Abstract
Streptococcus mutans strain GS-5 produces a two-peptide lantibiotic, Smb, which displays inhibitory activity against a broad spectrum of bacteria, including other streptococci. For inhibition, lantibiotics must recognize specific receptor molecules present on the sensitive bacterial cells. However, so far no such receptor proteins have been identified for any lantibiotics. In this study, using a powerful transposon mutagenesis approach, we have identified in Streptococcus pyogenes a gene that exhibits a receptor-like function for Smb. The protein encoded by that gene, which we named LsrS, is a membrane protein belonging to the CAAX protease family. We also found that nisin, a monopeptide lantibiotic, requires LsrS for its optimum inhibitory activity. However, we found that LsrS is not required for inhibition by haloduracin and galolacticin, both of which are two-peptide lantibiotics closely related to Smb. LsrS appears to be a well-conserved protein that is present in many streptococci, including S. mutans. Inactivation of SMU.662, an LsrS homolog, in S. mutans strains UA159 and V403 rendered the cells refractory to Smb-mediated killing. Furthermore, overexpression of LsrS in S. mutans created cells more susceptible to Smb. Although LsrS and its homolog contain the CAAX protease domain, we demonstrate that inactivation of the putative active sites on the LsrS protein has no effect on its receptor-like function. This is the first report describing a highly conserved membrane protein that displays a receptor-like function for lantibiotics.
INTRODUCTION
Lantibiotics are a group of ribosomally synthesized small peptides containing bactericidal or bacteriostatic activity. These peptides are posttranslationally modified at multiple residues (1–4). In general, the lantibiotic synthesis operon encodes various enzymes that dehydrate most of the serine and threonine residues to dehydroalanine (Dha) and dehydrobutyrine (Dhb), respectively. When cysteine residues are present in the vicinity, Dha and Dhb can form thioether-linked lanthionine and 3-methyllanthionine bridges, respectively. Occasionally, Dha, Dhb, and other modified residues can be present as unlinked residues (for reviews, see references 2 and 5 to 8). On the basis of the biochemical activities of the modifying enzymes, lantibiotics are grouped into three classes (2). Lantibiotics that belong to class I include nisin, streptin, and Pep5 and are modified by two enzymes, LanB and LanC (8, 9). Class II lantibiotics are generally globular peptides, with the prototype class II lantibiotics, mersacidin and cinnamycin, which are modified by a single enzyme, often being referred to as LanM-type enzymes. Class II lantibiotics also include two-component lantibiotics (such as lacticin 3147, plantaracin W, and haloduracin), and their antimicrobial activity requires the synergistic interaction of both peptides (10–12). Class III lantibiotics, such as SapT and SapB, constitute an emerging group of lantibiotics that mainly have morphogenetic functions and display very limited antibacterial activities (8, 13–15).
On the basis of their modes of action, lantibiotics can also be classified into several categories. Lantibiotics such as Pep5 directly target the bacterial membrane to form pores, with pore formation leading to the release of ions and molecules from the sensitive bacteria, eventually leading to cell death (16). Other lantibiotics, such as mersacidin and nukacin ISK-1, bind to lipid II and thereby inhibit peptidoglycan biosynthesis in the target bacteria, a mechanism similar to that of vancomycin, which also binds to lipid II (1, 17). Lantibiotics belonging to the next category function by a complex double mode-of-action mechanism where they inhibit cell wall biosynthesis by binding to lipid II molecules as well as forming pores in bacterial membranes. Both functions can often be combined into a single polypeptide, as in nisin and epidermin (18, 19). However, a combination of two functionally specialized peptides, known as two-peptide lantibiotics, is required for the activity. Two-peptide lantibiotics contain a globular α-peptide with homology to mersacidin that binds to lipid II and an elongated β-peptide that forms a complex with the α-peptide-bound lipid II complex. Subsequently, the β-peptide forms a pore by inserting inside the bacterial membrane (20, 21).
The lactic acid bacteria, such as enterococci, lactococci, and streptococci, secrete a wide range of lantibiotics with variable spectra of inhibition (4, 6, 22–25). Among the lantibiotics, nisin, which is secreted by lactococci, is one of the most well studied and widely used lantibiotics (22, 23). Nisin has a broad inhibitory spectrum and can inhibit several Gram-positive bacteria, including Staphylococcus aureus, Listeria monocytogenes, and a variety of streptococci and enterococci (26, 27). Furthermore, nisin can inhibit Bacillus spore outgrowth and germination (28). Among the two-peptide lantibiotics, lacticin 3147, which is also secreted by some strains of lactococci, inhibits many Gram-positive bacteria, including L. monocytogenes, S. aureus, and Clostridium difficile, in addition to streptococci, enterococci, and mycobacteria (29, 30).
Streptococcus mutans, an oral lactic acid bacterium and a major causative agent of dental caries in humans, secretes several types of lantibiotics, commonly known as mutacins (31–38). Most of these lantibiotics, such as mutacins I, II, and III (mutacin 1140), are monopeptide and presumably function as nisin or mersacidin. The only two-peptide lantibiotic so far identified in S. mutans is Smb produced by GS-5 and some other strains (36, 39). Although the mutacin lantibiotics are widely present in S. mutans (38, 40), surprisingly, the first sequenced reference strain, strain UA159, does not encode any lantibiotic; it encodes only nonlantibiotics (41). It appears that S. mutans has acquired many mutacin-encoding genes by a horizontal gene transfer mechanism. For example, the strains that produce mutacin II contain the mut operon, which is inserted after the alanyl-tRNA synthetase (ats, SMU.650) in the corresponding UA159 genome (38). Likewise, the smb locus, which contains the genes necessary for Smb biosynthesis, appears to be integrated between SMU.1942 and the syl locus.
The frequency of the presence of the smb locus among various S. mutans clinical isolates has not been systematically studied. However, we recently showed that as many as 50% of the S. mutans isolates in our laboratory collection have genes encoding the smb locus (42). Although very little is known about the structure or the mode of action of Smb, primary sequences suggest that Smb is similar to lacticin 3147 and haloduracin (42). Smb also has a broad inhibitory spectrum. It can inhibit the growth of streptococci belonging to all six phylogenetic groups as well as lactococci and enterococci (42, 43). However, it appears that Smb may not inhibit Staphylococcus epidermidis or Bacillus subtilis (42, 43).
One of the streptococci that Smb very efficiently inhibits is the human pathogen Streptococcus pyogenes, also known as group A streptococcus (GAS). GAS causes a wide variety of diseases, including relatively mild and self-limiting infections of the throat and skin as well as life-threatening invasive diseases like septicemia, myositis, necrotizing fasciitis, and streptococcal toxic shock syndrome (for recent reviews, see references 44 and 45). Earlier observations suggested that S. pyogenes and other sensitive bacteria express cell surface molecules that can act as receptors (46, 47). These molecules are different from the surface polymers, such as group and type antigens (48). A study by Perry and Slade (47) suggests that a partially purified fraction of sonicated extracts of S. pyogenes can inhibit the lantibiotic activity produced by strain GS-5, presumably because a receptor-like molecule sequesters one or both of the peptides. In this study, we attempted to identify in S. pyogenes receptor molecules for Smb by using a transposon mutagenesis approach. We identified a previously uncharacterized membrane protein that exhibits a receptor-like function for Smb.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The strains used in this study are described in Table 1. Luria-Bertani (LB) medium was used for culturing Escherichia coli strain XL1-Blue, and when necessary, 100 μg/ml ampicillin or 100 μg/ml kanamycin was added to the medium. S. mutans and other streptococci were normally grown at 37°C in Todd-Hewitt medium (BBL, Becton, Dickinson) supplemented with 0.2% yeast extract (THY medium) under microaerophilic conditions. When necessary, 5 μg/ml erythromycin or 500 μg/ml kanamycin was included in the THY medium. All the streptococcal strains except S. pyogenes were transformed by means of natural transformation following a standard protocol with the addition of competence-stimulating peptides (49). For S. pyogenes, electrotransformation was carried out as previously described (50).
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Description | Reference or source |
|---|---|---|
| Strains | ||
| S. mutans | ||
| UA159 | Wild type, serotype c | 41 |
| IBSA99 | UA159 derivative, ΔSMU.662 Emr | This study |
| V403 | Wild type, serotype c | 75 |
| IBSA98 | V403 derivative, ΔSMU.662 Emr | This study |
| GS-5 | Wild type, serotype c, Smb producer | 39 |
| IBSA76 | GS-5 ΔsmbAB Emr, Smb nonproducer | 42 |
| GAS | ||
| JRS4 | Wild type, M6 serotype | 76 |
| IBSA66 | JRS4::ISS1 clone 1, insertion site not mapped, Emr | This study |
| IBSA67 | JRS4::ISS1 clone 4, insertion at codon position 8, Emr | This study |
| IBSA68 | JRS4::ISS1 clone 13, insertion at codon position 148, Emr | This study |
| IBSA70 | Plasmid-cured IBSA68, Ems | This study |
| Plasmids | ||
| pGEM-T Easy | Commercial TA cloning vector, Apr | Promega |
| pIB184-Km | E. coli-streptococcal shuttle plasmid, Kmr | 77 |
| pNM480 | E. coli vector for lacZ fusion, Apr | 55 |
| pIBM01 | pGEM-T Easy containing ermB gene, Apr Emr | 42 |
| pIBA35 | pIB184-Km containing SPy1384, Kmr | This study |
| pIBA39 | pNM480 with lacZ fused to SPy1384 at codon 75 (M75), Apr | This study |
| pIBA40 | pGEM-T Easy containing SPy1384, Apr | This study |
| pIBA41 | pNM480 with lacZ fused to SPy1384 at codon 149 (V149), Apr | This study |
| pIBA42 | pGEM-T Easy with the H178A mutation in SPy1384, Apr | This study |
| pIBA43 | pGEM-T Easy with the EE145/146AA mutations in SPy1384, Apr | This study |
| pIBA44 | pIB184-Km with the EE145/146AA mutations in SPy1384, Kmr | This study |
| pIBA45 | pIB184-Km with the H178A mutation in SPy1384, Kmr | This study |
Antibiotic sensitivity assay.
Disk diffusion assays were performed to evaluate the antibiotic susceptibilities of different S. pyogenes and S. mutans strains as described previously (51). Briefly, antibiotic disks (diameter, 6 mm; Becton, Dickinson) were placed on THY agar plates that were overlaid with 10 ml of THY soft agar containing 400 μl of freshly grown cultures. The plates were incubated overnight at 37°C under microaerophilic condition, and the zones of inhibition (ZOIs) were measured. For some chemicals, such as nisin and tunicamycin, stock solutions were prepared at the strengths indicated below and 20 μl was spotted directly or on an empty disk (diameter, 6 mm). See Table 3 for the antibiotics used in the present study.
TABLE 3.
Zones of inhibition by bacteriocins and antibiotics
| Compound or strain | ZOI (mm) for the following indicator strains: |
|
|---|---|---|
| JRS4/pIB184-Km | IBSA70/pIB184-Km | |
| Lantibiotics | ||
| Smba | 24 ± 1 | 18 ± 1 |
| Haloduracin | 15 ± 1 | 15 ± 1 |
| Galolacticin | 15 ± 1 | 16 ± 1 |
| Nisina | 18 ± 1 | 14 ± 1 |
| Cell wall antibioticsb | ||
| Amdinocillin (AMD10) | 18 ± 1 | 20 ± 1 |
| Bacitracin (B10) | 27 ± 2 | 27 ± 2 |
| Colistin (CL10) | 9 ± 1 | 9 ± 1 |
| Cycloserine (100 mg/ml) | 35 ± 2 | 37 ± 2 |
| Fosfomycin (F300) | 24 ± 2 | 24 ± 2 |
| Penicillin (P2) | 29 ± 1 | 30 ± 1 |
| Polymyxin B (PB300) | 12 ± 1 | 13 ± 1 |
| Tunicamycin (5 mg/ml)a | 19 ± 1 | 13 ± 1 |
| Vancomycin (V5) | 17 ± 1 | 17 ± 1 |
| Strains producing bacteriocin | ||
| UA159 | 18 ± 1 | 17 ± 1 |
| UA159::ΔnlmAB | 15 ± 1 | 15 ± 1 |
| UA159::ΔnlmC | 16 ± 1 | 16 ± 1 |
| UA159::ΔnlmABC | 14 ± 1 | 13 ± 1 |
ZOIs shown differ substantially from the wild-type value.
The strengths of the antibiotics are shown in parentheses.
Bacteriocin assay for ZOI determination.
GS-5 and its mutant derivatives were stabbed on THY agar plates and incubated under microaerophilic condition at 37°C overnight (52). After 16 to 20 h, the plates were overlaid with freshly grown indicator strain cultures mixed with soft agar. When the indicator strains contained plasmids, appropriate antibiotics were also included in the soft agar. The overlaid plates were again incubated overnight under the same conditions described above. The diameter of the clearing zone was measured afterwards. Assays were repeated at least twice with a minimum of two replicates.
Isolation of receptor mutants.
The procedure described by Maguin et al. (53) was used to generate insertion mutants of GAS. Briefly, strain JRS4 was electroporated with pGhost9::ISS1 and transformants were selected on THY agar containing erythromycin at 30°C. An overnight culture was made from a single colony transformed at 30°C with erythromycin. Cultures were diluted 100-fold in the same medium without antibiotics, grown for 2 h at 30°C, and then shifted to 37°C for 2.5 h to select for transposition events. This culture was then stored at −80°C with 20% glycerol and used as the transposon library. Strain GS-5 was stabbed on THY agar plates (about four to six stabs per plate) and incubated overnight under microaerophilic condition at 37°C (52). The stabbed plates were overlaid with the 100 μl of a library that was freshly revived in 500 μl THY medium. Colonies that appeared inside the zone of inhibition were inoculated in THY broth containing erythromycin at 37°C. The location of the inserted ISS1 element was identified by one of two methods. A template generated by self-ligation of HindIII-digested chromosomal DNA was subjected to inverse PCR by using primers ISS1Rout1 and ISS1Fout1. The PCR product was sequenced with primer ISS1Rout2 to identify the flanking sequences. The insertion sequences were identified by comparison to the serotype M1 (strain SF370) and serotype M6 (strain MGAS10394) genome sequences.
Curing of integrated pGhost9:ISS1.
S. pyogenes cells carrying chromosomally inserted pGhost9:ISS1 were subjected to multiple growth cycles in liquid THY medium at permissive and nonpermissive temperatures in the absence of antibiotic to induce plasmid DNA excision. For each growth cycle, a saturated culture grown at 37°C was diluted 1,000-fold in fresh THY medium, followed by incubation at 30°C for 16 h. After 16 h, the cells were diluted and plated on THY agar. Colonies were then replica patched onto THY agar with or without erythromycin to determine the efficiency of plasmid excision and to isolate a pGhost9:ISS1-cured strain. Erythromycin-sensitive colonies were confirmed to have lost the plasmid sequence by PCR with primers homologous to the flanking regions.
Construction of SMU.662 deletion mutant.
SMU.662 was deleted by fusion PCR as previously described (51). Briefly, ∼0.5-kb upstream (up) and downstream (dn) flanking regions were separately amplified with the primer sets FSN662upF/FSN662upR and FSN662dnF/FSN662dnR using S. mutans UA159 chromosomal DNA as the template (Table 2). An erythromycin-resistant (Emr) cassette was amplified from pIBM01 with the primers NcoI-Kan-D7-F and PstI-Kan-D7-R, and overlapping fusion PCR was carried out with equal amounts of each PCR product with the primers FSN662upF and FSN662dnR. The amplified products were purified and transformed into S. mutans UA159 and V403 to generate strains IBSA99 and IBSA98, respectively. A gene replacement event on the chromosomal DNA isolated from strains IBSA98 and IBSA99 was confirmed by PCR.
TABLE 2.
Oligonucleotides used in this study
| Name | Sequence (5′ to 3′) | Purpose |
|---|---|---|
| NewCBam-1384F | AGTGGATCCAGACAATTTTACCGTTAGCCTAAAAGG | SPy1384 complementation |
| NewCXho-1384R | GTTCTCGAGCCGAAGCTTTTTATTATATGACTCC | SPy1384 complementation |
| FSN662upF | CAATTTTACTTTGTTTTGTTTTTCTGCCAAGAAG | SMU.662 deletion |
| FSN662upR | CGGCCGCCATGGCGGCCGGGAGCAAGTGATAATAAAATCAGTCCAATAAC | SMU.662 deletion |
| FSN662dnF | CGCGGCCGCCTGCAGGTCGACCTACGGCGCTTTATTTCTTATTTATAGC | SMU.662 deletion |
| FSN662dnR | GGACATTGACAAAATGACTGGACTCTGACAAGACCTTGCC | SMU.662 deletion |
| NcoI-Kan-D7-F | CTCCCGGCCGCCATGGCGGCCGC | ermB amplification |
| PstI-Kan-D7-R | GGTCGACCTGCAGGCGGCCGCG | ermB amplification |
| EE145AABbv1F | GCTTTTATCGCTCCTATTATGGCAGCACTAGTCTTTAGAGGATTTCCTATG | EE146/146AA mutation |
| EE145AABbv1R | CATAGGAAATCCTCTAAAGACTAGTGCTGCCATAATAGGAGCGATAAAAGC | EE146/146AA mutation |
| H178ABbv1F | CTTGTTTTTGCTTTACCAGCAGCCACCAATAGTGTTGAA | H178A mutation |
| H178ABbv1R | TTCAACACTATTGGTGGCTGCTGGTAAAGCAAAAACAAG | H178A mutation |
| pJRSF | TAAGGCTATTGGTGTTTATGGC | LacZ fusion (upstream) |
| M74LZHindR | CCTAAGCTTCCATTTTTTGCTGTTTAATAAAAGTGTCTTGCTTAGC | LacZ fusion (downstream) |
| V148LzHindR | CCTAAGCTTCGACTAGTTCTTCCATAATAGGAGCGATAAAAGCTAT | LacZ fusion (downstream) |
Construction of plasmids for complementation.
A PCR fragment containing the entire SPy1384-coding region plus a 36-bp upstream sequence (containing the ribosome binding site) was amplified from GAS strain JRS4 genomic DNA by using the primers NewCBam-1384F and NewCXho-1384R, which introduced a unique BamHI site at the 5′ end and a unique XhoI site at the 3′ end, respectively. The resulting ∼0.7-kb fragment was digested with BamHI plus XhoI and ligated into BamHI-XhoI-digested pIB184-Km (54), a kanamycin-resistant (Kmr) shuttle plasmid that replicates in streptococci and contains the P23 promoter from lactococcal phage (pOri23), to create pIBA35. This plasmid and the pIB184-Km vector were introduced into various streptococci.
Site-directed mutagenesis of SPy1384.
For site-directed mutagenesis of the putative CAAX protease domain, the coding region of SPy1384 was amplified by using the primers NewCBam-1384F and NewCXho-1384R and JRS4 genomic DNA as the template and inserted into the pGEM-T Easy vector by TA cloning, to generate pIBA40. Site-directed mutagenesis was performed using high-fidelity Pfu polymerase (QuikChange; Agilent Technologies) with mutagenic primers that encode either the EE145 and 146AA mutations or the H178M mutation with an additional recognition site for BbvI (to facilitate screening by restriction digestion) to create intermediate plasmids pIBA43 and pIBA42, respectively. The mutations were confirmed by sequencing. SPy1384 was then amplified from pIBA42 and pIBA43 with primers NewCBam-1384F and NewCXho-1384R, digested with BamHI plus XhoI, and ligated into BamHI-XhoI-digested pIB184-Km, to create pIBA45 and pIBA44, respectively. Sequencing of the entire coding region reconfirmed the mutations in these constructs. Plasmids pIBA44 and pIBA45 were transformed into S. pyogenes by electroporation and into S. mutans by natural transformation, as described above.
Construction of plasmids for topology studies.
Two fusion constructs were created with the help of an upstream primer (pJRSF) and a gene-specific primer that annealed within the coding region of SPy1384. Since the immediate upstream region of Spy1384 does not contain a promoter, we used pIBA35, in which SPy1384 is transcribed from the P23 promoter, as the template. The PCR products were purified and digested with HindIII and cloned into SmaI-HindIII-digested pNM480 reporter plasmid (55). The fusion junction in the constructs was confirmed by DNA sequencing. LacZ activity was assessed on an LB agar plate by hydrolysis of X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside).
RESULTS
Identification of a receptor gene in S. pyogenes.
A previous study indicated that S. pyogenes might encode some cell surface proteins that function as a receptor for the lantibiotic Smb (47). We wanted to identify the genes that encode those putative receptor molecules. Toward this end, we used the insertion sequence ISS1 because it randomly inserts into the genome of Gram-positive bacteria, including various streptococci, and because it rarely inserts itself into the same cell more than once (53, 56–59). We introduced this transposon into JRS4, an M6 serotype strain, on pGhost9::ISS1, a plasmid whose replication is temperature sensitive (60). An Emr transformant containing pGhost9::ISS1 was grown overnight at 30°C, and Emr colonies containing the transposon were isolated at 37°C. We reasoned that inactivation of a receptor molecule on GAS would produce a strain that would be recalcitrant to Smb-mediated inhibition. We plated a transposon library on THY agar plates that had previously been stabbed with S. mutans strain GS-5, which produces the lantibiotic Smb. While most of the stabbed GS-5 produced clear zones of inhibition (ZOIs) with diameters of 24 ± 1 mm, each of three stabbed cultures produced ZOIs with a single colony that grew inside the halo. Using an inverse PCR method, as described in Materials and Methods, we attempted to identify the ISS1 insertion sites in these survivor mutants that grew inside the halo. Two of the insertion sites were located within the SPy1384 gene (GAS M1 serotype strain SF370 is the reference strain), while the insertion site could not be determined for the third mutant.
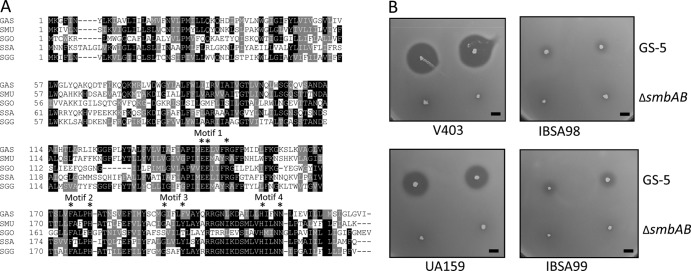
SPy1384 encodes a polypeptide of 231 residues; the ISS1 insertions occurred in this gene at codon positions 8 and 148 (Fig. 1A). We renamed this gene lsrS, for lantibiotic Smb receptor-like function in streptococci. It appears that lsrS is the last gene of a three-gene operon. SPy1386, which encodes a putative transcriptional regulator protein (71 amino acids) with a helix-turn-helix (HTH) XRE family-like motif, and SPy1385, which encodes a hypothetical protein with a DUF3169 domain, are the two other genes in the operon. Just upstream of the operon is the alaS gene, which encodes alanyl-tRNA synthetase. An intergenic region of 259 bp lies between the alaS and SPy1386 loci. Analysis by BPROM (Softberry) software indicated the presence of a −35 box (TTGTCA) and a −10 box (TACAAT) within a position 250 bp upstream of the ATG start codon of SPy1386 (Fig. 1A).
FIG 1.
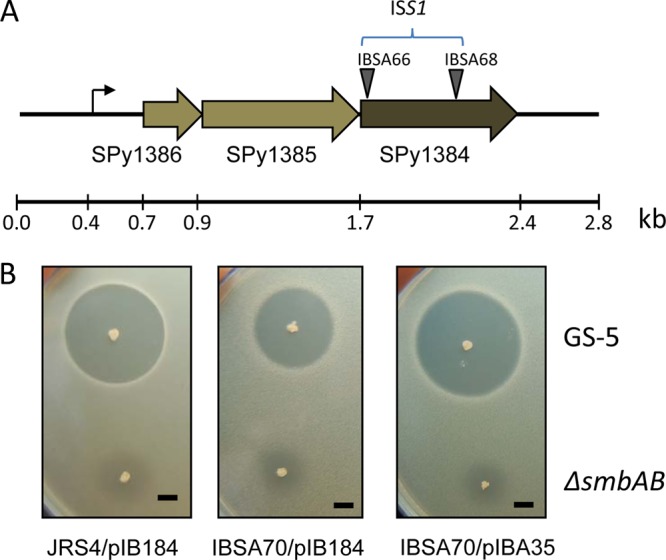
Isolation of a receptor protein for the Smb lantibiotic in S. pyogenes. (A) Genetic organization of the receptor locus (SPy1384). SPy1384 is the last gene of a three-gene operon. The first gene, SPy1386, encodes a putative transcription factor of the HTH XRE superfamily. The second gene, SPy1385, encodes a hypothetical membrane protein with the DUF3169 motif. The sites of the ISS1 insertions and their relative positions are shown. Bent arrow, putative promoter sequence. (B). Deferred antagonism assay for receptor activity. Bacterial cultures were stabbed on THY agar plate and incubated overnight at 37°C under microaerophilic conditions. The plates were then overlaid with soft agar containing the indicator strains. The ZOIs of the indicator strains were measured after overnight incubation. The observation is based on four separate experiments, and a representative area of interest is shown. The ZOI values for Smb-mediated inhibition are as follows: JRS4/pIB184-Km, 24 ± 1 mm; IBSA70/pIB184-Km, 18 ± 1 mm; and IBSA70/pIBA35, 25 ± 1 mm. Bars, 5 mm.
To confirm that lsrS indeed plays a role in Smb-mediated inhibition, we selected a mutant strain (IBSA68) in which ISS1 was inserted at the 148th codon. We generated a clean mutant derivative strain (IBSA70) by curing the integrated pGhost9::ISS1 plasmid from strain IBSA68 to create IBSA70. We also cloned the lsrS gene in plasmid pIB184-Km under the control of a heterologous promoter (P23) for complementation purposes. Both the vector plasmid (pIB184-Km) and the complementing plasmid (pIBA35) were introduced into IBSA70. The vector plasmid was also introduced into JRS4 for uniformity. These strains were then tested against GS-5 for sensitivity. As shown in Fig. 1B, IBSA70 carrying only the vector plasmid produced a ZOI with a diameter of 18 ± 1 mm, whereas JRS4 with the vector plasmid produced a ZOI with a diameter of 24 ± 1 mm, which was an approximately 40% reduction in the total area of inhibition (Fig. 1B). When we complemented IBSA70 with plasmid pIBA35 carrying lsrS, the ZOI became 25 ± 1 mm in diameter, suggesting that the observed reduction in ZOI is indeed due to the inactivation of lsrS.
LsrS plays a role in nisin and tunicamycin sensitivity.
Smb is a two-component lantibiotic, and like other lantibiotics, it is expected to interact with lipid II molecules. Because LsrS is involved in the optimum function of Smb to inhibit S. pyogenes, we wanted to test whether LsrS is also involved in inhibition by other two-component lantibiotics. For this purpose, we selected haloduracin, a well-characterized lantibiotic that targets lipid II, and galolacticin, which is produced by Streptococcus gallolyticus BAA2069 and which has sequence similarity to Smb (42). As shown in Fig. 2, the sensitivities of both the JRS4 and IBSA70 strains to the purified haloduracin and galolacticin were similar. This result suggests that LsrS is specific to Smb and does not recognize other two-component lantibiotics.
FIG 2.
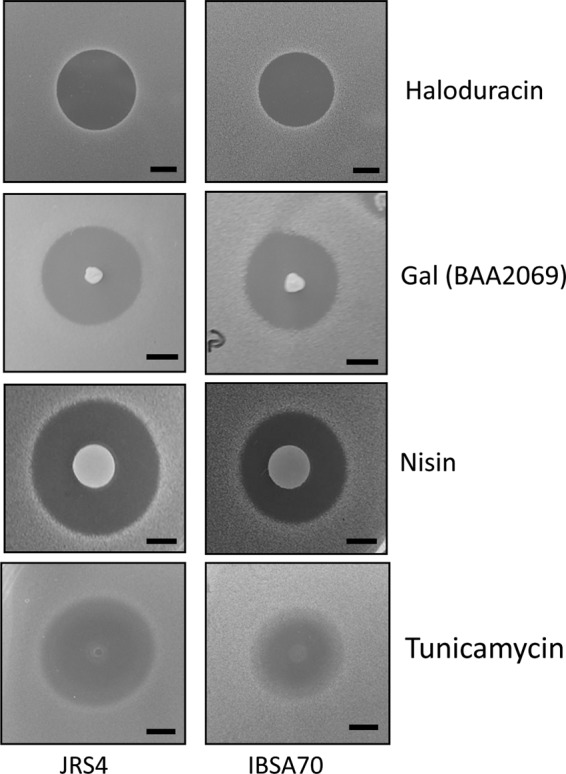
Sensitivity of the lsrS mutant to various antimicrobial agents. THY agar plates containing the indicator strains were either preseeded with a galolacticin (Gal)-producing strain (BAA2069) or spotted directly on the overlaid plates and incubated overnight at 37°C under microaerophilic conditions. Experiments were repeated at least three times, and representative areas of interest are shown. Both strains also contained the vector pIB184-Km. For actual values, refer to Table 3. Bars, 5 mm.
Nisin is a one-component lantibiotic that functions similarly to two-component lantibiotics; i.e., it binds to lipid II and inhibits cell wall biosynthesis as well as forms pores in the membrane. Therefore, we tested whether LsrS could display a receptor-like function for nisin. We observed that IBAS70 produced a ZOI with a diameter of 14 ± 1 mm, while wild-type strain JRS4 produced a ZOI with a diameter of 18 ± 1 mm, a net reduction in total area of about 40%. This indicates that, in addition to Smb, LsrS plays an important role, either directly or indirectly, for nisin recognition.
Since both Smb and nisin bind to lipid II and since LsrS is a putative membrane protein, we wanted to know whether inactivation of LsrS renders the cell sensitive to antibiotics that target cell wall biosynthesis. We tested antibiotics specific for lipid II synthesis, such as fosfomycin (which inhibits MurA), d-cycloserine (which inhibits d-Ala ligase), tunicamycin (which inhibits MraY), bacitracin (which blocks lipid carrier recycling), vancomycin (which blocks transglycosylation), and penicillin (which blocks transpeptidation). We also included polymyxin B and colistin, both of which target cell membranes. We found that among these antibiotics, only tunicamycin produced a 40% smaller halo in IBSA70 than JRS4 (Fig. 2). All other antibiotics produced similar halos in both strains (Table 3; data not shown).
LsrS homologs are present in other streptococci and function as Smb receptors.
Bioinformatics analysis suggests that LsrS belongs to COG1266, a highly conversed family predicted to encode a zinc-dependent CAAX prenyl metalloprotease domain. Furthermore, the C-terminal region of LsrS contains a domain called Abi (abortive infection; Pfam02517), which is a subfamily of CAAX proteases. A BLAST-P search showed that LsrS is present in all the sequenced S. pyogenes genomes. Furthermore, the search also fetched numerous streptococci, including many oral streptococci, with E values lower than −50. Surprisingly, it appears that all the sequenced S. mutans strains also encode an LsrS homolog protein with an E value of −61. The homolog in S. mutans UA159 is SMU.662, which showed 41% identity and 66% similarity with the LsrS sequence (Fig. 3A).
FIG 3.
Deletion of the lsrS homolog in S. mutans makes the strains resistant to Smb-mediated inhibition. (A). Multiple-sequence alignment of LsrS and its homolog from various streptococci. Sequences were aligned by use of the ClustalW program, and the degree of relatedness is displayed with the BoxShade program, where black and gray indicate identical and similar residues, respectively. Sequences were obtained from GenBank (accession numbers are in parentheses). The strains were S. pyogenes (GAS; NP_269484), S. mutans (SMU; NP_721090), Streptococcus gordonii (SGO; YP_001449790), S. sanguinis (SSA; YP_001034746), and S. gallolyticus (SGG; YP_004287423). The four conserved putative metalloprotease motifs along with the active-site residues (asterisks) are also indicated. (B). Deferred antagonism assay using two S. mutans isolates and their mutant derivatives. Assays were carried out with the GS-5 and ΔsmbAB strains as tester strains, as described in the legend to Fig. 1, and repeated at least four times. Bars, 5 mm.
To study whether SMU.662 could function as a receptor protein, we selected two S. mutans strains (UA159 and V403) that are sensitive to Smb (42). The entire SMU.662-coding region was replaced in these strains with an erythromycin resistance gene by fusion PCR, and these strains were tested for Smb-mediated inhibition. As shown in Fig. 3B, inactivation of SMU.662 nearly abolished the sensitivity toward the Smb lantibiotic in both strains. This finding suggests that SMU.662 indeed encodes a receptor-like function for Smb.
We also tested whether SMU.662 can be effective against haloduracin, galolacticin, and nisin. However, we found no difference between the wild-type strains and the strains in which SMU.662 was inactivated (data not shown). Thus, at least in S. mutans, SMU.662 is very specific and recognizes only Smb. Furthermore, we also observed that strains with the SMU.662 deletion were as susceptible to tunicamycin as their isogenic wild-type strains. Therefore, it seems that although SMU.662 recognizes Smb for lantibiotic activity, the LsrS protein in S. pyogenes might have additional functions that are absent in SMU.662.
Overexpression of LsrS in a heterologous host increases sensitivity.
It appears that Smb produced smaller ZOIs in S. mutans strains than in S. pyogenes strains. We speculated that SMU.662 might not function as efficiently as LsrS; therefore, we decided to overexpress LsrS in UA159. When we used UA159 containing pIBA35, we observed that the ZOI was increased about 2.5 times compared to that for UA159 containing the vector plasmid (Fig. 4). To rule out the possibility that the observed effect is not a strain-specific phenomenon, we also used strain V403 and observed the increased ZOI when LsrS was overexpressed. Taken together, these results suggest that LsrS can efficiently function in a heterologous host and overexpression can lead to increased sensitivity.
FIG 4.
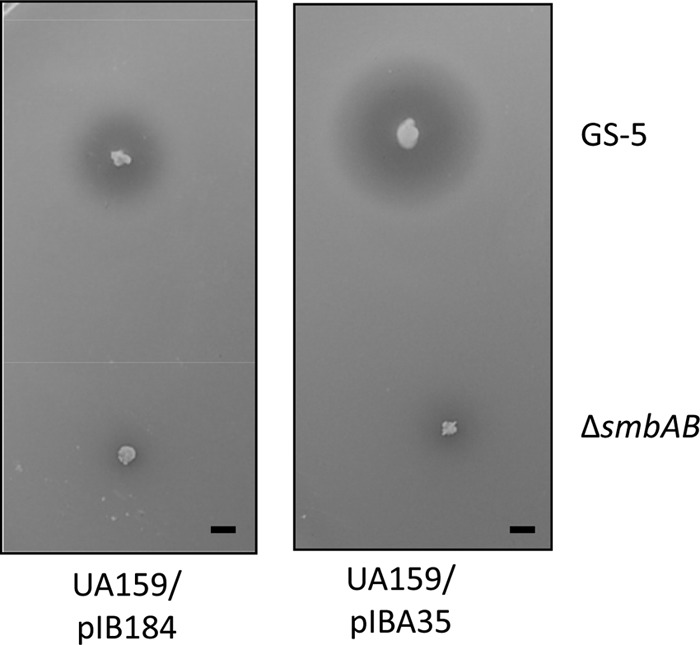
Overexpression of LsrS in S. mutans causes increased inhibition. Deferred antagonism assays were carried out with the GS-5 and ΔsmbAB strains as the tester strains and were performed as described in the legend to Fig. 1. These plates are representative of those from three independent assays. Bars, 5 mm.
Since LsrS-deficient S. pyogenes strains showed decreased sensitivity toward nisin and tunicamycin, we wanted to test whether overexpression of LsrS in S. mutans made the strain more sensitive to these compounds. We found that LsrS, when expressed in S. mutans, did not affect the sensitivity toward these reagents. This result indicates that in S. pyogenes, additional proteins which are absent in S. mutans are necessary for the observed LsrS functions.
Protease activity is not required for the receptor-like function.
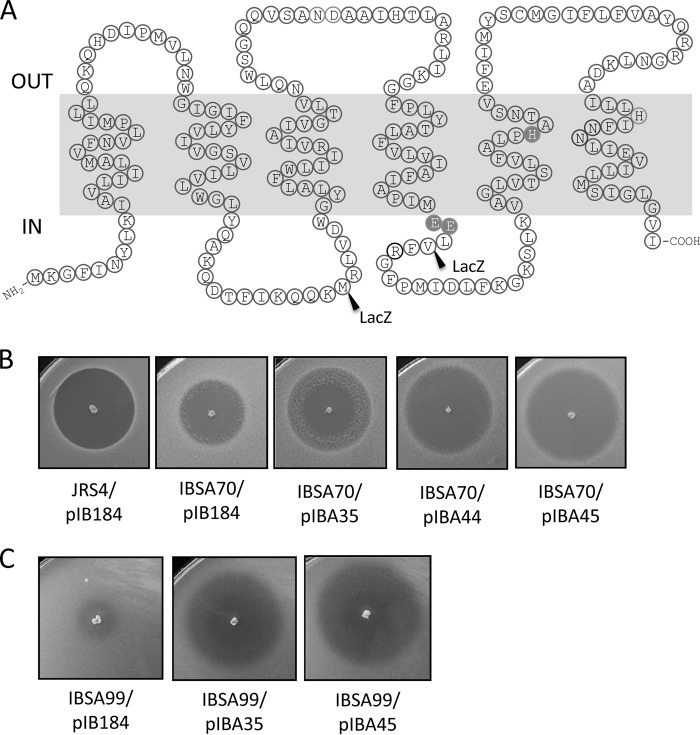
In eukaryotes, Abi domain-containing proteins are known to be involved in protein prenylation (61). These membrane proteases belong to the zinc metalloprotease family and cleave within CAAX of the substrate (A denotes an aliphatic residue, whereas X denotes one of several allowed residues that dictate the specificity of prenyltransferases). The Abi domain itself encodes four transmembrane helices (THs) with the conserved active-site residues. The catalytic glutamic acids in motif 1 and the histidines in motifs 2 and 4 are predicted to coordinate zinc ions (Fig. 3A). We used the TMpred and TopPred2 algorithms to determine the membrane topology of LsrS, and as shown in Fig. 5A, LsrS appears to contain six THs. We verified the orientations of TH3 and TH4 with the help of LacZ translational fusions at positions M74 and V148. E. coli XL1-Blue containing these translational fusions generated blue colonies on agar plates containing X-Gal, suggesting that the predicted TH orientations correlate with the experimentally verified ones.
FIG 5.
Putative protease activity is not required for LsrS activity. (A) Proposed transmembrane topology of LsrS. The hydrophobicity plots predicted from the TopPred2, TMpred, and TMHMM algorithms are similar. The predicted six putative transmembrane α helices are indicated. The positions (residues) for LacZ fusions are shown. The residues with a dark background are putative active sites for CAAX protease activity. (B and C) Site-directed mutations in the conserved active-site motifs do not affect LsrS receptor function in S. pyogenes (B) or S. mutans (C). Deferred antagonism assays were repeated at least three times, and representative areas of the plates are shown.
We then tested whether the protease-like mechanism of LsrS is necessary for the receptor function. For this, we replaced the conserved glutamic acids at positions 145 and 146 and the histidine residue at position 178 described to be critical for metalloprotease activity with alanine residues (resulting in EE145/146AA mutations in pIBA44 and the H178A mutation in pIBA45). As shown in Fig. 5B, neither of the mutations in the conserved active-site residues of LsrS had an effect on receptor activity in S. pyogenes or in S. mutans. Thus, the putative protease activity of LsrS is not necessary to exhibit receptor-like functions.
DISCUSSION
One of the noteworthy features of bacteriocins, specifically, lantibiotics, is that the peptides are highly potent and active in the nanomolar range. On the other hand, antimicrobial peptides produced by humans and animals (such as defensins and LL-37) are active in the micromolar range, a difference in concentration of 1,000-fold from that for bacteriocins (20, 62). It is assumed that the primary reason for this extreme potency is that bacteriocins recognize specific receptors on the target cells, while antimicrobial peptides of eukaryotic origin interact nonspecifically with their targets. This assumption was first validated by the identification of the mannose-phosphotransferase system (Man-PTS) as a receptor for some nonlantibiotics (class II) belonging to pediocin-like bacteriocins of subclass IIa (63) and also for some non-pediocin-like linear bacteriocins of subclass IId, such as lactococcins A and B (64). Subsequently, another sugar transporter, a maltose-ABC transporter, was also found to be required in target cells for sensitivity to garvicin ML, a circular bacteriocin belonging to subclass IIc (65). Furthermore, very recently, Uzelac et al. (66) identified a membrane-bound Zn-dependent metalloprotease in Lactococcus lactis that seems to act as a receptor for yet another nonlantibiotic, LsbB, produced by some strains of L. lactis. So far, no receptor molecules have been identified for lantibiotic peptides, including nisin, one of the most extensively studied lantibiotics. In the present study, we report the discovery of a new protein, LsrS, employed by Smb to target sensitive strains.
The locus that encodes lsrS is organized in a three-gene operon and has been found to be present in all S. pyogenes strains sequenced. A BLAST-P search with the LsrS sequence as a query against the S. pyogenes genomes did not return the sequence of any other protein, suggesting that LsrS does not have any paralogs. The inactivation of lsrS generated about 40% reductions in ZOIs but did not completely abolish the sensitivity to Smb. This indicates that in S. pyogenes, LsrS may not be the only protein with a receptor-like function. Since no other LsrS paralogs are present in S. pyogenes, we speculate that Smb utilizes as receptors other molecules unrelated to LsrS to inhibit this organism. Along this line, it is noteworthy that Perry and Slade (47) first isolated from S. pyogenes strain E14 (a sensitive strain) an inhibitory factor with a molecular mass of 93 kDa that neutralizes bacteriocins produced by GS-5. Soon after, Franker (46) isolated another factor of 74 kDa from S. pyogenes strain MJP-2 (which is also sensitive to GS-5) that also demonstrates inhibitory activity against GS-5. The exact identities of both factors are not known, and we speculate that these factors might act as a receptor for Smb.
We observed that LsrS has a receptor-like activity for Smb and not for other closely related two-peptide lantibiotics, such as haloduracin and galolacticin (Fig. 2). This was surprising to us, since the immunity protein SmbFT can recognize all three lantibiotics. While haloduracin and galolacticin are structurally similar to Smb, several differences in their sequences also exist. We speculate that some critical residues that are present in either one or both of the components of Smb might be important for peptide-receptor interactions and those critical residues are absent in haloduracin and galolacticin. Since these two lantibiotics inhibit S. pyogenes very efficiently, they might utilize other cell surface molecules as receptors.
In contrast, we found that LsrS facilitates nisin activity. This was also surprising to us, since nisin, which is a monopeptide lantibiotic, has very little sequence or structural similarity with Smb. Furthermore, the mechanism of inhibition by nisin and two-peptide lantibiotics is different. In the case of two-peptide lantibiotics, the α-peptide component interacts with lipid II, most likely in a manner that involves the mersacidin-like binding motif, and forms a complex. The β-peptide then binds to the α-peptide/lipid II complex and adopts a transmembrane conformation to form a defined pore. Although LsrS displays a receptor-like activity for both nisin and Smb, the molecular mechanism might be very different. It is possible that an accessory protein acts as the primary receptor for nisin and the function of LsrS is to enhance or stabilize the interaction. We speculate this because, when we overexpressed LsrS in S. mutans, it did not enhance the nisin-mediated inhibition and enhanced only the Smb-mediated inhibition (Fig. 4; data not shown). Since in S. pyogenes the LsrS-encoding gene is genetically linked to SPy1385, it is possible that SPy1385 might be involved in nisin recognition. SPy1385 is a hypothetical protein that is present in all the sequenced S. pyogenes strains that encode LsrS. In fact, the entire operon is very highly conserved in S. pyogenes strains and other pyogenic streptococci. When we performed a BLAST-P search, we did not find any Spy1385 homolog in S. mutans, strengthening our hypothesis. Furthermore, SPy1385 contains six transmembrane helices (data not shown), and thus, it also appears to be a membrane protein. We also found that the pyogenic group of streptococci is more sensitive to nisin than the mutans group (data not shown). Thus, we believe that for the pyogenic group of streptococci, both LsrS and SPy1385 play an important role for nisin-mediated inhibition.
In addition to forming a pore in the membrane, both nisin and Smb interfere with cell wall biosynthesis. Therefore, we tested the susceptibility of LsrS-deficient strains to various antibiotics that target enzymatic steps leading to lipid II biosynthesis and pathways leading to cell wall formation after lipid II biosynthesis. To our surprise, except for tunicamycin, we did not find any significant differences in sensitivity to any other antibiotics, including vancomycin, which also binds to lipid II (unpublished data). Thus, we speculate that LsrS has no negative effect on overall cell wall synthesis. However, we found that LsrS-deficient S. pyogenes strains were significantly resistant to the action of tunicamycin. The chemical composition of tunicamycin is complex, and it contains uracil, N-acetylglycosamine, an 11-carbon aminodialdose called tunicamine, and a fatty acid linked to the amino group. Tunicamycin inhibits the enzymatic activity of MraY, the phospho-MurNAc-pentapeptide translocase that catalyzes the synthesis of lipid I in the conserved pathway for peptidoglycan biosynthesis. Since MraY is also a transmembrane protein, it is possible that LsrS, either alone or in combination with other proteins, interferes with MraY activity in S. pyogenes. Hence, in the absence of LsrS, the enzymatic activity of MraY is enhanced. Alternatively, LsrS itself acts as a receptor for tunicamycin. We believe that the latter possibility is unlikely because when we overexpressed LsrS in S. mutans, we did not observe any change in tunicamycin sensitivity (data not shown).
In S. pyogenes, LsrS is encoded by a three-gene operon (Fig. 1). Our bioinformatics searches found that the entire operon is present in all strains of S. pyogenes that have been sequenced. We also found that this operon is present in some of isolates of Streptococcus anginosus, Streptococcus constellatus, Streptococcus dysgalactiae, Streptococcus pneumoniae, and Streptococcus suis. Since these streptococci are pathogenic, we speculate that in addition to a receptor-like function for Smb, the genes within this operon might play a role in virulence. Additional experiments are required to determine the true role of the genes in this operon.
While the operon that encodes LsrS is present in a few streptococci, a BLAST-P search with the LsrS sequence as the query yielded several additional streptococci with an E value of −35 or less. The streptococci that we found were S. gallolyticus, Streptococcus intermedius, S. mutans, and Streptococcus sanguinis. Apart from streptococci, the only other organism that we found was Carnobacterium sp. strain 17-4, a lactic acid bacterium often associated with seafood and dairy products. However, LsrS showed the highest degree of homology (E value, −60) to SMU.662 and its counterpart encoded by various S. mutans strains. This was surprising to us, since Smb is also secreted by S. mutans. We showed that SMU.662 alone could function as a receptor for Smb and deletion of SMU.662 makes the strains insensitive to Smb. Two recent large-scale genome sequencing studies have indicated that SMU.662 is a part of the core genome (67, 68). Furthermore, the upstream region (SMU.651 to SMU.658) and the downstream region (SMU.681 to SMU.687) appeared to be genomic islands and are present in some but not all S. mutans strains (69). Whether the primary function for SMU.662 is to act as a receptor for lantibiotics or whether it plays a role in other biological processes remains to be evaluated.
LsrS is a member of a highly conversed protein family with a putative CAAX prenyl protease domain. This family, which was recently renamed CPBP (CAAX protease and bacteriocin-processing enzymes), encompasses more than 5,000 proteins (70). Members of the CPBP family are involved in diverse biological functions. For example, Kjos et al. have shown that SkkI functions as a bacteriocin immunity protein for sakacin secreted by Lactobacillus plantarum (71). These authors have also shown that protease activity is necessary for the immunity function. In S. pneumoniae, PcnO is both necessary for the production of bacteriocin Pnc and involved in immunity against Pnc (72). The exact mechanism by which PncO regulates bacteriocin production or mediates immunity is currently unknown. CPBP proteins have also been shown to be involved in the expression of surface proteins containing the YSIRK signal peptide, as in the case of Staphylococcus aureus Spd proteins (73). Recently, Firon and colleagues have shown that in S. agalactiae, a CPBP protein, Abx, forms a signaling complex with the histidine kinase CovS and regulates expression of virulence factors (74). The number of CPBP-family proteins varies greatly depending on the organism. For example, while S. pyogenes encodes only 2 or 3 CPBP-family proteins (depending on the isolates), some streptococci, such as S. sanguinis, contain as many as 21 CAAX-family proteins, and the roles of most of these proteins remain largely unknown. In this study, we added another role for a CPBP family protein to the growing list of functions.
ACKNOWLEDGMENTS
We thank Wilfred A. van der Donk, UIC, for kindly providing us purified haloduracin.
This work was supported in part by an NIH-NIDCR grant (DE22660) to I.B.
Footnotes
Published ahead of print 7 February 2014
REFERENCES
- 1.Islam MR, Nishie M, Nagao J, Zendo T, Keller S, Nakayama J, Kohda D, Sahl HG, Sonomoto K. 2012. Ring A of nukacin ISK-1: a lipid II-binding motif for type-A(II) lantibiotic. J. Am. Chem. Soc. 134:3687–3690. 10.1021/ja300007h [DOI] [PubMed] [Google Scholar]
- 2.Willey JM, van der Donk WA. 2007. Lantibiotics: peptides of diverse structure and function. Annu. Rev. Microbiol. 61:477–501. 10.1146/annurev.micro.61.080706.093501 [DOI] [PubMed] [Google Scholar]
- 3.Guder A, Wiedemann I, Sahl HG. 2000. Posttranslationally modified bacteriocins—the lantibiotics. Biopolymers 55:62–73. [DOI] [PubMed] [Google Scholar]
- 4.Bierbaum G, Sahl HG. 2009. Lantibiotics: mode of action, biosynthesis and bioengineering. Curr. Pharm. Biotechnol. 10:2–18. 10.2174/138920109787048616 [DOI] [PubMed] [Google Scholar]
- 5.Chatterjee C, Paul M, Xie L, van der Donk WA. 2005. Biosynthesis and mode of action of lantibiotics. Chem. Rev. 105:633–684. 10.1021/cr030105v [DOI] [PubMed] [Google Scholar]
- 6.Diep DB, Nes IF. 2002. Ribosomally synthesized antibacterial peptides in Gram-positive bacteria. Curr. Drug Targets 3:107–122. 10.2174/1389450024605409 [DOI] [PubMed] [Google Scholar]
- 7.Woodyer RD, Li G, Zhao H, van der Donk WA. 2007. New insight into the mechanism of methyl transfer during the biosynthesis of fosfomycin. Chem. Commun. (Camb.), p 359–361. 10.1039/B614678C [DOI] [PubMed] [Google Scholar]
- 8.Knerr PJ, van der Donk WA. 2012. Discovery, biosynthesis, and engineering of lantipeptides. Annu. Rev. Biochem. 81:479–505. 10.1146/annurev-biochem-060110-113521 [DOI] [PubMed] [Google Scholar]
- 9.Wescombe PA, Tagg JR. 2003. Purification and characterization of streptin, a type A1 lantibiotic produced by Streptococcus pyogenes. Appl. Environ. Microbiol. 69:2737–2747. 10.1128/AEM.69.5.2737-2747.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McClerren AL, Cooper LE, Quan C, Thomas PM, Kelleher NL, van der Donk WA. 2006. Discovery and in vitro biosynthesis of haloduracin, a two-component lantibiotic. Proc. Natl. Acad. Sci. U. S. A. 103:17243–17248. 10.1073/pnas.0606088103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McAuliffe O, Hill C, Ross RP. 2000. Each peptide of the two-component lantibiotic lacticin 3147 requires a separate modification enzyme for activity. Microbiology 146(Pt 9):2147–2154 [DOI] [PubMed] [Google Scholar]
- 12.Holo H, Jeknic Z, Daeschel M, Stevanovic S, Nes IF. 2001. Plantaricin W from Lactobacillus plantarum belongs to a new family of two-peptide lantibiotics. Microbiology 147:643–651 [DOI] [PubMed] [Google Scholar]
- 13.Kodani S, Lodato MA, Durrant MC, Picart F, Willey JM. 2005. SapT, a lanthionine-containing peptide involved in aerial hyphae formation in the streptomycetes. Mol. Microbiol. 58:1368–1380. 10.1111/j.1365-2958.2005.04921.x [DOI] [PubMed] [Google Scholar]
- 14.Tillotson RD, Wosten HA, Richter M, Willey JM. 1998. A surface active protein involved in aerial hyphae formation in the filamentous fungus Schizophillum commune restores the capacity of a bald mutant of the filamentous bacterium Streptomyces coelicolor to erect aerial structures. Mol. Microbiol. 30:595–602. 10.1046/j.1365-2958.1998.01093.x [DOI] [PubMed] [Google Scholar]
- 15.Voller GH, Krawczyk JM, Pesic A, Krawczyk B, Nachtigall J, Sussmuth RD. 2012. Characterization of new class III lantibiotics—erythreapeptin, avermipeptin and griseopeptin from Saccharopolyspora erythraea, Streptomyces avermitilis and Streptomyces griseus demonstrates stepwise N-terminal leader processing. Chembiochem 13:1174–1183. 10.1002/cbic.201200118 [DOI] [PubMed] [Google Scholar]
- 16.Sahl HG. 1985. Influence of the staphylococcin-like peptide Pep 5 on membrane potential of bacterial cells and cytoplasmic membrane vesicles. J. Bacteriol. 162:833–836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brotz H, Bierbaum G, Leopold K, Reynolds PE, Sahl HG. 1998. The lantibiotic mersacidin inhibits peptidoglycan synthesis by targeting lipid II. Antimicrob. Agents Chemother. 42:154–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wiedemann I, Benz R, Sahl HG. 2004. Lipid II-mediated pore formation by the peptide antibiotic nisin: a black lipid membrane study. J. Bacteriol. 186:3259–3261. 10.1128/JB.186.10.3259-3261.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wiedemann I, Breukink E, van Kraaij C, Kuipers OP, Bierbaum G, de Kruijff B, Sahl HG. 2001. Specific binding of nisin to the peptidoglycan precursor lipid II combines pore formation and inhibition of cell wall biosynthesis for potent antibiotic activity. J. Biol. Chem. 276:1772–1779. 10.1074/jbc.M006770200 [DOI] [PubMed] [Google Scholar]
- 20.Morgan SM, O'Connor PM, Cotter PD, Ross RP, Hill C. 2005. Sequential actions of the two component peptides of the lantibiotic lacticin 3147 explain its antimicrobial activity at nanomolar concentrations. Antimicrob. Agents Chemother. 49:2606–2611. 10.1128/AAC.49.7.2606-2611.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wiedemann I, Bottiger T, Bonelli RR, Wiese A, Hagge SO, Gutsmann T, Seydel U, Deegan L, Hill C, Ross P, Sahl HG. 2006. The mode of action of the lantibiotic lacticin 3147—a complex mechanism involving specific interaction of two peptides and the cell wall precursor lipid II. Mol. Microbiol. 61:285–296. 10.1111/j.1365-2958.2006.05223.x [DOI] [PubMed] [Google Scholar]
- 22.Cotter PD, Hill C, Ross RP. 2005. Bacteriocins: developing innate immunity for food. Nat. Rev. Microbiol. 3:777–788. 10.1038/nrmicro1273 [DOI] [PubMed] [Google Scholar]
- 23.Cotter PD, Hill C, Ross RP. 2005. Bacterial lantibiotics: strategies to improve therapeutic potential. Curr. Protein Pept. Sci. 6:61–75. 10.2174/1389203053027584 [DOI] [PubMed] [Google Scholar]
- 24.Heng NCK, Wescombe PA, Burton JP, Jack RW, Tagg JR. 2007. The diversity of bacteriocins in Gram-positive bacteria, p 45–92 In Riley MA, Chavan MA. (ed), Bacteriocins: ecology and evolution. Springer-Verlag, Berlin, Germany [Google Scholar]
- 25.Nes IF, Diep DB, Holo H. 2007. Bacteriocin diversity in Streptococcus and Enterococcus. J. Bacteriol. 189:1189–1198. 10.1128/JB.01254-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thomas LV, Wimpenny JW. 1996. Investigation of the effect of combined variations in temperature, pH, and NaCl concentration on nisin inhibition of Listeria monocytogenes and Staphylococcus aureus. Appl. Environ. Microbiol. 62:2006–2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Field D, Connor PM, Cotter PD, Hill C, Ross RP. 2008. The generation of nisin variants with enhanced activity against specific gram-positive pathogens. Mol. Microbiol. 69:218–230. 10.1111/j.1365-2958.2008.06279.x [DOI] [PubMed] [Google Scholar]
- 28.Gut IM, Prouty AM, Ballard JD, van der Donk WA, Blanke SR. 2008. Inhibition of Bacillus anthracis spore outgrowth by nisin. Antimicrob. Agents Chemother. 52:4281–4288. 10.1128/AAC.00625-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Carroll J, Draper LA, O'Connor PM, Coffey A, Hill C, Ross RP, Cotter PD, O'Mahony J. 2010. Comparison of the activities of the lantibiotics nisin and lacticin 3147 against clinically significant mycobacteria. Int. J. Antimicrob. Agents 36:132–136. 10.1016/j.ijantimicag.2010.03.029 [DOI] [PubMed] [Google Scholar]
- 30.Rea MC, Clayton E, O'Connor PM, Shanahan F, Kiely B, Ross RP, Hill C. 2007. Antimicrobial activity of lacticin 3147 against clinical Clostridium difficile strains. J. Med. Microbiol. 56:940–946. 10.1099/jmm.0.47085-0 [DOI] [PubMed] [Google Scholar]
- 31.Hillman JD, Novak J, Sagura E, Gutierrez JA, Brooks TA, Crowley PJ, Hess M, Azizi A, Leung K, Cvitkovitch D, Bleiweis AS. 1998. Genetic and biochemical analysis of mutacin 1140, a lantibiotic from Streptococcus mutans. Infect. Immun. 66:2743–2749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Qi F, Chen P, Caufield PW. 1999. Purification of mutacin III from group III Streptococcus mutans UA787 and genetic analyses of mutacin III biosynthesis genes. Appl. Environ. Microbiol. 65:3880–3887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Qi F, Chen P, Caufield PW. 1999. Functional analyses of the promoters in the lantibiotic mutacin II biosynthetic locus in Streptococcus mutans. Appl. Environ. Microbiol. 65:652–658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Qi F, Chen P, Caufield PW. 2000. Purification and biochemical characterization of mutacin I from the group I strain of Streptococcus mutans, CH43, and genetic analysis of mutacin I biosynthesis genes. Appl. Environ. Microbiol. 66:3221–3229. 10.1128/AEM.66.8.3221-3229.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tsang P, Merritt J, Nguyen T, Shi W, Qi F. 2005. Identification of genes associated with mutacin I production in Streptococcus mutans using random insertional mutagenesis. Microbiology 151:3947–3955. 10.1099/mic.0.28221-0 [DOI] [PubMed] [Google Scholar]
- 36.Yonezawa H, Kuramitsu HK. 2005. Genetic analysis of a unique bacteriocin, Smb, produced by Streptococcus mutans GS5. Antimicrob. Agents Chemother. 49:541–548. 10.1128/AAC.49.2.541-548.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mota-Meira M, Lacroix C, LaPointe G, Lavoie MC. 1997. Purification and structure of mutacin B-Ny266: a new lantibiotic produced by Streptococcus mutans. FEBS Lett. 410:275–279. 10.1016/S0014-5793(97)00425-0 [DOI] [PubMed] [Google Scholar]
- 38.Robson CL, Wescombe PA, Klesse NA, Tagg JR. 2007. Isolation and partial characterization of the Streptococcus mutans type AII lantibiotic mutacin K8. Microbiology 153:1631–1641. 10.1099/mic.0.2006/003756-0 [DOI] [PubMed] [Google Scholar]
- 39.Biswas S, Biswas I. 2012. Complete genome sequence of Streptococcus mutans GS-5, a serotype c strain. J. Bacteriol. 194:4787–4788. 10.1128/JB.01106-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kamiya RU, Hofling JF, Goncalves RB. 2008. Frequency and expression of mutacin biosynthesis genes in isolates of Streptococcus mutans with different mutacin-producing phenotypes. J. Med. Microbiol. 57:626–635. 10.1099/jmm.0.47749-0 [DOI] [PubMed] [Google Scholar]
- 41.Ajdic D, McShan WM, McLaughlin RE, Savic G, Chang J, Carson MB, Primeaux C, Tian R, Kenton S, Jia H, Lin S, Qian Y, Li S, Zhu H, Najar F, Lai H, White J, Roe BA, Ferretti JJ. 2002. Genome sequence of Streptococcus mutans UA159, a cariogenic dental pathogen. Proc. Natl. Acad. Sci. U. S. A. 99:14434–14439. 10.1073/pnas.172501299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Biswas S, Biswas I. 2013. SmbFT, a putative ABC transporter complex, confers protection against the lantibiotic Smb in streptococci. J. Bacteriol. 195:5592–5601. 10.1128/JB.01060-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Petersen FC, Fimland G, Scheie AA. 2006. Purification and functional studies of a potent modified quorum-sensing peptide and a two-peptide bacteriocin in Streptococcus mutans. Mol. Microbiol. 61:1322–1334. 10.1111/j.1365-2958.2006.05312.x [DOI] [PubMed] [Google Scholar]
- 44.Henningham A, Barnett TC, Maamary PG, Walker MJ. 2012. Pathogenesis of group A streptococcal infections. Discov. Med. 13:329–342 [PubMed] [Google Scholar]
- 45.Ralph AP, Carapetis JR. 2013. Group A streptococcal diseases and their global burden. Curr. Top. Microbiol. Immunol. 368:1–27. 10.1007/82_2012_280 [DOI] [PubMed] [Google Scholar]
- 46.Franker CK. 1980. Mutational loss of sensitivity to mutacin GS-5 in Streptococcus pyogenes: characterization of a mutant deficient in receptor protein. Antimicrob. Agents Chemother. 17:151–156. 10.1128/AAC.17.2.151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Perry D, Slade HD. 1978. Isolation and characterization of a Streptococcus mutans bacteriocin inhibitor from Streptococcus pyogenes. Infect. Immun. 20:578–580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Paul D, Slade HD. 1975. Production and properties of an extracellular bacteriocin from Streptococcus mutans bacteriocidal for group A and other streptococci. Infect. Immun. 12:1375–1385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Biswas I, Drake L, Johnson S, Thielen D. 2007. Unmarked gene modification in Streptococcus mutans by a cotransformation strategy with a thermosensitive plasmid. Biotechniques 42:487–490. 10.2144/000112414 [DOI] [PubMed] [Google Scholar]
- 50.Biswas I, Jha JK, Fromm N. 2008. Shuttle expression plasmids for genetic studies in Streptococcus mutans. Microbiology 154:2275–2282. 10.1099/mic.0.2008/019265-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Biswas S, Biswas I. 2011. Role of VltAB, an ABC transporter complex, in viologen tolerance in Streptococcus mutans. Antimicrob. Agents Chemother. 55:1460–1469. 10.1128/AAC.01094-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Banas JA, Biswas S, Zhu M. 2011. Effects of DNA methylation on expression of virulence genes in Streptococcus mutans. Appl. Environ. Microbiol. 77:7236–7242. 10.1128/AEM.00543-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Maguin E, Prevost H, Ehrlich SD, Gruss A. 1996. Efficient insertional mutagenesis in lactococci and other gram-positive bacteria. J. Bacteriol. 178:931–935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hossain MS, Biswas I. 2012. An extracellular protease, SepM, generates functional competence-stimulating peptide in Streptococcus mutans UA159. J. Bacteriol. 194:5886–5896. 10.1128/JB.01381-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Minton NP. 1984. Improved plasmid vectors for the isolation of translational lac gene fusions. Gene 31:269–273. 10.1016/0378-1119(84)90220-8 [DOI] [PubMed] [Google Scholar]
- 56.Biswas I, Scott JR. 2003. Identification of rocA, a positive regulator of covR expression in the group A streptococcus. J. Bacteriol. 185:3081–3090. 10.1128/JB.185.10.3081-3090.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Spellerberg B, Pohl B, Haase G, Martin S, Weber-Heynemann J, Lutticken R. 1999. Identification of genetic determinants for the hemolytic activity of Streptococcus agalactiae by ISS1 transposition. J. Bacteriol. 181:3212–3219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ward PN, Holden MT, Leigh JA, Lennard N, Bignell A, Barron A, Clark L, Quail MA, Woodward J, Barrell BG, Egan SA, Field TR, Maskell D, Kehoe M, Dowson CG, Chanter N, Whatmore AM, Bentley SD, Parkhill J. 2009. Evidence for niche adaptation in the genome of the bovine pathogen Streptococcus uberis. BMC Genomics 10:54. 10.1186/1471-2164-10-54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Thibessard A, Fernandez A, Gintz B, Decaris B, Leblond-Bourget N. 2002. Transposition of pGh9:ISS1 is random and efficient in Streptococcus thermophilus CNRZ368. Can. J. Microbiol. 48:473–478. 10.1139/w02-038 [DOI] [PubMed] [Google Scholar]
- 60.Maguin E, Duwat P, Hege T, Ehrlich D, Gruss A. 1992. New thermosensitive plasmid for gram-positive bacteria. J. Bacteriol. 174:5633–5638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pei J, Grishin NV. 2001. Type II CAAX prenyl endopeptidases belong to a novel superfamily of putative membrane-bound metalloproteases. Trends Biochem. Sci. 26:275–277. 10.1016/S0968-0004(01)01813-8 [DOI] [PubMed] [Google Scholar]
- 62.Cotter PD, Ross RP, Hill C. 2013. Bacteriocins—a viable alternative to antibiotics? Nat. Rev. Microbiol. 11:95–105. 10.1038/nrmicro2937 [DOI] [PubMed] [Google Scholar]
- 63.Diep DB, Skaugen M, Salehian Z, Holo H, Nes IF. 2007. Common mechanisms of target cell recognition and immunity for class II bacteriocins. Proc. Natl. Acad. Sci. U. S. A. 104:2384–2389. 10.1073/pnas.0608775104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kjos M, Nes IF, Diep DB. 2009. Class II one-peptide bacteriocins target a phylogenetically defined subgroup of mannose phosphotransferase systems on sensitive cells. Microbiology 155:2949–2961. 10.1099/mic.0.030015-0 [DOI] [PubMed] [Google Scholar]
- 65.Gabrielsen C, Brede DA, Hernandez PE, Nes IF, Diep DB. 2012. The maltose ABC transporter in Lactococcus lactis facilitates high-level sensitivity to the circular bacteriocin garvicin ML. Antimicrob. Agents Chemother. 56:2908–2915. 10.1128/AAC.00314-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Uzelac G, Kojic M, Lozo J, Aleksandrzak-Piekarczyk T, Gabrielsen C, Kristensen T, Nes IF, Diep DB, Topisirovic L. 2013. A Zn-dependent metallopeptidase is responsible for sensitivity to LsbB, a class II leaderless bacteriocin of Lactococcus lactis subsp. lactis BGMN1-5. J. Bacteriol. 195:5614–5621. 10.1128/JB.00859-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cornejo OE, Lefebure T, Bitar PD, Lang P, Richards VP, Eilertson K, Do T, Beighton D, Zeng L, Ahn SJ, Burne RA, Siepel A, Bustamante CD, Stanhope MJ. 2013. Evolutionary and population genomics of the cavity causing bacteria Streptococcus mutans. Mol. Biol. Evol. 30:881–893. 10.1093/molbev/mss278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Song L, Sudhakar P, Wang W, Conrads G, Brock A, Sun J, Wagner-Dobler I, Zeng AP. 2012. A genome-wide study of two-component signal transduction systems in eight newly sequenced mutans streptococci strains. BMC Genomics 13:128. 10.1186/1471-2164-13-128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Waterhouse JC, Russell RR. 2006. Dispensable genes and foreign DNA in Streptococcus mutans. Microbiology 152:1777–1788. 10.1099/mic.0.28647-0 [DOI] [PubMed] [Google Scholar]
- 70.Pei J, Mitchell DA, Dixon JE, Grishin NV. 2011. Expansion of type II CAAX proteases reveals evolutionary origin of gamma-secretase subunit APH-1. J. Mol. Biol. 410:18–26. 10.1016/j.jmb.2011.04.066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kjos M, Snipen L, Salehian Z, Nes IF, Diep DB. 2010. The Abi proteins and their involvement in bacteriocin self-immunity. J. Bacteriol. 192:2068–2076. 10.1128/JB.01553-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lux T, Nuhn M, Hakenbeck R, Reichmann P. 2007. Diversity of bacteriocins and activity spectrum in Streptococcus pneumoniae. J. Bacteriol. 189:7741–7751. 10.1128/JB.00474-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Frankel MB, Wojcik BM, DeDent AC, Missiakas DM, Schneewind O. 2010. ABI domain-containing proteins contribute to surface protein display and cell division in Staphylococcus aureus. Mol. Microbiol. 78:238–252. 10.1111/j.1365-2958.2010.07334.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Firon A, Tazi A, Da Cunha V, Brinster S, Sauvage E, Dramsi S, Golenbock DT, Glaser P, Poyart C, Trieu-Cuot P. 2013. The Abi-domain protein Abx1 interacts with the CovS histidine kinase to control virulence gene expression in group B streptococcus. PLoS Pathog. 9:e1003179. 10.1371/journal.ppat.1003179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Munro C, Michalek SM, Macrina FL. 1991. Cariogenicity of Streptococcus mutans V403 glucosyltransferase and fructosyltransferase mutants constructed by allelic exchange. Infect. Immun. 59:2316–2323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Scott JR, Guenthner PC, Malone LM, Fischetti VA. 1986. Conversion of an M− group A streptococcus to M+ by transfer of a plasmid containing an M6 gene. J. Exp. Med. 164:1641–1651. 10.1084/jem.164.5.1641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hossain MS, Biswas I. 2011. Mutacins from Streptococcus mutans UA159 are active against multiple streptococcal species. Appl. Environ. Microbiol. 77:2428–2434. 10.1128/AEM.02320-10 [DOI] [PMC free article] [PubMed] [Google Scholar]