrows. The small black rectangles at the ends of the element represent the 60-bp direct repeats that contain the site-specific recombination sites in the left and right attachment sites, attL and attR. (B and C) Various deletions of ICEBs1 used in this study. The thin horizontal lines below the map of ICEBs1 represent regions that are present, and the open spaces represent regions that are missing. (B) Δ(rapI-phrI) contains an insertion of kan (not shown). (C) This construct is contained at thrC and was used to complement various cwlT mutations in ICEBs1 in the normal attachment site. Δ(yddI-attR) deletes all ICEBs1 genes to the right of cwlT and contains an insertion of tet (not shown). (Figure adapted from PLoS Genetics [56].)
The ICEBs1 gene cwlT (named for cell wall lytic; previously yddH) encodes a bifunctional cell wall hydrolase (Fig. 2) capable of degrading peptidoglycan (9). Peptidoglycan is the major component of the bacterial cell wall and is composed of long carbohydrate chains of alternating amino sugars, N-acetylglucosamine and N-acetylmuramic acid, cross-linked by short peptide chains (10–12). In B. subtilis, the cell wall is approximately 40 to 50 nm thick (13, 14), and the genome encodes a complement of >30 hydrolases that digest the various covalent bonds in the cell wall peptidoglycan to facilitate processes such as growth, separation of cells after division, and mother cell lysis during sporulation (12, 13).
FIG 2.
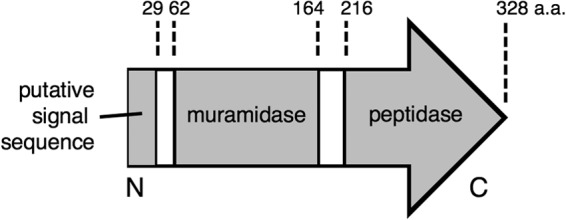
Features of CwlT. CwlT has 328 amino acids (a.a.). The putative signal sequence (amino acids 1 to 29), muramidase domain (amino acids 62 to 164), peptidase domain (amino acids 216 to 328), and N and C termini are indicated. The putative lipid attachment residue is a cysteine at amino acid 23 (not shown). This figure is based on data from reference 9.
Peptidoglycan hydrolases are widespread in mobile genetic elements and are often found associated with type IV secretion systems (T4SS) involved in conjugation (15–19). One of the best characterized of the type IV secretion systems is the VirB/D4 system from Agrobacterium tumefaciens. This system is composed of a large multiprotein channel that spans the cell envelope and mediates the secretion of conjugative DNA and associated proteins. It is generally assumed that the hydrolases cause localized degradation of the cell wall to allow the assembly of the large secretion apparatus. However, relatively little is known about their function in conjugation. Hydrolases in Gram-negative organisms tend to have one hydrolytic domain and are usually not essential for conjugation (20–22). Hydrolases from conjugative systems in Gram-positive organisms typically have two or more catalytic domains, and conjugation is significantly reduced or eliminated in mutants (23–25).
Cell wall hydrolases from B. subtilis phage and conjugative elements typically have multiple domains (9, 26). CwlT has two domains for peptidoglycan hydrolysis, and each has been characterized biochemically (9). The N-terminal domain is an N-acetylmuramidase (muramidase) that cleaves the linkage between N-acetylmuramic acid and N-acetylglucosamine. The C-terminal endopeptidase (peptidase) domain cleaves the bond between d-γ-glutamate and meso-diaminopimelic acid (9).
We found that cwlT is required for conjugation of ICEBs1. Using mutations in each of the two domains (Fig. 2), we found that the muramidase function is essential and that the peptidase function is important but partially dispensable for ICEBs1 conjugation. We found that the signal sequence involved in secretion of CwlT is critical for its function in conjugation. It was previously predicted that CwlT might be a lipoprotein (9, 27). We found that alteration of the putative lipid anchor site in CwlT had no effect on conjugation, indicating that if CwlT is a lipoprotein, lipid attachment is likely not required for CwlT function. We also analyzed whether CwlT functions were needed for ICEBs1 to function in Bacillus anthracis, whose cell wall is modified and resistant to hydrolysis by CwlT (and lysozyme). Our results indicate that CwlT activity is essential for ICEBs1 to transfer into and/or out of B. subtilis and B. anthracis.
MATERIALS AND METHODS
Strains and alleles.
B. subtilis and B. anthracis strains used are listed in Table 1. Standard techniques were used for cloning and strain construction (28). Some alleles related to ICEBs1 were previously described and are summarized below. Donor strains contained a derivative of ICEBs1 that contains a deletion of rapI-phrI and insertion of a kan cassette, Δ(rapI-phrI)::kan (4, 29–31) (Fig. 1B). rapI was overexpressed from Pxyl-rapI (Pxyl is the xylose-inducible promoter) integrated into amyE, amyE::{(Pxyl-rapI) spc} (29) to induce ICEBs1 gene expression and excision in donor cells. ICEBs10 indicates that the strain is cured of ICEBs1. B. subtilis recipients were streptomycin resistant due to the spontaneous streptomycin-resistant allele str84, most likely in rpsL (30), and this was used as a counterselective marker in mating experiments.
TABLE 1.
Bacillus strains used in this study
| Strain | Relevant genotypea [reference(s)] |
|---|---|
| B. subtilis strains | |
| JH642 | trpC2 pheA1 (contains wild-type ICEBs1) |
| CAL85 | ICEBs10 (cured of ICEBs1) str84 [30] |
| CAL229 | thrC::{mls ICEBs1 Δ(rapI-phrI)::kan} |
| MMB970 | Δ(rapI-phrI)342::kan amyE::{(Pxyl-rapI) spc} |
| TD19 | Δ(rapI-phrI)342::kan ΔcwlT19 amyE::{(Pxyl-rapI) spc} |
| TD37 | Δ(rapI-phrI)342::kan ΔcwlT19 thrC11::{mls ICEBs1Δ(yddI-attR::tet)} amyE::{(Pxyl-rapI) spc} |
| TD46 | Δ(rapI-phrI)342::kan cwlT-E87Q amyE::{(Pxyl-rapI) spc} |
| TD48 | Δ(rapI-phrI)342::kan cwlT-C237A amyE::{(Pxyl-rapI) spc} |
| TD50 | Δ(rapI-phrI)342::kan cwlT-E87Q-C237A amyE::{(Pxyl-rapI) spc} |
| TD52 | Δ(rapI-phrI)342::kan cwlT-C237A amyE::{(Pxyl-rapI) spc} thrC11::{mls ICEBs1 Δ(yddI-attR)::tet} |
| TD57 | Δ(rapI-phrI)342::kan cwlT-E87Q-C237A amyE::{(Pxyl-rapI) spc} thrC11::{mls ICEBs1 Δ(yddI-attR)::tet} |
| TD62 | Δ(rapI-phrI)342::kan cwlT-E87Q amyE::{(Pxyl-rapI) spc} thrC11::{mls ICEBs1Δ(yddI-attR)::tet} |
| TD123 | Δ(rapI-phrI)342::kan cwlTΔ(1-29) amyE::{(Pxyl-rapI) spc} |
| TD221 | Δ(rapI-phrI)342::kan cwlT-C27A amyE::{(Pxyl-rapI) spc} |
| TD319 | Δ(rapI-phrI)342::kan cwlTΔ(207-327) amyE::{(Pxyl-rapI) spc} |
| TD321 | Δ(rapI-phrI)342::kan cwlTΔ(207-327) amyE::{(Pxyl-rapI) spc} thrC11::{mls ICEBs1 Δ(yddI-attR)::tet} |
| B. anthracis strains | |
| UM44-1C9 | (AG1924) str ind (plasmid-free strain) [4, 36] |
| JMA921 | str ind nal |
| CAL2257 | str ind nal pBS42 (Cmr) |
| TD230 | ICEBs1 Δ(rapI-phrI)342::kan str ind nal |
| TD322 | ICEBs1 Δ(rapI-phrI)342::kan cwlT-E87Q str ind nal |
| TD324 | ICEBs1 Δ(rapI-phrI)342::kan cwlT-C237A str ind nal |
| TD326 | ICEBs1 Δ(rapI-phrI)342::kan cwlT-E87Q-C237A str ind nal |
All B. subtilis strains are derived from strain JH642 (54) and contain trpC2 and pheA1 (not shown). Unless otherwise indicated, all B. subtilis strains contain ICEBs1 integrated at its normal attachment site in trnS-leu2. B. anthracis plasmid-free strain UM44-19C (55) is streptomycin resistant (str), requires indole (ind) or tryptophan for growth, and was the parent for other B. anthracis strains. B. anthracis strains do not contain ICEBs1 unless otherwise indicated.
(i) Deletion of cwlT.
The ΔcwlT19 allele is an unmarked deletion that removes cwlT entirely and fuses the stop codon of conG (upstream of cwlT) to the intergenic region upstream of yddI. A 2.1-kb fragment of ICEBs1 DNA with the ΔcwlT19 allele was obtained by the splice overlap extension PCR method (32, 33) and cloned into the EcoRI and BamHI sites of pEX44 (34), a chloramphenicol-resistant vector containing the Escherichia coli lacZ gene. The resulting plasmid, pTD6, was used to introduce ΔcwlT19 into the chromosome of B. subtilis MMB970 by first integrating by single crossover and then screening for loss of the plasmid by virtue of loss of the lacZ gene and then testing by PCR for introduction of the indicated allele, essentially as described previously (31).
(ii) Modification of muramidase and peptidase domains.
Mutations in the muramidase and peptidase domains were created using a strategy similar to that for ΔcwlT. cwlT-E87Q contains a missense mutation at position 87 of cwlT, converting a glutamate codon to a glutamine codon. cwlT-C237A contains a missense mutation at position 237, converting a cysteine codon to an alanine codon. cwlT-E87Q-C237A contains both of these mutations. cwlTΔ(207-329) is a deletion of the entire peptidase domain, consisting of a fusion of the first 206 codons of cwlT to its stop codon (Fig. 2). DNA fragments (∼1.2 kb) containing one or both of these mutations were constructed and cloned into pCAL1422 by isothermal assembly (35) to yield pTD8 (cwlT-E87Q), pTD9 (cwlT-C237A), pTD10 (cwlT-E87Q-C237A), and pTD310 (cwlTΔ207-329). These plasmids were used to introduce their respective alleles into the chromosome of ΔcwlT19 strain TD19 as described for pEX44 above. Mutations in cwlT were then confirmed by sequencing appropriate PCR products from genomic DNA.
(iii) Modifications to cwlT signal sequence.
cwlTΔ1-29 contains a deletion of the first 29 codons of cwlT and introduces a start codon at the beginning of the truncated gene (Fig. 2). cwlT-C23A contains a missense mutation that removes the putative lipoprotein anchoring site by converting the cysteine codon at position 23 to an alanine codon. These mutations were introduced into B. subtilis MMB970 with pCAL1422-derived plasmids pTD95 (cwlTΔ1-29), pTD99 (cwlTspoVD1-32), and pTD116 (cwlT-C23A) as described above.
(iv) Construction of ICEBs1-cwlT at thrC.
To test for complementation of various cwlT mutants, we provided wild-type cwlT from an ectopic copy of ICEBs1 integrated at thrC (29, 31). We found that complementation required expression of cwlT along with the upstream genes, similar to findings with complementation of other ICEBs1 mutants (29). As discussed previously, we suspect that this has to do with some type of coupling, perhaps translational, between expression of many of the ICEBs1 genes (29). A complementation construct, thrC11::{mls ICEBs1Δ(yddI-attR::tet)} (Fig. 1C), was created by starting with B. subtilis CAL229, which contains the entire ICEBs1 integrated into an attachment site (attB) placed at thrC and marked with macrolide-lincosamide-streptogramin (mls) resistance. Genes downstream from cwlT were deleted, and a tetracycline resistance cassette was inserted, analogous to previously described alleles (31), yielding strain TD11. Transformation with chromosomal DNA from strain TD11 was used to introduce the complementation construct to other strains.
(v) Construction of donor and recipient B. anthracis strains.
In mating experiments, counterselection for B. anthracis recipients was with either chloramphenicol or nalidixic acid. Chloramphenicol resistance in B. anthracis was from the plasmid pBS42, introduced into B. anthracis strain UM44-1C9 (AG1924) (4, 36) by ICEBs1-mediated mobilization from B. subtilis strain CAL1394 (37). Nalidixic acid resistance was due to a spontaneous mutation (4). ICEBs1 elements with mutations in cwlT were introduced into B. anthracis via conjugation from B. subtilis donors harboring a wild-type cwlT allele at an exogenous chromosomal locus, complementing the loss of cwlT function and allowing transfer. B. anthracis strains TD322 (cwlT-E87Q), TD324 (cwlT-C237A), and TD326 (cwlT-E87Q-C237A) were created by conjugation of ICEBs1 from B. subtilis strains TD62, TD52, and TD57, respectively.
(vi) Construction of cwlT overexpression plasmids.
Plasmids for the overproduction of CwlT and both mutant and wild-type versions of the peptidase domain of CwlT were constructed similarly to those previously described (9). Overproduction of full-length CwlT in E. coli caused rapid cell lysis. However, deletion of the N-terminal 29 amino acids prevented lysis in E. coli, and this deletion was used to express CwlT for purification. In contrast, overproduction of full-length CwlT in B. subtilis had no obvious effect on cell growth or viability, perhaps indicating that activation of CwlT might be regulated. A fragment of cwlT containing codons 30 to 329 (with an N-terminal initiation codon) was amplified by PCR and cloned into pET21b (Novagen) digested with NdeI and HindIII, placing a hexahistidine tag (His6) at the C terminus of the CwlT protein (CwlT-His6). This yielded pTD3, which was used for overexpression of CwlT-His6. For expression of the peptidase domain, a fragment encoding amino acids 207 to 329 of cwlT was amplified by PCR either from strain AG174 (wild-type cwlT) or TD48 (cwlT-C237A), and cloned into pET28a (Novagen) digested with NdeI and HindIII, placing a hexahistidine tag at the N terminus of the protein. This yielded plasmids pTD106 (His6-CwlT-Pep) and pTD107 (His6-CwlT-PepC237A).
Media and growth conditions.
Cells were grown at 37°C with agitation in LB medium (28, 38) as indicated. Antibiotics were used at the following concentrations: ampicillin (100 μg/ml), chloramphenicol (5 μg/ml), kanamycin (5 μg/ml for B. subtilis; 25 μg/ml for E. coli), spectinomycin (100 μg/ml), streptomycin (100 μg/ml), and nalidixic acid (40 μg/ml). Erythromycin and lincomycin were used together (0.5 and 12/5 μg/ml, respectively) to select for macrolide-lincosamide-streptogramin B (MLS) resistance. Isopropyl-β-d-thiogalactopyranoside (IPTG) (Sigma) was used at a final concentration of 1 mM.
Mating assays.
Matings were performed essentially as previously described (4). Briefly, donor and recipient cells were grown in LB. Expression of ICEBs1 genes was achieved in one of two ways: either by production of the activator RapI from the xylose-inducible promoter Pxyl or by activation of the SOS response by the addition of the DNA-damaging agent mitomycin C (MMC) (4). For activation of the Pxyl promoter, xylose (1%) was added to donor cells in mid-exponential growth (optical density at 600 nm [OD600] of ∼0.2) to induce expression of Pxyl-rapI. For mitomycin C induction, 1 μg/ml mitomycin C (Sigma) was added to donor cells in mid-exponential growth (OD600 of ∼0.5). After 1 h of induction, approximately equal numbers of donor and recipient cells were mixed and filtered onto sterile nitrocellulose filters. When the cultures were induced with MMC, the filter was then washed with 25 ml of LB to minimize exposure of recipients to MMC in the donor culture.
The filters were placed on plates comprised of Spizizen minimal salts (28) and 1.5% agar for 3 h. Cells were collected from the filter and spread on selective plates. Transconjugants were identified, and mating frequencies were calculated per donor cell. The reported transfer frequencies are means (± standard errors of the means) of at least two independent biological replicates. In mating experiments induced by MMC, donor CFU was determined prior to the addition of MMC, as it can cause a drop in cell viability.
Purification of CwlT proteins.
Plasmids pTD3 (CwlT-His6), pTD106 (His6-CwlT-Pep), and pTD107 (His6-CwlT-PepC237A) were introduced into E. coli strain BL21-A1 (Invitrogen), generating strains TD103, TD106, and TD107 for expression of the different alleles of cwlT. Cells were grown in LB containing 100 μg/ml ampicillin (pTD3) or 25 μg/ml kanamycin (pTD106 and pTD107), shaking at 37°C. At an OD600 of ∼0.7 to 0.9, l-arabinose (final concentration of 0.2%) and IPTG (final concentration of 1 mM) were added to induce expression of the T7 polymerase and derepress expression of cwlT. Cells were collected after 2 h of induction and pelleted by centrifugation. Cell pellets were stored at −80°C until needed.
For purification of CwlT, the cell pellet was thawed on ice, resuspended in 0.2 volume of lysis buffer (50 mM NaH2PO4, 300 mM NaCl, 10 mM imidazole [pH 8.0]) and lysed by the addition (final concentration of 1×) of CelLytic B (Sigma) and by sonication (microtip, 50% power) on ice by four 20-s pulses. The lysate was incubated with DNase I (10 μg/ml) for 30 min on ice, and the supernatant was separated by centrifugation at 14,000 × g at 4°C for 20 min. CwlT-His6, His6-CwlT-Pep, and His6-CwlT-PepC237A were purified by nickel-nitrilotriacetic acid (Ni-NTA) column chromatography (Qiagen) according to the manufacturer's protocol for batch purification under native conditions.
Elution fractions were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). The fractions containing more than ∼95% CwlT were pooled and exchanged into storage buffer (50 mM NaH2PO4, 300 mM NaCl, 1 mM dithiothreitol [DTT] [pH 7.4]) using PD-10 desalting columns (GE Healthcare). The protein concentration was determined by using the Bradford protein assay kit (Bio-Rad), glycerol was added to 25%, and protein was stored at −80°C. CwlT was often unstable (degraded) after storage, so most assays were done with freshly purified protein.
Activity of CwlT against B. subtilis and B. anthracis.
Cells (B. anthracis or B. subtilis) were grown to mid-exponential phase in LB liquid medium at 37°C with shaking. Purified CwlT-His was added to the growing culture in final concentrations ranging from 1 μg/ml to 1 mg/ml. The cultures were incubated for 20 min, and then the cells were plated to determine the number of CFU. Survival percentage was determined by comparison to a culture to which no CwlT was added.
Preparation of cell walls.
Cell walls from B. subtilis and B. anthracis were prepared essentially as described previously (9, 39, 40). Briefly, cells were harvested from cultures (2 liters) in mid-exponential growth phase, resuspended in cold phosphate-buffered saline (PBS) (40 ml), and disrupted by sonication (microtip, 50% power) by 15 30-s pulses. After low-speed centrifugation (1,500 × g, 10 min) to remove unbroken cells, the crude cell wall was pelleted at 27,000 × g for 5 min at 4°C, suspended in 20 ml of a 4% (wt/vol) sodium dodecyl sulfate solution, and put in a boiling water bath for 20 min. The pellets were washed three times with warm deionized water (to prevent precipitation of SDS), two times with 1 M NaCl, and four more times with deionized water. After each of the last four washes, the sample was first spun at low speed (1500 × g, 5 min) to separate whole cells and other contaminating material from the cell wall fraction, which was then pelleted by spinning at 27,000 × g for 5 min.
Determination of hydrolytic activities of CwlT proteins.
Hydrolytic activities were determined essentially as described previously (9). Reactions were performed in 50 mM morpholinepropanesulfonic acid (MOPS)-NaOH buffer (pH 6.5) at 32°C with 1-mg/ml B. subtilis or B. anthracis cell wall preparations. Proteins were added to a final concentration of 10 μg/ml (CwlT-His6) or 5 μg/ml (His6-CwlT-Pep and His6-CwlT-PepC237A), and the reaction mixture was agitated constantly to maintain the cell walls in suspension. The turbidity of the reaction was monitored at 540 nm using a spectrophotometer (Genesys 10 Bio; Thermo Corporation).
Polyacrylamide gels and zymography.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis and zymography were performed as previously described (38, 41). For zymography, approximately 1 μg of various purified CwlT proteins were electrophoresed through a 12% polyacrylamide gel containing cell wall preparations (∼1 mg/ml) from B. subtilis or B. anthracis. Following electrophoresis, gels were soaked in deionized water for 30 min and then transferred into renaturation buffer (25 mM Tris-HCl, 1% Triton X-100 [pH 7.2]) at 30°C overnight with gentle agitation. After incubation, the gels were rinsed with deionized water, stained with 0.1% methylene blue in 0.01% KOH for 3 h, and destained with deionized water. Hydrolytic activity appeared as zones of clearing in the blue background of the stained cell walls.
Western blot analysis.
Samples were collected from cultures after 3 h of induction of ICEBs1 expression. Cells were pelleted and stored at −80°C. Pellets were thawed and resuspended in buffer (10 mM Tris, 10 mM EDTA [pH 7]) containing 0.1 mg/ml lysozyme and the protease inhibitor 4-(2-aminoethyl) benzenesulfonyl fluoride hydrochloride (AEBSF) at 1 mM. The volume of buffer used to resuspend each sample of cells was adjusted to the optical density at 600 nm in order to normalize the concentration of proteins in each sample. Resuspended cells were incubated at 37°C for 30 min, SDS sample buffer was added, and samples were heated at 100°C for 10 min, followed by centrifugation to remove insoluble material.
Proteins were separated by SDS-PAGE on 12% polyacrylamide gels and transferred to an Immobilon polyvinylidene difluoride (PVDF) membrane (Millipore) using a Trans-blot semidry electroblot transfer apparatus (Bio-Rad). The membranes were blocked in Odyssey blocking buffer (Li-Cor Biosciences) for 1 h and then incubated in a 1:5,000 dilution of anti-CwlT rabbit polyclonal antisera (made commercially by Covance using CwlT-His6 protein purified from E. coli) in Odyssey blocking buffer with 0.2% Tween for 1 h, and washed several times in phosphate-buffered saline, pH 7.8, with 0.1% Tween 20. The membranes were then incubated with 1:5,000 goat anti-rabbit IRDye 800 CW conjugate (Li-Cor) in Odyssey blocking buffer, 0.2% Tween, and 0.01% SDS for 1 h and washed several times in PBS with 0.1% Tween. Signals were detected using the Odyssey infrared imaging system (Li-Cor) according to the manufacturer's protocols, and the gel image was desaturated and inverted using Adobe Photoshop.
RESULTS AND DISCUSSION
CwlT is required for horizontal transfer of ICEBs1.
We constructed a deletion of cwlT (ΔcwlT19) in ICEBs1 (Materials and Methods) and tested for the ability of ICEBs1ΔcwlT to function in conjugation. The conjugation efficiency of wild-type (cwlT+) ICEBs1 was ∼5% transconjugants per donor (Table 2, line 1), similar to frequencies described previously (4). In contrast, there was no detectable transfer (≤5 × 10−5%) of ICEBs1ΔcwlT (Table 2, line 2). The mutant phenotype was largely complemented by expression of wild-type cwlT and all upstream ICEBs1 genes (Fig. 1) from an ectopic locus (Table 2, line 3). We were unable to complement ICEBs1ΔcwlT by expressing cwlT alone at an exogenous locus (data not shown). We suspect that proper expression of cwlT requires coupling to expression of the upstream genes, similar to what has been observed with other ICEBs1 mutants (29). The complementation results indicate that the defect in conjugation was due predominantly to loss of cwlT and not an unexpected effect on downstream genes or a site in ICEBs1 and that the ICEBs1-encoded cell wall hydrolase CwlT is indispensable for conjugation.
TABLE 2.
Effects of cwlT mutations on transfer of ICEBs1 from B. subtilis
| Line | Relevant genotype of donora (strain) | Mating efficiencyb |
|---|---|---|
| 1 | WT cwlT (MMB970) | 5.9 × 10−2 ± 1.2 × 10−2 |
| 2 | ΔcwlT19 (TD19) | <5 × 10−7 |
| 3 | ΔcwlT19 thrC11::ICEBs1ΔyddI-attR (TD37) | 6.6 × 10−2 ± 6.4 × 10−2 |
| 4 | cwlT-E87Q (muramidase mutant) (TD46) | <5 × 10−7 |
| 5 | cwlT-E87Q thrC11::ICEBs1Δ(yddI-attR) (TD62) | 6.0 × 10−2 ± 1.1 × 10−2 |
| 6 | cwlT-C237A (peptidase mutant) (TD48) | 5.3 × 10−5 ± 3.0 × 10−5 |
| 7 | cwlT-C237A thrC11::ICEBs1Δ(yddI-attR) (TD52) | 4.4 × 10−2 ± 6.0 × 10−3 |
| 8 | cwlTΔ(207-329) (deletion of peptidase domain) (TD319) | 3.0 × 10−5 ± 7.6 × 10−6 |
| 9 | cwlTΔ(207-329) thrC11::ICEBs1Δ(yddI-attR) (TD321) | 1.8 × 10−2 ± 2.1 × 10−2 |
| 10 | cwlTΔ(1-29) (TD123) | <6 × 10−7 |
| 11 | cwlT-C23A (TD221) | 6.1 × 10−2 ± 2.8 × 10−2 |
All donor strains contain Δ(rapI-phrI)::kan in ICEBs1 and Pxyl-rapI (not shown) and the indicated cwlT allele. WT, wild type.
The recipient in each conjugation experiment was B. subtilis CAL85 (streptomycin resistant). Mating efficiencies were calculated from the number of kanamycin-resistant, streptomycin-resistant transconjugants per initial donor (± standard error of the mean). The cells were grown in LB medium at 37°C, and expression of RapI (Pxyl-rapI) in donors was induced by the addition of xylose for 1 h. Mating mixtures were incubated at 37°C for 3 h on filters (Materials and Methods).
Our results with cwlT contrast with those for cell wall hydrolases from Gram-negative conjugative elements and are consistent with recent findings in Gram-positive organisms. In Gram-negative bacteria, loss of the element-encoded hydrolase reduces, but does not eliminate, conjugative transfer. For example, deletion of virB1 from the A. tumefaciens Ti plasmid (20), gene 19 of R1 (21), and traL of pKM101 (22) results in an approximately 10- to 100-fold reduction in conjugative transfer. In contrast, loss of the hydrolase from Gram-positive conjugative elements causes either a complete elimination in transfer or a more severe reduction than that observed for conjugative elements from Gram-negative bacteria. For example, loss of the hydrolase TcpG from pCW3 in Clostridium perfringens causes an approximately 1,000-fold decrease in conjugation (25), and loss of TraG from pIP501 (23) or PrgK from pCF10 (24) in Enterococcus faecalis causes complete elimination (>105-fold) of transfer.
We suspect that the apparently greater contribution to conjugation by the element-encoded hydrolases in Gram-positive bacteria than that in Gram-negative bacteria is partly due to the thicker cell wall. Consistent with the increased hydrolytic requirement, many hydrolases associated with Gram-positive mobile elements have multiple hydrolytic domains. Like CwlT, TraG (from pIP501) is predicted to contain both muramidase and peptidase function (23). PrgK (from pCF10) contains three hydrolytic domains: two muramidases and one peptidase (24). It has been suggested that the peptidase domains are important in assisting digestion of highly cross-linked Gram-positive cell wall (19, 42, 43).
Partial requirement for some hydrolases in conjugation might be due to redundant functions in the host. Many hydrolases have a high degree of cross-functionality (13, 18, 44). That is, there can be redundancy, and the loss of one hydrolase is masked by the presence of others. For the conjugative elements, we suspect that the partial requirement for hydrolases in either Gram-positive or Gram-negative bacteria could be due to the activities of host hydrolases or hydrolases from other resident mobile elements (25, 45–47). For CwlT of ICEBs1 and the essential hydrolases from other mobile elements, it appears that the host hydrolases are not capable of providing sufficient function to allow any detectable conjugative transfer.
Different effects of muramidase and peptidase mutants of CwlT.
CwlT contains two peptidoglycan hydrolytic domains, a muramidase and a peptidase (9). To determine their respective contributions to ICEBs1 transfer, we made mutations in each of the two domains of CwlT and assayed for effects on the conjugation efficiency of ICEBs1. Our findings indicate that the muramidase is essential and the peptidase is partly dispensable for the function of CwlT in conjugation.
Muramidase activity is abolished by a previously characterized cwlT-E87Q mutation that alters the catalytic site of the muramidase domain (9). We introduced this mutation into cwlT in ICEBs1. There was no detectable transfer of the ICEBs1 cwlT-E87Q mutant (Table 2, line 4), indicating that muramidase activity is required for transfer of ICEBs1. The levels of CwlT-E87Q protein accumulation appear comparable to those of wild-type CwlT, as shown on Western blots (Fig. 3), indicating that the mutant protein was accumulating to normal levels. The defect in conjugation was due to the cwlT-E87Q mutation and not to an unexpected effect on downstream genes because the mutant phenotype was fully complemented by exogenous expression of wild-type cwlT and the upstream ICEBs1 genes (Table 2, line 5).
FIG 3.
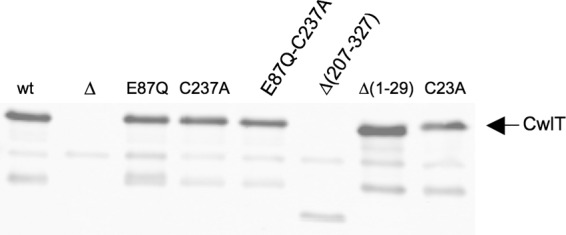
Accumulation of wild-type and mutant CwlT proteins. Western blots of cell extracts 3 h after induction of ICEBs1 by overproduction of RapI. The arrow on the right indicates full-length CwlT. The relevant cwlT allele or change (B. subtilis strain shown in the parentheses) is indicated above each lane as follows: wt, wild-type cwlT (MMB970); ΔcwlT (TD19); cwlT-E87Q (TD46); cwlT-C237A (TD48); cwlT-E87Q-C237A (TD50); cwlTΔ(207-329) (TD319); cwlTΔ(1-29) (TD123); and cwlT-C23A (TD221). Blots were probed with anti-CwlT antiserum (Materials and Methods).
To investigate the role of the peptidase domain, we constructed a point mutation in cwlT that changes its putative catalytic cysteine (48, 49), cwlT-C237A. We used two assays to verify that the mutant protein was defective in enzymatic function: a quantitative kinetic assay to measure the rate at which CwlT degraded purified peptidoglycan and a zymography assay to detect hydrolase activity in purified proteins or cell lysates (41). We purified both wild-type and mutant peptidase fragments of CwlT separate from the muramidase domain (9). There was no detectable hydrolytic activity in the C237A mutant peptidase fragment by either kinetic assay or by zymography (data not shown).
We introduced the cwlT-C237A mutation into ICEBs1 and tested for effects on conjugation. This mutant had a conjugation efficiency of ∼5 × 10−3% transconjugants per donor (Table 2, line 6), approximately 1,000-fold less than that of the wild type. The levels of CwlT-C237A protein accumulation were comparable to those of wild-type CwlT, as shown on Western blots (Fig. 3), again indicating that the mutant fragment was accumulating to normal levels. The defect in conjugation was due to the cwlT mutation and not an unexpected effect on downstream genes because the mutant phenotype was fully complemented by exogenous expression of wild-type cwlT and the upstream ICEBs1 genes (Table 2, line 7).
The conjugation efficiency of the cwlT-C237A peptidase mutant (∼5 × 10−3%) was significantly and reproducibly greater than that of the muramidase mutant (<5 × 10−5%). We were concerned that the cwlT-C237A mutation might not fully eliminate the peptidase activity in vivo and that the conjugation detected could be a result of residual peptidase activity. To test this, we constructed an allele that deletes the peptidase domain, cwlTΔ(207-329), leaving the signal sequence and the muramidase domain (Fig. 2). The muramidase and peptidase domains have been shown to maintain robust enzymatic function when separated and purified as fragments (9). The deletion of the peptidase domain was introduced into cwlT in ICEBs1. The conjugation efficiency of ICEBs1 cwlTΔ207-329 was ∼3.0 × 10−3% (Table 2, line 8), approximately 1,000-fold below that of wild-type ICEBs1, and similar to that of the cwlT-C237A mutant (Table 2, line 6). Again, the conjugation defect was fully complemented by expression of wild-type cwlT and the upstream ICEBs1 genes (Table 2, line 9). These results indicate that the peptidase is partly dispensable for conjugation efficiency. They further indicate that the cysteine at amino acid 237 is required for peptidase activity. Based on comparisons to other peptidases, C237 is likely in the active site, and histidine at amino acid 290 and the asparagine at amino acid 302 are also likely required for peptidase activity (48, 49). Together, our results indicate that the muramidase function is absolutely required and that the peptidase function is partially required for transfer of ICEBs1.
CwlT is similar to other hydrolases from well-characterized conjugative elements in Gram-positive bacteria (Tn916, pIP501, pCW3, and pCF10). The cell wall hydrolases from these elements have or are predicted to have two catalytic domains, a muramidase and a peptidase. Many other putative two-domain hydrolases are found in uncharacterized mobile elements from Gram-positive hosts. Hydrolases in Gram-negative conjugative elements appear to have only a single muramidase domain, and the peptidase domain appears to be a unique addition to hydrolases from Gram-positive systems. Some phage enzymes from Gram-positive hosts share a similar domain structure, and it has been suggested that the peptidase domains are important in assisting digestion of highly cross-linked Gram-positive cell wall (42, 43). Our results with ICEBs1 are consistent with this suggestion. In ICEBs1, the muramidase function of CwlT is essential, which is consistent with the observation that such activity is conserved in conjugative systems in both Gram-negative and Gram-positive organisms. The peptidase is partly dispensable, perhaps due to partial redundancy with host peptidases (see above).
The putative N-terminal signal sequence is, but the putative lipid attachment site is not, needed for CwlT function.
Subcellular localization plays an essential role in the regulation of many hydrolases. CwlT contains a putative N-terminal signal sequence, residues 1 to 29 (Fig. 2) that may determine its localization, though predictions of this region's function are discrepant. Different methods have predicted it to be either a lipoprotein signal sequence (9, 27) or a stable transmembrane domain (50, 51).
To determine whether the putative signal sequence of CwlT in B. subtilis is required for conjugation, we deleted codons 1 to 29 of cwlT [cwlTΔ(1-29)], removing the putative signal sequence. There was no detectable transfer of ICEBs1 cwlTΔ1-29 (Table 2, line 10), indicating that this region of CwlT is important for function. These results are consistent with the notion that CwlT is a secreted protein.
The cwlT gene product contains an FVLC motif at amino acids 20 to 23, which was identified as a putative lipobox, a conserved sequence in lipoproteins (27). The cysteine in this motif is required for lipid attachment in bona fide lipoproteins. We changed the cysteine at amino acid 23 to alanine (cwlT-C23A) and found that there was no detectable change in conjugation efficiency (Table 2, line 11). This result indicates that if CwlT were a lipoprotein, then a lipid attachment at cysteine 23 would not be required for CwlT function. Alternatively, and more likely, CwlT is not a lipoprotein, although we have not tested this directly.
The amount of each of the mutant proteins was analyzed by Western blotting and was indistinguishable from that of wild-type CwlT (Fig. 3). Together, our results indicate that the putative signal sequence of CwlT is needed for CwlT function but that the putative lipid attachment site is not. Preliminary results indicate that CwlT accumulates in culture supernatant (data not shown) and that some of it is found associated with the cell (Fig. 3).
CwlT can hydrolyze B. subtilis but not B. anthracis peptidoglycan in vitro.
ICEBs1 is capable of transferring from B. subtilis to B. anthracis (4). However, the cell wall of B. anthracis is different from that of B. subtilis, and we found that CwlT cannot degrade purified B. anthracis peptidoglycan. The glycan strands from the cell wall of B. anthracis differ from those of B. subtilis in two ways: B. anthracis glycan chains are O-acetylated and N-deacetylated. Both of these modifications confer lysozyme resistance to B. anthracis, and they might also cause resistance to the muramidase activity of CwlT. In addition, although the peptides of B. subtilis and B. anthracis peptidoglycan have the same amino acid sequence, in B. subtilis, the carboxyl group of meso-diaminopimelic acid (m-DAP) is amidated (52). This modification is not found in B. anthracis (11).
We purified CwlT and tested for degradation of cell wall material from B. subtilis and B. anthracis. As expected, CwlT was able to degrade cell wall from B. subtilis, but not that from B. anthracis (Fig. 4). We mixed 1.5 nmol of CwlT with 5 mg of purified B. subtilis cell wall and monitored the change in turbidity of the solution over time (Fig. 4). There was a rapid drop in turbidity within 5 min, indicating that the B. subtilis cell wall was degraded. In a similar reaction with the B. anthracis cell wall, there was little or no change in turbidity in 20 min (Fig. 4), indicating that the B. anthracis cell wall was resistant to degradation by CwlT. To be sure that the preparation of peptidoglycan from B. anthracis did not contain an inhibitor of CwlT activity, we mixed the peptidoglycan from B. anthracis with that from B. subtilis. In this mixed peptidoglycan, CwlT was able to degrade about half of the material present (Fig. 4), indicating that CwlT activity is not inhibited by anything in the peptidoglycan preparation from B. anthracis. We also found that there was no detectable degradation of the B. anthracis cell wall by CwlT in a polyacrylamide gel using zymography (data not shown), consistent with the results in solution.
FIG 4.
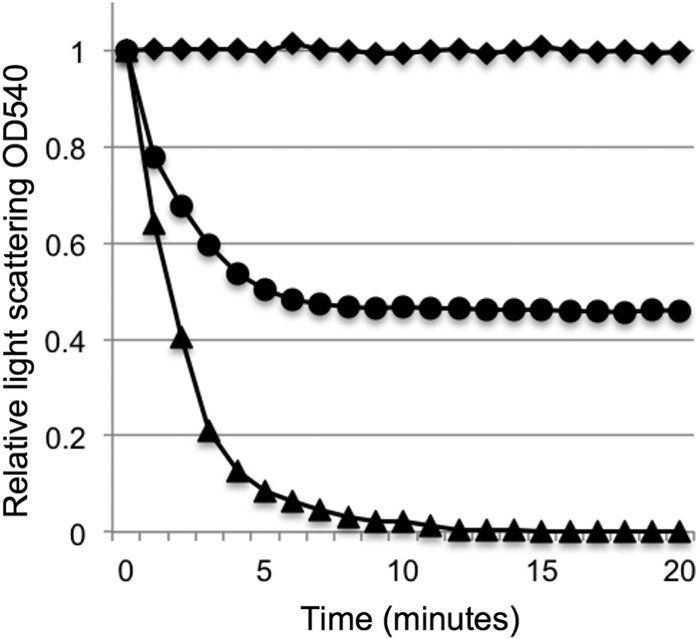
CwlT degrades purified cell wall peptidoglycan from B. subtilis but not B. anthracis. Cell wall lytic activity of CwlT on peptidoglycan from B. subtilis (triangles), B. anthracis (diamonds), or a 1:1 mix of both types (circles). CwlT-His (10 μg/ml) was mixed with approximately 1.0 mg/ml of purified peptidoglycan, and the turbidity of the reaction was monitored at 540 nm (Materials and Methods). OD540, optical density at 540 nm.
CwlT is required for ICEBs1 mating from B. subtilis into B. anthracis.
B. anthracis was a very effective recipient of ICEBs1, even though its cell wall was not degraded by CwlT. ICEBs1 was able to transfer from B. subtilis into B. anthracis with an efficiency of ∼3% transconjugants per donor, virtually indistinguishable from that of transfer from B. subtilis to B. subtilis (Table 3). Like transfer of ICEBs1 from B. subtilis to B. subtilis, transfer to B. anthracis was also dependent on cwlT. Both the muramidase mutant and the peptidase mutant were defective in transfer from B. subtilis to B. anthracis (Table 3). Because the peptidoglycan of B. anthracis is different from that of B. subtilis and was not digested by CwlT, these results could indicate that CwlT is needed to act on the cell wall of the donor, in this case B. subtilis, and not that of the recipient. However, subsequent experiments showed that CwlT is also needed for ICEBs1 to transfer from B. anthracis donors.
TABLE 3.
cwlT is required for ICEBs1 transfer from B. subtilis into B. anthracis
| Relevant genotype of donora (strain) | Mating efficiency with the following recipientb: |
|
|---|---|---|
| B. subtilis CAL85 | B. anthracis JMA921 | |
| Wild type (MMB970) | 5.5 × 10−2 ± 1.2 × 10−2 | 3.2 × 10−2 ± 5.9 × 10−3 |
| cwlT-E87Q (muramidase mutant) (TD46) | <6 × 10−7 | <6 × 10−7 |
| cwlT-C237A (peptidase mutant) (TD48) | 2.9 × 10−5 ± 9.2 × 10−4 | 4.3 × 10−5 ± 1.0 × 10−5 |
All donor strains are B. subtilis and contain ICEBs1 with Δ(rapI-phrI)::kan and Pxyl-rapI (not shown) and the indicated cwlT allele.
The efficiency of transfer of ICEBs1 from the indicated donor strain into either recipient strain CAL85 (B. subtilis) or JMA921 (B. anthracis) was calculated from the number of transconjugants per initial donor. The mating mixtures were incubated on filters at 37°C for 3 h.
ICEBs1 can transfer out of B. anthracis into B. subtilis and B. anthracis.
We found that ICEBs1 could transfer out of B. anthracis into both B. subtilis and B. anthracis with similar efficiencies (Table 4). We used the DNA-damaging agent mitomycin C to induce ICEBs1 in B. anthracis. Mitomycin C induces ICEBs1 in B. subtilis, although less efficiently than overproduction of RapI (4). The addition of mitomycin C to either B. subtilis or B. anthracis donors caused induction of ICEBs1 and enabled transfer to either B. subtilis or B. anthracis (Table 4). These results were somewhat surprising, since CwlT appeared incapable of degrading the B. anthracis cell wall (Fig. 4).
TABLE 4.
cwlT is required for mitomycin C-induced transfer of ICEBs1 from B. anthracis
| Donor strain (relevant genotype)a | Mating efficiency with the following recipientb: |
|
|---|---|---|
| B. subtilis CAL419 | B. anthracis CAL2257 | |
| B. subtilis IRN342 (cwlT+) | 1.2 × 10−3 ± 7.1 × 10−4 | 6.8 × 10−3 ± 1.1 × 10−4 |
| B. anthracis strains | ||
| TD230 (cwlT+) | 7.0 × 10−4 ± 4.2 × 10−4 | 1.5 × 10−4 ± 8.0 × 10−5 |
| TD322 (cwlT-E87Q) | <3.6 × 10−7 | <5.0 × 10−7 |
| TD324 (cwlT-C237A) | <3.8 × 10−7 | <5.0 × 10−7 |
| TD326 (cwlT-E87Q-C237A) | <8.4 × 10−7 | <5.1 × 10−7 |
All donor strains contained ICEBs1 with Δ(rapI-phrI)::kan (not shown) and the indicated cwlT allele. ICEBs1 was induced by the addition of mitomycin C for 1 h.
The mating efficiency from the indicated donor strain into either recipient strain CAL419 (B. subtilis) or CAL2257 (B. anthracis) was calculated from the number of transconjugants per initial donor. The cells were grown in LB medium at 37°C, and ICEBs1 was induced by the addition of mitomycin C for 1 h (Materials and Methods). The mating mixtures were incubated on filters at 37°C for 3 h.
CwlT is required for ICEBs1 mating from B. anthracis into B. subtilis and B. anthracis.
It seemed possible that CwlT was not needed for ICEBs1 function in B. anthracis and that other factors (perhaps cell wall hydrolases) in the B. anthracis donor strain might bypass the need for cwlT. For example, mitomycin C treatment induces a DNA damage response and the induction of many genes, some of which are in phage or prophage elements that contain their own hydrolytic enzymes that could substitute for CwlT (53).
We found that cwlT was needed for transfer of ICEBs1 from B. anthracis even after treatment with mitomycin C. We transferred ICEBs1 cwlT mutants from B. subtilis into B. anthracis. This was done by complementing the cwlT mutants with a wild-type cwlT in trans in the B. subtilis donor strains (Materials and Methods). We then used the B. anthracis strains with the ICEBs1 cwlT mutants as donors in conjugation experiments with either B. subtilis or B. anthracis as the recipient (Table 4). When ICEBs1 was induced with mitomycin C, no transfer was detected from either B. subtilis or B. anthracis donors containing the cwlT-E87Q, cwlT-C237A, or cwlT-E87Q-C237A allele (Table 4). These results demonstrate that cwlT is needed for transfer from B. anthracis, that both enzymatic activities are required for transfer, and that the requirement for cwlT is not bypassed by treatment with mitomycin C.
Exogenous CwlT causes lysis of B. subtilis and B. anthracis.
We found it puzzling that cwlT appeared to be required for transfer out of an organism with cell wall peptidoglycan that was resistant to its activity. We were interested in examining whether CwlT might exhibit different activity on growing cell walls in vivo than what we observed on purified peptidoglycan in vitro. To test this, we added purified CwlT to B. anthracis and B. subtilis cells growing in LB medium and measured its effect on cell viability. Despite their differences in cell wall composition and lysozyme resistance, both species were killed by CwlT. The addition of 100 μg/ml of CwlT for 20 min caused an approximately 500- to 1,000-fold drop in CFU of both B. anthracis and B. subtilis. These results indicate that CwlT was able to kill both B. subtilis and B. anthracis, most likely by causing at least minimal degradation of the cell wall. The amount of peptidoglycan hydrolysis by CwlT that is needed for cell lysis is probably much less than that needed for detection of hydrolysis in vitro. In B. anthracis, the cell wall is first assembled in an unmodified form that resembles that of B. subtilis. Following the initial synthesis, N-deacetylases and O-acetylases introduce modifications during peptidoglycan maturation (10, 11). Our results indicate that CwlT may act on newly synthesized peptidoglycan before it is fully modified.
Summary and model for CwlT activity.
We found that the putative signal sequence on CwlT is essential for ICEBs1 conjugation, but the putative lipid attachment site (cysteine at residue 23) is not. More importantly, we found that the peptidase activity of CwlT is important but not essential, whereas the muramidase activity is essential for conjugation. Surprisingly, we found that CwlT was needed for ICEBs1 to function in B. anthracis, whose mature cell wall is resistant to degradation by CwlT. We interpret these results to indicate that CwlT can act before full maturation of the cell wall, and this expands the range of organisms in which ICEBs1 can function. We suspect that analogous cell wall hydrolyases from other conjugative elements function similarly.
Our findings that CwlT is required for conjugation of ICEBs1 are consistent with recent results on cell wall hydrolases encoded by genes on Gram-positive conjugative plasmids (23–25). CwlT-mediated digestion likely causes local alteration of the peptidoglycan meshwork to allow assembly of the conjugation machinery. It is unknown what other ICEBs1-encoded proteins associate with CwlT, though in the Gram-positive conjugative plasmid pIP501, the cell wall hydrolase associates with the coupling protein, a putative ATPase, and a membrane-associated conjugation protein, indicating that it may be playing a role in recruitment of these proteins and in the assembly of the conjugation machinery (19). CwlT may play a similar role, and it would be interesting to determine whether CwlT affects localization or assembly of components of the ICEBs1 conjugation machinery.
ACKNOWLEDGMENTS
We thank C. Lee for help with some of the B. anthracis experiments, useful discussions, and comments on the manuscript and J. Thomas for helpful discussions.
Research reported here is based upon work supported by the National Science Foundation Graduate Research Fellowship Program under grant 1122374 and by the National Institute of General Medical Sciences of the National Institutes of Health under award R01GM050895.
Any opinions, findings, and conclusions or recommendations expressed in this report are those of the authors and do not necessarily reflect the views of the National Science Foundation or the National Institutes of Health.
Footnotes
Published ahead of print 14 February 2014
REFERENCES
- 1.Burrus V, Waldor MK. 2004. Shaping bacterial genomes with integrative and conjugative elements. Res. Microbiol. 155:376–386. 10.1016/j.resmic.2004.01.012 [DOI] [PubMed] [Google Scholar]
- 2.Wozniak RA, Waldor MK. 2010. Integrative and conjugative elements: mosaic mobile genetic elements enabling dynamic lateral gene flow. Nat. Rev. Microbiol. 8:552–563. 10.1038/nrmicro2382 [DOI] [PubMed] [Google Scholar]
- 3.Guglielmini J, Quintais L, Garcillan-Barcia MP, de la Cruz F, Rocha EP. 2011. The repertoire of ICE in prokaryotes underscores the unity, diversity, and ubiquity of conjugation. PLoS Genet. 7:e1002222. 10.1371/journal.pgen.1002222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Auchtung JM, Lee CA, Monson RE, Lehman AP, Grossman AD. 2005. Regulation of a Bacillus subtilis mobile genetic element by intercellular signaling and the global DNA damage response. Proc. Natl. Acad. Sci. U. S. A. 102:12554–12559. 10.1073/pnas.0505835102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Burrus V, Pavlovic G, Decaris B, Guedon G. 2002. The ICESt1 element of Streptococcus thermophilus belongs to a large family of integrative and conjugative elements that exchange modules and change their specificity of integration. Plasmid 48:77–97. 10.1016/S0147-619X(02)00102-6 [DOI] [PubMed] [Google Scholar]
- 6.Earl AM, Losick R, Kolter R. 2007. Bacillus subtilis genome diversity. J. Bacteriol. 189:1163–1170. 10.1128/JB.01343-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Auchtung JM, Lee CA, Garrison KL, Grossman AD. 2007. Identification and characterization of the immunity repressor (ImmR) that controls the mobile genetic element ICEBs1 of Bacillus subtilis. Mol. Microbiol. 64:1515–1528. 10.1111/j.1365-2958.2007.05748.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bose B, Auchtung JM, Lee CA, Grossman AD. 2008. A conserved anti-repressor controls horizontal gene transfer by proteolysis. Mol. Microbiol. 70:570–582. 10.1111/j.1365-2958.2008.06414.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fukushima T, Kitajima T, Yamaguchi H, Ouyang Q, Furuhata K, Yamamoto H, Shida T, Sekiguchi J. 2008. Identification and characterization of novel cell wall hydrolase CwlT: a two-domain autolysin exhibiting N-acetylmuramidase and dl-endopeptidase activities. J. Biol. Chem. 283:11117–11125. 10.1074/jbc.M706626200 [DOI] [PubMed] [Google Scholar]
- 10.Vollmer W. 2008. Structural variation in the glycan strands of bacterial peptidoglycan. FEMS Microbiol. Rev. 32:287–306. 10.1111/j.1574-6976.2007.00088.x [DOI] [PubMed] [Google Scholar]
- 11.Vollmer W, Blanot D, de Pedro MA. 2008. Peptidoglycan structure and architecture. FEMS Microbiol. Rev. 32:149–167. 10.1111/j.1574-6976.2007.00094.x [DOI] [PubMed] [Google Scholar]
- 12.Foster SJ, Popham D. 2002. Structure and synthesis of cell wall, spore cortex, teichoic acids, S-layers, and capsules, p 21–41 In Sonenshein AL, Hoch JA, Losick R. (ed), Bacillus subtilis and its closest relatives: from genes to cells. ASM Press, Washington, DC [Google Scholar]
- 13.Smith TJ, Blackman SA, Foster SJ. 2000. Autolysins of Bacillus subtilis: multiple enzymes with multiple functions. Microbiology 146:249–262 [DOI] [PubMed] [Google Scholar]
- 14.Hayhurst EJ, Kailas L, Hobbs JK, Foster SJ. 2008. Cell wall peptidoglycan architecture in Bacillus subtilis. Proc. Natl. Acad. Sci. U. S. A. 105:14603–14608. 10.1073/pnas.0804138105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Alvarez-Martinez CE, Christie PJ. 2009. Biological diversity of prokaryotic type IV secretion systems. Microbiol. Mol. Biol. Rev. 73:775–808. 10.1128/MMBR.00023-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Koraimann G. 2003. Lytic transglycosylases in macromolecular transport systems of Gram-negative bacteria. Cell. Mol. Life Sci. 60:2371–2388. 10.1007/s00018-003-3056-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Scheurwater EM, Burrows LL. 2011. Maintaining network security: how macromolecular structures cross the peptidoglycan layer. FEMS Microbiol. Lett. 318:1–9. 10.1111/j.1574-6968.2011.02228.x [DOI] [PubMed] [Google Scholar]
- 18.Zahrl D, Wagner M, Bischof K, Bayer M, Zavecz B, Beranek A, Ruckenstuhl C, Zarfel GE, Koraimann G. 2005. Peptidoglycan degradation by specialized lytic transglycosylases associated with type III and type IV secretion systems. Microbiology 151:3455–3467. 10.1099/mic.0.28141-0 [DOI] [PubMed] [Google Scholar]
- 19.Abajy MY, Kopec J, Schiwon K, Burzynski M, Doring M, Bohn C, Grohmann E. 2007. A type IV-secretion-like system is required for conjugative DNA transport of broad-host-range plasmid pIP501 in Gram-positive bacteria. J. Bacteriol. 189:2487–2496. 10.1128/JB.01491-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Berger BR, Christie PJ. 1994. Genetic complementation analysis of the Agrobacterium tumefaciens virB operon: virB2 through virB11 are essential virulence genes. J. Bacteriol. 176:3646–3660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bayer M, Eferl R, Zellnig G, Teferle K, Dijkstra A, Koraimann G, Hogenauer G. 1995. Gene 19 of plasmid R1 is required for both efficient conjugative DNA transfer and bacteriophage R17 infection. J. Bacteriol. 177:4279–4288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Winans SC, Walker GC. 1985. Conjugal transfer system of the IncN plasmid pKM101. J. Bacteriol. 161:402–410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Arends K, Celik EK, Probst I, Goessweiner-Mohr N, Fercher C, Grumet L, Soellue C, Abajy MY, Sakinc T, Broszat M, Schiwon K, Koraimann G, Keller W, Grohmann E. 2013. TraG encoded by the pIP501 type IV secretion system is a two-domain peptidoglycan-degrading enzyme essential for conjugative transfer. J. Bacteriol. 195:4436–4444. 10.1128/JB.02263-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Laverde Gomez JA, Bhatty M, Christie PJ. 2014. PrgK, a multidomain peptidoglycan hydrolase, is essential for conjugative transfer of the pheromone-responsive plasmid pCF10. J. Bacteriol. 196:527–539. 10.1128/JB.00950-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bantwal R, Bannam TL, Porter CJ, Quinsey NS, Lyras D, Adams V, Rood JI. 2012. The peptidoglycan hydrolase TcpG is required for efficient conjugative transfer of pCW3 in Clostridium perfringens. Plasmid 67:139–147. 10.1016/j.plasmid.2011.12.016 [DOI] [PubMed] [Google Scholar]
- 26.Sudiarta IP, Fukushima T, Sekiguchi J. 2010. Bacillus subtilis CwlP of the SP-β prophage has two novel peptidoglycan hydrolase domains, muramidase and cross-linkage digesting dd-endopeptidase. J. Biol. Chem. 285:41232–41243. 10.1074/jbc.M110.156273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tjalsma H, Bolhuis A, Jongbloed JD, Bron S, van Dijl JM. 2000. Signal peptide-dependent protein transport in Bacillus subtilis: a genome-based survey of the secretome. Microbiol. Mol. Biol. Rev. 64:515–547. 10.1128/MMBR.64.3.515-547.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Harwood CR, Cutting SM. 1990. Molecular biological methods for Bacillus. John Wiley & Sons, Chichester, United Kingdom [Google Scholar]
- 29.Berkmen MB, Lee CA, Loveday EK, Grossman AD. 2010. Polar positioning of a conjugation protein from the integrative and conjugative element ICEBs1 of Bacillus subtilis. J. Bacteriol. 192:38–45. 10.1128/JB.00860-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee CA, Auchtung JM, Monson RE, Grossman AD. 2007. Identification and characterization of int (integrase), xis (excisionase) and chromosomal attachment sites of the integrative and conjugative element ICEBs1 of Bacillus subtilis. Mol. Microbiol. 66:1356–1369 [DOI] [PubMed] [Google Scholar]
- 31.Lee CA, Grossman AD. 2007. Identification of the origin of transfer (oriT) and DNA relaxase required for conjugation of the integrative and conjugative element ICEBs1 of Bacillus subtilis. J. Bacteriol. 189:7254–7261. 10.1128/JB.00932-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ho SN, Hunt HD, Horton RM, Pullen JK, Pease LR. 1989. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene 77:51–59. 10.1016/0378-1119(89)90358-2 [DOI] [PubMed] [Google Scholar]
- 33.Horton RM, Hunt HD, Ho SN, Pullen JK, Pease LR. 1989. Engineering hybrid genes without the use of restriction enzymes: gene splicing by overlap extension. Gene 77:61–68. 10.1016/0378-1119(89)90359-4 [DOI] [PubMed] [Google Scholar]
- 34.Comella N, Grossman AD. 2005. Conservation of genes and processes controlled by the quorum response in bacteria: characterization of genes controlled by the quorum sensing transcription factor ComA in Bacillus subtilis. Mol. Microbiol. 57:1159–1174. 10.1111/j.1365-2958.2005.04749.x [DOI] [PubMed] [Google Scholar]
- 35.Gibson DG, Young L, Chuang RY, Venter JC, Hutchison CA, III, Smith HO. 2009. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods 6:343–345. 10.1038/nmeth.1318 [DOI] [PubMed] [Google Scholar]
- 36.Hoffmaster AR, Koehler TM. 1997. The anthrax toxin activator gene atxA is associated with CO2-enhanced non-toxin gene expression in Bacillus anthracis. Infect. Immun. 65:3091–3099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee CA, Thomas J, Grossman AD. 2012. The Bacillus subtilis conjugative transposon ICEBs1 mobilizes plasmids lacking dedicated mobilization functions. J. Bacteriol. 194:3165–3172. 10.1128/JB.00301-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sambrook J, Russell DW. 2001. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 39.Kuroda A, Sekiguchi J. 1990. Cloning, sequencing and genetic mapping of a Bacillus subtilis cell wall hydrolase gene. J. Gen. Microbiol. 136:2209–2216. 10.1099/00221287-136-11-2209 [DOI] [PubMed] [Google Scholar]
- 40.Fein JE, Rogers HJ. 1976. Autolytic enzyme-deficient mutants of Bacillus subtilis 168. J. Bacteriol. 127:1427–1442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Leclerc D, Asselin A. 1989. Detection of bacterial cell wall hydrolases after denaturing polyacrylamide gel electrophoresis. Can. J. Microbiol. 35:749–753. 10.1139/m89-125 [DOI] [PubMed] [Google Scholar]
- 42.Navarre WW, Ton-That H, Faull KF, Schneewind O. 1999. Multiple enzymatic activities of the murein hydrolase from staphylococcal phage phi11. Identification of a d-alanyl-glycine endopeptidase activity. J. Biol. Chem. 274:15847–15856 [DOI] [PubMed] [Google Scholar]
- 43.Payne KM, Hatfull GF. 2012. Mycobacteriophage endolysins: diverse and modular enzymes with multiple catalytic activities. PLoS One 7:e34052. 10.1371/journal.pone.0034052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vollmer W, Joris B, Charlier P, Foster S. 2008. Bacterial peptidoglycan (murein) hydrolases. FEMS Microbiol. Rev. 32:259–286. 10.1111/j.1574-6976.2007.00099.x [DOI] [PubMed] [Google Scholar]
- 45.Hoppner C, Liu Z, Domke N, Binns AN, Baron C. 2004. VirB1 orthologs from Brucella suis and pKM101 complement defects of the lytic transglycosylase required for efficient type IV secretion from Agrobacterium tumefaciens. J. Bacteriol. 186:1415–1422. 10.1128/JB.186.5.1415-1422.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zupan J, Hackworth CA, Aguilar J, Ward D, Zambryski P. 2007. VirB1* promotes T-pilus formation in the vir-type IV secretion system of Agrobacterium tumefaciens. J. Bacteriol. 189:6551–6563. 10.1128/JB.00480-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Baron C, Llosa M, Zhou S, Zambryski PC. 1997. VirB1, a component of the T-complex transfer machinery of Agrobacterium tumefaciens, is processed to a C-terminal secreted product, VirB1. J. Bacteriol. 179:1203–1210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Anantharaman V, Aravind L. 2003. Evolutionary history, structural features and biochemical diversity of the NlpC/P60 superfamily of enzymes. Genome Biol. 4:R11. 10.1186/gb-2003-4-2-r11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Xu Q, Chiu HJ, Farr CL, Jaroszewski L, Knuth MW, Miller MD, Lesley SA, Godzik A, Elsliger MA, Deacon AM, Wilson IA. 2014. Structures of a bifunctional cell wall hydrolase CwlT containing a novel bacterial lysozyme and an NlpC/P60 dl-endopeptidase. J. Mol. Biol. 426:169–184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Juncker AS, Willenbrock H, Von Heijne G, Brunak S, Nielsen H, Krogh A. 2003. Prediction of lipoprotein signal peptides in Gram-negative bacteria. Protein Sci. 12:1652–1662. 10.1110/ps.0303703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kall L, Krogh A, Sonnhammer EL. 2004. A combined transmembrane topology and signal peptide prediction method. J. Mol. Biol. 338:1027–1036. 10.1016/j.jmb.2004.03.016 [DOI] [PubMed] [Google Scholar]
- 52.Atrih A, Bacher G, Allmaier G, Williamson MP, Foster SJ. 1999. Analysis of peptidoglycan structure from vegetative cells of Bacillus subtilis 168 and role of PBP 5 in peptidoglycan maturation. J. Bacteriol. 181:3956–3966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sozhamannan S, Chute MD, McAfee FD, Fouts DE, Akmal A, Galloway DR, Mateczun A, Baillie LW, Read TD. 2006. The Bacillus anthracis chromosome contains four conserved, excision-proficient, putative prophages. BMC Microbiol. 6:34. 10.1186/1471-2180-6-34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Perego M, Spiegelman GB, Hoch JA. 1988. Structure of the gene for the transition state regulator, abrB: regulator synthesis is controlled by the spo0A sporulation gene in Bacillus subtilis. Mol. Microbiol. 2:689–699. 10.1111/j.1365-2958.1988.tb00079.x [DOI] [PubMed] [Google Scholar]
- 55.Koehler TM, Thorne CB. 1987. Bacillus subtilis (natto) plasmid pLS20 mediates interspecies plasmid transfer. J. Bacteriol. 169:5271–5278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Menard KL, Grossman AD. 2013. Selective pressures to maintain attachment site specificity of integrative and conjugative elements. PLoS Genet. 9:e1003623. 10.1371/journal.pgen.1003623 [DOI] [PMC free article] [PubMed] [Google Scholar]


