Abstract
Staphylococcus aureus small-colony variants (SCVs) are implicated in chronic and relapsing infections that are difficult to diagnose and treat. Despite many years of study, the underlying molecular mechanisms and virulence effect of the small-colony phenotype remain incompletely understood. We sequenced the genomes of five S. aureus SCV strains recovered from human patients and discovered previously unidentified nonsynonymous point mutations in three genes encoding proteins in the menadione biosynthesis pathway. Analysis of genetic revertants and complementation with wild-type alleles confirmed that these mutations caused the SCV phenotype and decreased virulence for mice.
INTRODUCTION
Staphylococcus aureus is a leading cause of bacterial infections globally and has posed serious infection control problems since the emergence of methicillin-resistant S. aureus (MRSA) strains in 1961 (1). MRSA strains are now the leading cause of death by an infectious agent in the United States, with a mortality rate of approximately 20% (2). MRSA and methicillin-susceptible S. aureus (MSSA) strains cause a variety of diseases, including superficial skin and soft tissue infections and more severe diseases such as bacteremia, pneumonia, endocarditis, and osteomyelitis (3). S. aureus can also be a commensal organism, asymptomatically colonizing approximately 30% of healthy individuals (4), which can lead to subsequent infection (5). Strains of antibiotic-resistant S. aureus have spread worldwide, resulting in an endemic population of MRSA in many industrialized nations.
An ongoing diagnostic and treatment problem in S. aureus infections is due to a subpopulation of small-colony variants (SCVs) found in patients with persistent, antibiotic-resistant, and recurrent infections (6–8). Although SCVs were described >100 years ago (9), it was not until approximately 20 years ago that investigators began to study the biological and pathogenic traits of these organisms in earnest (7). SCVs have been recovered from patients with endocarditis, pneumonia, soft tissue infections, osteomyelitis, and severe bacteremia (reviewed in reference 10). The frequency of SCV recovery from clinical specimens ranges from 1 to 30% (reviewed in reference 11). A distinct characteristic of S. aureus SCV infections is their ability to persist in the presence of aggressive antimicrobial therapy. SCVs constitute a slow-growing, auxotrophic subpopulation with atypical colony morphology and physiology. For example, they produce small, nonpigmented, and nonhemolytic colonies with decreased coagulase activity and an unstable colony phenotype. In addition, they have altered drug resistance profiles, such as an increased resistance to aminoglycosides, and thus are particularly difficult to detect and treat (reviewed in references 11 and 12). SCV strains have significant clinical importance and in some cases have been shown to persist for >50 years (7). These naturally occurring variants have a survival advantage in their ability to persist within eukaryotic cells, thereby protecting them from host defenses and antibiotics (reviewed in reference 13).
The atypical morphology, slow growth, and abnormal biochemical characteristics of SCVs often lead to misidentification by clinical laboratories (14). An additional confounding attribute is the instability of the SCV phenotype, as the variants may revert to a normal phenotype (reviewed in reference 13). Three major categories of SCVs have been found in clinical isolates, namely, electron transport-defective strains that are auxotrophs for menadione, hemin, or thiamine. Auxotrophy for menadione and hemin makes the bacteria unable to synthesize menaquinone and cytochromes, respectively. This often results from mutations in genes coding for enzymes involved in the biosynthesis of these two molecules. Thiamine auxotrophs are rarely identified in human infections and are considered a subtype of menadione-dependent strains. As a consequence of auxotrophy, specific nutritional supplementation and prolonged culture enhance recovery from patient specimens (15). Many of the typical SCV characteristics (e.g., low growth rate, lack of pigmentation, increased aminoglycoside resistance, and decreased alpha-toxin production) can be linked to electron transport deficiencies (reviewed in reference 11). Most studies investigating SCVs have used laboratory strains containing stable mutations in the menD or hemB gene, resulting in menadione- and hemin-auxotrophic strains, respectively, the two most common SCV phenotypes (16–18).
Here we report genetic analyses of five SCV strains cultured from clinically important human infections. The strains are auxotrophic for menadione, and each strain has a single-nucleotide polymorphism (SNP) in one of the menadione biosynthesis genes, including menC, menE, or menF. Genetic complementation of the mutant alleles restored menadione auxotrophy, wild-type colony morphology and growth characteristics, and strain virulence for mice.
MATERIALS AND METHODS
Bacterial strains, growth conditions, and growth curves.
Clinical isolates were recovered from hospitalized patients in Houston, TX. Five strains with small-colony morphology were analyzed (Table 1). Strain LA County (LAC) was used as a reference strain because it is a well-characterized USA300 community-acquired organism whose genome has been sequenced (19). The SCV isolates were grown on Columbia sheep blood agar, Trypticase soy agar (TSA), and Mueller-Hinton agar for 24 to 72 h at 37°C in 5% CO2. The revertant and plasmid-complemented strains were grown on the same media for 12 to 16 h at 37°C. Liquid cultures were grown in Trypticase soy broth (TSB) at 37°C with shaking at 250 rpm. The five SCV strains produce nonpigmented, slow-growing microcolonies. Menadione (Acros Organics) was used, when indicated, at 1 μg/ml, and chloramphenicol was added at 20 μg/ml.
TABLE 1.
S. aureus strains used in this study
| Strain | Phenotype, auxotrophism | Diagnosis | Patient treatment(s) | Gene with SNP | Nucleotide change | Amino acid change |
|---|---|---|---|---|---|---|
| LAC | Wild type, none | |||||
| 221SCV | SCV, menadione | Abscess | Clindamycin | menE | G762A | L254F |
| 221REV | Wild type | Wild-type menE | ||||
| 377SCV | SCV, menadione | Bacteremia | Vancomycin | |||
| Piperacillin | menC | G625A | A209V | |||
| Tazobactam | ||||||
| Gentamicin | menF | G382A | G128E | |||
| 377COM | Wild type | psk265menCa (wild type) | G625A | A209V | ||
| G382A | G128E | |||||
| 453SCV | SCV, menadione | Osteomyelitis | Vancomycin | menC | G514A | A172V |
| Nafcillin | ||||||
| Gentamicin | ||||||
| 453COM | Wild type | psk265menC (wild type) | G514A | A172V | ||
| 726SCV | SCV, menadione | Osteomyelitis | Ciprofloxacin, clindamycin | menC | G625A | A209V |
| menF | G382A | G128E | ||||
| 726COM | Wild type | psk265menC (wild type) | G625A | A209V | ||
| G382A | G128E | |||||
| 927SCV | SCV, menadione | Abscess | Clindamycin, trimethoprim-sulfamethoxazole | menF | A594G | I198M |
| 927REV | Wild-type menF |
psk265 contains the wild-type menC allele from strain LAC (menC accession number SAU300_1735 from the NCBI whole-genome database).
For growth curve experiments, fresh colonies from each strain were picked from TSA plates and incubated overnight in 5 ml of TSB at 37°C with shaking. Cultures grown overnight were diluted to an optical density at 600 nm (OD600) of 0.02 in 50 ml of fresh TSB and incubated at 37°C with shaking. The OD600 was measured hourly for 8 h.
Genome sequencing and data analysis.
Methods for genome sequencing and data processing and analysis were described previously (20). The sequencing reads were compared to the LAC reference genome to identify genetic polymorphisms (19).
Targeted men gene sequencing.
S. aureus strains were grown overnight on TSA. Genomic DNA was extracted by alkaline-boiling lysis (21). Sanger sequencing was used to determine the men gene alleles by using previously described methods (20). The primer pairs used (5′ to 3′) were menC1 (CAGCAAGCAAGCATTCACAT) and menC2 (TCCAGGTATCAGTGACGCAG), menE1 (GCGTTACATTCCCCGAAATA) and menE2 (AGATGATGTGGCAACAATGGGT), and menF1 (AGTGAAATGGATGGCTACGG) and menF2 (AATTCGCTGCTGCTGTACCT). Electropherograms were visually inspected by using Sequencher 4.7 (Gene Codes).
Selection of fast-growing (wild-type) revertant colonies.
Small colonies were picked from fresh TSA plates, grown in TSB with serial passage daily for 3 to 10 days, and then plated and screened for fast-growing colonies after 14 h of incubation at 37°C. Wild-type-sized colonies were picked, purified by restreaking, and stored in TSB with 60% glycerol at −80°C. The menE and menF genes were sequenced by Sanger sequencing using primer pair menE1 and menE2 and primer pair menF1 and menF2, respectively, with an Applied Biosystems 3730 capillary sequencer. Fast-growing colonies that were derived from strains 221 and 927 and were confirmed to have a wild-type (revertant) allele in menE or menF were designated strains 221REV and 927REV, respectively.
Construction of genetically complemented strains.
cis-acting elements that are required for complete menE and menC gene expression, including an upstream putative ribosomal binding site and promoter region, were amplified with primers menE2 and ID4 (5′-ATGTCATCCGCTTCATAAAGG-3′). The 2.7-kb fragment of the PCR product was purified with the QIAquick gel extraction kit, ligated into the PCR 2.1-TOPO vector (Invitrogen, Carlsbad, CA), and transformed into competent Escherichia coli TOP10 cells (Invitrogen). A staphylococcal origin of replication was introduced by cloning plasmid pSK265 into the unique BamHI site on PCR 2.1-TOPO, and the construct was moved into S. aureus RN4220 by electroporation (22). Phage 80α was used to transduce the constructs from RN4220 into strains 377SCV, 453SCV, and 726SCV. The final constructs were designated 377COM, 453COM, and 726COM.
Determination of gentamicin MIC.
The MIC of gentamicin was determined with Etest strips (0.016 to 256 μg/ml gentamicin) (bioMérieux SA) according to the manufacturer's instructions.
Hemolysis assay.
Hemolytic activity was determined spectrophotometrically by measuring the release of hemoglobin from rabbit red blood cells (RBCs) (23). Briefly, bacteria were grown overnight in TSB at 37°C with shaking at 250 rpm. The cells were pelleted by centrifugation, and the supernatant was discarded. The cells were suspended in phosphate-buffered saline (PBS) containing 1 mM CaCl and 0.5 mM Mg, incubated for 1 h, and collected by centrifugation, and the supernatant was filter sterilized. Rabbit RBCs were washed three times in PBS with 0.85% NaCl to remove free hemoglobin. The washed RBCs were diluted to 1.4% in PBS containing 0.2% (wt/vol) bovine serum albumin. The filtered bacterial supernatants were added to an equal volume of the 1.4% RBC solution and incubated for 1 h at 37°C. A PBS control was used for spontaneous lysis, and Staphylococcus epidermidis was used as a nonhemolytic control. The cells were then pelleted for 10 s at maximum speed in a desktop microcentrifuge. The released hemoglobin was measured as the A540 with a spectrophotometer. Standard dilutions were made for comparison; deionized water was added to an equal volume of RBCs to represent 100% hemolysis (24). Additional standards were made for comparison (75%, 50%, 25%, 12.5%, and 6.25%). The results shown are the means ± standard deviations of at least three replicate experiments.
Mouse virulence experiments.
Virulence of SCV (454SCV) and isogenic complemented (453com) strains were compared with a mouse model of bacteremia. Fifteen 5- to 6-week-old outbred immunocompetent female CD1 mice (Harlan Laboratories) were randomly assigned to a strain treatment group and inoculated intraperitoneally with 1 × 107 CFU. Bacterial doses were prepared at the time of inoculation from previously quantified frozen stocks, and the actual dose was confirmed by CFU analysis. Near mortality was determined by observation. Data were graphically displayed as a Kaplan-Meier survival curve and analyzed with the log-rank test (Prism 6; GraphPad Software Inc., La Jolla, CA). All mouse experiments were approved by the Institutional Animal Care and Use Committee, Houston Methodist Research Institute (protocol AUP-1010-0022).
RESULTS
Isolation of S. aureus SCVs.
We screened ∼1,000 S. aureus strains cultured from normally sterile sites for a SCV phenotype on blood agar plates. Five SCV strains were identified among these organisms (Table 1). Two methicillin-resistant strains (221SCV and 927SCV) caused abscesses in community-acquired infections occurring in patients with no underlying illness or risk factors (community-acquired MRSA [CA-MRSA]). The other three strains were from hospital-associated S. aureus (HA-SA) infections, two of which were MRSA strains (strains 377SCV and 453SCV) and one of which was an MSSA strain (strain 726SCV). Both HA-MRSA-infected patients were treated with an antibiotic regimen that included gentamicin, an agent known to select for SCVs (25). The three patients with HA-SA infections had underlying medical conditions and had previously been hospitalized. All five strains were of the USA300 clonotype.
The five SCV strains formed colonies that were approximately 1/10 the diameter of normal S. aureus colonies after 24 h of incubation at 37°C (Fig. 1A and B). The colonies were white, versus the yellow-gold colonies of strain LAC and many other S. aureus strains (data not shown).
FIG 1.
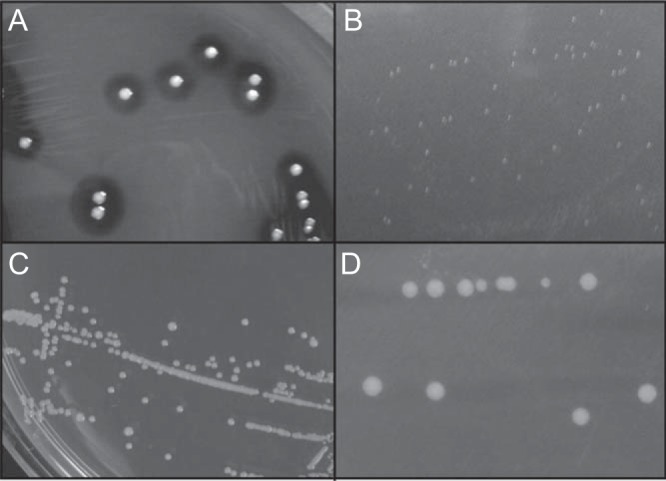
Colony morphology on TSA or blood agar plates. (A) S. aureus wild-type strain LAC grown on blood agar plates for 14 h; (B) pinpoint SCV colonies on TSA after 24 h of growth; (C) SCV colonies on TSA supplemented with 1 μg/ml menadione; (D) plasmid-complemented SCV strain 453COM grown on TSA.
Identification of single-nucleotide polymorphisms in the men genes of the SCV strains.
As a first step toward determining the genetic mechanism underlying the SCV phenotype in these five strains, the genome of each strain was sequenced. Inasmuch as mutations in hemB, menB, menD, or thyA have been reported to cause menadione, hemin, or thymine auxotrophy and SCV phenotypes in S. aureus (7, 17), we initially analyzed the genome sequences for polymorphisms in genes implicated in menadione (men), hemin (hem), or thymidine (thy) biosynthesis. Relative to the reference genome of strain LAC (19), all five SCV variants had nonsynonymous (amino-acid-altering) SNPs in one of the men genes. In contrast, no polymorphisms that have been associated with the SCV phenotype were present in the hem or thy gene (Table 1). Three strains each had a different SNP in one of the nine genes implicated in menadione biosynthesis. Strain 221SCV had an SNP in menE at nucleotide 762, resulting in a L254F replacement. Strain 453SCV had an SNP in menC at nucleotide 514 that resulted in an A172V change, and the menF gene in strain 927SCV had an SNP that produced an I198M replacement. Strains 377SCV and 726SCV each had one SNP in menC that resulted in an A209V change and a second SNP in menF that produced a G128E replacement (Table 1). No other polymorphisms that could be readily hypothesized to cause SCV variants and menadione, hemin, or thymine auxotrophy were identified in the five genomes.
We next tested the hypothesis that the identified single-amino-acid replacements caused the SCV phenotype. SCVs are known to be unstable, so we first screened for wild-type revertants by growth on TSB after serial in vitro passage for 3 to 10 days (see Materials and Methods). Two strains (221SCV and 927SCV) produced growth revertants (designated 221REV and 927REV) at rates of 1.76 × 10−6 and 1.21 × 10−6 per cell per generation, respectively, as assessed by fluctuation analysis (22). These rates are consistent with expected values for the reversion of a point mutation and are similar to other data in the literature reported for reversion of SCV strains (26, 27). Sanger DNA sequencing confirmed that the revertant strains had a wild-type allele of the cognate men gene.
As in vitro passage yielded revertant colonies only for two of the five SCV strains, we introduced the wild-type allele of either the menE or menC gene into the remaining three strains to create 377COM, 726COM, and 453COM. Sanger sequencing confirmed the presence of a wild-type copy of the cognate mutant gene.
Although strains 377SCV and 726SCV had SNPs in both menC and menF, we began our analysis by complementing the menC gene. The SNP in the menC gene changes an amino acid located directly between two invariant amino acids responsible for Mg cofactor binding (28).
Strain 453SCV contains an SNP in the menC gene that changes an amino acid located within the absolutely conserved DAN (Asp-Ala-Asn) motif found in the o-succinylbenzoate synthase family of proteins, of which MenC is a member. The alanine residue of this motif is required to maintain the position of the Mg+ cofactor-binding pocket (28). In strain 453SCV, this critical alanine is replaced by valine (A172V), which may explain the stability of the SCV phenotype in strain 453SCV. MenE belongs to the o-succinylbenzoate–coenzyme A (CoA) ligase family of proteins. Members of this family are responsible for catalyzing two reactions and share conserved catalytic domain structures (29). The SNP in the menE gene of strain 221SCV results in a L254F replacement located directly adjacent to a conserved functional domain of the protein. This amino acid change may alter the catalytic site, leading to menadione auxotrophism and the observed SCV phenotype.
Using the five clinical SCV strains and their wild-type revertant or plasmid-complemented counterparts, we sought to characterize the functional consequences of these naturally occurring mutations. Our results demonstrate that correction of the men gene mutation causes reversion of the small-colony phenotype and restores wild-type growth, hemolytic activity, and antibiotic susceptibility.
Growth of bacteria.
To test the hypothesis that the SCV phenotype in these five strains was related to menadione auxotrophy, we grew the strains on TSA supplemented with menadione. The results (Fig. 1C) showed that wild-type colony morphology was restored by growth in the presence of exogenous menadione. To further test this hypothesis, we grew plasmid-complemented cells on TSA lacking supplemental menadione. Consistent with our hypothesis, genetic complementation of SCV strains with a plasmid containing a wild-type men gene restored wild-type colony morphology, including size and gold pigmentation (Fig. 1D). Similarly, growth of SCV strains in liquid medium (TSB) supplemented with 1 μg/ml menadione or with a plasmid containing the cognate wild-type men gene restored growth to a wild-type phenotype (Fig. 2A and B).
FIG 2.
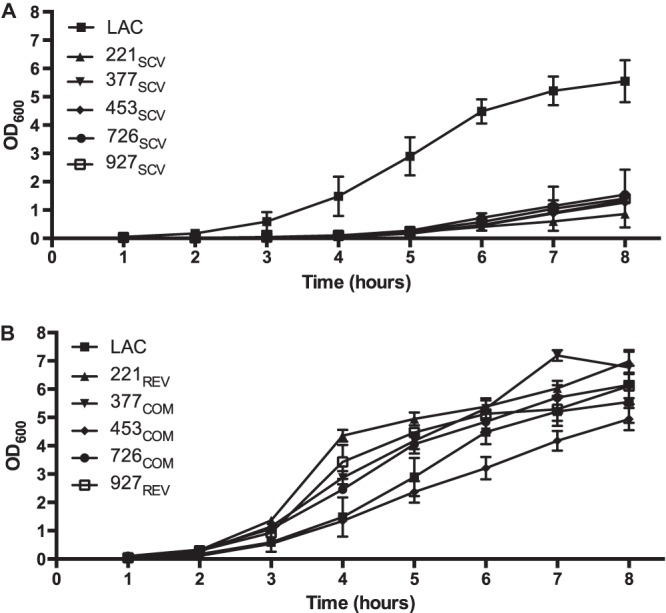
Growth curves of SCVs and revertant or genetically complemented strains. (A) Growth curves of control strain LAC and the five clinical SCV strains; (B) growth curves of the plasmid-complemented strains and revertant strains 927REV and 221REV. Growth curves were performed with TSB. Data are means ± standard deviations of at least 3 independent experiments.
Hemolysis of red blood cells.
A key characteristic phenotype of S. aureus SCVs is a reduced level of alpha-toxin production, which is believed to enhance persistence in nonprofessional phagocytes (24). Consistent with other SCV strains reported in the literature, these five strains had greatly reduced alpha-toxin production, as assayed by hemolysis of red blood cells (Fig. 3). To establish that this reduction in hemolysis was due to the mutations in the men genes, bacteria were grown in TSB supplemented with 1 μg/ml menadione (Fig. 3, black bars). The addition of exogenous menadione significantly increased hemolysis of red blood cells compared to the SCV strains grown in the absence of menadione (P < 0.05). The natural revertant strains 221REV and 927REV and the plasmid-complemented strains 377COM, 453COM, and 726COM (Fig. 3, white bars) caused significantly more RBC hemolysis (P < 0.0001) than their SCV parental strains. These results show that reversion of this mutation restores the amount of hemolysis to that of a wild-type strain, indicating that the men gene mutations are responsible for the small-colony phenotypes.
FIG 3.
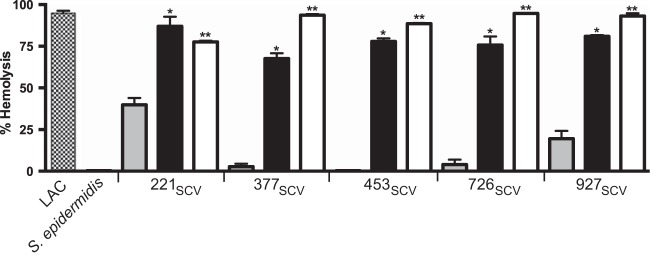
Hemolysis of red blood cells. Hemolysis of rabbit erythrocytes was measured spectrophotometrically and compared with lysed erythrocyte standards to calculate percent hemolysis. The tests were performed by using each SCV strain (gray bars), the SCV strains grown in media supplemented with 1 μg/ml menadione (black bars), and the SCV strains with wild-type men genes (white bars). LAC was included as a wild-type positive control; S. epidermidis does not produce alpha-toxin (negative control). A single asterisk indicates a significant difference in red blood cell hemolysis between the menadione-supplemented strain and the matching SCV strain. A double asterisk indicates a significant difference in percent hemolysis between the complemented or revertant strain and the corresponding SCV strain (P < 0.05 by t test). Data are the means ± standard deviations of at least 3 independent experiments.
Gentamicin resistance.
SCV S. aureus strains have altered antibiotic resistance profiles that are thought to be caused by interruptions in electron transport and low growth rates. Menadione-auxotrophic SCVs have defective electron transport chains that result in the decreased uptake of aminoglycosides such as gentamicin, thereby causing increased antibiotic resistance (30). Gentamicin-resistant SCVs are important pathogens repeatedly recovered from cystic fibrosis patients (31) and patients with chronic staphylococcal bone infections (32). Consistent with previous reports, our five clinical strains had increased resistance to gentamicin compared to the control strain LAC (Fig. 4). Genetically complemented strains 377COM, 453COM, and 726COM and natural revertant strains 221REV and 927REV had MICs comparable to that of the wild-type strain (Fig. 4C and D). Additionally, menadione added exogenously to the SCV strains resulted in an MIC of gentamicin that was identical to that for the wild-type strain (data not shown). Thus, these results are consistent with the idea that mutations in the menadione biosynthesis genes responsible for the SCV phenotype also result in increased gentamicin resistance.
FIG 4.
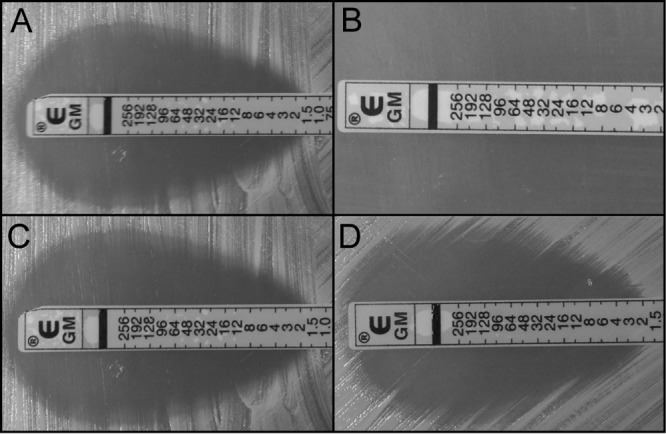
Gentamicin susceptibility of various strains. (A) USA300 LAC; (B) SCV strain 776SCV; (C) plasmid-complemented strain 453COM; (D) natural revertant strain 927REV.
SCV strain 453SCV is less virulent for mice than genetically complemented strain 453COM in an invasive infection model.
To test the hypothesis that S. aureus SCV strains have altered virulence, we compared strain 453SCV with the isogenic genetically complemented strain 453COM in a mouse model of bacteremia. The use of strain 453 in these experiments was an arbitrary choice. Consistent with the hypothesis of altered virulence, strain 453SCV was significantly less virulent than the genetically complemented strain 453COM (Fig. 5).
FIG 5.
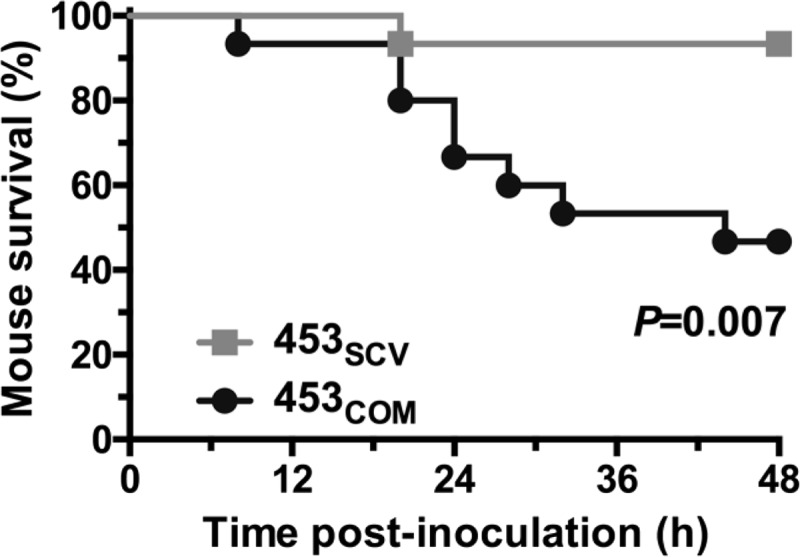
Virulence in mice. Fifteen outbred CD1 mice were infected intraperitoneally with 1 × 107 CFU of each indicated strain. Data are shown as Kaplan-Meier survival over time, with P values calculated with the log-rank test.
DISCUSSION
In this work, we identified the genetic basis of the SCV phenotype observed among five naturally occurring clinical strains of S. aureus cultured from normally sterile sites. These five strains were isolated from community-acquired and hospital-associated infections, included both MRSA and MSSA, and caused multiple infection types. All five strains had classic SCV phenotypes, including pinpoint colonies, lack of pigmentation, slow growth, reduced alpha-toxin production, and increased antibiotic resistance. Full-genome sequencing led to the discovery that these strains contained mutations in the men genes menC, menE, and menF. We showed a reversal of the SCV phenotype through restoration of a wild-type copy of the mutant men gene containing the point mutations or mutational reversion or through the addition of exogenous menadione. In aggregate, the data provide compelling evidence that mutations in menC, menE, or menF are the direct cause of menadione-auxotrophic SCVs in these clinical isolates of S. aureus.
Traditional experiments focused on identifying compounds that restored wild-type growth to small colonies, leading to the discovery that many SCVs are auxotrophic for menadione or hemin (7, 9, 33). The failure of SCVs to produce menadione or hemin results in interrupted electron transport and decreased ATP synthesis. Many of the phenotypes associated with SCVs, such as slow growth, decreased pigmentation, and resistance to aminoglycosides, can be linked to disrupted respiration and energy production (34, 35). In the majority of Gram-positive bacteria, menaquinone is the only quinone in the electron transport chain, making the enzymes that synthesize this compound promising drug targets for multidrug-resistant pathogens such as S. aureus and Mycobacterium tuberculosis (34). In this regard, we note that growth inhibition has been reported for compounds that target the gene products of menA (34, 36), menD (16), menE (18), and menB (37). The initial findings for this new class of antimicrobial molecules highlight the importance of the work presented here. Given the relative lack of success over the last 10 years in identifying new compounds to treat multidrug-resistant pathogens such as S. aureus (38), the menaquinone biosynthesis pathway may prove to be a fruitful area of focus to selectively target prokaryotes while preventing side effects to eukaryotes, which lack menaquinone.
ACKNOWLEDGMENTS
This work was supported by the Houston Methodist Hospital. A.E.R. was supported by NIH grant 5R01AI080688-04.
We thank Concepcion Cantu for technical assistance.
Footnotes
Published ahead of print 22 January 2014
REFERENCES
- 1.Jevons M. 1961. “Celbenin”-resistant staphylococci. Br. Med. J. i:124–125. 10.1136/bmj.1.5219.124-a [DOI] [Google Scholar]
- 2.Klevens RM, Morrison MA, Nadle J, Petit S, Gershman K, Ray S, Harrison LH, Lynfield R, Dumyati G, Townes JM, Craig AS, Zell ER, Fosheim GE, McDougal LK, Carey RB, Fridkin SK, Active Bacterial Core Surveillance MRSA Investigators 2007. Invasive methicillin-resistant Staphylococcus aureus infections in the United States. JAMA 298:1763–1771. 10.1001/jama.298.15.1763 [DOI] [PubMed] [Google Scholar]
- 3.Diekema DJ, Pfaller MA, Schmitz FJ, Smayevsky J, Bell J, Jones RN, Beach M, Sentry Participants Group 2001. Survey of infections due to Staphylococcus species: frequency of occurrence and antimicrobial susceptibility of isolates collected in the United States, Canada, Latin America, Europe, and the Western Pacific region for the SENTRY Antimicrobial Surveillance Program, 1997–1999. Clin. Infect. Dis. 32(Suppl 2):S114–S132. 10.1086/320184 [DOI] [PubMed] [Google Scholar]
- 4.Gorwitz RJ, Kruszon-Moran D, McAllister SK, McQuillan G, McDougal LK, Fosheim GE, Jensen BJ, Killgore G, Tenover FC, Kuehnert MJ. 2008. Changes in the prevalence of nasal colonization with Staphylococcus aureus in the United States, 2001-2004. J. Infect. Dis. 197:1226–1234. 10.1086/533494 [DOI] [PubMed] [Google Scholar]
- 5.von Eiff C, Becker K, Machka K, Stammer H, Peters G. 2001. Nasal carriage as a source of Staphylococcus aureus bacteremia. N. Engl. J. Med. 344:11–16. 10.1056/NEJM200101043440102 [DOI] [PubMed] [Google Scholar]
- 6.Kahl B, Herrmann M, Everding AS, Koch HG, Becker K, Harms E, Proctor RA, Peters G. 1998. Persistent infection with small colony variant strains of Staphylococcus aureus in patients with cystic fibrosis. J. Infect. Dis. 177:1023–1029. 10.1086/515238 [DOI] [PubMed] [Google Scholar]
- 7.Proctor RA, van Langevelde P, Kristjansson M, Maslow JN, Arbeit RD. 1995. Persistent and relapsing infections associated with small-colony variants of Staphylococcus aureus. Clin. Infect. Dis. 20:95–102. 10.1093/clinids/20.1.95 [DOI] [PubMed] [Google Scholar]
- 8.von Eiff C, Vaudaux P, Kahl BC, Lew D, Emler S, Schmidt A, Peters G, Proctor RA. 1999. Bloodstream infections caused by small-colony variants of coagulase-negative staphylococci following pacemaker implantation. Clin. Infect. Dis. 29:932–934. 10.1086/520462 [DOI] [PubMed] [Google Scholar]
- 9.Proctor RA, Peters G. 1998. Small colony variants in staphylococcal infections: diagnostic and therapeutic implications. Clin. Infect. Dis. 27:419–422. 10.1086/514706 [DOI] [PubMed] [Google Scholar]
- 10.McNamara PJ, Proctor RA. 2000. Staphylococcus aureus small colony variants, electron transport and persistent infections. Int. J. Antimicrob. Agents 14:117–122. 10.1016/S0924-8579(99)00170-3 [DOI] [PubMed] [Google Scholar]
- 11.Proctor RA, von Eiff C, Kahl BC, Becker K, McNamara P, Herrmann M, Peters G. 2006. Small colony variants: a pathogenic form of bacteria that facilitates persistent and recurrent infections. Nat. Rev. Microbiol. 4:295–305. 10.1038/nrmicro1384 [DOI] [PubMed] [Google Scholar]
- 12.Sendi P, Proctor RA. 2009. Staphylococcus aureus as an intracellular pathogen: the role of small colony variants. Trends Microbiol. 17:54–58. 10.1016/j.tim.2008.11.004 [DOI] [PubMed] [Google Scholar]
- 13.Melter O, Radojevic B. 2010. Small colony variants of Staphylococcus aureus—review. Folia Microbiol. (Praha) 55:548–558. 10.1007/s12223-010-0089-3 [DOI] [PubMed] [Google Scholar]
- 14.Kahl B, von Eiff C, Hermann M, Peters G, Proctor RA. 1996. Staphylococcal small colony variants present a challenge to clinicians and clinical microbiologists. Antimicrob. Infect. Dis. Newsl. 15:59–63. 10.1016/S1069-417X(00)80009-6 [DOI] [Google Scholar]
- 15.Garcia LG, Lemaire S, Kahl BC, Becker K, Proctor RA, Denis O, Tulkens PM, Van Bambeke F. 2013. Antibiotic activity against small-colony variants of Staphylococcus aureus: review of in vitro, animal and clinical data. J. Antimicrob. Chemother. 68:1455–1464. 10.1093/jac/dkt072 [DOI] [PubMed] [Google Scholar]
- 16.Fang M, Macova A, Hanson KL, Kos J, Palmer DR. 2011. Using substrate analogues to probe the kinetic mechanism and active site of Escherichia coli MenD. Biochemistry 50:8712–8721. 10.1021/bi201202n [DOI] [PubMed] [Google Scholar]
- 17.Lannergard J, von Eiff C, Sander G, Cordes T, Seggewiss J, Peters G, Proctor RA, Becker K, Hughes D. 2008. Identification of the genetic basis for clinical menadione-auxotrophic small-colony variant isolates of Staphylococcus aureus. Antimicrob. Agents Chemother. 52:4017–4022. 10.1128/AAC.00668-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lu X, Zhang H, Tonge PJ, Tan DS. 2008. Mechanism-based inhibitors of MenE, an acyl-CoA synthetase involved in bacterial menaquinone biosynthesis. Bioorg. Med. Chem. Lett. 18:5963–5966. 10.1016/j.bmcl.2008.07.130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Diep BA, Gill SR, Chang RF, Phan TH, Chen JH, Davidson MG, Lin F, Lin J, Carleton HA, Mongodin EF, Sensabaugh GF, Perdreau-Remington F. 2006. Complete genome sequence of USA300, an epidemic clone of community-acquired meticillin-resistant Staphylococcus aureus. Lancet 367:731–739. 10.1016/S0140-6736(06)68231-7 [DOI] [PubMed] [Google Scholar]
- 20.Shea PR, Beres SB, Flores AR, Ewbank AL, Gonzalez-Lugo JH, Martagon-Rosado AJ, Martinez-Gutierrez JC, Rehman HA, Serrano-Gonzalez M, Fittipaldi N, Ayers SD, Webb P, Willey BM, Low DE, Musser JM. 2011. Distinct signatures of diversifying selection revealed by genome analysis of respiratory tract and invasive bacterial populations. Proc. Natl. Acad. Sci. U. S. A. 108:5039–5044. 10.1073/pnas.1016282108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hartas J, Hibble M, Sriprakash KS. 1998. Simplification of a locus-specific DNA typing method (Vir typing) for Streptococcus pyogenes. J. Clin. Microbiol. 36:1428–1429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Finan JE, Rosato AE, Dickinson TM, Ko D, Archer GL. 2002. Conversion of oxacillin-resistant staphylococci from heterotypic to homotypic resistance expression. Antimicrob. Agents Chemother. 46:24–30. 10.1128/AAC.46.1.24-30.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bernheimer AW. 1988. Assay of hemolytic toxins. Methods Enzymol. 165:213–217. 10.1016/S0076-6879(88)65033-6 [DOI] [PubMed] [Google Scholar]
- 24.Balwit JM, van Langevelde P, Vann JM, Proctor RA. 1994. Gentamicin-resistant menadione and hemin auxotrophic Staphylococcus aureus persist within cultured endothelial cells. J. Infect. Dis. 170:1033–1037. 10.1093/infdis/170.4.1033 [DOI] [PubMed] [Google Scholar]
- 25.von Eiff C, Bettin D, Proctor RA, Rolauffs B, Lindner N, Winkelmann W, Peters G. 1997. Recovery of small colony variants of Staphylococcus aureus following gentamicin bead placement for osteomyelitis. Clin. Infect. Dis. 25:1250–1251. 10.1086/516962 [DOI] [PubMed] [Google Scholar]
- 26.Maple PA, Hamilton-Miller JM, Brumfitt W. 1991. Differing activities of quinolones against ciprofloxacin-susceptible and ciprofloxacin-resistant, methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 35:345–350. 10.1128/AAC.35.2.345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schmitz FJ, Fluit AC, Hafner D, Beeck A, Perdikouli M, Boos M, Scheuring S, Verhoef J, Kohrer K, Von Eiff C. 2000. Development of resistance to ciprofloxacin, rifampin, and mupirocin in methicillin-susceptible and -resistant Staphylococcus aureus isolates. Antimicrob. Agents Chemother. 44:3229–3231. 10.1128/AAC.44.11.3229-3231.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thompson TB, Garrett JB, Taylor EA, Meganathan R, Gerlt JA, Rayment I. 2000. Evolution of enzymatic activity in the enolase superfamily: structure of o-succinylbenzoate synthase from Escherichia coli in complex with Mg2+ and o-succinylbenzoate. Biochemistry 39:10662–10676. 10.1021/bi000855o [DOI] [PubMed] [Google Scholar]
- 29.Gulick AM, Starai VJ, Horswill AR, Homick KM, Escalante-Semerena JC. 2003. The 1.75 A crystal structure of acetyl-CoA synthetase bound to adenosine-5′-propylphosphate and coenzyme A. Biochemistry 42:2866–2873. 10.1021/bi0271603 [DOI] [PubMed] [Google Scholar]
- 30.von Eiff C. 2008. Staphylococcus aureus small colony variants: a challenge to microbiologists and clinicians. Int. J. Antimicrob. Agents 31:507–510. 10.1016/j.ijantimicag.2007.10.026 [DOI] [PubMed] [Google Scholar]
- 31.Proctor RA, Kahl B, von Eiff C, Vaudaux PE, Lew DP, Peters G. 1998. Staphylococcal small colony variants have novel mechanisms for antibiotic resistance. Clin. Infect. Dis. 27(Suppl 1):S68–S74 [DOI] [PubMed] [Google Scholar]
- 32.von Eiff C, Peters G, Becker K. 2006. The small colony variant (SCV) concept—the role of staphylococcal SCVs in persistent infections. Injury 37(Suppl 2):S26–S33. 10.1016/j.injury.2006.04.006 [DOI] [PubMed] [Google Scholar]
- 33.Proctor RA, Balwit JM, Vesga O. 1994. Variant subpopulations of Staphylococcus aureus as cause of persistent and recurrent infections. Infect. Agents Dis. 3:302–312 [PubMed] [Google Scholar]
- 34.Kurosu M, Narayanasamy P, Biswas K, Dhiman R, Crick DC. 2007. Discovery of 1,4-dihydroxy-2-naphthoate prenyltransferase inhibitors: new drug leads for multidrug-resistant gram-positive pathogens. J. Med. Chem. 50:3973–3975. 10.1021/jm070638m [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.von Eiff C, Heilmann C, Proctor RA, Woltz C, Peters G, Gotz F. 1997. A site-directed Staphylococcus aureus hemB mutant is a small-colony variant which persists intracellularly. J. Bacteriol. 179:4706–4712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kurosu M, Crick DC. 2009. MenA is a promising drug target for developing novel lead molecules to combat Mycobacterium tuberculosis. Med. Chem. 5:197–207. 10.2174/157340609787582882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Debnath J, Siricilla S, Wan B, Crick DC, Lenaerts AJ, Franzblau SG, Kurosu M. 2012. Discovery of selective menaquinone biosynthesis inhibitors against Mycobacterium tuberculosis. J. Med. Chem. 55:3739–3755. 10.1021/jm201608g [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Payne DJ, Gwynn MN, Holmes DJ, Pompliano DL. 2007. Drugs for bad bugs: confronting the challenges of antibacterial discovery. Nat. Rev. Drug Discov. 6:29–40. 10.1038/nrd2201 [DOI] [PubMed] [Google Scholar]


