Abstract
Biofilm formation has been associated with bacterial pathogenesis, such as nosocomial and chronic infections, as the resistance of biofilms to environmental stresses has increased. Clostridium perfringens is a Gram-positive spore-forming anaerobic pathogen. This organism survives antibiotic treatment through the formation of biofilms or spores, but the environmental and regulatory factors involved in the biofilm formation remain unclear. Here, we observed that temperature regulates C. perfringens biofilm morphology. At 37°C, C. perfringens adhered to the substrate surface and formed a flat, thin biofilm, herein referred to as adhered biofilm. However, at 25°C, this bacterium did not adhere and produced a threadlike extracellular matrix, forming a viscous, thick biofilm, herein referred to as pellicle biofilm. Pellicle biofilm formation requires the sporulation master regulator, Spo0A, and the toxin regulator, CtrAB, and is enhanced in the absence of the global repressor, AbrB. These transcriptional regulator genes are regulated by each other and temperature. Adhered-biofilm formation requires AbrB and pilA2, which encodes a component of type IV pili (TFP). TFP expression was activated at 37°C and regulated through Spo0A, AbrB, and CtrAB. These results indicate that the morphology of C. perfringens biofilm is dependent on temperature through the differential production of extracellular matrix and the activity of TFP. Moreover, pellicle biofilm formation is involved in sporulation and toxin production. Here, we demonstrated that clostridial biofilm formation is closely associated with sporulation and that the morphological change of the biofilms could play an important role in the pathogenesis of this organism.
INTRODUCTION
Clostridium perfringens is a Gram-positive spore-forming anaerobic bacterium that causes gas gangrene, food-borne and non-food-borne poisoning, and antibiotic-associated diarrhea (1–3). Although this organism is an obligate anaerobe, C. perfringens is widely distributed throughout many environments, such as soil, sewage, and the intestine of humans and animals, since this organism produces endospores. Bacterial endospores are highly resistant to various stresses, such as oxygen, starvation, UV irradiation, and drying. However, the process of sporulation consumes a lot of energy and does not enable rapid adaption to environmental change. Recently, it has been shown that C. perfringens produces biofilms with increasing resistance to oxygen and antibiotics (4). The biofilms can form more quickly than endospores, and the cells in biofilms can respond more quickly to the environment, since endospores are in a highly dormant state, suggesting that biofilm formation, as well as sporulation, is involved in stress resistance and the pathogenesis of this organism.
Biofilm is a multicellular community in which the cells are surrounded by an extracellular matrix and possess phenotypes different from those of planktonic cells, such as resistance to extracellular stresses and specialized gene expression (5). Most bacteria grow in multicellular aggregates and form biofilms. Moreover, due to the resistance of these cells to antibiotics, microbial biofilm formation is involved in the pathogenesis of diseases such as nosocomial and chronic infections by this bacterium (6). C. perfringens causes antibiotic-associated diarrhea (AAD) (2) and survives the intestinal environment through biofilm formation and/or sporulation even when antibiotics are administered to the patient.
The regulatory cascade of sporulation and biofilm formation is shared among spore-forming bacilli, and the biofilm is an optimal environment for sporulation (7, 8). Bacillus spp. and Clostridium spp. share a regulatory cascade for sporulation. The sporulation master regulator Spo0A is required for sporulation and efficient biofilm formation in some Bacillus spp. and Clostridium spp. (9–11). Therefore, sporulation and biofilm formation would be relevant in Clostridium and probably regulated through a common regulatory cascade. However, in other clostridia, except C. difficile, this possibility has not been confirmed (12).
Varga et al. observed C. perfringens biofilm formation, showing that type IV pili and glucose concentration are involved in maximal biofilm formation on the substrate surface (4). However, most regulatory and environmental factors involved in C. perfringens biofilm growth remain unknown, and knowledge of these factors is important to understand the regulation of biofilm formation in Clostridium species. In the present study, we observed that C. perfringens forms biofilms with different structures in response to different temperatures. Specifically, at 37°C, cells adhered densely to the substrate surface, forming a flat, thin biofilm. However, at 25°C, most of the cells did not adhere to the surface, and they formed a viscous, thick, and pelliclelike biofilm. Moreover, the sporulation transcription factors Spo0A and AbrB and the toxin regulator CtrAB are required for the regulation of type IV pili and the production of an extracellular matrix, which contribute to the differing morphologies of biofilms in response to temperature. Therefore, the structural changes of C. perfringens biofilms are involved in sporulation and toxin production and are likely associated with the pathogenesis of this organism.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The strains of C. perfringens used in this study are listed in Table 1. All strains were cultured under anaerobic conditions using an Anaeropack (Mitsubishi Gas Chemical Co., Inc., Tokyo, Japan) at 25 or 37°C in Gifu anaerobic medium (GAM) broth (Nissui Co. Japan) with or without 50 μg/ml of erythromycin. Plasmid vector-harboring strains were cultured in GAM broth containing 10 μg/ml chloramphenicol.
TABLE 1.
Bacterial strains used in this study
| Strain | Relevant characteristic(s) | Reference or source |
|---|---|---|
| 13 | Wild type | 13 |
| HN13 | galKT in-frame deletion mutant of strain 13 | 15 |
| NO16 | ctrAB operon null mutant of strain 13; Emr | 14 |
| NO23 | spo0A mutant of strain 13; Emr | This study |
| NO24 | abrB null mutant of strain 13; Emr | This study |
| NO25 | pilA2 in-frame deletion mutant of HN13 | This study |
| NO33 | ctrAB mutant of NO25; Emr | This study |
Oligonucleotides.
Table S1 in the supplemental material lists the oligonucleotide primers used in this study.
Mutant strain construction.
The spo0A and abrB mutant strains (NO23 and NO24, respectively) were constructed using double-crossover homologous recombination as previously described (14). The upstream and downstream regions of the disrupting genes were PCR amplified using primers NOB-0428/NOB-0429 or NOB-0436/NOB-0437 and NOB-0495/NOB-0496 or NOB-0438NOB-0439 from the genomic DNA of C. perfringens and subsequently digested with EcoRI and BamHI, respectively. The ermBP gene, amplified from pJIR418 using primers NOB-0234/NOB-0240, was digested with EcoRI and BamHI. To construct the mutant strains NO23 and NO24, the DNA fragments amplified from the ligation product of the upstream and downstream regions and the ermBP fragments, amplified using primers NOB-0428/NOB-0496 or NOB-0436/NOB0439, were introduced into C. perfringens strain 13 through electroporation. The transformants were selected on brain heart infusion (BHI) sheep blood agar containing 50 μg/ml of erythromycin. To construct the pilA2 mutant strain, we used an in-frame deletion system as previously described (15). The upstream and downstream regions of the pilA2 gene were amplified using primers NOB-0576/NOB-0577 and NOB-0578/NOB-0579 and digested with SalI/EcoRI and EcoRI/BamHI, respectively. These fragments were cloned into the SalI/BamHI site of pCM-GALK, and the resulting plasmid was introduced into C. perfringens HN13 through electroporation. Each mutant was confirmed through PCR and DNA sequencing.
Northern blot analysis.
Total RNA was extracted from C. perfringens derivatives grown at 37°C in GAM broth and used for Northern blot analysis as previously described (16), with digoxigenin (DIG)-labeled DNA probes generated by using DIG-High Prime according to the manufacturer's instructions (17). The template DNA for the toxin gene-specific probes was PCR amplified from strain 13 genomic DNA using the primers described in Abe et al. (18).
Western blot analysis.
C. perfringens strain 13, harboring the CtrA and CtrB-His6 expression vector pCPE6 (14), was cultured at 37°C in GAM broth for 2, 3, or 4 h. The cells were harvested through centrifugation and washed with sterile phosphate-buffered saline (PBS). Whole-cell lysate was extracted using glass beads, and the CtrB-His6 proteins in the lysate were detected through Western blotting as previously described (14).
Biofilm assay.
C. perfringens was cultured overnight in PGY medium (3% proteose peptone no. 3, 2% glucose, 1% yeast extract, and 0.1% sodium thioglycolate) and diluted 1:50 into 1 or 3 ml of GAM broth or TY-G1 medium (3% tryptone, 2% yeast extract, 1% glucose, and 0.1% sodium thioglycolate) in 24- or 6-well polystyrene plates, and the cells were subsequently incubated under anaerobic conditions for 1 to 2 days. After incubation, the supernatants were removed, and the wells were washed once with sterile phosphate-buffered saline (PBS). The surface-associated cells were stained with 0.1% crystal violet for 10 min, followed by being washed once with sterile PBS. To quantify the amount of surface-adhered cells, each well was incubated with 1 ml of 99.5% ethanol for 10 min, and subsequently, the liquid was transferred to fresh 96-well plates. The optical density at 570 nm (OD570) of each well was measured using an Ultramark microplate reader (Bio-Rad).
Visualization of biofilms.
C. perfringens biofilms were formed in 35-mm glass-based dishes (Iwaki) and monitored using confocal reflection microscopy (CRM) (19, 20). A Carl Zeiss LSM710 laser scanning microscope equipped with a 63×/1.4 numerical aperture Plan-Apochromat objective (Carl Zeiss, Jena, Germany) was used to capture the confocal microscopy images. The cells were irradiated with a 514-nm argon laser, and 500- to 530-nm reflected light was used to visualize the cell localization and attachment to the substrate surface in biofilms. The images were processed using ZEN and Imaris software (Carl Zeiss, Jena, Germany).
SEM of biofilms.
A total of 4 ml of GAM broth in 6-well plates (Iwaki) containing 12-mm coverslips (Fisher) precoated with polylysine was inoculated with 80 μl of overnight culture. After incubation for 2 days at 25 or 37°C, the coverslips were transferred to a fresh 24-well plate, and the cells attached to the coverslips were fixed in 2.5% glutaraldehyde–10 mM sodium phosphate (pH 7.5) overnight. The samples were washed with 10 mM sodium phosphate (pH 7.5) twice and dehydrated in 50%, 70%, 90%, and 99.5% ethanol. Subsequently, the ethanol was replaced with 50% ethanol–50% isoamyl acetate and 100% isoamyl acetate. The coverslips were mounted onto aluminum stubs, dried using a critical-point dryer (HCP-2; Hitachi Ltd., Japan), and subsequently sputter coated with platinum using an E-1030 ion sputtering machine (Hitachi Ltd., Japan). The biofilms were observed using a scanning electron microscope (SEM) (HITACH-S-4200) (Hitachi Ltd., Japan).
RESULTS
C. perfringens forms differently structured biofilms in response to temperature.
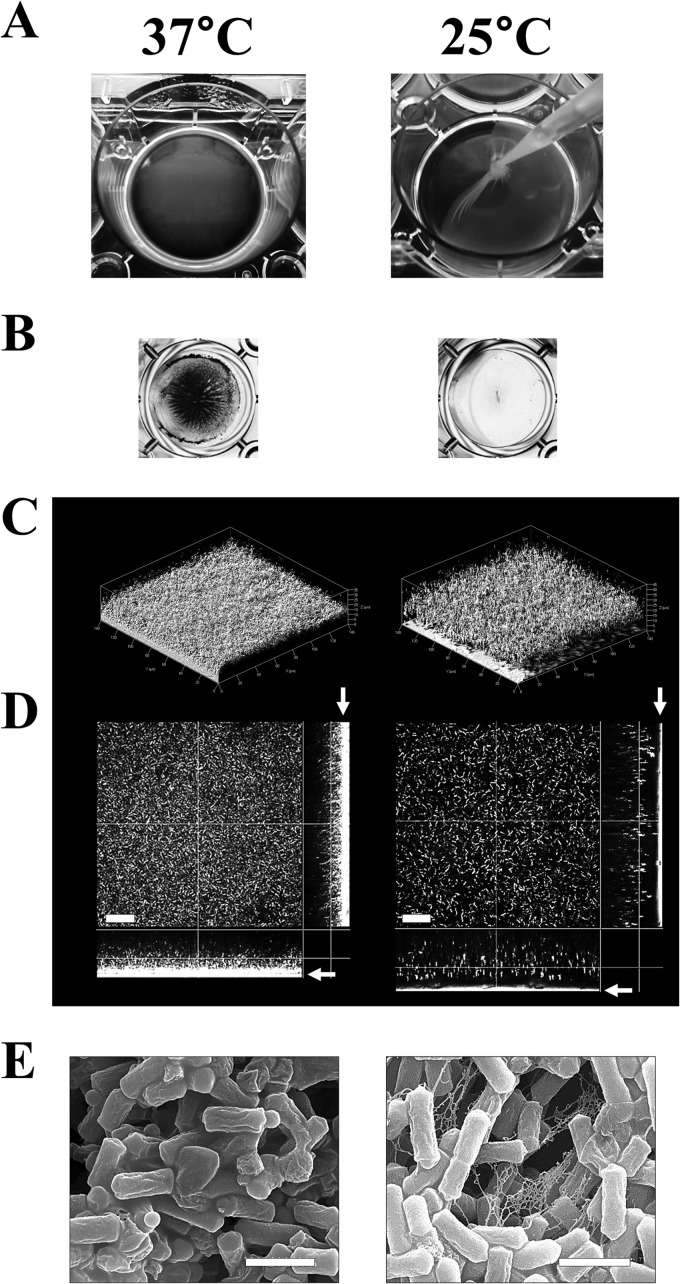
Most pathogens regulate virulence factors in response to temperature change for adaptation to the conditions in the host tissue and for efficient infection (21, 22). Here, we compared C. perfringens biofilms grown at 25 and 37°C. These temperatures resemble the environments outside or inside the host, such as in soil or in the intestine, respectively. C. perfringens formed biofilms that adhered to the substrate surface at 37°C, as previously reported (Fig. 1A, left) (4). The adhered cells were stained with crystal violet and easily visualized (Fig. 1B, left). However, C. perfringens formed thick, pelliclelike, viscous biofilms on the bottom of the well at 25°C, although under this condition, the cells did not adhere to the surface and were not detected through crystal violet staining because the biofilm structure was strongly connected and easily removed through pipette aspiration (Fig. 1A and B, right). This result suggests that at 25°C, the adherence of C. perfringens to the surface decreases and the biofilm morphology is dramatically changed. Hereinafter, we refer to the biofilms formed at 37°C as “adhered biofilm” and those formed at 25°C as “pellicle biofilm.” Confocal laser microscopy was used to visualize the cells in the biofilms, revealing that at 37°C, the surface of the glass-based dishes were densely covered with adhered biofilms, with many cells attached to the surface (Fig. 1C and D, left). This observation is consistent with the results of a previous report (4). However, the pellicle biofilm formed at 25°C did not contact the surface, and the density of the cells in this biofilm was lower than that in the adhered biofilm (Fig. 1C and D, right). These observations suggest that the cells at 25°C do not adhere to the surface and that they form biofilms with a structure different than that of biofilms at 37°C, indicating that pellicle biofilms predominantly contain extracellular matrix. Indeed, SEM imaging revealed threadlike extracellular materials between the cells in the pellicle biofilm but not in the adhered biofilm. Threadlike structures are detected in biofilms of various organisms, such as Bacillus subtilis and Pseudomonas aeruginosa (23, 24), suggesting that the threadlike material is an extracellular polymeric substance (EPS) of the pellicle biofilm and that temperature affects the specific EPS production (Fig. 1E).
FIG 1.
C. perfringens forms biofilms with different structures in response to temperature. (A) Biofilms were formed at 37°C (left) or 25°C (right). The edges of pellicle biofilms, formed at the bottom of wells, were picked through gentle pipette aspiration. (B to E) The biofilms were visualized through crystal violet staining (B), confocal reflection microscopy (C and D), and scanning electron microscopy (E). Three-dimensional projection (C) and orthometric images (D) show a field of 140 by 140 μm. The arrows indicate the substrate surface (D). The bars indicate 20 μm (D) or 2 μm (E).
DNA and protein are components of the extracellular matrix in pellicle biofilm.
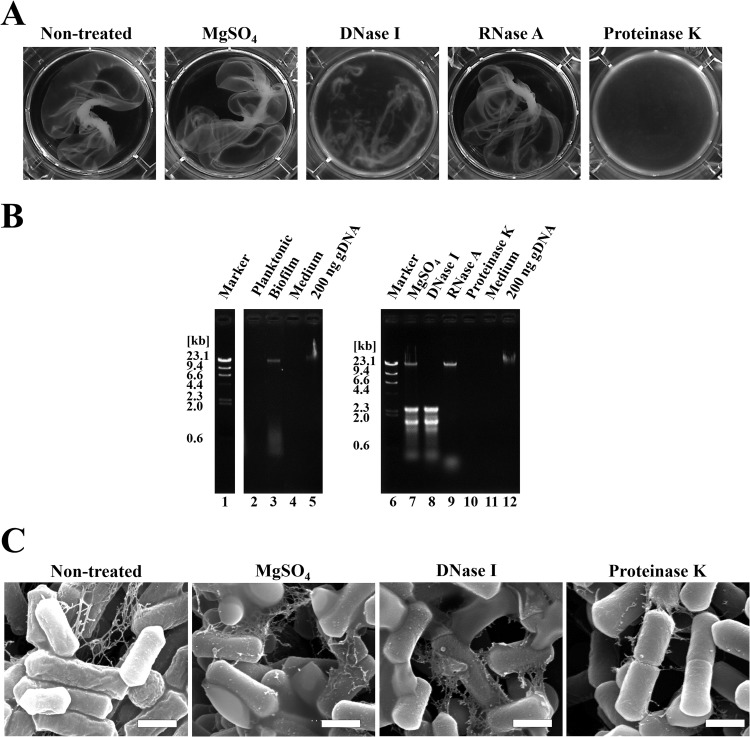
Next, we focused on the formation of a novel biofilm structure, the pellicle biofilm that is primarily comprised of extracellular matrix. To identify the extracellular matrix components of pellicle biofilm, we added enzymes that degrade extracellular matrices to the culture for pellicle biofilm formation. Figure 2A shows the pellicle biofilm after gentle agitation. The addition of DNase I or proteinase K at the start of the culture made the pellicle biofilm structure fragile, but RNase A addition had no apparent effect (Fig. 2A). Therefore, extracellular DNA and proteins are the components of pellicle biofilms. Biofilms of various organisms contain extracellular DNA (eDNA), and eDNA could be the primary structural component of biofilms and used as an adhesion factor (5, 25, 26). We isolated the extracellular nucleic acids from pellicle biofilm and analyzed these components using agarose gel electrophoresis. A 23-kb DNA fragment was observed in the biofilm fraction but not in the planktonic cell sample, indicating that pellicle biofilm contains DNA as a matrix component (Fig. 2B, lanes 2 to 5). Because 23S and 16S rRNA were detected, magnesium sulfate, which is required for DNase I activity, was added to the culture because magnesium divalent cations could stabilize extracellular RNAs (eRNAs) (Fig. 2B, lanes 6 to 10). However, the addition of magnesium sulfate did not affect the structure of pellicle biofilm (Fig. 2A; also data not shown). Under this condition, the addition of DNase I and RNase A completely degraded eDNA and eRNA, respectively. Furthermore, proteinase K treatment decreased both eDNA and eRNA. These results indicate that long eDNA is a component of pellicle biofilm and important for the formation of the viscous film structure. However, although SEM imaging showed that DNase I or proteinase K treatment decreased the amount of extracellular materials around the cells, threadlike strands remained (Fig. 2C).
FIG 2.
Treatment with DNase I or proteinase K affects the pellicle biofilm structure. (A) A concentration of 10 mM MgSO4, 1 mg/ml DNase I, 1 mg/ml RNase A, or 0.2 mg/ml proteinase K was added to the medium prior to culturing for 2 days. The biofilm formed in the medium was gently agitated to make pellicle biofilm more visible. (B) Extracellular nucleic acids extracted from the biofilms were dissolved in 0.8% agarose. Each lane contains equivalent amounts of the culture obtained at an OD600 of 0.2. gDNA, genomic DNA. (C) Scanning electron microscopy images of the biofilm treated with DNase I or proteinase K. The bars indicate 1 μm.
Transcriptional regulators necessary for biofilm formation.
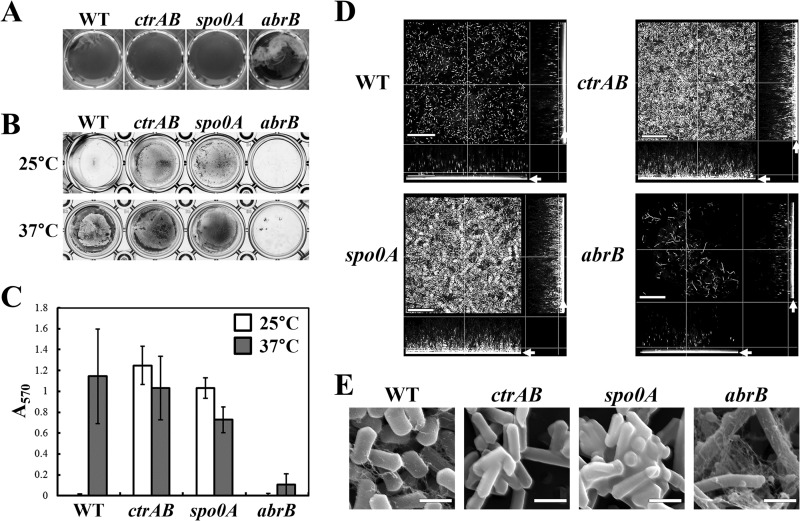
The Gram-positive aerobic spore-forming bacterium B. subtilis forms robust biofilms, and the regulatory cascade of this biofilm formation has been well studied (27). The master regulator of sporulation, Spo0A, and the global repressor, AbrB, control the biofilm formation in B. subtilis positively and negatively, respectively (8, 28). C. perfringens contains homologs of the spo0A and abrB genes, and Spo0A is also necessary for efficient sporulation in C. perfringens, similar to B. subtilis (10). In addition, we recently reported a protein complex of CtrA and CtrB (clostridial toxin regulator; formerly CPE1447-CPE1446) as a novel toxin regulator (14). The ctrAB complex is widely conserved in clostridia and regulates many genes, including those encoding toxins, suggesting that ctrAB is important for clostridial metabolism. Therefore, we examined biofilm formation in spo0A, abrB, and ctrAB mutant strains. As shown in Fig. 3A, spo0A and ctrAB mutants did not form pellicle biofilms, and instead, these mutants formed adhered biofilms at 25°C (Fig. 3B). However, the adhered-biofilm formation at 37°C was not significantly affected in these mutants, as demonstrated through microtiter plate crystal violet assay (Fig. 3B and C). The abrB mutant forms thick pellicle biofilms at 25°C (Fig. 3A), and the efficiency of the adhered-biofilm formation in this mutant was significantly lower than that in wild-type strains (Fig. 3B and C). Confocal laser microscopy also indicated that the ctrAB and spo0A mutant cells adhered to the surface and formed dense populations at 25°C (Fig. 3D). We used SEM to observe the extracellular matrix of the biofilms formed by spo0A, abrB, and ctrAB mutants at 25°C. We observed that spo0A and ctrAB mutants showed no production of threadlike EPS, but the abrB mutant featured robust production of EPS (Fig. 3E). Complementation of spo0A or ctrAB restored pellicle biofilm formation (see Fig. S1 in the supplemental material). These results suggest that spo0A, abrB, and ctrAB are involved in cell adherence and EPS production. Thus, Spo0A and CtrAB are required for pellicle biofilm formation and AbrB is required for adhered-biofilm formation.
FIG 3.
Multiple transcriptional regulators affect biofilm formation in C. perfringens. (A) Pellicle biofilm formation at 25°C by ctrAB, spo0A, or abrB mutant strains. The cells were incubated at 25°C for 2 days. The edges of pellicle biofilms formed at the bottom of wells were picked and flipped through gentle pipette aspiration. (B) Adhered-biofilm formation of the mutant strains. The cells were cultured at 25 or 37°C for 2 days and stained with crystal violet. (C) Quantification of the adhered-biofilm biomass. The means and standard deviations of the OD570 values of three independent experiments are shown. (D) Confocal microscopy images of the mutant strain biofilms formed at 25°C. The arrows indicate the substrate surface. (E) Scanning electron microscopy of the mutant cells in the biofilms formed at 25°C. The bars indicate 30 μm (D) or 2 μm (E).
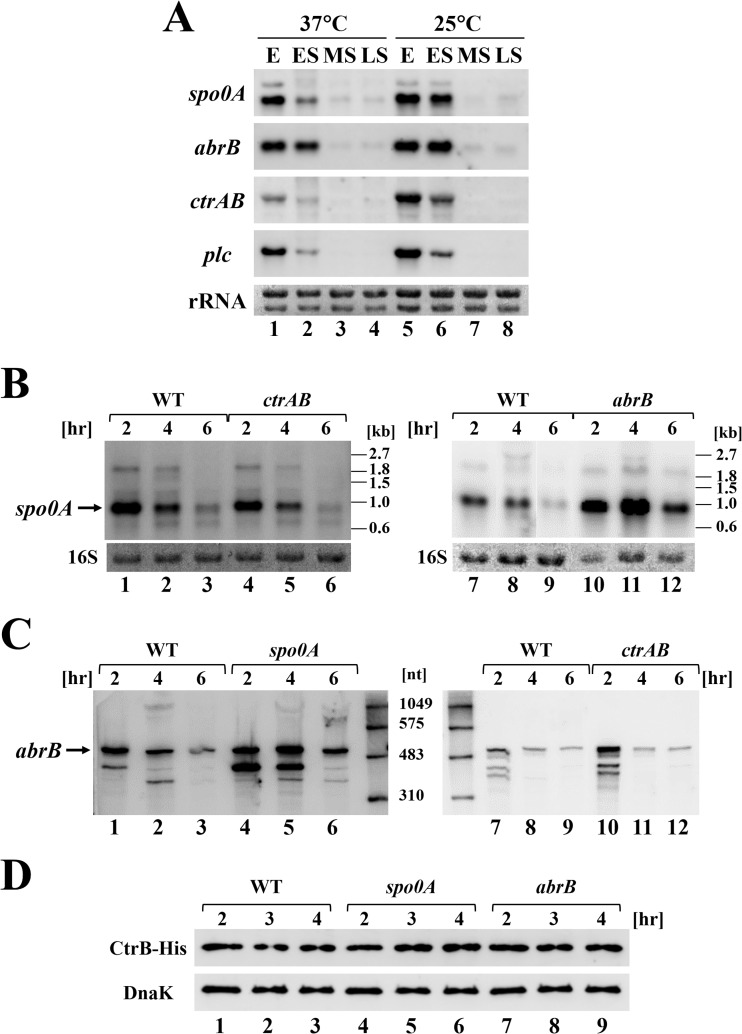
The morphological change of biofilms in response to temperature requires the transcriptional regulators Spo0A, AbrB, and CtrAB. Thus, we compared the expression of genes encoding these transcriptional regulators at 25°C and 37°C and performed a Northern blot analysis using wild-type C. perfringens (Fig. 4A). The plc gene encoding the alpha-toxin, phospholipase C, is highly expressed at the exponential phase and activated at a lower temperature, and this toxin was used as an intrinsic control (18, 29). The expression of spo0A and ctrAB also peaked at the exponential phase, and these genes were more highly activated at 25°C than at 37°C, implying that transcription of spo0A and ctrAB is activated at low temperatures and, subsequently, morphological changes of the biofilm are induced (Fig. 4A). In contrast, temperature did not affect the expression of abrB (Fig. 4A). We also examined whether these transcriptional regulators control each other's expression. Northern blot analysis of the mutant strains showed that spo0A expression was derepressed in the abrB mutant but was not affected in the ctrAB mutant (Fig. 4B). abrB expression was derepressed in both spo0A and ctrAB mutants (Fig. 4C). These results suggest that the expression of spo0A and abrB is repressed by AbrB and Spo0A, respectively, as has been observed in B. subtilis (28, 30). Moreover, CtrAB could repress abrB transcription in C. perfringens. We also tested whether ctrAB expression is affected by spo0A and abrB mutations by using Western blot analysis of CtrB-His6 expressed from the pCPE6 vector, in which ctrA and ctrB-His6 genes are transcribed from the ctrAB native promoter. However, the amount of CtrB-His6 in spo0A or abrB mutants was not changed compared with the amount in the wild-type strain (Fig. 4D). Moreover, we analyzed chromosomally encoded ctrAB expression by Northern blotting, and the amount of ctrAB mRNA was not significantly different in the mutants (data not shown). These data suggest that ctrAB expression is not regulated through Spo0A and AbrB. Therefore, these transcriptional regulators control the expression of the other genes and could affect biofilm formation.
FIG 4.
Expression of transcriptional regulator genes involved in biofilm formation. (A to C) Northern blot analyses of spo0A, abrB, and ctrAB. 23S and 16S rRNAs stained with methylene blue are presented as loading controls at the bottom as indicated. In each lane, 2 μg of total RNA was loaded. (A) Wild-type C. perfringens was cultured at 37 or 25°C and harvested at the exponential (E; OD600 of 1.5 to 2.0), early stationary (ES; OD600 of 4.5 to 5.0), mid-stationary (MS; 2 and 8 h after OD600 reached 4.5 at 37 and 25°C, respectively), and late-stationary (LS; 4 and 16 h after OD600 reached 4.5 at 37 and 25°C, respectively) phases. (B) spo0A expression in wild-type and abrB mutant cells cultured at 37°C for 2, 4, or 6 h, corresponding to the exponential, early stationary, and mid-stationary phase, respectively. (C) abrB expression in wild-type and spo0A mutant cells cultured at 37°C for 2, 4, or 6 h, corresponding to the exponential, early stationary, and mid-stationary phase, respectively. nt, nucleotides. (D) Western blot analysis of CtrB-His6 proteins using the anti-His tag antibody. Wild-type C. perfringens and spo0A and abrB mutant strains harboring the CtrA and His-tagged CtrB expression vector were cultured at 37°C for 2, 3, and 4 h, corresponding to the mid-exponential, late-exponential, and early stationary phase, respectively. DnaK proteins were detected using the anti-DnaK antibody and are indicated as a loading control at the bottom.
TFP component PilA2 is activated at 37°C.
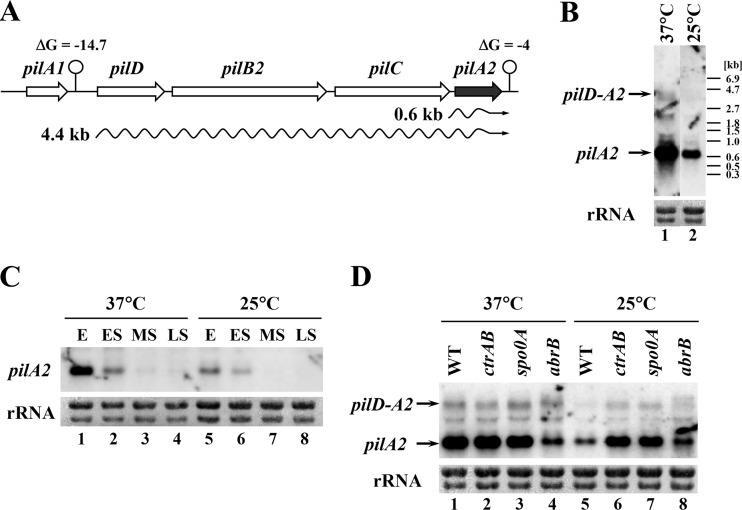
As shown by the results described above, the adhesive property of C. perfringens was controlled through temperature. It had been reported that type IV pili (TFP) are required for the maturation of adhered biofilm and adherence to myoblast cells (4, 31, 32), suggesting that the temperature-dependent regulation of the adhesive property is associated with the TFP machinery. First, we analyzed the TFP biosynthesis gene expression at different temperatures. Three gene clusters encoding putative TFP biosynthesis proteins were identified in the C. perfringens genome (31). We performed Northern blot analysis using DNA probes specific for these genes. The pilT-ftsA-ftsZ mRNA expression was not significantly changed at different temperatures, and pilB-pilC-CPE1842-CPE1841 mRNA was undetectable on Northern blots due to low expression levels (data not shown). Although PilT and PilC were required for gliding motility and adherence to eukaryotic cells in C. perfringens, neither gene was drastically regulated through temperature. The 4.4- and 0.6-kb transcripts were detected on the Northern blot using the DNA probe specific for the pilA2 gene that is located in the residual gene cluster encoding TFP biosynthesis proteins (Fig. 5A and B). Because putative intrinsic transcriptional terminators were predicted downstream from pilA1 and pilA2 (ΔG = −14.7 and −4, respectively) (33), these 4.4- and 0.6-kb transcripts would, respectively, represent pilD-pilB2-pilC2-pilA2 polycistronic and pilA2 monocistronic mRNA. The expression peaked at the exponential phase and was activated at 37°C, suggesting that pilA2 expression was regulated through temperature (Fig. 5C). Moreover, the pilA2 expression was higher in the ctrAB and spo0A mutant strains at 25°C and was repressed in the abrB mutant strain compared with its level in the wild-type strain (Fig. 5D). Thus, the amount of the pilA2 mRNA corresponded to the efficiency of adherence to the substrate surface and the biomass of the adhered biofilm (Fig. 3B and 5D). These results suggest that temperature-dependent pilA2 regulation requires ctrAB and spo0A and basal pilA2 expression requires abrB.
FIG 5.
pilA2 expression is regulated through temperature and multiple transcriptional factors. (A) Schematic of the type IV pilus biosynthesis operon containing the pilA2 gene. The predicted intrinsic transcriptional terminators and ΔG values are indicated. The arrows represent transcriptional units. (B to D) Northern blot analyses of the pilA2 gene. 23S and 16S rRNAs stained with methylene blue are represented as loading controls at the bottom of the gels. In each lane, 2 μg of total RNA was loaded. (B) Expression of pilD-pilA2 polycistronic and pilA2 monocistronic mRNA at 25 or 37°C. (C) pilA2 expression in wild-type C. perfringens grown to the exponential (E; OD600 of 1.5 to 2.0), early stationary (ES; OD600 of 4.5 to 5.0), mid-stationary (MS; 2 and 8 h after OD600 reached 4.5 at 25 and 37°C, respectively), and late-stationary (LS; 4 and 16 h after OD600 reached 4.5 at 25 and 37°C, respectively) phases. (D) pilA2 expression in ctrAB, spo0A, and abrB mutants grown to the mid-exponential phase at 25 or 37°C.
PilA2 is required for cell adherence to the substrate surface and adhered-biofilm formation.
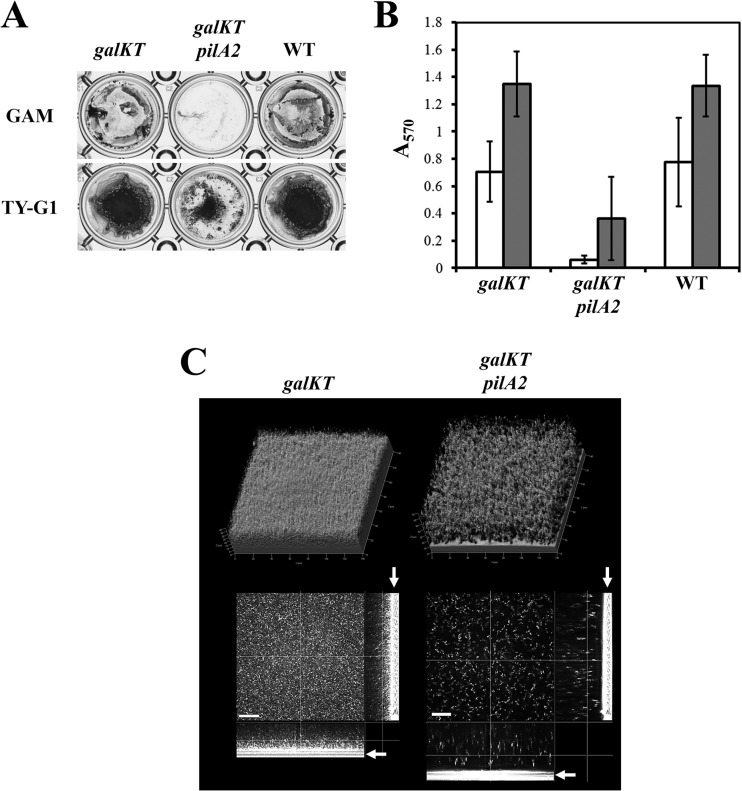
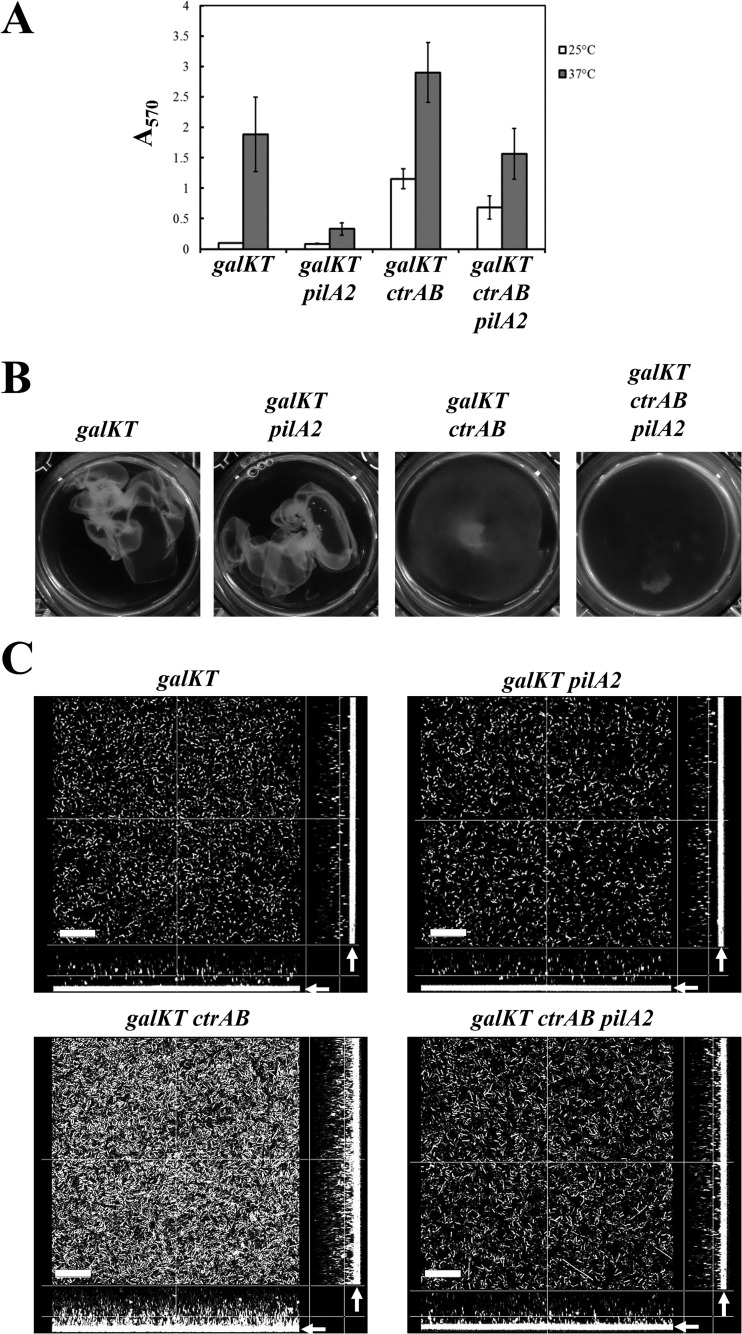
The reduction of pilA2 expression and cell adherence to the substrate surface at 25°C suggest that changes in the adhesive property and biofilm morphology of C. perfringens occur through the regulation of pilA2 expression in response to temperature. To examine this idea, we constructed a pilA2 mutant strain and analyzed whether pilA2 is required for adherence to the substrate surface and adhered-biofilm formation. We used an in-frame deletion system to construct the mutant according to the method of Nariya et al. (15). Subsequently, the strains were cultured in GAM or TY-G1 at 37°C. The wild-type and HN13 (parent strain of the pilA2 mutant; a galKT disruptant) strains showed comparable formation of adhered biofilms, but the pilA2 mutant could not form adhered biofilms in GAM and showed poor formation of adhered biofilms in TY-G1 compared with the adhered-biofilm formation of the parent strain (Fig. 6A and B). In addition, confocal microscopy revealed that the pilA2 mutant strain did not adhere to the glass surface in GAM broth (Fig. 6C). The ctrAB mutant could not form pellicle biofilm but did form adhered biofilm at 25°C. We constructed a ctrAB pilA2 double mutant and analyzed biofilm formation using this mutant. The ctrAB mutant of HN13 also formed adhered biofilm at 25°C, and a reduction in the biofilm biomass was observed for the ctrAB pilA2 double mutant compared with the biofilm mass of the ctrAB mutant (Fig. 7A). The ctrAB pilA2 mutant could not form pellicle biofilm, and the number of cells adhered to the substrate surface was drastically decreased in the confocal microscopy image (Fig. 7B and C). These results indicate that the pilA2 gene, encoding the main component of TFP, pilin, is necessary for adherence to the substrate surface and the formation of adhered biofilm in C. perfringens.
FIG 6.
PilA2 is required for adhered-biofilm formation. (A) Adhered-biofilm formation of the pilA2 mutant strain. C. perfringens was cultured at 37°C in GAM or TY-G1 broth for 2 days, and the adhered cells were stained with crystal violet. (B) Quantification of the adhered-biofilm biomass. The means and standard deviations of the OD570 values of three independent experiments are shown. (C) Confocal laser microscopy images of the galKT or galKT pilA2 mutant strain biofilms formed at 37°C in GAM broth. The bars indicate 20 μm. The arrows indicate the substrate surface.
FIG 7.
PilA2 affects cell adherence in the ctrAB mutant. (A) Adhered-biofilm formation of the mutant strains. C. perfringens was cultured at 25 or 37°C in GAM broth for 2 days, and the biomass of the adhered biofilm was quantified through staining with crystal violet. The means and standard deviations of the OD570 values of a representative experiment are shown. The experiments were repeated at least three times. (B) Pellicle biofilm formation of the mutant strains at 25°C. The cells were incubated at 25°C for 2 days. The photographs show the pellicle biofilm after picking by gentle pipette aspiration. (C) Confocal laser microscopy images of the galKT or galKT pilA2 mutant strain biofilms formed at 37°C in GAM broth. The bars indicate 30 μm. The arrows indicate the substrate surface.
DISCUSSION
We observed that the C. perfringens biofilm structure is drastically different at 25°C than at 37°C. At 37°C, C. perfringens adhered to the substrate surface and formed a flat, thin, adhered biofilm in which the cells were densely packed. However, at 25°C, the cells did not adhere to the surface and precipitated to the bottom of the well, forming a thick, viscous, pellicle biofilm in which the density of the cells was low. These results suggest that the pellicle biofilm comprises extracellular matrices. Indeed, SEM imaging of adhered and pellicle biofilms indicated that the threadlike extracellular structures were only observed in pellicle biofilms and were comprised of a low-temperature-specific EPS produced in the process of C. perfringens biofilm formation. C. perfringens grows more rapidly at 37°C than 25°C, and it might be possible that biofilm morphology is caused by differences in cell density and/or physiological status. However, we observed no pellicle biofilm formation when cells were cultured at 37°C for 12 h or 24 h, which are shorter incubation times than used for the biofilm formation assay whose results are shown in Fig. 1 (data not shown). In addition, adhered biofilm never formed at 25°C even if the incubation was prolonged for 8 days (data not shown). Thus, we concluded that pellicle biofilm formation is not an early stage of adhered, dense biofilm formation and that the differences in the morphology of biofilms formed by C. perfringens are dependent on temperature. We therefore suggest that C. perfringens regulates adherence and EPS production through changes in the biofilm structure in response to temperature.
The biofilm component EPS is important for biofilm architecture, protection, and energy storage (5). Treatment with DNase I or proteinase K destabilized the pellicle biofilm structure, suggesting that extracellular DNA or proteins are components of pellicle biofilms. It has been reported that C. difficile biofilm formation was reduced through DNase I and proteinase K treatment and that the biofilm contained extracellular DNA and proteins (34). Thus, extracellular nucleic acids and proteins could be important for biofilm formation in Clostridium species, similar to other bacterial species. However, although DNase I or proteinase K treatment makes the C. perfringens pellicle biofilm fragile, it does not completely inhibit biofilm formation. SEM revealed that DNase I or proteinase K could not digest the threadlike EPS, which is specifically produced at lower temperatures and would also be a main component of the pellicle biofilm. Therefore, threadlike EPS could not be protein or DNA. Threadlike EPS in the biofilms formed by other species have also been analyzed using SEM. Extracellular polysaccharide and poly-γ-glutamate in B. subtilis and Pel extracellular polysaccharide in P. aeruginosa have been detected as threadlike structures and are the components of the biofilms formed by these organisms (23, 24). These observations imply that the EPS in the pellicle biofilm also contains extracellular polysaccharide.
In general, adherence to the substrate surface is an important step in surface-associated biofilm formation, and TFP is involved in cell attachment and biofilm formation in various species (35–37). It has been reported that the maturation of C. perfringens surface-associated biofilm formation requires type IV pili (TFP) and the transcriptional regulator CcpA (4). TFP is involved in gliding motility and cell attachment of C. perfringens and is an important factor for biofilm formation and pathogenesis in many bacteria (31, 32, 38). We observed that the expression of pilA2, which encodes pilin, a major component of TFP, is activated at 37°C but not at 25°C. This result suggested that pilA2 regulation is thermodependent and TFP activity is controlled through temperature. In addition, adhered-biofilm formation requires pilA2, indicating that the thermoregulation of pilA2 expression is important to control the biofilm morphology. We detected pilD-pilC2-pilB2-pilA2 polycistronic and pilA2 monocistronic mRNA using Northern blot analysis, and the amount of the latter transcript was markedly affected by temperature. This effect suggests that the transcription from a promoter upstream from the pilA2 gene is regulated through temperature, but no sequence corresponding to a promoter has been identified. In addition, we cloned a DNA fragment containing the pilA2 gene and 200 bp of its upstream sequence into the Escherichia coli-C. perfringens shuttle vector and introduced the resultant plasmid into C. perfringens. No pilA2 transcript or PilA2 protein was derived from the plasmid, suggesting that pilA2 monocistronic mRNA was processed from the pilD-pilC2-pilB2-pilA2 polycistronic mRNA and posttranscriptional regulation was involved in the expression of pilA2 (data not shown).
We identified the transcriptional regulators Spo0A, AbrB, and CtrAB as essential factors for biofilm formation in C. perfringens. In addition, we revealed that threadlike-EPS production and cell adherence to the surface were regulated by these global transcriptional factors. In bacilli, the regulatory cascade of sporulation is involved in biofilm development (8). Spo0A is an essential factor for biofilm formation and sporulation in bacilli. In C. perfringens, spo0A is also required for sporulation (10), and the spo0A mutant could form adhered biofilm but not pellicle biofilm (Fig. 3). Therefore, it was suggested that pellicle biofilm formation is associated with sporulation in C. perfringens. In C. difficile, the biofilm formation of the spo0A mutant was decreased compared with that of the wild-type strain (12, 34). Thus, there might be a deep connection between biofilm formation and sporulation in clostridia.
AbrB is a global repressor involved in myriad biological processes, such as antibiotic production, sporulation, and biofilm formation in B. subtilis (28). The abrB gene is conserved in Bacillus and Clostridium species and is widely investigated in Bacillus but not in Clostridium species. In the present study, we observed that AbrB is also important for cell adherence to the substrate surface and adhered-biofilm formation (Fig. 3). In addition, AbrB negatively regulates spo0A expression and vice versa (Fig. 4B and C). The expression of abrB is directly regulated through Spo0A in B. subtilis, and we identified the 0A box, the consensus sequence of the Spo0A binding site, in the C. perfringens abrB promoter region (39; also data not shown). In addition, abrB regulates spo0A expression through the repression of sigH, which encodes the stationary-phase sigma factor in B. subtilis (28, 30). We confirmed that sigH expression is derepressed in the C. perfringens abrB mutant (data not shown). Therefore, Spo0A and AbrB directly or indirectly reciprocally regulate each other in C. perfringens. The abrB mutant produces abundant threadlike EPS and forms a robust pellicle biofilm (Fig. 3). We also observed cell elongation and aggregation of the abrB mutant, and the exogenous expression of abrB from the plasmid vector restored the phenotype (see Fig. S2 in the supplemental material). The dramatic phenotype of the abrB mutant suggested that the AbrB repressor possesses a pivotal biological role in C. perfringens.
We previously identified a CtrAB protein complex as a global toxin regulator and observed that ctrAB genes are widely conserved in Clostridium species (14). The results obtained in the present study revealed that the protein complex regulator also regulates the formation and morphology of the biofilm. The deletion of the ctrAB genes reduced abrB expression, and ctrAB mRNA accumulated to a greater extent at 25°C than at 37°C (Fig. 4A). Therefore, the CtrAB protein complex might be a critical determinant of biofilm morphology, and the homologous proteins identified in other Clostridium species might also be important for biofilm formation.
It has been reported that toxin gene expression was altered in the ctrAB mutant (14). Moreover, we analyzed the toxin gene expression in the spo0A or abrB mutant and observed altered expression of several toxin genes through Northern blot analysis (see Fig. S3 in the supplemental material). These results suggest that the morphological changes of the biofilm in response to temperature were associated with toxin gene regulation. Indeed, it has been reported that the expression of the phospholipase C gene, plc, was activated at lower temperatures through changes in the affinity of RNA polymerase for poly(A) tracts upstream from the plc promoter (40). Moreover, we observed that the sporulation factors Spo0A and AbrB are involved in biofilm morphology and that the CtrAB toxin regulator also controls biofilm formation, suggesting that the morphological changes of the biofilm could be involved in the toxin production and virulence of this organism.
Temperature is an important factor for biofilm formation, and numerous studies have indicated that temperature affects the amount of biofilm in Escherichia coli, Legionella pneumophila, Listeria monocytogenes, and B. cereus (41–44). In C. perfringens, temperature drastically affects biofilm morphology, potentially reflecting the temperature-dependent regulation of EPS production and cell adhesion. We suggest that temperature-regulated biofilm formation is a strategy for adaptation to the environment and appropriate infection, as temperature is an environmental signal that alternates between the outside and inside of the mammalian host. After infection, C. perfringens spores are germinated and grown at 37°C, and these cells produce TFP and adhere to the epithelial cells. Subsequently, they secrete a number of toxins that degrade the host cell. However, when the host dies or C. perfringens cells are secreted from the host, these organisms might sense the shift to lower temperatures and produce extracellular matrices to form the pellicle biofilm. Under this condition, these cells are more likely to be exposed to oxygen. Thus, the pellicle biofilm, which predominantly contains the extracellular matrix, might provide increased resistance to external environments, thereby facilitating survival. Therefore, the morphological changes of biofilms in C. perfringens could be involved in the pathogenesis of these organisms. However, the mechanisms underlying the regulatory cascade in C. perfringens biofilm formation remain unknown, and further investigation is needed.
Supplementary Material
ACKNOWLEDGMENTS
This work was financially supported through the Advanced Low Carbon Technology Research and Development Program (ALCA) and the Core Research for Evolutional Science and Technology (CREST) of Japan Science and Technology Agency (JST). N. Obana was supported through Research Fellowships from the Japan Society for the Promotion of Science for Young Scientists.
Footnotes
Published ahead of print 7 February 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.01444-13.
REFERENCES
- 1.Rood JI, Lyristis M. 1995. Regulation of extracellular toxin production in Clostridium perfringens. Trends Microbiol. 3:192–196. 10.1016/S0966-842X(00)88919-2 [DOI] [PubMed] [Google Scholar]
- 2.Modi N, Wilcox MH. 2001. Evidence for antibiotic induced Clostridium perfringens diarrhoea. J. Clin. Pathol. 54:748–751. 10.1136/jcp.54.10.748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stevens DL, Aldape MJ, Bryant AE. 2012. Life-threatening clostridial infections. Anaerobe 18:254–259. 10.1016/j.anaerobe.2011.11.001 [DOI] [PubMed] [Google Scholar]
- 4.Varga JJ, Therit B, Melville SB. 2008. Type IV pili and the CcpA protein are needed for maximal biofilm formation by the gram-positive anaerobic pathogen Clostridium perfringens. Infect. Immun. 76:4944–4951. 10.1128/IAI.00692-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Flemming H-C, Wingender J. 2010. The biofilm matrix. Nat. Rev. Microbiol. 8:623–633. 10.1038/nrmicro2415 [DOI] [PubMed] [Google Scholar]
- 6.Høiby N, Bjarnsholt T, Givskov M, Molin S, Ciofu O. 2010. Antibiotic resistance of bacterial biofilms. Int. J. Antimicrob. Agents 35:322–332. 10.1016/j.ijantimicag.2009.12.011 [DOI] [PubMed] [Google Scholar]
- 7.Branda SS, González-Pastor JE, Ben-Yehuda S, Losick R, Kolter R. 2001. Fruiting body formation by Bacillus subtilis. Proc. Natl. Acad. Sci. U. S. A. 98:11621–11626. 10.1073/pnas.191384198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hamon M, Lazazzera B. 2001. The sporulation transcription factor Spo0A is required for biofilm development in Bacillus subtilis. Mol. Microbiol. 42:1199–1209. 10.1046/j.1365-2958.2001.02709.x [DOI] [PubMed] [Google Scholar]
- 9.Steiner E, Dago AE, Young DI, Heap JT, Minton NP, Hoch JA, Young M. 2011. Multiple orphan histidine kinases interact directly with Spo0A to control the initiation of endospore formation in Clostridium acetobutylicum. Mol. Microbiol. 80:641–654. 10.1111/j.1365-2958.2011.07608.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huang I-H, Waters M, Grau RR, Sarker MR. 2004. Disruption of the gene (spo0A) encoding sporulation transcription factor blocks endospore formation and enterotoxin production in enterotoxigenic Clostridium perfringens type A. FEMS Microbiol. Lett. 233:233–240. 10.1111/j.1574-6968.2004.tb09487.x [DOI] [PubMed] [Google Scholar]
- 11.Rosenbusch KE, Bakker D, Kuijper EJ, Smits WK. 2012. C. difficile 630Δerm Spo0A regulates sporulation, but does not contribute to toxin production, by direct high-affinity binding to target DNA. PLoS One 7:e48608. 10.1371/journal.pone.0048608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dawson LF, Valiente E, Faulds-Pain A, Donahue EH, Wren BW. 2012. Characterisation of Clostridium difficile biofilm formation, a role for Spo0A. PLoS One 7:e50527. 10.1371/journal.pone.0050527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shimizu T, Ohtani K, Hirakawa H, Ohshima K, Yamashita A, Shiba T, Ogasawara N, Hattori M, Kuhara S, Hayashi H. 2002. Complete genome sequence of Clostridium perfringens, an anaerobic flesh-eater. Proc. Natl. Acad. Sci. U. S. A. 99:996–1001. 10.1073/pnas.022493799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Obana N, Nakamura K. 2011. A novel toxin regulator, the CPE1446-CPE1447 protein heteromeric complex, controls toxin genes in Clostridium perfringens. J. Bacteriol. 193:4417–4424. 10.1128/JB.00262-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nariya H, Miyata S, Suzuki M, Tamai E, Okabe A. 2011. Development and application of a method for counterselectable in-frame deletion in Clostridium perfringens. Appl. Environ. Microbiol. 77:1375–1382. 10.1128/AEM.01572-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Obana N, Nomura N, Nakamura K. 2013. Structural requirement in Clostridium perfringens collagenase mRNA 5′ leader sequence for translational induction through small RNA-mRNA base pairing. J. Bacteriol. 195:2937–2946. 10.1128/JB.00148-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Roche. 2008. DIG application manual for filter hybridization. Roche Diagnostics GmbH, Freiburg, Germany: https://www.roche-applied-science.com/wcsstore/RASCatalogAssetStore/Articles/05353149001_08.08.pdf [Google Scholar]
- 18.Abe K, Obana N, Nakamura K. 2010. Effects of depletion of RNA-binding protein Tex on the expression of toxin genes in Clostridium perfringens. Biosci. Biotechnol. Biochem. 74:1564–1571. 10.1271/bbb.100135 [DOI] [PubMed] [Google Scholar]
- 19.Yawata Y, Toda K, Setoyama E, Fukuda J, Suzuki H, Uchiyama H, Nomura N. 2010. Monitoring biofilm development in a microfluidic device using modified confocal reflection microscopy. J. Biosci. Bioeng. 110:377–380. 10.1016/j.jbiosc.2010.04.002 [DOI] [PubMed] [Google Scholar]
- 20.Inaba T, Ichihara T, Yawata Y, Toyofuku M, Uchiyama H, Nomura N. 2013. Three-dimensional visualization of mixed species biofilm formation together with its substratum. Microbiol. Immunol. 57:589–593. 10.1111/1348-0421.12064 [DOI] [PubMed] [Google Scholar]
- 21.Shapiro RS, Cowen LE. 2012. Thermal control of microbial development and virulence: molecular mechanisms of microbial temperature sensing. mBio 3(5):e00238–12. 10.1128/mBio.00238-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Konkel ME, Tilly K. 2000. Temperature-regulated expression of bacterial virulence genes. Microbes Infect. 2:157–166. 10.1016/S1286-4579(00)00272-0 [DOI] [PubMed] [Google Scholar]
- 23.Morikawa M, Kagihiro S, Haruki M, Takano K, Branda S, Kolter R, Kanaya S. 2006. Biofilm formation by a Bacillus subtilis strain that produces gamma-polyglutamate. Microbiology 152:2801–2807. 10.1099/mic.0.29060-0 [DOI] [PubMed] [Google Scholar]
- 24.Friedman L, Kolter R. 2003. Genes involved in matrix formation in Pseudomonas aeruginosa PA14 biofilms. Mol. Microbiol. 51:675–690. 10.1046/j.1365-2958.2003.03877.x [DOI] [PubMed] [Google Scholar]
- 25.Vilain S, Pretorius JM, Theron J, Brözel VS. 2009. DNA as an adhesin: Bacillus cereus requires extracellular DNA to form biofilms. Appl. Environ. Microbiol. 75:2861–2868. 10.1128/AEM.01317-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Whitchurch CB, Tolker-Nielsen T, Ragas PC, Mattick JS. 2002. Extracellular DNA required for bacterial biofilm formation. Science 295:1487. 10.1126/science.295.5559.1487 [DOI] [PubMed] [Google Scholar]
- 27.Vlamakis H, Chai Y, Beauregard P, Losick R, Kolter R. 2013. Sticking together: building a biofilm the Bacillus subtilis way. Nat. Rev. Microbiol. 11:157–168. 10.1038/nrmicro2960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hamon M, Stanley NR, Britton R, Grossman AD, Lazazzera B. 2004. Identification of AbrB-regulated genes involved in biofilm formation by Bacillus subtilis. Mol. Microbiol. 52:847–860. 10.1111/j.1365-2958.2004.04023.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Katayama S, Matsushita O, Jung CM, Minami J, Okabe A. 1999. Promoter upstream bent DNA activates the transcription of the Clostridium perfringens phospholipase C gene in a low temperature-dependent manner. EMBO J. 18:3442–3450. 10.1093/emboj/18.12.3442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jordan S, Rietkötter E, Strauch MA, Kalamorz F, Butcher BG, Helmann JD, Mascher T. 2007. LiaRS-dependent gene expression is embedded in transition state regulation in Bacillus subtilis. Microbiology 153:2530–2540. 10.1099/mic.0.2007/006817-0 [DOI] [PubMed] [Google Scholar]
- 31.Varga JJ, Nguyen V, O'Brien DK, Rodgers K, Walker R, Melville SB. 2006. Type IV pili-dependent gliding motility in the Gram-positive pathogen Clostridium perfringens and other clostridia. Mol. Microbiol. 62:680–694. 10.1111/j.1365-2958.2006.05414.x [DOI] [PubMed] [Google Scholar]
- 32.Rodgers K, Arvidson CG, Melville S. 2011. Expression of a Clostridium perfringens type IV pilin by Neisseria gonorrhoeae mediates adherence to muscle cells. Infect. Immun. 79:3096–3105. 10.1128/IAI.00909-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.de Hoon MJL, Makita Y, Nakai K, Miyano S. 2005. Prediction of transcriptional terminators in Bacillus subtilis and related species. PLoS Comput. Biol. 1:e25. 10.1371/journal.pcbi.0010025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ðapa T, Leuzzi R, Ng YK, Baban ST, Adamo R, Kuehne SA, Scarselli M, Minton NP, Serruto D, Unnikrishnan M. 2013. Multiple factors modulate biofilm formation by the anaerobic pathogen Clostridium difficile. J. Bacteriol. 195:545–555. 10.1128/JB.01980-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nallapareddy S, Singh K. 2006. Endocarditis and biofilm-associated pili of Enterococcus faecalis. J. Clin. Invest. 116:2799–2807. 10.1172/JCI29021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Manetti AG, Zingaretti C, Falugi F, Capo S, Bombaci M, Bagnoli F, Gambellini G, Bensi G, Mora M, Edwards AM, Musser JM, Graviss EA, Telford JL, Grandi G, Margarit I. 2007. Streptococcus pyogenes pili promote pharyngeal cell adhesion and biofilm formation. Mol. Microbiol. 64:968–983. 10.1111/j.1365-2958.2007.05704.x [DOI] [PubMed] [Google Scholar]
- 37.Watnick PI, Fullner KJ, Kolter R. 1999. A role for the mannose-sensitive hemagglutinin in biofilm formation by Vibrio cholerae El Tor. J. Bacteriol. 181:3606–3609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Danne C, Dramsi S. 2012. Pili of gram-positive bacteria: roles in host colonization. Res. Microbiol. 163:645–658. 10.1016/j.resmic.2012.10.012 [DOI] [PubMed] [Google Scholar]
- 39.Strauch M, Webb V, Spiegelman G, Hoch JA. 1990. The SpoOA protein of Bacillus subtilis is a repressor of the abrB gene. Proc. Natl. Acad. Sci. U. S. A. 87:1801–1805. 10.1073/pnas.87.5.1801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Katayama S, Matsushita O, Tamai E, Miyata S, Okabe A. 2001. Phased A-tracts bind to the alpha subunit of RNA polymerase with increased affinity at low temperature. FEBS Lett. 509:235–238. 10.1016/S0014-5793(01)03148-9 [DOI] [PubMed] [Google Scholar]
- 41.Weiss-Muszkat M, Shakh D, Zhou Y, Pinto R, Belausov E, Chapman MR, Sela S. 2010. Biofilm formation by and multicellular behavior of Escherichia coli O55:H7, an atypical enteropathogenic strain. Appl. Environ. Microbiol. 76:1545–1554. 10.1128/AEM.01395-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Piao Z, Sze CC, Barysheva O, Iida K, Yoshida S. 2006. Temperature-regulated formation of mycelial mat-like biofilms by Legionella pneumophila. Appl. Environ. Microbiol. 72:1613–1622. 10.1128/AEM.72.2.1613-1622.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lemon KP, Freitag NE, Kolter R. 2010. The virulence regulator PrfA promotes biofilm formation by Listeria monocytogenes. J. Bacteriol. 192:3969–3976. 10.1128/JB.00179-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wijman JG, de Leeuw PP, Moezelaar R, Zwietering MH, Abee T. 2007. Air-liquid interface biofilms of Bacillus cereus: formation, sporulation, and dispersion. Appl. Environ. Microbiol. 73:1481–1488. 10.1128/AEM.01781-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.