Abstract
Comparative genomics have shown that 5% of Synechococcus elongatus PCC 7942 genes are of probable proteobacterial origin. To investigate the role of interphylum conjugation in cyanobacterial gene acquisition, we tested the ability of a set of prototype proteobacterial conjugative plasmids (RP4, pKM101, R388, R64, and F) to transfer DNA from Escherichia coli to S. elongatus. A series of BioBrick-compatible, mobilizable shuttle vectors was developed. These vectors were based on the putative origin of replication of the Synechococcus resident plasmid pANL. Not only broad-host-range plasmids, such as RP4 and R388, but also narrower-host-range plasmids, such as pKM101, all encoding MPFT-type IV secretion systems, were able to transfer plasmid DNA from E. coli to S. elongatus by conjugation. Neither MPFF nor MPFI could be used as interphylum DNA delivery agents. Reciprocally, pANL-derived cointegrates could be introduced in E. coli by electroporation, where they conferred a functional phenotype. These results suggest the existence of potentially ample channels of gene flow between proteobacteria and cyanobacteria and point to MPFT-based interphylum conjugation as a potential mechanism to explain the proteobacterial origin of a majority of S. elongatus xenologous genes.
INTRODUCTION
Horizontal gene transfer (HGT) is an outstanding player of bacterial evolution (1). Among classical HGT mechanisms, natural transformation was demonstrated in several cyanobacteria, including Synechococcus elongatus PCC 7942 (referred to here as Se7942) (2), Synechococcus sp. strain PCC 7002 (3), and Synechocystis sp. strain PCC 6803 (4), while conjugative transfer among Anabaena strains was also reported (5). Although no experimental evidence for transduction has been reported, several marine phages that contain photosynthetic genes have been detected (6, 7). This fact could indicate that photosynthetic genes are also mobilized by transduction. Comparative genomic analysis of the Se7942 and other cyanobacterial genomes identified xenologous genes based on the combination of multiple approaches: best BLAST hit out of cyanobacteria, absence of the ubiquitous octanucleotide HIP1 motif (for highly iterated palindrome 1) (8), differences in codon usage, GC index and trinucleotide skews (9). Based on these criteria, a majority of these genes (162 out of 253) probably originated from the phylum proteobacteria. These data suggest that functional mechanisms of HGT must exist, to provide a genetic bridge between phyla proteobacteria and cyanobacteria.
Conjugation has been used as a tool for introducing shuttle vectors from Escherichia coli to both pluricellular (several strains of heterocyst-forming Anabaena [10], non-heterocyst-forming Leptolyngbya sp. strain BL0902 [11], akinetes, hormogonia, and heterocyst-forming Fischerella muscicola PCC 7414, and Chlorogloeopsis fritschii PCC 6912 [12]) and unicellular (Se7942 [13, 14], several strains of marine Synechococcus [60], Prochlorococcus strain MIT9313 [15], and Synechocystis sp. strain PCC 6803 [13]) cyanobacteria. These shuttle vectors were either based on the mobilization of the promiscuous plasmid RSF1010 or in a ColE1-like origin of transfer (16). Although conjugative plasmids of several incompatibility groups were tested as helpers to mobilize these shuttle vectors, only IncP1-MOBP11 plasmids, such as RP4 and R751, were successful at transferring DNA from E. coli to cyanobacteria (17). Thus, up to now, mobilizable shuttle vectors have relied on IncP1 helper plasmids to be transferred to cyanobacterial recipients by conjugation (14, 16).
With few exceptions, proteobacterial conjugative plasmids can be grouped into five MOB families (MOBP, MOBF, MOBQ, MOBC, and MOBH) and three mating-pair formation types (MPFT, MPFF, and MPFI) (18). Natural combinations MOBP11-MPFT (present in IncP1α plasmid RP4), MOBF11-MPFT (present in IncN plasmid pKM101 and IncW plasmid R388), MOBP12-MPFI (present in IncIα plasmid R64), and MOBF12-MPFF (present in IncFI plasmid F) were tested here to investigate the range of proteobacterial conjugative systems able to conjugate DNA to cyanobacteria. These plasmids were used as helpers to mobilize a series of BioBrick-compatible shuttle vectors containing a cognate MOB from E. coli to Se7942. Such vectors were helpful to define the functional replicon of plasmid pANL. The conjugation results showed that all tested MPFT plasmids, regardless of their MOB type, were proficient for delivering DNA to cyanobacteria by conjugation, suggesting that plasmid conjugation from proteobacteria has contributed to the composition and evolution of Se7942 genome.
MATERIALS AND METHODS
Strains and culture conditions.
Strains used are detailed in Table 1. The original Se7942 strain we used was already cured of the endogenous plasmid pANS. Se7942 was cultured at 30°C in BG11 medium (19) by bubbling 1% CO2 with continuous light at 60 μmol of photons m−2 s−1. Leptolyngbya PCC 7410, Anabaena variabilis ATCC 29413, Plectonema boryanum, Nostoc punctiforme PCC 73102, and Nostoc punctiforme ATCC 29133 were cultured in BG11 at 25°C using 20 μmol of photons m−2 s−1, 10 rpm, and atmospheric CO2 conditions. The E. coli strains used were BW27783, DH5α, and β2150. They were grown at 37°C under shaking in Luria-Bertani medium (LB). Strain β2150 was supplemented with 30 μM diaminopimelic acid (DAP30). The antibiotics used for selecting cyanobacteria were neomycin at 5 or 25 μg/ml (Neo5 or Neo25), chloramphenicol at 5 or 10 μg/ml (Cm5 or Cm10), and streptomycin at 10 or 50 μg/ml (Sm10 or Sm50). The antibiotics used for selecting E. coli were kanamycin at 50 μg/ml (Km50), rifampin at 50 μg/ml (Rif50), chloramphenicol at 25 μg/ml (Cm25), nalidixic acid at 20 μg/ml (Nx20), and streptomycin at 300 μg/ml (Sm300).
TABLE 1.
Bacterial strains used in this study
| Strain | Description and/or relevant characteristicsa | Source or referenceb |
|---|---|---|
| β2150 | ΔdapA::(erm-pir) thrB1004 pro thi strA hsdS lacZΔM15 (F′ lacZΔM15 lacIq traD36 proA+ proB+); Emr Smr | 39 |
| BW27783 | lacIq rrnB3 ΔlacZ4787 hsdR514 DE(araBAD)567 DE(rhaBAD)568 DE(araFGH) Φ(ΔaraEp PCP8-araE); Nxr | 52 |
| DH5α | F− endA1 glnV44 thi-1 recA1 relA1 gyrA96 deoR nupG Φ80dlacZΔM15 Δ(lacZYA-argF)U169, hsdR17(rK− mK+) λ−; Nxr | 53 |
| Synechococcus elongatus PCC 7942 | Wild-type strain lacking plasmid pANS (NC_007604 plus NC_004073); also known as Anacystis nidulans R2-SPc; classified into the cyanobacterial section I | 54; PCC |
| GRPS1 | S. elongatus PCC 7942 with a mutation in rps12-R43; Smr | 55 |
| Leptolyngbya sp. strain PCC 7410 | Wild-type strain; classified into the cyanobacterial section III | PCC |
| Plectonema boryanum | Wild-type strain; classified into the cyanobacterial section III | PCC |
| Anabaena variabilis ATCC 29413 | Wild-type strain (NC_007413 plus NC_007410 plus NC_007411 plus NC_007412); classified into the cyanobacterial section IV | ATCC |
| Nostoc punctiforme PCC 73102 | Wild-type strain (NC_010628 plus NC_010631 plus NC_010632 + NC_010630 plus NC_010633 plus NC_010629); classified into cyanobacterial section IV | PCC |
| Nostoc punctiforme ATCC 29133 | Wild-type strain; classified into the cyanobacterial section IV | ATCC |
Emr, erythromycin resistance; Nxr, nalidixic acid resistance; Smr, streptomycin resistance.
PCC, Pasteur Culture Collection; ATCC, American Type Culture Collection.
Construction of vectors.
The plasmids and oligonucleotides are given in Table 2 and Table S1 in the supplemental material, respectively. The steps for the construction of the shuttle vectors are depicted in Fig. S1 in the supplemental material, while dislodging vectors are described in Fig. S2 and S3 in the supplemental material. Details on the construction procedures are summarized in the supplemental material.
TABLE 2.
Plasmids used in this study
| Plasmid | Descriptiona | Source or reference |
|---|---|---|
| pRL443 | Kms RP4 derivative; Apr Tcr | 51 |
| R388 | Sur Tpr | 56 |
| R64drd11 | Smr Tcr | 57 |
| pKM101 | Apr | 58 |
| pOX38 | F derivative; Cmr | 59 |
| pSB1K3 | Rep(pMB8); Kmr; backbone for BioBrick parts cloning | http://parts.igem.org/Part:pSB1K3 |
| pSB1C3 | Rep(pMB8); Cmr; backbone for BioBrick parts cloning | http://parts.igem.org/Part:pSB1C3 |
| pEXR91 | Rep(pMB8); Apr Kmr; containing gene rps12 under the promoter of the psbAI gene | 25 |
| pDEP5 | pSB1K3::(reppANL); Kmr | This study; see also Fig. S1 |
| pDEP6 | pDEP11::(reppANL); Kmr | This study; see also Fig. 2 and S1 |
| pDEP7 | pDEP12::(reppANL); Kmr | This study; see also Fig. 2 and S1 |
| pDEP8 | pDEP13::(reppANL); Kmr | This study; see also Fig. 2 and S1 |
| pDEP9 | pDEP14::(reppANL); Kmr | This study; see also Fig. 2 and S1 |
| pDEP10 | pDEP15::(reppANL); Kmr | This study; see also Fig. 2 and S1 |
| pDEP11 | pSB1K3::(oriTRP4); Kmr | This study; see also Fig. S1 |
| pDEP12 | pSB1K3::(oriTpKM101); Kmr | This study; see also Fig. S1 |
| pDEP13 | pSB1K3::(oriTR388); Kmr | This study; see also Fig. S1 |
| pDEP14 | pSB1K3::(oriTR64); Kmr | This study; see also Fig. S1 |
| pDEP15 | pSB1K3::(oriTF); Kmr | This study; see also Fig. S1 |
| pDEP16 | pSB1K3::(sepA1); Kmr | This study; see also Fig. S2 |
| pDEP17 | pSB1K3::(sepA2); Kmr | This study; see also Fig. S2 |
| pDEP18 | pSB1K3::(Ptac); Kmr | This study; see also Fig. S2 |
| pDEP19 | pDEP17::(sepA1); Kmr | This study; see also Fig. S2 |
| pDEP20 | pDEP19::(Ptac); Kmr | This study; see also Fig. S2 |
| pDEP21 | pDEP6::(Ptac-sepA1-sepA2); Kmr | This study; see also Fig. 1 and S2 |
| pDEP22 | pSB1K3::(gap2-3); Kmr | This study; see also Fig. S2 |
| pDEP23 | pDEP21::(gap2-3); Kmr | This study; see also Fig. S2 |
| pDEP24 | pSB1K3::(HS1); Kmr | This study; see also Fig. S3 |
| pDEP25 | pSB1K3::(HS2); Kmr | This study; see also Fig. S3 |
| pDEP26 | pSB1K3::(cat); Kmr Cmr | This study; see also Fig. S3 |
| pDEP27 | pSB1K3::(rsp12); Kmr | This study; see also Fig. S3 |
| pDEP28 | pDEP27::(cat); Kmr Cmr | This study; see also Fig. S3 |
| pDEP29 | pDEP25::(cat-rsp12); Kmr Cmr | This study; see also Fig. S3 |
| pDEP30 | pDEP29::(HS1); Kmr Cmr | This study; see also Fig. S3 |
| pDEP31 | pDEP6-pANL cointegrate; Kmr | This study; see also Fig. 3 |
| pDEP32 | pANL in which the maintenance region comprised between HS1 and HS2 has been replaced by rps12-cat; Kmr Cmr | This study; see also Fig. S4 |
Cmr, chloramphenicol resistance; Tcr, tetracycline resistance; Apr, ampicillin resistance; Smr, streptomycin resistance; Kmr, kanamycin resistance; Tpr, trimethoprim resistance; Sur, sulfonamide resistance.
Conjugation assays between E. coli and cyanobacteria and between E. coli.
Biparental assays were used to conjugate DNA from E. coli to cyanobacteria. They were performed at 30°C for Se7942 and at 25°C for other cyanobacterial genera. For each conjugation, a Se7942 culture sample equivalent to 15 μg of chlorophyll (around 6 × 108 cyanobacterial cells/μg of chlorophyll) was mixed with 100 μl of serial dilutions of a 109-cells/ml E. coli culture. Conjugative mixtures were placed on top of a nitrocellulose filter that was in turn placed on a BG11 plate supplemented with 5% LB plus DAP30 (LB-DAP30) for 1 h in the dark. The conjugative mixture was incubated for 24 h in the presence of light (60 μmol of photons m−2 s−1). Filters were later changed to fresh BG11 plates, incubated for an additional 24-h period, and finally transferred to BG11-Neo25 under the same conditions. Transconjugant colonies became visible after 7 to 14 days of incubation. Conjugation between E. coli cells was performed as previously described (20). Strains β2150 or BW27783 were used as donors, while DH5α was used as a recipient strain. Conjugation frequencies were expressed as the number of transconjugants per donor cell and calculated as described previously (21).
Natural transformation.
Se7942 was transformed with plasmid pDEP30 according to a protocol described earlier (22). Transformation mixtures were deposited onto nitrocellulose filters (Millipore) and incubated in BG11 plates at 30°C with continuous light for 24 h. Transformants were selected in BG11-Cm10.
Dislodging assays and analysis of cyanobacterial transconjugant colonies.
Dislodging vectors pDEP21 and pDEP23 were introduced in Se7942 by conjugation from E. coli using RP4 as helper plasmid. Individual transconjugant Se7942 colonies, carrying either pDEP21 or pDEP23, were grown in 250 ml of BG11-Neo5. Once cultures reached an optical density at 750 nm (OD750) of 2, 1.0 ml was transferred to 250 ml of BG11-Neo5. Serial dilutions were repeated for 42 or 64 generations of growth (for pDEP23 or pDEP21, respectively), when the axenic condition of these cultures was confirmed. The presence or absence of pANL was checked by PCR using the primer pairs 31/32, 33/34, and/or 35/36 when appropriate.
Transconjugant colonies of Se7942 strain GRPS1 carrying pDEP23 were serially replicated in BG11-Neo5 plates to remove E. coli cells. Axenic cultures of GRPS1 carrying pDEP23 were transformed with pDEP30 plasmid DNA. The latter plasmid contains a Synechocystis sp. strain PCC 6803 gene rps12 under the control of the psbA1 gene promoter and a cat gene flanked by two 1-kb fragments located at both sides of the pANL maintenance region (see Materials and Methods and Fig. S3 in the supplemental material). Thus, a double crossover between pANL and pDEP30 should remove pANL maintenance region. Transformant colonies were grown in BG11-Cm5 plates to improve the segregation of the mutation, which was checked by PCR (30 cycles of 94°C for 60 s, 50°C for 30 s, and 72°C for 30 s) using the primer pairs 37/38 and 39/40 (see Fig. S4 in the supplemental material). The colonies were grown in liquid medium BG11-Neo5-Sm10 to favor displacement of the pANL derivative lacking the maintenance region (pDEP32) by pDEP23. Individual colonies recovered from BG11-Neo5-Sm50 plates were analyzed by PCR (30 cycles of 94°C for 60 s, 50°C for 30 s, and 72°C for 30 s) using the primer pairs 33/34 and 35/36 to check for the absence of the pANL derivative.
To analyze the cyanobacterial transconjugants, single colonies were streaked out twice in BG11-Neo5 plates to remove E. coli donors. Transconjugants were then grown in BG11-Neo5 up to an OD750 of 1 to 1.5. The axenic condition of these cultures was tested in LB-DAP30. Plasmid DNA was isolated using GenElute TM Plasmid Miniprep kit (Sigma-Aldrich) and used to transform E. coli DH5α by electroporation. Kanamycin-resistant (Kmr) E. coli transformants were analyzed by electrophoresis of plasmid DNAs with EcoRI and PstI in 1% agarose gel run in 0.5× TBE buffer (44.5 mM Tris borate, 44.5 mM boric acid, 1 mM EDTA [pH 8.2 to 8.4]). Gels were stained with Real-Safe (Real) and developed in a Gel Doc Imager (Bio-Rad).
Chromate resistance test.
E. coli DH5α was independently transformed with cointegrate pDEP31 or vector pDEP6 by electroporation. Saturated cultures from single transformant colonies, grown in LB-Km50, were used to inoculate 96-well plates containing 150 μl of LB-Km50 per well and different concentrations of K2CrO4. Plates were incubated at 37°C. Bacterial growth was monitored based on the OD600 in a Victor3 plate reader (Perkin-Elmer). Generation times were calculated as ln(2)/k, where k represents the growth rate and corresponds to the slope of the exponential growth phase (three experiments, eight replicas per experiment).
RESULTS
Dislodging vectors to displace the indigenous Se7942 plasmid pANL.
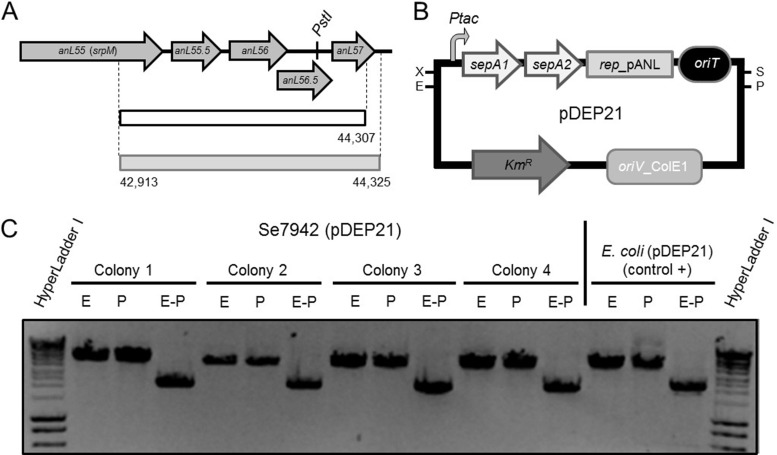
The 46.3-kb pANL plasmid, indigenous to Se7942, could potentially interfere with the conjugation or stability of other plasmids, hence our interest in attempting pANL plasmid curing. Several shuttle plasmids were built, all based on a 1,395-bp DNA segment containing pANL putative origin of replication (23). The replication region (here named rep_pANL) that was cloned to construct the pDEP vector series included an additional 18-bp fragment to complete the coding region of gene anL57 (Fig. 1A).
FIG 1.
pDEP21 autoreplicates in Se7942. (A) Genetic organization of the putative replication region of pANL. The replication sequence described previously (23) is indicated by a white box outlined in black, while the segment used in the present study as rep_pANL is indicated by a gray box outlined in gray. (B) Genetic map of the dislodging vector pDEP21. (C) Plasmid DNA recovered from Se7942(pDEP21) cultured in liquid BG11-Neo5 for 64 generations was restricted with EcoRI (E) and/or PstI (P). Plasmid DNA directly extracted from E. coli DH5α(pDEP21) without any passage through Se7942 was used as a control in the restriction analysis.
pDEP21 was built as a dislodging vector containing the pANL replication region in order to remove plasmid pANL from Se7942 by vectorial plasmid incompatibility under selective pressure (24) (Fig. 1B and see Fig. S2 in the supplemental material). Since plasmid pANL encodes two toxin-antitoxin systems, both pANL antitoxin genes (sepA1 and sepA2) were also included in pDEP21 to avoid killing pANL segregants. The dislodging plasmid pDEP21 also contains oriT_RP4 to allow conjugation from E. coli to cyanobacteria. Transconjugant colonies were subcultured in liquid BG11-Neo5 for 64 generations. Ten individual colonies were analyzed by PCR using primers that specifically hybridized to pANL and not to pDEP21. They all rendered amplicons congruent with the presence of pANL. In addition, plasmid DNA isolated from four independent colonies, transformed into E. coli and subjected to restriction analysis, showed no differences in restriction pattern between the recovered plasmid DNAs and the original dislodging vector pEDP21 (Fig. 1C), indicating that it could be autonomously maintained in Se7942 but was not able to completely displace pANL.
In another attempt to remove pANL, regions named gap2 and gap3, described as essential in an earlier study (23), were incorporated into the dislodging vector, thus producing pDEP23 (see Fig. S2 in the supplemental material). This vector was introduced in Se7942 by conjugation. Transconjugant colonies were cultured for 42 generations in BG11-Neo5 and plated out again, and plasmid DNA was extracted from single colonies. Restriction analysis showed a cointegrate between plasmids pDEP23 and pANL. No instance of pANL curing was detected.
A third attempt to remove pANL was carried out by deleting its maintenance region, previously described to be essential for stable carriage of pANL in Se7942 (23). This region is composed of two toxin-antitoxin system cassettes (sepA1-sepT1 and sepA2-sepT2), a set of partition genes (parA and parB), and an orf encoding a putative nucleotidyltransferase (anL30). Deletion of this pANL segment (coordinates 20984 to 24487 under GenBank accession no. AF441790) was carried out by homologous recombination in the Se7942 strain GRPS1, which is used for the construction of gene-replacement mutants (25). Plasmid pDEP30 (see Fig. S3 in the supplemental material) was introduced into strain GRPS1(pDEP23). The flanking areas of the pANL maintenance region surround genes rps12 and cat in pDEP30, which confer a dominant streptomycin-sensitive (Sms), chloramphenicol-resistant (Cmr) phenotype. Deletion of the maintenance region was confirmed by PCR (see Fig. S4 in the supplemental material), and the deleted pANL derivative was named pDEP32. To favor pDEP23 in the competition with pDEP32, GRPS1 was grown in Neo25-Sm50. No GRPS1 colonies free of pDEP32 were detected. All Smr neomycin-resistant (Neor) colonies tested contained a cointegrate between pDEP23 and pDEP32 (data not shown). In conclusion, all attempts at curing pANL failed, suggesting that this plasmid contains some genes that are essential for Se7942 viability under the tested conditions.
Mobilization of shuttle vectors from E. coli to Se7942 using different prototype proteobacterial conjugative plasmids.
To test for the ability of different MPF systems to transfer plasmid DNA by conjugation to cyanobacteria, we tested the mobilization of a series of plasmid oriTs by their cognate conjugative plasmids from E. coli to Se7942.
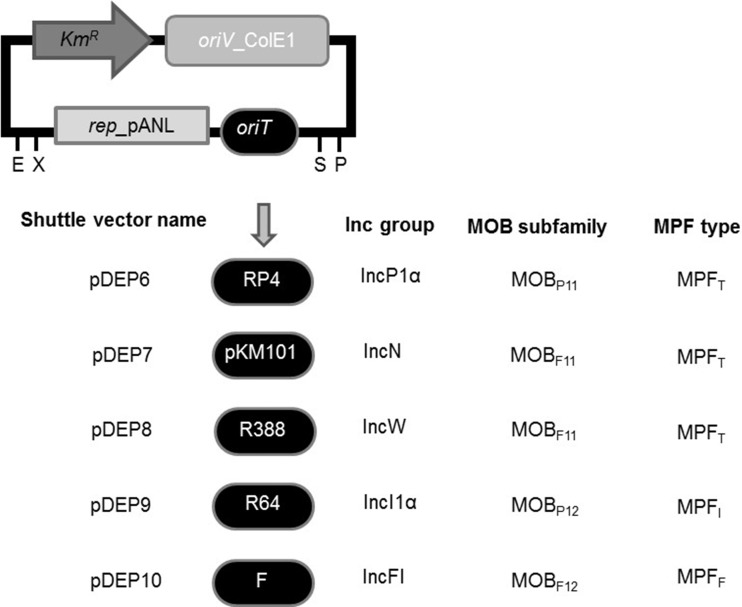
Five prototype conjugative plasmids were tested (RP4, pKM101, R388, R64 and F) for transfer from E. coli to Se7942 (Fig. 2). They represent five frequently found incompatibility groups (Inc) in gammaproteobacteria, including four MOB subfamilies and three MPF types, thus comprising a representation of the diversity of proteobacterial mobility systems (18). First, mobilization of the shuttle vectors was tested by E. coli intraspecies crosses. As shown in Table 3 (column 3), all shuttle vectors were mobilized by their cognate conjugative plasmid between E. coli strains at frequencies of roughly 10−1 transconjugants/donor cell, indicating that the helper plasmids were efficient in promoting mobilization of their cognate mobilizable vectors. When mobilization of the same shuttle vectors was tested, but using Se7942 as a recipient (Table 3, column 4), vectors containing the oriTs of RP4, pKM101, and R388 produced cyanobacterial Neor transconjugants. RP4, pKM101, and R388 transconjugants were obtained at frequencies of ∼10−3 in the mobilization of their cognate pDEP vectors. On the other hand, plasmids R64 and F showed conjugation frequencies undistinguishable from background levels. The temperature conditions used for Se7942 growth and mating were critical, since pDEP6 mobilization drastically dropped to background levels when the mating temperature was shifted from 30°C to 35, 37, or 40°C (data not shown). To rule out natural transformation as the cause of Se7942 Neor colonies, a control assay was carried out by repeating the conjugation experiment, using as the donor an E. coli strain containing vector pDEP5. Since pDEP5 is devoid of oriT, transfer of its Neor marker can occur only by natural transformation. The results shown in Table 3 (column 5) indicate that natural transformation was extremely inefficient under these conditions, occurring at frequencies not exceeding 10−8. This in turn suggests that the number of transconjugants obtained using pDEP9 and pDEP10 as shuttle vectors (respectively mobilized by R64 and F) was within the range of natural transformation efficiency.
FIG 2.
Shuttle vector series. Mobilizable vectors are based on plasmid pSB1K3. All contain the rep region of plasmid pANL. Each one includes the origin of transfer (oriT) from a different prototype conjugative plasmid for which the Inc group, relaxase MOB family, and MPF type are indicated.
TABLE 3.
Conjugative frequencies between E. coli and from E. coli to Se7942
| Plasmid contained in donor straina | Inc MOB MPF typeb | Mobilization frequencyc: |
||
|---|---|---|---|---|
| Between E. coli strains | From E. coli to Se7942 | Of ΔoriT derivativesd | ||
| RP4+pDEP6e | IncP1α MOBP11 MPFT | 4.8 × 10−1 (8.6 × 10−2–2.7) | 2.2 × 10−3 (5.0 × 10−4–1 × 10−2) | 5 × 10−9 (9 × 10−10–2 × 10−8) |
| pKM101+pDEP7 | IncN MOBF11 MPFT | 5.2 × 10−1 (3.1 × 10−1–8.4 × 10−1) | 1.2 × 10−3 (4.5 × 10−4–3.3 × 10−3) | 2 × 10−9 (6 × 10−10–1 × 10−8) |
| R388+pDEP8 | IncW MOBF11 MPFT | 5.0 × 10−2 (1.9 × 10−2–1.3 × 10−1) | 8.9 × 10−3 (2.5 × 10−3–3.1 × 10−2) | 1 × 10−8 (6 × 10−9 –2 × 10−8) |
| R64drd11+pDEP9 | IncI1α MOBP12 MPFI | 8.5 × 10−2 (6.1 × 10−2–1.2 × 10−1) | <10−9 | 4 × 10−10 (8 × 10−11–2 × 10−9) |
| F+pDEP10f | IncFI MOBF12 MPFF | 4.8 × 10−1 (3.8 × 10−1–5.9 × 10−1) | <10−9 | 1 × 10−9 (3 × 10−10–7 × 10−9) |
Donor strains were derivatives of E. coli strain β2150 containing the plasmids shown in the first column. Strain BW27783 was used as the donor in the case of plasmid R388, since β2150 was inhibitory to R388 conjugation, due to an inhibitory effect of the integrated F-plasmid (39; our unpublished results).
For a description of MOB and MPF types, see reference 18.
Mobilization frequencies are the average of at least six experiments. They were calculated by the log conversion of frequencies (number of transconjugants per donor cell) to obtain the means and standard deviations (indicated in parentheses), which are expressed as the anti-log of the calculated figures.
The same conjugation experiment from E. coli to Se7942 was conducted using the helper plasmids in combination with plasmid pDEP5, a nonmobilizable vector, to determine the background level of Neor Se7942 colonies that could arise in the mating experiments due to natural transformation. Transformation frequencies were calculated as the number of transforming cells/number of donor cells.
pRL443, an RP4 derivative sensitive to kanamycin, was used as a helper plasmid.
pOX38, a Cmr F derivative, was used as a helper plasmid.
Finally, experiments to mobilize pDEP6 (the vector containing oriT_RP4) to other cyanobacterial genera using RP4 as a helper plasmid were carried out using a diversity of cyanobacterial recipients: Leptolyngbya PCC 7410, Anabaena variabilis ATCC 29413, Plectonema boryanum, Nostoc punctiforme PCC 7310, and Nostoc punctiforme ATCC 29133. No transconjugants were obtained in any of these cases (data not shown). Since RP4 could mobilize RSF1010 to different cyanobacteria (16), it can be assumed that plasmid pANL (or its derivatives) cannot replicate in those cyanobacteria.
Analysis of pEDP6 in Se7942 transconjugants.
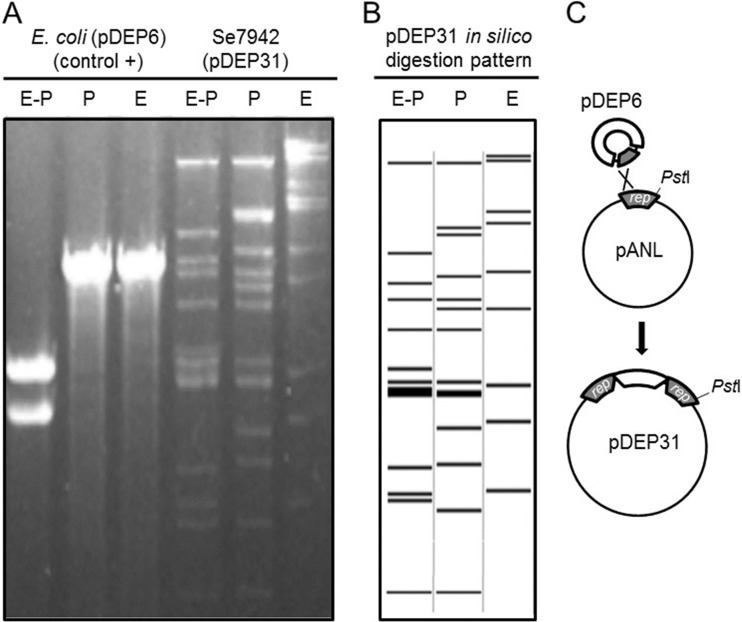
To test whether pDEP6 could be maintained as an autonomous replicon in Se7942, plasmid DNA was extracted from four transconjugant colonies, transformed to E. coli to amplify the amount of plasmid DNA, and analyzed with restriction enzymes. All plasmid preparations recovered from Se7942 rendered the same restriction pattern. It was different from the original pDEP6 plasmid and consistent with it being a cointegrate (named pDEP31), formed by homologous recombination between pDEP6 and pANL rep regions, as shown in Fig. 3. Since pDEP31 contains the sulfate/chromate uptake operon of pANL (srp operon [23, 26]), it should result in increased chromate sensitivity of E. coli cells containing cointegrate pDEP31. This was proven by analysis of chromate sensitivity (see Fig. S5 in the supplemental material), which showed increased sensitivity to increasing amounts of chromate in DH5α (pEDP31) with respect to the control strain of DH5α (pDEP6). This result further indicates that the srp operon of Se7942 is adequately expressed in E. coli.
FIG 3.
Analysis of transconjugant plasmid DNAs produced by pDEP6 conjugation to Se7942. The figure shows the electrophoretic analysis of DNA bands obtained by restriction analysis of transconjugant plasmid DNA. (A) Plasmid DNA recovered from Se7942(pDEP6) transconjugants (i.e., pEDP31) was digested with EcoRI (E) and/or PstI (P) and developed by 1% agarose gel electrophoresis. Plasmid pDEP6 DNA (isolated from E. coli DH5α) was used as a control in the restriction analysis. (B) In silico restriction analysis of pDEP6-pANL cointegrate pDEP31 assuming a single crossover across the pANL rep region. (C) Schematic representation of the single crossover leading to pDEP31 formation, according to the results shown in panels A and B. The position of the single PstI site present in pANL replication region, which is not present in pDEP6, helps to determine the direction of the pDEP6 insertion.
DISCUSSION
The main purposes of the present study were to determine which of the main conjugative systems from proteobacteria were able to transfer DNA to cyanobacteria by conjugation, to compare their relative efficiencies, and to optimize the conjugation protocol. The finding that all MPFT-type plasmids were efficient donors is probably the most relevant result. It indicates that conjugation of proteobacterial plasmids is a probable source of xenologous genes in Se7942, as suggested previously (9).
For a start, and in order to optimize Se7942 as a conjugation recipient, we attempted to remove the indigenous pANL plasmid from Se7942 by using a plasmid incompatibility strategy. This technique is based in the fact that, when two plasmids carrying the same origin of replication coexist in a cell, they become unstable due to interactions between their replication machineries. Vectorial incompatibility (one of the plasmids is lost with higher probability than the other [24]) was previously exploited to cure native plasmids from Agrobacterium tumefaciens (27, 28), Bacillus anthracis (29), or Yersinia pestis (30), among many other examples. To cure pANL, two dislodging vectors were built: pDEP21 and pDEP23. They contain the proposed minimal replication region of plasmid pANL, defined as discussed in the supplemental material. When pDEP21 or pDEP23 were mobilized to Se7942 by RP4, pANL was not cured. Not even a pANL derivative lacking its maintenance region could be displaced by the dislodging vectors by applying selection to favor them in the competition. Thus, either the native pANL contains one or more essential genes that we did not include in our constructs, or the origin of replication described earlier (23) is incomplete, and some additional functions provided by pANL are indispensable for replication or maintenance of the dislodging vector. DNA extraction of transconjugant colonies showed that, in some cases, the pDEP derivative was still an autonomous replicon (Fig. 1), while in others it formed a cointegrate with pANL (Fig. 3). This result could occur if the incoming plasmid is unstable (by incompatibility) but cannot completely dislodge the resident plasmid, as discussed above. In our experiments, the restriction pattern of plasmid pDEP21 was not altered, indicating that it can remain as an autonomous plasmid in Se7942. On the other hand, plasmids pDEP6 and pDEP23, which contain the same rep_pANL as pDEP21, were always found forming cointegrates with pANL. Their different behavior remains unexplained. In any case, pANL could not be dislodged so all conjugation experiments were carried out in a Se7942 containing pANL.
Interphylum conjugation from E. coli to cyanobacteria is carried out in the laboratory solely by using an RP4-based helper plasmid (10, 16). Conjugation has been used as an alternative to transformation for the insertion of foreign genes in the Se7942 chromosome (22, 31). Mobilizable shuttle vectors were based either on plasmid pBR322 oriT (while MOB and MPF functions were provided in trans by a ColE1-like plasmid and RP4, respectively) (32), or on RSF1010, which is mobilized by RP4 (31). pBR322-based vectors were used to study whether plasmids other than RP4 supported mobilization to Anabaena strains M-131 or PCC 7120 (17). Other IncP1 plasmids could mobilize such vectors, while IncW plasmids could not. It should be pointed out that the transfer efficiency of a given mobilizable plasmid depends on the conjugative plasmid used as helper, because the relaxosome provided by the mobilizable plasmid should make appropriate contacts with the coupling protein and the mating apparatus provided by the conjugative plasmid. For example, RP4 mobilizes ColE1 and RSF1010 between E. coli strains 2 and 4 log more efficiently, respectively, than the IncW plasmid R388 (33). Thus, it is not surprising that IncW plasmids were unable to sustain conjugation to Anabaena in the referred conditions (17).
In the present study we used plasmids RP4, pKM101, R388, R64, and F, which represent the diversity of conjugative systems in proteobacteria (18), as potential donors in conjugation from proteobacteria to cyanobacteria. The relevant shuttle vectors contained always the same oriT as the conjugative plasmid used as helper, to avoid the above-mentioned inefficiencies due to heterologous interactions. This fact allows attention to be focused on the relative proficiency of each MPF type, not being distracted by the interactions between MPF and MOB modules. Actual transfer of a conjugative plasmid is an indication of two capabilities: first, its ability to invade the host population and, second, its efficiency of replicating in that host. Even in the absence of replication, the invasive plasmid DNA can recombine with the host genome (given the appropriate recombination sites), thus integrating within the chromosome and becoming an integrative and conjugative element (34). The host range and therefore the promiscuity of a plasmid can be inferred from sequence data (35). This type of analysis established that IncP1 plasmids (such as RP4) show a broad host range, in accordance with the fact that RP4 conjugates to cyanobacteria (10) and even to yeast (36). The same analysis indicated that IncW (R388) plasmids have a conjugative range perhaps broader than IncP, IncN (pKM101) plasmids have an intermediate range, and IncI (R64) or IncF (F) plasmids are narrow host range. Ample experimental evidence confirms these assumptions (37, 38).
Results shown in Table 3 indicate that all three conjugative plasmids with an MPFT-type transport channel (i.e., RP4, pKM101, and R388) were able to achieve interphylum conjugation from E. coli to Se7942. The other two plasmids, F and R64, which contain MPFF- or MPFI-type conjugation systems (Fig. 2), could not introduce DNA in Se7942. These results suggest that different MPF types exhibit different abilities to conjugate to cyanobacteria. All transfer systems are able to transfer DNA to a much wider range of recipients than the replication ability of the vector plasmids (hence, suicide vectors used for gene delivery by conjugation [39, 40] or the induction of SOS response toward invading DNA [41]). Thus, our results suggest that neither MPFF nor MPFI could deliver DNA to cyanobacteria, whereas all three MPFT plasmids tested, irrespective of their MOB type, could do it. Moreover, the fact that pANL-based conjugation can occur through a variety of conjugative systems from E. coli to Se7942, but not to other cyanobacteria, suggests that pANL has a narrow replication host range.
What is special about MPFT? It is known that MPFT, MPFI, and MPFF share homology in some protein components but also contain specific components that are signatures for each group (18). Thus, one possibility is that these differences could be responsible for the increased promiscuity of MPFT-type elements. Curiously, transkingdom transfer from bacteria to yeast and plants has only been reported for MPFT conjugal types (42–44). There is scarce information on factors that determine the range of potential recipients of a given mating system. TraN, one of the MPFF components of plasmid F, interacts with the outer membrane protein OmpA. This interaction results in stabilization of conjugation partners and is necessary for efficient mobilization (45). Adhesin PilV of the thin pilus encoded by plasmid R64 specifically interacts with the lipopolysaccharide of the recipient cells, determining recipient specificity (46, 47). Receptors similar to OmpA or specific lipopolysaccharide components might be lacking in cyanobacteria and thus prevent the interphylum conjugation of MPFF (F) or MPFI (R64) plasmids, this being a second alternative to explain MPFT enhanced promiscuity. In any case, our results clearly demonstrate that conjugation to cyanobacteria is not limited to IncP1 plasmids but involves many MPFT plasmids. These results broaden the number of conjugative systems that can be used for the genetic manipulation of cyanobacteria and explain the origin of Se7942 xenologous genes.
But genetic exchange between proteobacteria and cyanobacteria could occur in both directions. In fact, E. coli cells harboring a cointegrate between the shuttle vector pDEP6 and the endogenous Se7942 plasmid pANL, which contains the complete sulfur-regulated region of pANL, exhibited increased generation time in the presence of chromate (see Fig. S5 in the supplemental material), as occurred in Se7942 (26). When encoded out of the context of the sulfur-regulated gene cluster, gene srpC conferred chromate resistance to E. coli (48). On the contrary, a Se7942 srpC deletion mutant exhibited a lower doubling time than the wild-type Se7942 in the presence of chromate (26). The reproduction of a Se7942 phenotype in E. coli as a consequence of the presence of the pANL genome suggests that the genetic flow between proteobacteria and cyanobacteria could be bidirectional.
Finally, it should be emphasized that several parameters of the conjugation protocol were optimized during the course of the present study to maximize conjugation frequencies to cyanobacteria. First, conjugation worked best at 30°C, with few or no transconjugants obtained at higher temperatures (35, 37, and 40°C were tested), although Se7942 growth rate is maximal at 41°C. Second, E. coli strain β2150 was used as a donor. This strain is auxotrophic for DAP, which helped killing donor E. coli cells during the selection regime. Third, several donor/recipient ratios were assayed for each mating experiment in order to obtain countable colonies in one of each series of filters. Other barriers to transphylum conjugation might exist that affect the efficiency of transphylum conjugation, including restriction/modification systems and CRISPRs, among others. Se7942 contains both (49, 50; our unpublished data). In our conjugation assays, the incoming plasmid DNAs were not methylated for Se7942. It would be interesting to know whether an appropriate modification, such as that devised for RP4-based conjugation to Anabaena, which was based on the methylases that protect DNA against restriction by AvaI, AvaII, and AvaIII (51), would result in still higher conjugation frequencies.
Supplementary Material
ACKNOWLEDGMENTS
Work in the FdlC laboratory was financed by the Spanish Ministry of Economy and Competitivity (MINECO) grant BFU2011-26608 and the European Seventh Framework Program (282004/FP7-HEALTH-2011-2.3.1-2). M.S.-M. is received a Ph.D. fellowship from MINECO. M.P.G.-B. received a JAE-Doc contract from CSIC, cofinanced by the European Science Foundation.
We thank Paula Tamagnini for allowing D.E. to conduct some conjugation experiments in her laboratory, the Pasteur Culture Collection (Institute Pasteur, Paris, France) for supplying Se7942, and K. Takahama for kindly sharing Se7942 strain GRPS1.
Footnotes
Published ahead of print 7 February 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.01464-13.
REFERENCES
- 1.de la Cruz F, Davies J. 2000. Horizontal gene transfer and the origin of species: lessons from bacteria. Trends Microbiol. 8:128–133. 10.1016/S0966-842X(00)01703-0 [DOI] [PubMed] [Google Scholar]
- 2.Shestakov SV, Khyen NT. 1970. Evidence for genetic transformation in blue-green alga Anacystis nidulans. Mol. Gen. Genet. 107:372–375. 10.1007/BF00441199 [DOI] [PubMed] [Google Scholar]
- 3.Stevens SE, Porter RD. 1980. Transformation in Agmenellum quadruplicatum. Proc. Natl. Acad. Sci. U. S. A. 77:6052–6056. 10.1073/pnas.77.10.6052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Grigorieva G, Shestakov S. 1982. Transformation in the cyanobacterium Synechocystis Sp6803. FEMS Microbiol. Lett. 13:367–370. 10.1111/j.1574-6968.1982.tb08289.x [DOI] [Google Scholar]
- 5.Muro-Pastor AM, Kuritz T, Flores E, Herrero A, Wolk CP. 1994. Transfer of a genetic marker from a megaplasmid of Anabaena sp. strain PCC 7120 to a megaplasmid of a different Anabaena strain. J. Bacteriol. 176:1093–1098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mann NH, Clokie MR, Millard A, Cook A, Wilson WH, Wheatley PJ, Letarov A, Krisch HM. 2005. The genome of S-PM2, a “photosynthetic” T4-type bacteriophage that infects marine Synechococcus strains. J. Bacteriol. 187:3188–3200. 10.1128/JB.187.9.3188-3200.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Weigele PR, Pope WH, Pedulla ML, Houtz JM, Smith AL, Conway JF, King J, Hatfull GF, Lawrence JG, Hendrix RW. 2007. Genomic and structural analysis of Syn9, a cyanophage infecting marine Prochlorococcus and Synechococcus. Environ. Microbiol. 9:1675–1695. 10.1111/j.1462-2920.2007.01285.x [DOI] [PubMed] [Google Scholar]
- 8.Delaye L, Moya A. 2011. Abundance and distribution of the highly iterated palindrome 1 (HIP1) among prokaryotes. Mobile Genet. Elements 1:159–168. 10.4161/mge.1.3.18300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Delaye L, Gonzalez-Domenech CM, Garcillan-Barcia MP, Pereto J, de la Cruz F, Moya A. 2011. Blueprint for a minimal photoautotrophic cell: conserved and variable genes in Synechococcus elongatus PCC 7942. BMC Genomics 12:25. 10.1186/1471-2164-12-25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wolk CP, Vonshak A, Kehoe P, Elhai J. 1984. Construction of shuttle vectors capable of conjugative transfer from Escherichia coli to nitrogen-fixing filamentous cyanobacteria. Proc. Natl. Acad. Sci. U. S. A. 81:1561–1565. 10.1073/pnas.81.5.1561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Taton A, Lis E, Adin DM, Dong G, Cookson S, Kay SA, Golden SS, Golden JW. 2012. Gene transfer in Leptolyngbya sp. strain BL0902, a cyanobacterium suitable for production of biomass and bioproducts. PLoS One 7:e30901. 10.1371/journal.pone.0030901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stucken K, Ilhan J, Roettger M, Dagan T, Martin WF. 2012. Transformation and conjugal transfer of foreign genes into the filamentous multicellular cyanobacteria (subsection V) Fischerella and Chlorogloeopsis. Curr. Microbiol. 65:552–560. 10.1007/s00284-012-0193-5 [DOI] [PubMed] [Google Scholar]
- 13.Marraccini P, Bulteau S, Cassier-Chauvat C, Mermet-Bouvier P, Chauvat F. 1993. A conjugative plasmid vector for promoter analysis in several cyanobacteria of the genera Synechococcus and Synechocystis. Plant Mol. Biol. 23:905–909. 10.1007/BF00021546 [DOI] [PubMed] [Google Scholar]
- 14.Tsinoremas NF, Kutach AK, Strayer CA, Golden SS. 1994. Efficient gene transfer in Synechococcus sp. strains PCC 7942 and PCC 6301 by interspecies conjugation and chromosomal recombination. J. Bacteriol. 176:6764–6768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tolonen AC, Liszt GB, Hess WR. 2006. Genetic manipulation of Prochlorococcus strain MIT9313: green fluorescent protein expression from an RSF1010 plasmid and Tn5 transposition. Appl. Environ. Microbiol. 72:7607–7613. 10.1128/AEM.02034-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Koksharova OA, Wolk CP. 2002. Genetic tools for cyanobacteria. Appl. Microbiol. Biotechnol. 58:123–137. 10.1007/s00253-001-0864-9 [DOI] [PubMed] [Google Scholar]
- 17.Thiel T, Wolk CP. 1987. Conjugal transfer of plasmids to cyanobacteria. Methods Enzymol. 153:232–243. 10.1016/0076-6879(87)53056-7 [DOI] [PubMed] [Google Scholar]
- 18.Smillie C, Garcillan-Barcia MP, Francia MV, Rocha EP, de la Cruz F. 2010. Mobility of plasmids. Microbiol. Mol. Biol. Rev. 74:434–452. 10.1128/MMBR.00020-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rippka R, Deruelles J, Waterbury JB, Herdman M, Stanier RY. 1979. Generic assignments, strain histories, and properties of pure cultures of cyanobacteria. J. Gen. Microbiol. 111:1–61. 10.1099/00221287-111-1-1 [DOI] [Google Scholar]
- 20.del Campo I, Ruiz R, Cuevas A, Revilla C, Vielva L, de la Cruz F. 2012. Determination of conjugation rates on solid surfaces. Plasmid 67:174–182. 10.1016/j.plasmid.2012.01.008 [DOI] [PubMed] [Google Scholar]
- 21.Garcillan-Barcia MP, Jurado P, Gonzalez-Perez B, Moncalian G, Fernandez LA, de la Cruz F. 2007. Conjugative transfer can be inhibited by blocking relaxase activity within recipient cells with intrabodies. Mol. Microbiol. 63:404–416. 10.1111/j.1365-2958.2006.05523.x [DOI] [PubMed] [Google Scholar]
- 22.Golden SS, Sherman LA. 1984. Optimal conditions for genetic transformation of the cyanobacterium Anacystis nidulans R2. J. Bacteriol. 158:36–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen Y, Holtman CK, Magnuson RD, Youderian PA, Golden SS. 2008. The complete sequence and functional analysis of pANL, the large plasmid of the unicellular freshwater cyanobacterium Synechococcus elongatus PCC 7942. Plasmid 59:176–192. 10.1016/j.plasmid.2008.01.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Novick RP. 1987. Plasmid incompatibility. Microbiol. Rev. 51:381–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Takahama K, Matsuoka M, Nagahama K, Ogawa T. 2004. High-frequency gene replacement in cyanobacteria using a heterologous rps12 gene. Plant Cell Physiol. 45:333–339. 10.1093/pcp/pch041 [DOI] [PubMed] [Google Scholar]
- 26.Nicholson ML, Laudenbach DE. 1995. Genes encoded on a cyanobacterial plasmid are transcriptionally regulated by sulfur availability and CysR. J. Bacteriol. 177:2143–2150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Uraji M, Suzuki K, Yoshida K. 2001. A new method for construction of Ti plasmid-less strains in Agrobacterium tumefaciens. Nucleic Acids Symp. Ser. 1:173–174. 10.1093/nass/1.1.173 [DOI] [PubMed] [Google Scholar]
- 28.Uraji M, Suzuki K, Yoshida K. 2002. A novel plasmid curing method using incompatibility of plant-pathogenic Ti plasmids in Agrobacterium tumefaciens. Genes Genet. Syst. 77:1–9. 10.1266/ggs.77.1 [DOI] [PubMed] [Google Scholar]
- 29.Liu X, Wang D, Wang H, Feng E, Zhu L. 2012. Curing of plasmid pXO1 from Bacillus anthracis using plasmid incompatibility. PLoS One 7:e29875. 10.1371/journal.pone.0029875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ni B, Du Z, Guo Z, Zhang Y, Yang R. 2008. Curing of four different plasmids in Yersinia pestis using plasmid incompatibility. Lett. Appl. Microbiol. 47:235–240. 10.1111/j.1472-765X.2008.02426.x [DOI] [PubMed] [Google Scholar]
- 31.Muhlenhoff U, Chauvat F. 1996. Gene transfer and manipulation in the thermophilic cyanobacterium Synechococcus elongatus. Mol. Gen. Genet. 252:93–100. 10.1007/BF02173209 [DOI] [PubMed] [Google Scholar]
- 32.Golden SS, Sherman LA. 1983. A hybrid plasmid is a stable cloning vector for the cyanobacterium Anacystis nidulans R2. J. Bacteriol. 155:966–972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cabezon E, Sastre JI, de la Cruz F. 1997. Genetic evidence of a coupling role for the TraG protein family in bacterial conjugation. Mol. Gen. Genet. 254:400–406. 10.1007/s004380050432 [DOI] [PubMed] [Google Scholar]
- 34.Guglielmini J, Quintais L, Garcillan-Barcia MP, de la Cruz F, Rocha EP. 2011. The repertoire of ICE in prokaryotes underscores the unity, diversity, and ubiquity of conjugation. PLoS Genet. 7:e1002222. 10.1371/journal.pgen.1002222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Suzuki H, Yano H, Brown CJ, Top EM. 2010. Predicting plasmid promiscuity based on genomic signature. J. Bacteriol. 192:6045–6055. 10.1128/JB.00277-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Heinemann JA, Sprague GF., Jr 1989. Bacterial conjugative plasmids mobilize DNA transfer between bacteria and yeast. Nature 340:205–209. 10.1038/340205a0 [DOI] [PubMed] [Google Scholar]
- 37.Mazodier P, Davies J. 1991. Gene transfer between distantly related bacteria. Annu. Rev. Genet. 25:147–171. 10.1146/annurev.ge.25.120191.001051 [DOI] [PubMed] [Google Scholar]
- 38.Kues U, Stahl U. 1989. Replication of plasmids in gram-negative bacteria. Microbiol. Rev. 53:491–516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Demarre G, Guerout AM, Matsumoto-Mashimo C, Rowe-Magnus DA, Marliere P, Mazel D. 2005. A new family of mobilizable suicide plasmids based on broad-host-range R388 plasmid (IncW) and RP4 plasmid (IncPα) conjugative machineries and their cognate Escherichia coli host strains. Res. Microbiol. 156:245–255. 10.1016/j.resmic.2004.09.007 [DOI] [PubMed] [Google Scholar]
- 40.Smorawinska M, Szuplewska M, Zaleski P, Wawrzyniak P, Maj A, Plucienniczak A, Bartosik D. 2012. Mobilizable narrow host range plasmids as natural suicide vectors enabling horizontal gene transfer among distantly related bacterial species. FEMS Microbiol. Lett. 326:76–82. 10.1111/j.1574-6968.2011.02432.x [DOI] [PubMed] [Google Scholar]
- 41.Baharoglu Z, Bikard D, Mazel D. 2010. Conjugative DNA transfer induces the bacterial SOS response and promotes antibiotic resistance development through integron activation. PLoS Genet. 6:e1001165. 10.1371/journal.pgen.1001165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bates S, Cashmore AM, Wilkins BM. 1998. IncP plasmids are unusually effective in mediating conjugation of Escherichia coli and Saccharomyces cerevisiae: involvement of the tra2 mating system. J. Bacteriol. 180:6538–6543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zambryski P, Tempe J, Schell J. 1989. Transfer and function of T-DNA genes from agrobacterium Ti and Ri plasmids in plants. Cell 56:193–201. 10.1016/0092-8674(89)90892-1 [DOI] [PubMed] [Google Scholar]
- 44.Cascales E, Christie PJ. 2003. The versatile bacterial type IV secretion systems. Nat. Rev. Microbiol. 1:137–149. 10.1038/nrmicro753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Klimke WA, Frost LS. 1998. Genetic analysis of the role of the transfer gene, traN, of the F and R100-1 plasmids in mating pair stabilization during conjugation. J. Bacteriol. 180:4036–4043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ishiwa A, Komano T. 2003. Thin pilus PilV adhesins of plasmid R64 recognize specific structures of the lipopolysaccharide molecules of recipient cells. J. Bacteriol. 185:5192–5199. 10.1128/JB.185.17.5192-5199.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ishiwa A, Komano T. 2004. PilV adhesins of plasmid R64 thin pili specifically bind to the lipopolysaccharides of recipient cells. J. Mol. Biol. 343:615–625. 10.1016/j.jmb.2004.08.059 [DOI] [PubMed] [Google Scholar]
- 48.Aguilar-Barajas E, Jeronimo-Rodriguez P, Ramirez-Diaz MI, Rensing C, Cervantes C. 2011. The ChrA homologue from a sulfur-regulated gene cluster in cyanobacterial plasmid pANL confers chromate resistance. World J. Microbiol. Biotechnol. 28:865–869 [DOI] [PubMed] [Google Scholar]
- 49.Miyake M, Kotani H, Asada Y. 1992. Isolation and identification of restriction endonuclease, SelI from a cyanobacterium, Synechococcus elongatus. Nucleic Acids Res. 20:2605. 10.1093/nar/20.10.2605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cai F, Axen SD, Kerfeld CA. 2013. Evidence for the widespread distribution of CRISPR-Cas system in the phylum Cyanobacteria. RNA Biol. 10:687–693. 10.4161/rna.24571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Elhai J, Vepritskiy A, Muro-Pastor AM, Flores E, Wolk CP. 1997. Reduction of conjugal transfer efficiency by three restriction activities of Anabaena sp. strain PCC 7120. J. Bacteriol. 179:1998–2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Khlebnikov A, Datsenko KA, Skaug T, Wanner BL, Keasling JD. 2001. Homogeneous expression of the P(BAD) promoter in Escherichia coli by constitutive expression of the low-affinity high-capacity AraE transporter. Microbiology 147:3241–3247 [DOI] [PubMed] [Google Scholar]
- 53.Grant SG, Jessee J, Bloom FR, Hanahan D. 1990. Differential plasmid rescue from transgenic mouse DNAs into Escherichia coli methylation-restriction mutants. Proc. Natl. Acad. Sci. U. S. A. 87:4645–4649. 10.1073/pnas.87.12.4645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kuhlemeier CJ, Thomas AA, van der Ende A, van Leen RW, Borrias WE, van den Hondel CA, van Arkel GA. 1983. A host-vector system for gene cloning in the cyanobacterium Anacystis nidulans R2. Plasmid 10:156–163. 10.1016/0147-619X(83)90068-9 [DOI] [PubMed] [Google Scholar]
- 55.Matsuoka M, Takahama K, Ogawa T. 2001. Gene replacement in cyanobacteria mediated by a dominant streptomycin-sensitive rps12 gene that allows selection of mutants free from drug resistance markers. Microbiology 147:2077–2087 [DOI] [PubMed] [Google Scholar]
- 56.Datta N, Hedges RW. 1972. R factors identified in Paris, some conferring gentamicin resistance, constitute a new compatibility group. Ann. Inst. Pasteur (Paris) 123:849–852 [PubMed] [Google Scholar]
- 57.Komano T, Yoshida T, Narahara K, Furuya N. 2000. The transfer region of IncI1 plasmid R64: similarities between R64 tra and Legionella icm/dot genes. Mol. Microbiol. 35:1348–1359 [DOI] [PubMed] [Google Scholar]
- 58.Langer PJ, Walker GC. 1981. Restriction endonuclease cleavage map of pKM101: relationship to parental plasmid R46. Mol. Gen. Genet. 182:268–272. 10.1007/BF00269669 [DOI] [PubMed] [Google Scholar]
- 59.Chandler M, Galas DJ. 1983. Cointegrate formation mediated by Tn9. II. Activity of IS1 is modulated by external DNA sequences. J. Mol. Biol. 170:61–91 [DOI] [PubMed] [Google Scholar]
- 60.Brahamsha B. 1996. A genetic manipulation system for oceanic cyanobacteria of the genus Synechococcus. Appl. Environ. Microbiol. 62:1747–1751 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.