Abstract
We performed a genomewide analysis using a next-generation sequencer to investigate the effect of pulmonary surfactant on gene expression in Staphylococcus aureus, a clinically important opportunistic pathogen. RNA sequence (RNA-seq) analysis of bacterial transcripts at late log phase revealed 142 genes that were upregulated >2-fold following the addition of pulmonary surfactant to the culture medium. Among these genes, we confirmed by quantitative reverse transcription-PCR analysis that mRNA amounts for genes encoding ESAT-6 secretion system C (EssC), an unknown hypothetical protein (NWMN_0246; also called pulmonary surfactant-inducible factor A [PsiA] in this study), and hemolysin gamma subunit B (HlgB) were increased 3- to 10-fold by the surfactant treatment. Among the major constituents of pulmonary surfactant, i.e., phospholipids and palmitate, only palmitate, which is the most abundant fatty acid in the pulmonary surfactant and a known antibacterial substance, stimulated the expression of these three genes. Moreover, these genes were also induced by supplementing the culture with detergents. The induction of gene expression by surfactant or palmitate was not observed in a disruption mutant of the sigB gene, which encodes an alternative sigma factor involved in bacterial stress responses. Furthermore, each disruption mutant of the essC, psiA, and hlgB genes showed attenuation of both survival in the lung and host-killing ability in a murine pneumonia model. These findings suggest that S. aureus resists membrane stress caused by free fatty acids present in the pulmonary surfactant through the regulation of virulence gene expression, which contributes to its pathogenesis within the lungs of the host animal.
INTRODUCTION
Pathogenic bacteria infecting host animals sense environmental changes and alter the expression of various genes to exert their virulence. Recently, the virulence genes required for Staphylococcus aureus to cause sepsis in infected hosts were identified by studying the global alterations of the gene expression patterns induced by serum and blood (1). Pathogens that induce pneumonia, however, are inhaled through the nasal cavities and transferred through the bronchus to the lung, followed by colonization on alveolar lumens and alveolar epithelial cells (2). Bacteria infecting the lungs are then exposed to pulmonary surfactant, which covers the alveolar surfaces. Pulmonary surfactant is a secretion comprising mostly lipids (90% of dry weight) that is produced by alveolar epithelial cells to maintain the surface tension of the alveoli (3). The major lipid constituents of surfactant are phosphatidylcholine (80% of the dry weight of total lipids), phosphatidylglycerol (10%), and free fatty acids (10%) (3). Approximately 60% of the phosphatidylcholine is dipalmitoyl-phosphatidylcholine (DPPC), and the other 40% is phosphatidylcholines containing unsaturated fatty acids, such as 1-palmitoyl-2-oleoyl-phosphatidylcholine (POPC). Palmitate is the most abundant free fatty acid in the pulmonary surfactant. When bacteria are exposed to such a lipid-rich environment, distinctive responses different from those in bacteria circulating in the bloodstream or infecting other tissues are likely to be induced, which may contribute to virulence in host lungs. Clarification of the mechanisms underlying the bacterial responses to pulmonary surfactant is important for enhancing our understanding of lung infections and overcoming pneumonia. The effects of pulmonary surfactant on gene expression in pathogenic bacteria have not yet been studied at the molecular level.
Staphylococcus aureus is an opportunistic pathogen that causes sepsis and pneumonia. The recent emergence of S. aureus strains resistant to several antibiotics, including methicillin, has led to serious clinical issues. Moreover, some of these resistant strains, community-associated methicillin-resistant S. aureus (CA-MRSA), have acquired higher virulence potentials than those of hospital-associated methicillin-resistant S. aureus (HA-MRSA) (4, 5). Our research group has investigated various aspects of the virulence mechanisms of S. aureus, e.g., identification and characterization of novel virulence genes (6); investigation of the involvement of the staphylococcal cassette chromosome mec element (SCCmec), a mobile genetic element that provides methicillin resistance, in pathogenesis (7, 8); analysis of activation mechanisms of innate immunity by use of insects as animal models (9–13); and searching for new antibiotics with novel chemical structures (14, 15). These studies have provided some insights into sepsis and have contributed to the establishment of some drug therapies. Here we performed a genomewide analysis of gene expression in S. aureus exposed to pulmonary surfactant to determine the virulence mechanism of lung-infecting S. aureus.
MATERIALS AND METHODS
Ethics statement.
This study was performed in strict accordance with the recommendations of the Fundamental Guidelines for Proper Conduct of Animal Experiments and Related Activities in Academic Research Institutions (16) under the jurisdiction of the Ministry of Education, Culture, Sports, Science, and Technology of Japan. All mouse protocols followed the regulations for animal care and use of the University of Tokyo and were approved by the Animal Use Committee at the Graduate School of Pharmaceutical Science at the University of Tokyo (approval number 24-51).
Bacteria, animals, and reagents.
S. aureus strains Newman, RN4220, MW2, and FRP3757 were aerobically cultured in tryptic soy broth (TSB) (Becton, Dickinson and Co.) at 37°C for 18 to 24 h with vigorous shaking. Escherichia coli JM109 was used as a host for plasmids in DNA manipulations. C57BL/6JJc1 mice were obtained from CLEA Japan, Inc. Surfacten, a pulmonary surfactant extracted from cow lungs, was purchased from the Mitsubishi Tanabe Pharma Corporation and suspended in saline. Bovine calf serum was purchased from SAFC Biosciences and was heat inactivated at 50°C for 30 min before use. DPPC and POPC were purchased from Wako Pure Chemical Industries and the NOF Corporation, respectively. 1-Palmitoyl-2-oleoyl-phosphatidylglycerol (POPG) and sodium palmitate were purchased from Sigma-Aldrich. Oleic acid sodium salt and detergents, such as sodium dodecyl sulfate (SDS), Triton X-100, and polyoxyethylene sorbitan monolaurate (Tween 20), were purchased from Nacalai Tesque.
RNA sequence (RNA-seq) analysis by use of a next-generation sequencer.
TSB medium (5 ml) supplied with 0.3 mg/ml of Surfacten or 10% calf serum was inoculated with 50 μl of an overnight culture of S. aureus Newman. The culture was incubated at 37°C to an optical density at 600 nm (OD600) of 1.0 ± 0.1, and then RNAprotect bacterial reagent (Qiagen) was added at twice the culture volume. After 5 min of incubation at room temperature, the cells were washed in 0.5 ml of phosphate-buffered saline (PBS). Each pellet was suspended in 180 μl Tris-EDTA (TE) buffer (pH 8.0) supplied with 20 μl of 10-mg/ml lysostaphin, and samples were incubated for 30 min at room temperature. RNA extraction was performed with an RNeasy minikit (Qiagen) according to the manufacturer's instructions.
RNA-seq analysis was conducted basically following the manufacturer's instructions. Briefly, the RNA-seq template was prepared using a TruSeq RNA sample preparation kit (version 5; Illumina), omitting the poly(A) selection procedure. The double-stranded PCR products were purified and size fractionated using a bead-mediated method with AMPure (Ambion). Sequencing was conducted on a HiSeq2000 platform (Illumina), using a TruSeq RNA sample prep kit, version 2, and a TruSeq SBS kit, version 3-HS. At least 20 million sequences of 36-base single-end reads were generated per sample. The obtained data were analyzed by CLC Genomics Workbench software, version 6.0.2 (CLC Bio, Aarhus, Denmark) (for details, see Table S1 in the supplemental material). The number of reads per kilobase of exon per million mapped reads (rpkm) for each gene was compared between surfactant- or serum-treated groups and the nontreated group.
Quantitative RT-PCR analysis.
RNAs were extracted from 1- to 5-ml cultures supplied with Surfacten, serum, lipids, or detergents as described above. The genomic DNA was degraded with RQ1 RNase-free DNase (Promega). cDNAs were synthesized by TaqMan reverse transcription (RT) reagents (Applied Biosystems) according to the manufacturer's instructions. Sequences of primers used in this study are described in Table S2 in the supplemental material. cDNAs were mixed with primers and FastStart Universal SYBR green master (ROX) mix (Roche), and the analysis was performed using a Step One Plus real-time PCR system (Applied Biosystems). The expression level of each gene was normalized to that of 16S rRNA, which is widely used as an internal control (7, 17). We confirmed that the amount of 16S rRNA did not vary significantly under our experimental conditions. The induction ratio relative to that of the nontreated group was also calculated.
Lipid extraction.
Lipids were extracted from cow lungs by the Bligh-Dyer method, as follows. Cow lungs (90 g) in PBS were homogenized by use of a food processor. Approximately 100 ml of the tissue homogenate was mixed with 250 ml of methanol and 125 ml of chloroform (volume ratio of 0.8:2:1), followed by stirring for 30 min. The solution was filtered through filter paper (Toyo Roshi Kaisha, Ltd.), and 125 ml each of chloroform and water was added with rigorous mixing. The sample was centrifuged at 1,000 × g for 15 min, and the lower (organic) and upper (water) phases were collected and evaporated. The obtained solid materials were suspended in saline before use.
Measurement of promoter activity.
Construction of strains harboring luciferase-encoding plasmids and measurement of luciferase activity were performed essentially as described previously (7). The predicted promoter regions of essC and hlgB were cloned from the genomic DNA of the Newman strain. Because the hlgB gene is suggested to be transcribed as part of an operon with the hlgC gene (18), the promoter region of hlgC was cloned for analysis of the hlgB gene. The PCR primers used for cloning of the essC and hlgC promoters are listed in Table S2 in the supplemental material. The amplified fragments (550 bp and 441 bp) were digested with both EcoRI and KpnI and then ligated with the pluc vector, a modified pND50 plasmid (19) that contains a functional ribosomal binding site and the translational start codon of luc in all reading frames. The constructed plasmids (pluc-hlgBCpro and pluc-essCpro) were first introduced into the S. aureus RN4220 laboratory strain by electroporation (20) and then transduced into the S. aureus Newman strain by using phage 80α (21).
Transductants of strain Newman harboring either the pluc-hlgBCpro or pluc-essCpro plasmid were grown in TSB medium supplied with 12.5 μg/ml chloramphenicol. Each overnight culture (10 µl) was inoculated into 1 ml of TSB medium in the presence of palmitate (0.1 mg/ml) or Surfacten (0.3 mg/ml). The culture was incubated at 37°C to an OD600 of 0.7, and then the cells were collected by centrifugation. Immediately after discarding the supernatant, pellets were frozen in liquid nitrogen. Cells were suspended in 50 μl PBS and mixed with 0.5 ml lysis buffer (25 mmol/liter KH2PO4 [pH 7.8], 10 μg/ml lysostaphin, 0.04% Triton X-100, 0.1 mmol/liter dithiothreitol, and Complete proteinase inhibitor cocktail [Roche]). After incubation at room temperature for 30 min, the samples were centrifuged. The supernatant was diluted 10-fold with the lysis buffer, and then 100 μl of each sample was mixed with 100 μl substrate solution from a luciferase assay system (Promega). The luciferase activities were measured with a luminometer (Lumat LB 9507; EG&G Berthold). Protein concentrations were determined using the Coomassie Plus protein assay reagent (Thermo) with bovine serum albumin as a standard, and the luminescence units were normalized to the protein amounts.
Construction of disruption mutants of S. aureus.
Disruption mutants of the two-component system regulators (Δagr, ΔarlS, and ΔsaeS), with Newman as the parent strain, were constructed in a previous study (22). Disruptions of the sigB (NWMN_1970), essC (NWMN_0223), psiA (NWMN_0246), and hlgB (NWMN_2320) genes in the Newman strain were performed essentially as described previously (6, 7, 22). Briefly, internal regions within open reading frames of the sigB (+33 to +538; the first nucleotide of the start codon is numbered +1, and the length of the target region is 506 bp), essC (+109 to +1044; 936 bp), and hlgB (+25 to +862; 838 bp) genes were amplified by PCR and cloned into an S. aureus integration vector, pSF151 (23). The constructed plasmids were transferred into S. aureus RN4220 by electroporation, and the integrated regions containing the Kmr gene were transduced into Newman by use of phage 80α as described above, resulting in disrupted mutants of the sigB, essC, and hlgB genes (designated ΔsigB, ΔessC, and ΔhlgB mutants). For the psiA gene, with a relatively short coding region (399 bp), the upstream (−1126 to −241; 898 bp) and downstream (+584 to +1403; 820 bp) regions of the start codon were amplified by a PCR using degenerative primers to create linker sequences at the ends. The Kmr gene of the pSF151 vector with linker sequences was amplified separately. The DNA fragments of the upstream region, the Kmr gene, and the downstream region were linked in this order by a PCR using these fragments as templates, the forward primer of the upstream region, and the reverse primer of the downstream region. The obtained DNA (3 kb) was ligated with SmaI-digested pKOR3A, a vector (containing the Cmr gene as a marker) for allelic replacement in S. aureus (24). The constructed plasmid was introduced into S. aureus RN4220 by electroporation, and transformants were grown overnight in TSB medium containing 50 μg/ml kanamycin. The cultures were diluted and spread on TSB agar plates containing 50 μg/ml kanamycin, and the plates were incubated overnight at 43°C. The temperature-resistant cells were cultured in TSB medium, and the overnight cultures were diluted and spread on TSB agar containing 50 μg/ml kanamycin and 1.5 μg/ml anhydrotetracycline. Among colonies of a variety of sizes, relatively large colonies were selected and spread on two plates, containing either 50 μg/ml kanamycin or 12.5 μg/ml chloramphenicol. Cells resistant to kanamycin and susceptible to chloramphenicol were selected as disrupted mutants of the psiA gene in an RN4220 background (RNΔpsiA mutant). The integrated region was transferred from the RNΔpsiA strain to Newman by phage transduction, resulting in a disrupted mutant of the psiA gene in the Newman background (NMΔpsiA mutant). In the following sections, the NMΔpsiA strain is referred to as the ΔpsiA strain. The nucleotide sequences of primers used for gene disruption are listed in Table S2 in the supplemental material. The disruption of each target gene in the mutants was confirmed by Southern blotting.
Construction of complementation strains of S. aureus.
Complementation of the sigB, essC, psiA, and hlgB genes in disruption mutants of each gene was performed essentially as described previously (6, 7, 22). For sigB, the promoter region of the upstream rsbV gene (NWMN_1972) (241 bp) and the coding region of the sigB gene (851 bp) were amplified separately. Two PCR products were sequentially ligated with a pHY300E vector, resulting in pHYsigB. Similarly, for essC, the promoter region of the upstream essB gene (NWMN_0222) (457 bp) and the coding region of the essC gene (4,506 bp) were amplified separately and sequentially cloned, resulting in pHYessC. For psiA, the promoter and coding regions (638 bp) were amplified together and cloned into the vector, resulting in pHYpsiA. For hlgB, the promoter of the upstream hlgC gene (NWMN_2319) and the coding regions of both hlgC and hlgB (2,318 bp) were amplified together and cloned, resulting in pHYhlgB. Constructed plasmids were first introduced into S. aureus RN4220 by electroporation and then transferred to each mutant with the S. aureus Newman background by phage transduction as described above. Transductants showing resistance to both 50 μg/ml kanamycin (12.5 µg/ml chloramphenicol for the ΔsigB/pHYsigB strain) and 10 μg/ml erythromycin were selected as complementation strains. The nucleotide sequences of the primers used for complementation are listed in Table S2 in the supplemental material.
Mouse infection experiments.
Female mice (6 to 8 weeks of age) were anesthetized by intraperitoneal injection of pentobarbital (Nembutal). TSB medium (50 ml) was added to 0.5 ml of overnight cultures, and bacteria were grown to log phase (OD600 = 0.5 to 0.7). Bacteria suspended in PBS (30 μl; 1 × 108 CFU) were spotted into the nostrils of the mice. Survival rates of the mice were monitored. To measure amounts of viable bacteria in the infected lungs, sublethal doses (3 × 107 CFU) of bacterial suspensions were administered to mice. After 2 days, the mice were anesthetized and the lungs were dissected. The diluted homogenates were spread on mannitol salt agar, and yellow colonies were counted. Survival data were plotted using Prism 5 (GraphPad Software, Inc.), and statistically significant differences between the survival curves were analyzed by the log rank test.
RESULTS
Global analysis of gene expression profiles of S. aureus altered by pulmonary surfactant.
First, we analyzed the gene expression alterations of S. aureus induced by pulmonary surfactant and serum. S. aureus Newman was grown in TSB supplied with either calf-derived pulmonary surfactant or calf serum, and total RNAs were extracted from cells collected at late log phase (OD600 = 1). The growth curves for S. aureus treated with either serum or surfactant were similar (see Fig. S1 in the supplemental material). RNA-seq analysis by use of a next-generation analyzer revealed 142 and 259 genes whose transcript levels were increased by surfactant and serum, respectively (Fig. 1). Among them, 49 genes were upregulated by both surfactant and serum (Fig. 1). Recent studies demonstrated that genes involved in iron acquisition, such as the isd, sir, and sbn families, were upregulated in S. aureus grown in human or calf serum (1, 25). Under our experimental conditions, the expression levels of isdABEFG, sirAB, and sbnABCDEFHI increased from 4- to 34-fold in the presence of calf serum (see Table S3 in the supplemental material). On the other hand, more than half of the genes upregulated by serum were not induced by surfactant, and vice versa. Therefore, the patterns of gene induction by both host factors differed markedly. The RNA-seq data obtained by the next-generation sequencer are shown in Table S4 in the supplemental material.
FIG 1.
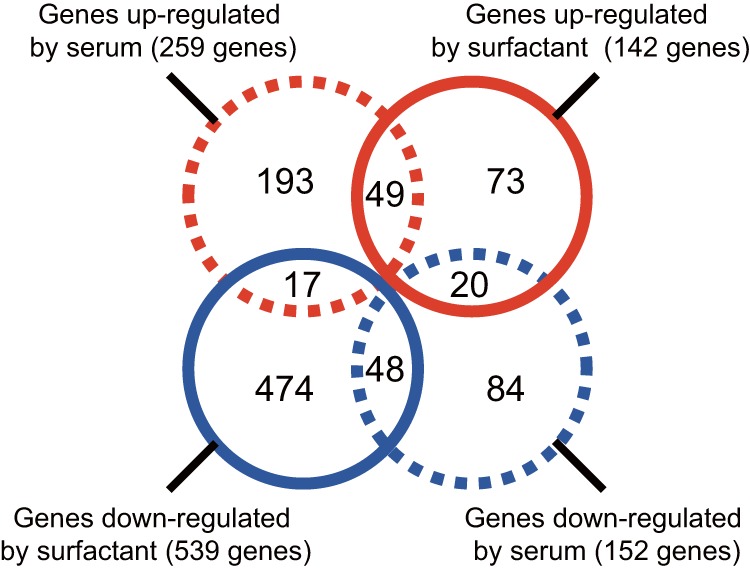
Numbers of S. aureus genes with altered expression in cultures with serum or pulmonary surfactant. S. aureus Newman was aerobically cultured in TSB medium containing 10% calf serum or 0.3 mg/ml Surfacten, and total RNAs extracted from bacteria at an OD600 of 1 were subjected to RNA-seq analysis using a next-generation sequencer. Circles written with dashed and solid lines indicate the results for samples treated with serum and surfactant, respectively. Numbers in blue circles show downregulated genes, and those in red circles indicate upregulated genes for each treatment.
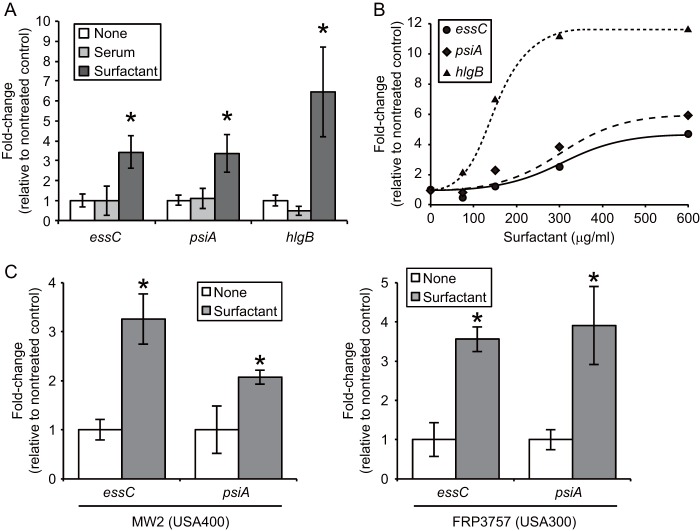
We then performed another genomewide analysis by using a next-generation sequencer on RNAs extracted at the early log phase (OD600 = 0.3) from bacteria grown in the presence of surfactant or serum. Among the 43 genes whose transcript levels were increased >4-fold at an OD600 of 1 in the presence of surfactant, half of them showed a surfactant-dependent increase at an OD600 of 0.3 (Table 1). Next, we performed quantitative RT-PCR analysis of the genes that were upregulated by the pulmonary surfactant. We confirmed that mRNA levels of genes encoding ESAT-6 secretion system C (EssC), the unknown factor NWMN_0246, and hemolysin gamma subunit B (HlgB) were increased 3- to 10-fold when S. aureus cells were incubated in TSB medium supplied with the pulmonary surfactant (Fig. 2A). On the other hand, serum treatment failed to increase the mRNA amounts for these three genes in S. aureus (Fig. 2A). The upregulation of these genes depended on the concentration of the surfactant added to the medium (Fig. 2B). Among these genes, we named the NWMN_0246 gene, which had not been characterized in S. aureus, pulmonary surfactant-inducible factor A (psiA). The predicted sequence of the PsiA protein contained an NfeD-like C-terminal partner-binding domain (see Fig. S2 in the supplemental material). Next, we examined whether induction of the above-mentioned genes was observed in CA-MRSA strains, clinically isolated MRSA strains that are highly virulent, as well as in strain Newman. Two CA-MRSA strains, MW2 (USA400) (26) and FRP3757 (USA300) (27), were cultured in TSB medium containing the pulmonary surfactant, and RNAs extracted at an OD600 of 1 were subjected to quantitative RT-PCR analysis. As observed in the Newman strain, the mRNA quantities for essC and psiA were increased by the surfactant treatment (Fig. 2C). On the other hand, the hlgB mRNA levels in MW2 and FRP3757 were <1/10-fold that in Newman, and the ratios of surfactant-dependent induction of hlgB mRNA were 1.3- and 1.4-fold, respectively. Recent studies demonstrated that expression of the hlgB gene is dependent on the SaeRS system (28) and that the Newman strain possesses a point mutation that leads to constitutive expression of the saeS gene (29). Based on these reports, the hlgB gene is assumed to be highly expressed in S. aureus Newman, which would account for our data showing that the expression level of the hlgB gene in S. aureus Newman was higher than those in clinical isolates.
TABLE 1.
Genes whose expression levels were altered by serum or pulmonary surfactanta
| Gene no. | Gene product | Gene expression |
|||
|---|---|---|---|---|---|
| Surfactant |
Serum |
||||
| OD = 1 | OD = 0.3 | OD = 1 | OD = 0.3 | ||
| NWMN_0093 | Truncated IS200 transposase family | ++ | ++ | +/− | +/− |
| NWMN_0095 | CapA (capsular polysaccharide synthesis enzyme) | ++ | + | + | +/− |
| NWMN_0112 | Hypothetical protein | ++ | − | ++ | +/− |
| NWMN_0207 | Hypothetical protein | ++ | +/− | ++ | + |
| NWMN_0221 | EssA (ESAT-6 secretion system) | ++ | ++ | + | +/− |
| NWMN_0223 | EssC (ESAT-6 secretion system) | ++ | + | +/− | +/− |
| NWMN_0224 | EsaC (ESAT-6 secretion system) | ++ | + | +/− | +/− |
| NWMN_0225 | EsxB (ESAT-6-like protein) | ++ | + | +/− | +/− |
| NWMN_0232 | Hypothetical protein | ++ | + | +/− | +/− |
| NWMN_0233 | Hypothetical protein | ++ | +/− | ++ | − |
| NWMN_0245 | Hypothetical protein | ++ | ++ | − | − |
| NWMN_0246 | Hypothetical protein | ++ | +/− | − | − |
| NWMN_0360 | Hypothetical protein | ++ | − | +/− | +/− |
| NWMN_0429 | Sle1 (N-acetylmuramoyl-l-alanine amidase) | ++ | + | +/− | − |
| NWMN_0405 | Truncated staphylococcal tandem lipoprotein | ++ | + | ++ | +/− |
| NWMN_0658 | Hypothetical protein | ++ | +/− | ++ | +/− |
| NWMN_0765 | Hypothetical protein | ++ | +/− | ++ | +/− |
| NWMN_0902 | Hypothetical protein | ++ | +/− | ++ | +/− |
| NWMN_0909 | Hypothetical protein | ++ | +/− | ND | − |
| NWMN_1059 | Hypothetical protein | ++ | +/− | ND | +/− |
| NWMN_1106 | Putative transposase | ++ | +/− | ND | − |
| NWMN_1196 | Hypothetical protein | ++ | +/− | ++ | − |
| NWMN_1275 | 4-Oxalocrotonate tautomerase | ++ | − | ++ | − |
| NWMN_1554 | Hypothetical protein | ++ | +/− | +/− | − |
| NWMN_1716 | BsaA2 (lantibiotic precursor) | ++ | − | ND | ND |
| NWMN_1745 | Hypothetical protein | ++ | − | +/− | − |
| NWMN_1776 | Hypothetical protein | ++ | + | ++ | +/− |
| NWMN_1861 | Hypothetical protein | ++ | ++ | ++ | + |
| NWMN_1898 | Phage portal protein | ++ | +/− | − | + |
| NWMN_2074 | Hypothetical protein | ++ | +/− | ++ | − |
| NWMN_2188 | UreA (urease subunit) | ++ | + | ND | − |
| NWMN_2189 | UreB (urease subunit) | ++ | + | + | − |
| NWMN_2320 | HlgB (gamma hemolysin component) | ++ | + | +/− | +/− |
| NWMN_2328 | ABC transporter ATP-binding protein | ++ | +/− | + | +/− |
| NWMN_2417 | Hypothetical protein | ++ | +/− | ++ | +/− |
| NWMN_2492 | Transcriptional regulator, TetR family | ++ | − | +/− | +/− |
| NWMN_2549 | Hypothetical protein | ++ | ++ | +/− | +/− |
| NWMN_2550 | Hypothetical protein | ++ | + | − | − |
| NWMN_2551 | Asp1 (accessory secretory protein) | ++ | + | +/− | − |
| NWMN_2552 | SecY (preprotein translocase subunit) | ++ | + | +/− | − |
S. aureus Newman was cultured in TSB medium supplied with 0.3 mg/ml Surfacten to an OD600 of 1. The 43 genes listed were identified by RNA-seq analysis to be upregulated >4-fold following surfactant treatment. Alterations in transcript levels observed in RNA-seq data for S. aureus incubated with either Surfacten or calf serum to an OD600 of 1 or 0.3 are also indicated. ++, upregulated >4-fold; +, upregulated 2- to 4-fold; +/−, altered 0.5- to 2-fold; −, downregulated >2-fold; ND, not detected.
FIG 2.
Pulmonary surfactant-dependent induction of S. aureus essC, psiA, and hlgB genes. (A) S. aureus Newman was cultured in TSB medium containing either 10% serum or 0.3 mg/ml Surfacten to an OD600 of 1. Amounts of essC, psiA, and hlgB mRNAs were determined by quantitative RT-PCR. Each value was normalized to 16S rRNA, and the ratio of the surfactant-treated to the nontreated group was calculated. Data represent means ± standard deviations (SD) for 4 experiments. For each gene, statistical differences compared with the nontreated group were analyzed by one-way analysis of variance (ANOVA) with Dunnett's multiple-comparison test (*, P < 0. 01). (B) Dose dependency of pulmonary surfactant for the induction of S. aureus gene expression. RNAs were collected from bacteria at an OD600 of 1 and subjected to quantitative RT-PCR analysis. The expression level of each gene was normalized to that of 16S rRNA, and the induction ratio relative to the nontreated group is indicated. (C) Induction of gene expression by pulmonary surfactant in CA-MRSA. MW2 (USA400) and FRP3757 (USA300) were cultured in the presence of 0.3 mg/ml Surfacten to an OD600 of 1, and quantitative RT-PCR analysis of each gene was performed. Data represent means ± SD for 3 or 4 experiments. Statistical differences for each gene compared with nontreated groups were analyzed by Student's t test (*, P < 0.05).
Molecular mechanisms of gene induction in S. aureus by pulmonary surfactant.
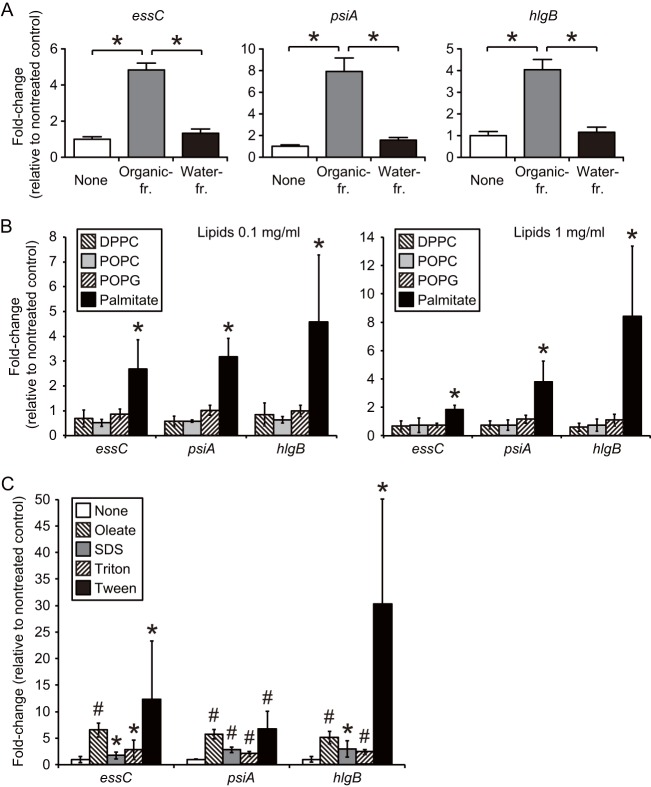
Pulmonary surfactant is a lipid-rich secreted material; 90% of its weight comprises lipids, and the remaining 10% contains proteins (3). We thus hypothesized that lipids, the major constituents of pulmonary surfactant, had gene expression-inducing activities in S. aureus. Lipid fractions from either the pulmonary surfactant (see Fig. S3 in the supplemental material) or cow lung homogenates (Fig. 3A) induced the upregulation of essC, psiA, and hlgB in S. aureus >2-fold. In contrast, the activity in the water fraction of lung extracts was much lower than that in the organic solvent fraction (Fig. 3A). We next examined the effects of the major phospholipids and fatty acids in the pulmonary surfactant, such as DPPC, POPC, POPG, and palmitate, on gene induction in S. aureus. S. aureus Newman was cultured to an OD600 of 1 in TSB medium supplied with each lipid at 0.1 or 1 mg/ml, and RNAs were extracted. The amounts of mRNA for the three genes (essC, psiA, and hlgB) induced by surfactant treatment were measured by quantitative RT-PCR. Neither DPPC, POPC, nor POPG, the major phospholipids in surfactant, affected the expression levels of the three genes (Fig. 3B). On the other hand, supplementation with palmitate increased the mRNA levels of the three genes 2- to 8-fold (Fig. 3B). Moreover, like palmitate, oleate promoted the expression of the three genes (Fig. 3C). These free fatty acids have detergent-like activities, and the growth of S. aureus is inhibited when these fatty acids are supplied at high concentrations (30). Under our experimental conditions, the viability of S. aureus was partly affected by supplementation with palmitate: the numbers of CFU in log phase were 2- to 5-fold lower than those for the nontreated group (see Fig. S4 in the supplemental material). We therefore considered the possibility that reactions of detergent-like substances targeting the cell membrane stimulate gene expression. We examined the effects of various detergents on S. aureus gene expression and found that supplementation of the medium with SDS, Triton X-100, and Tween 20 led to increases in the mRNA levels of the three genes (Fig. 3C). Among these detergents, Tween 20 showed the highest induction ability (Fig. 3C).
FIG 3.
Induction of essC, psiA, and hlgB genes in S. aureus by pulmonary surfactant lipid components. (A) Effects of lipid fractions of cow lung homogenates on gene expression of S. aureus. Cow lung homogenates were fractionated by the Bligh-Dyer method to an organic phase (Organic-fr.) and a water phase (Water-fr.). S. aureus Newman was cultured in TSB medium supplied with either the organic or water phase to an OD600 of 1, and the expression levels of the essC, psiA, and hlgB genes relative to those of the nontreated group were determined by quantitative RT-PCR. Data represent means ± SD for 4 experiments. Statistical differences were analyzed by one-way ANOVA with Tukey's multiple-comparison test (*, P < 0.001). (B) Effects of lipids contained in pulmonary surfactant on gene expression of S. aureus. S. aureus Newman was cultured in TSB medium supplied with 0.1 (left) or 1 (right) mg/ml of either DPPC, POPC, POPG, or palmitate to an OD600 of 1. mRNA levels of the essC, psiA, and hlgB genes were measured by quantitative RT-PCR. The expression level of each gene was normalized with that of 16S rRNA, and the induction ratio relative to the nontreated group is indicated. Data represent means ± SD for 3 or 4 experiments. Statistical differences were analyzed by one-way ANOVA with Tukey's multiple-comparison test (*, P < 0.05). (C) Effects of oleate and detergents on gene expression of S. aureus. S. aureus Newman was cultured in TSB medium containing either 0.1 mg/ml oleate, 0.0003% SDS, 0.006% Triton X-100, or 0.006% Tween 20 to an OD600 of 1. Expression levels of the three genes relative to those in the nontreated group were determined by quantitative RT-PCR as described above. Data represent means ± SD for 7 experiments (*, P < 0.05; #, P < 0.005).
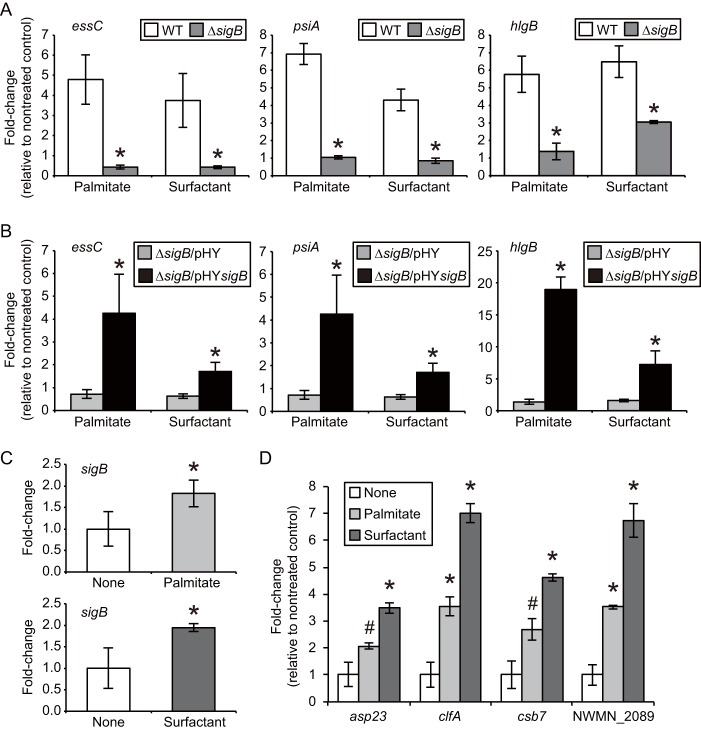
We further evaluated the molecular mechanisms of gene induction in S. aureus by pulmonary surfactant. To test whether pulmonary surfactant induced the upregulation of transcriptional activities of the essC and hlgB genes at the transcription initiation step, we performed promoter analysis of these genes. The promoter sequences of the essC and hlgB genes were cloned into a luciferase reporter plasmid, and the promoter activities were then assessed. Supplementation of the culture medium with palmitate or pulmonary surfactant increased the promoter activities of the essC and hlgB genes in S. aureus strains harboring the respective plasmids (Fig. 4). In contrast, serum treatment did not increase the essC and hlgB promoter activities (see Fig. S5 in the supplemental material), consistent with the gene induction results (Fig. 2A). Induction of some genes induced by damage to the cell membranes depends on the function of the sigB gene, which encodes a stress-responsive RNA polymerase subunit (31). We therefore examined the involvement of SigB in the upregulation of the essC, psiA, and hlgB genes. In contrast to the parent S. aureus strain, a disruption mutant of the sigB gene (ΔsigB mutant) did not show increases in the mRNA levels of the three genes, even in the presence of palmitate or pulmonary surfactant (Fig. 5A). The reduced ability to respond to palmitate and surfactant in the ΔsigB mutant was restored by complementation with the sigB gene (Fig. 5B). Moreover, the mRNA level of the sigB gene itself increased with S. aureus grown in medium supplemented with palmitate or surfactant (Fig. 5C). In addition, genes whose expression is regulated in a SigB-dependent pathway, such as asp23 (31–36), clfA (32, 37, 38), csb7 (31, 32, 39), and opuD2 (NWMN_2089; an ortholog of SACOL2176 in S. aureus strain COL) (31–34, 37), were also upregulated by treatment with palmitate or surfactant (Fig. 5D). These findings suggest that activation of the SigB pathway is involved in the gene induction by free fatty acids present in the pulmonary surfactant. There are a group of regulators of gene expression, called two-component systems, that sense and respond to environmental changes in bacteria. Among them, Agr (40), ArlRS (41), and SaeRS (42) are involved in sensing host environments and regulating virulence in pathogenic bacteria infecting host animals. The essC, psiA, and hlgB genes were still upregulated depending upon the pulmonary surfactant in the deletion mutant of each gene (see Fig. S6 in the supplemental material). These findings suggest that these two-component systems are not involved in the surfactant-dependent induction of those genes.
FIG 4.
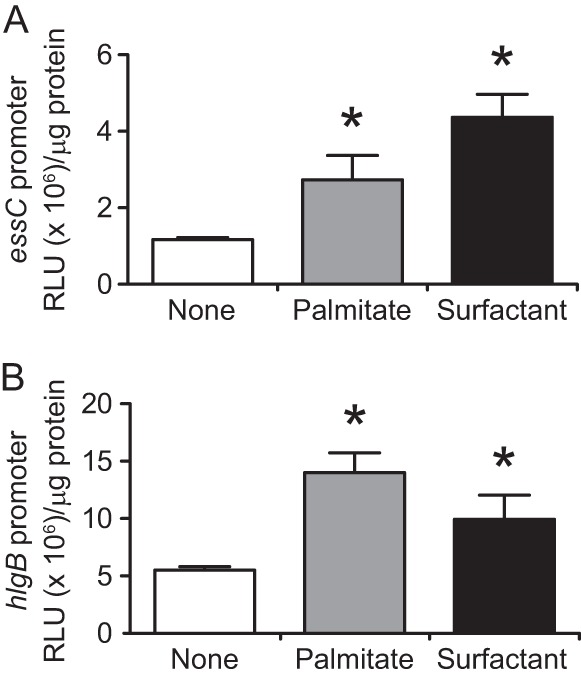
Palmitate and pulmonary surfactant upregulate essC and hlgB promoter activities. Promoter sequences of either the essC (A) or hlgB (B) gene were inserted into a reporter plasmid upstream of the luciferase gene. S. aureus Newman harboring each plasmid was cultured in TSB medium supplied with 0.1 mg/ml palmitate or 0.3 mg/ml Surfacten to an OD600 of 0.7. The luciferase activities were measured with a luminometer, and the luminescence units were normalized to the protein amounts. Data represent means and SD for 3 experiments. Statistical differences compared with the nontreated group were analyzed by one-way ANOVA with Dunnett's multiple-comparison test (*, P < 0.05). RLU, relative light units.
FIG 5.
Involvement of SigB in S. aureus gene induction by palmitate and pulmonary surfactant. (A) Requirement of the sigB gene in palmitate- and surfactant-dependent gene induction. S. aureus Newman as the parent strain (wild type [WT]) or a disruption mutant of the sigB gene (ΔsigB) was cultured in TSB medium containing 0.1 mg/ml palmitate or 0.3 mg/ml Surfacten to an OD600 of 1. Expression levels of the three genes relative to those of the nontreated group were determined by quantitative RT-PCR as described in the text. Data represent means ± SD for 3 experiments. Statistical differences between the WT and ΔsigB strains were analyzed by Student's t test (*, P < 0.05). (B) Complementation of phenotypes of the S. aureus sigB disruption mutant. Either a control plasmid (pHY300E) or a complementation plasmid containing an open reading frame of the sigB gene (pHYsigB) was introduced into the ΔsigB mutant, resulting in ΔsigB/pHY or ΔsigB/pHYsigB, respectively. Cells were cultured in the presence of 0.1 mg/ml palmitate or 0.3 mg/ml Surfacten to an OD600 of 1, and quantitative RT-PCR analysis of the three genes was performed as described in the text. Data represent means ± SD for 3 experiments. Statistical differences between the ΔsigB/pHY and ΔsigB/pHYsigB strains were analyzed by Student's t test (*, P < 0.05). (C) Induction of sigB gene expression in S. aureus by palmitate and pulmonary surfactant. S. aureus Newman was cultured in TSB medium supplied with either 0.1 mg/ml palmitate or 0.3 mg/ml Surfacten to an OD600 of 1. The expression level of the sigB gene relative to that for the nontreated group was determined by quantitative RT-PCR. Data represent means ± SD for 3 experiments. Statistical differences compared to the nontreated group were analyzed by Student's t test (*, P < 0.05). (D) Induction of SigB-regulated genes by palmitate and pulmonary surfactant. S. aureus Newman was cultured in TSB medium supplied with either 0.1 mg/ml palmitate or 0.3 mg/ml Surfacten to an OD600 of 1. Expression levels of the asp23, clfA, csb7, and NWMN_2089 genes relative to those for the nontreated group were determined by quantitative RT-PCR. Data represent means ± SD for 3 experiments. For each gene, statistical differences compared with the nontreated group were analyzed by one-way ANOVA with Dunnett's multiple-comparison test (#, P < 0.01; *, P < 0.001).
Involvement of pulmonary surfactant-inducible genes in pneumonia caused by S. aureus.
Pathogens invading host animals through the airway contact the pulmonary surfactant that covers the alveolar surfaces in the lungs. Responding to pulmonary surfactant and expressing an array of virulence factors in a distinct pattern seem to be important for bacteria to exert their pathogenicity in lung infections. We considered that the above surfactant-inducible genes contribute to virulence exertion of S. aureus in lung infection. To test this, we constructed S. aureus disruption mutants of each of the three surfactant-inducible genes that we found in the present study (ΔessC, ΔpsiA, and ΔhlgB mutants), together with complementation strains of each gene, and examined their virulence potentials in a mouse pneumonia model. The growth curves for these disruption mutants in TSB medium were indistinguishable from that of the parent strain (see Fig. S7 in the supplemental material). Suspensions of the parent or mutant strains were spotted into the nostrils of anesthetized mice, and the lungs were dissected 2 days later (see Fig. S8 in the supplemental material). Bacterial numbers were much smaller in mutants than in the parent strain (Fig. 6A) and strains complemented with each deleted gene (Fig. 6B). Moreover, all of these mutants showed delayed killing of mice compared to the parent and complementation strains (Fig. 6C). These findings suggest that the three genes induced by pulmonary surfactant are required for S. aureus virulence in animal lungs.
FIG 6.
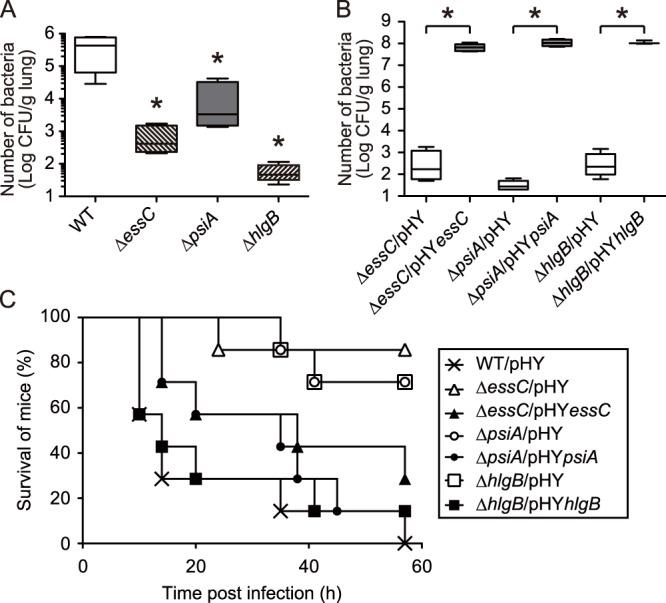
Involvement of essC, psiA, and hlgB genes in S. aureus virulence in a mouse pneumonia model. (A) Survival of S. aureus mutants in mouse lungs. Sublethal doses of either S. aureus Newman (WT) or a mutant strain (ΔessC, ΔpsiA, or ΔhlgB strain) were administered into the lungs of anesthetized mice through the nasal cavities. After 2 days, the lungs were dissected and numbers of viable bacteria in the homogenates were determined. Data represent means ± SD for 5 mice. Statistical differences compared with the WT were analyzed by one-way ANOVA with Dunnett's multiple-comparison test (*, P < 0.05). (B) Survival of S. aureus mutants and complementation strains in mouse lungs. Mice were infected with either S. aureus mutants harboring empty vectors (ΔessC/pHY, ΔpsiA/pHY, and ΔhlgB/pHY strains) or mutants harboring complementation vectors (ΔessC/pHYessC, ΔpsiA/pHYpsiA, and ΔhlgB/pHYhlgB strains). After 2 days, the numbers of viable bacteria in lung homogenates were determined. Data represent means ± SD for 3 to 6 mice. Statistical differences of the mutants compared with each complemented strain were analyzed by Student's t test (*, P < 0.001). (C) Host-killing ability in a mouse pneumonia model by mutant and complementation strains of surfactant-inducible genes. Bacterial cell suspensions of S. aureus Newman harboring an empty vector (×; WT/pHY), disruption mutants of each surfactant-inducible gene containing empty vectors (open symbols; ΔessC/pHY, ΔpsiA/pHY, and ΔhlgB/pHY strains), or mutants complemented with each gene (closed symbols; ΔessC/pHYessC, ΔpsiA/pHYpsiA, and ΔhlgB/pHYhlgB strains) were administered into the lungs of anesthetized mice through the nasal cavities (n = 7). Statistical differences between the wild type and each mutant or between each mutant and its complementation strain were determined to be significant (P < 0.05) by the log rank test.
DISCUSSION
Pathogens possess complex gene expression regulatory systems that are necessary for rapid adaptation to host environments that have not yet been clarified. Recently, several mammalian infection models, including those of pneumonia, skin abscess formation, bacteremia, and osteomyelitis, were established to investigate the virulence mechanism of S. aureus (4). S. aureus and other pathogenic bacteria show altered expression patterns of their virulence genes at different infection sites depending on the growth conditions within each tissue. Therefore, analyses of gene expression regulations under conditions that mimic host environments are important for determining bacterial pathogenesis. In the present study, we cultured S. aureus in the presence of pulmonary surfactant, a lipid-rich material secreted at infection sites such as lung alveoli, and we observed that an array of virulence genes were induced. Among them, the essC, psiA, and hlgB genes were upregulated by palmitate, a major fatty acid in surfactant, and were necessary for S. aureus virulence in lung infections (Fig. 7). Our results suggest that in vitro gene analysis studies following treatment with pulmonary surfactant would be useful for investigating the virulence mechanisms of pathogens that infect host lungs.
FIG 7.
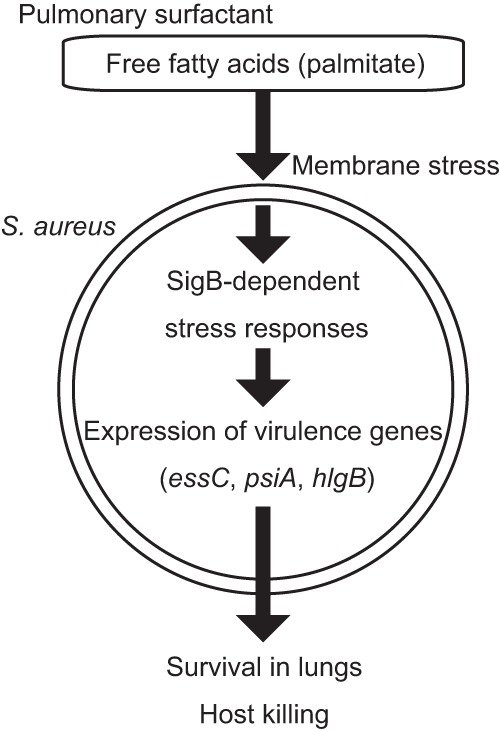
Proposed mechanism of S. aureus virulence gene induction by free fatty acids in pulmonary surfactant.
Mechanistic insights into the role of the three genes induced by pulmonary surfactant in S. aureus virulence.
The present study demonstrated that the essC, psiS, and hlgB gene expression levels were increased by treatment with surfactant. In this section, we discuss the relationships between our results and previous reports concerning the involvement of the three genes in pathogenesis.
The essC gene is a member of the ess cluster, a genomic region that contains the esxAB, esaABC, and essABC genes and is conserved in Gram-positive bacteria such as Listeria monocytogenes, Bacillus thuringiensis, and Mycobacterium tuberculosis (43). The ess genes encode components of the type VII secretion system, a recently identified Sec-independent secretion pathway of bacteria that is involved in virulence via the secretion of various toxins, including EsxA, EsxB, and EsaC (43–45). Interestingly, the amount of EsaC secreted into the blood increases when S. aureus infects host animals (43). Based on these reports, it is plausible that S. aureus possesses a system to sense the host environment and to thereby upregulate the expression of Ess factors. Validation of this hypothesis at the molecular level, however, has been inadequate. Here we demonstrated that the essC gene was induced by fatty acids in pulmonary surfactant and detergents. A previous report showed that the extracellular protein patterns of S. aureus are altered in response to detergents (29) and, together with our results, showed that the activation of the Ess secretion system is likely to be involved in membrane damage stress responses.
The NWMN_0246 gene, which we named psiA in this study, encodes a 132-amino-acid protein with an unknown function. The primary sequence contains an NfeD-like C-terminal partner-binding domain. Several NfeD-like proteins are found in prokaryotes, and some of them contribute to lipid metabolism (46). Thus, the psiA gene product might be involved in membrane repair responses in cells damaged by fatty acids and detergents. Our genomewide analysis showing that the psiA gene was not induced by serum suggests that this gene functions as an S. aureus virulence factor acting specifically in lung infections. We expect that further investigation of the potential role of PsiA in lipid metabolism could contribute to novel approaches toward reaching an understanding of pulmonary infections caused by S. aureus.
The hlgB gene encodes a component of hemolysin gamma (Hlg), a representative extracellular toxin of S. aureus. Hlg shows hemolytic and cytolytic activities against human monocytes, polymorphonuclear cells, and macrophages (47). Hlg and other related hemolysins are predicted to form transmembrane channel complexes on target cells, and the tertiary structure of the Hlg protomer is similar to those of hemolysin alpha (Hla) and leukocidin (48). Among these hemolysins, S. aureus Hla, Panton-Valentine leukocidin, and hemolysin beta (Hlb) contribute to evoking inflammation and host killing in experimental mammalian pneumonia models (49–51). On the other hand, Malachowa et al. reported only a modest contribution of Hlg to host killing by S. aureus in a sepsis model (1), and there has been little analysis of its virulence potential in a pneumonia model. Furthermore, the expression of hla and hlg genes is differentially regulated by the two-component systems Agr and Sae, respectively (28). Under our experimental conditions for surfactant treatment, the transcript levels of the hla and agr genes were not significantly affected, whereas the mRNA quantities of hlgB and hlgC were upregulated. Moreover, disruption of the hlgB gene in S. aureus attenuated its host-killing ability in a pneumonia model. These results suggest a possible role for Hlg as a major cytotoxin of S. aureus in lung infections. Because alveolar macrophages possessing high phagocytic capacities are likely to be present at or attracted to infected alveoli, promotion of Hlg production in response to pulmonary surfactant seems to be reasonable for S. aureus to avoid attacks by host immune cells at infection sites.
Molecular mechanisms of gene expression induction by fatty acid and detergents.
The two-component systems Agr and Sae, which are major regulators of virulence factors in pathogenic bacteria (17), are induced in S. aureus cultured in serum (25) and blood (1), respectively. In contrast to these reports, we found that neither the agr gene nor the sae gene was upregulated in the culture medium supplied with pulmonary surfactant. In addition, disruption of these two-component system genes did not suppress the gene expression stimulated by the surfactant. Montgomery and colleagues reported that mutations in the agr and sae genes attenuated the host-killing ability of S. aureus in a mouse pneumonia model (17). Our data do not conflict with the notion, based on this previous report, that Agr and Sae systems are required for S. aureus virulence; the robust impact of agr and sae mutations on the expression of numerous virulence genes likely masked the effects of the surfactant-inducible genes we focused on in the present study, leading to attenuated host killing as observed by Montgomery et al.
We then focused on SigB, an alternative sigma factor that contributes to various stress responses in several bacteria, as a candidate regulator of surfactant-dependent gene expression. In Bacillus subtilis, activation of SigB-dependent transcription of stress-responsive genes is induced by either heat shock or decline of the intracellular ATP level (52, 53). On the other hand, SigB of S. aureus differs from that of B. subtilis with regard to triggering stimuli for activation; while S. aureus SigB is stimulated by environmental stresses such as alkali, it is not activated by energy stressors (31). Under our experimental conditions, the pH values of the culture medium were not significantly changed (data not shown). Recently, unsaturated long fatty acids with antibacterial effects against S. aureus were demonstrated to increase the sigB mRNA level (54). Moreover, mutations in the sigB gene in S. aureus affect the expression profiles of numerous genes (32). These findings together with our results suggest that detergent-like palmitate contained in the pulmonary surfactant damages cell membranes, followed by activation of the stress-responsive regulator SigB and downstream expression of virulence genes (Fig. 7). Previous reports suggest that SigB contributes to S. aureus virulence in several infection models (55, 56). To our knowledge, however, there are no reports demonstrating that host-derived materials such as pulmonary surfactant induce SigB-mediated virulence gene expression in pathogenic bacteria. Although the transcriptional activity of B. subtilis SigB is controlled through interactions with Rsb family proteins (52), genes encoding these regulatory proteins (rsbRSTX) are missing in the genome of S. aureus (57, 58). For this reason, the SigB regulatory system of S. aureus is considered to differ from that of B. subtilis (59), and most of the activation pathways remain unclear. Precise mechanisms of host sensing via SigB in S. aureus under conditions of lung infections should be clarified in future studies.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by JSPS KAKENHI grants 24-11042 (grant-in-aid for JSPS fellows to K.I.), 24-10784 (grant-in-aid for JSPS fellows to T.A.), and 24689008 (grant-in-aid for young scientists [A] to H.H.) and by a Grant-in-Aid for Scientific Research on Innovative Areas, Genome Science, from the Ministry of Education, Culture, Sports, Science, and Technology of Japan.
Footnotes
Published ahead of print 22 January 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/IAI.01635-13.
REFERENCES
- 1.Malachowa N, Whitney AR, Kobayashi SD, Sturdevant DE, Kennedy AD, Braughton KR, Shabb DW, Diep BA, Chambers HF, Otto M, DeLeo FR. 2011. Global changes in Staphylococcus aureus gene expression in human blood. PLoS One 6:e18617. 10.1371/journal.pone.0018617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brouillette E, Grondin G, Shkreta L, Lacasse P, Talbot BG. 2003. In vivo and in vitro demonstration that Staphylococcus aureus is an intracellular pathogen in the presence or absence of fibronectin-binding proteins. Microb. Pathog. 35:159–168. 10.1016/S0882-4010(03)00112-8 [DOI] [PubMed] [Google Scholar]
- 3.Agassandian M, Mallampalli RK. 2013. Surfactant phospholipid metabolism. Biochim. Biophys. Acta 1831:612–625. 10.1016/j.bbalip.2012.09.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Otto M. 2010. Basis of virulence in community-associated methicillin-resistant Staphylococcus aureus. Annu. Rev. Microbiol. 64:143–162. 10.1146/annurev.micro.112408.134309 [DOI] [PubMed] [Google Scholar]
- 5.Li M, Du X, Villaruz AE, Diep BA, Wang D, Song Y, Tian Y, Hu J, Yu F, Lu Y, Otto M. 2012. MRSA epidemic linked to a quickly spreading colonization and virulence determinant. Nat. Med. 18:816–819. 10.1038/nm.2692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kaito C, Kurokawa K, Matsumoto Y, Terao Y, Kawabata S, Hamada S, Sekimizu K. 2005. Silkworm pathogenic bacteria infection model for identification of novel virulence genes. Mol. Microbiol. 56:934–944. 10.1111/j.1365-2958.2005.04596.x [DOI] [PubMed] [Google Scholar]
- 7.Kaito C, Saito Y, Nagano G, Ikuo M, Omae Y, Hanada Y, Han X, Kuwahara-Arai K, Hishinuma T, Baba T, Ito T, Hiramatsu K, Sekimizu K. 2011. Transcription and translation products of the cytolysin gene psm-mec on the mobile genetic element SCCmec regulate Staphylococcus aureus virulence. PLoS Pathog. 7:e1001267. 10.1371/journal.ppat.1001267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kaito C, Saito Y, Ikuo M, Omae Y, Mao H, Nagano G, Fujiyuki T, Numata S, Han X, Obata K, Hasegawa S, Yamaguchi H, Inokuchi K, Ito T, Hiramatsu K, Sekimizu K. 2013. Mobile genetic element SCCmec-encoded psm-mec RNA suppresses translation of agrA and attenuates MRSA virulence. PLoS Pathog. 9:e1003269. 10.1371/journal.ppat.1003269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ishii K, Hamamoto H, Kamimura M, Sekimizu K. 2008. Activation of the silkworm cytokine by bacterial and fungal cell wall components via a reactive oxygen species-triggered mechanism. J. Biol. Chem. 283:2185–2191. 10.1074/jbc.M705480200 [DOI] [PubMed] [Google Scholar]
- 10.Ishii K, Adachi T, Hamamoto H, Oonishi T, Kamimura M, Imamura K, Sekimizu K. 2013. Insect cytokine paralytic peptide activates innate immunity via nitric oxide production in the silkworm Bombyx mori. Dev. Comp. Immunol. 39:147–153. 10.1016/j.dci.2012.10.014 [DOI] [PubMed] [Google Scholar]
- 11.Ishii K, Hamamoto H, Kamimura M, Nakamura Y, Noda H, Imamura K, Mita K, Sekimizu K. 2010. Insect cytokine paralytic peptide (PP) induces cellular and humoral immune responses in the silkworm Bombyx mori. J. Biol. Chem. 285:28635–28642. 10.1074/jbc.M110.138446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ishii K, Hamamoto H, Imamura K, Adachi T, Shoji M, Nakayama K, Sekimizu K. 2010. Porphyromonas gingivalis peptidoglycans induce excessive activation of the innate immune system in silkworm larvae J. Biol. Chem. 285:33338–33347. 10.1074/jbc.M110.112987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ishii K, Adachi T, Imamura K, Takano S, Usui K, Suzuki K, Hamamoto H, Watanabe T, Sekimizu K. 2012. Serratia marcescens induces apoptotic cell death in host immune cells via a lipopolysaccharide- and flagella-dependent mechanism. J. Biol. Chem. 287:36582–36592. 10.1074/jbc.M112.399667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Paudel A, Hamamoto H, Kobayashi Y, Yokoshima S, Fukuyama T, Sekimizu K. 2012. Identification of novel deoxyribofuranosyl indole antimicrobial agents. J. Antibiot. (Tokyo) 65:53–57. 10.1038/ja.2011.110 [DOI] [PubMed] [Google Scholar]
- 15.Hamamoto H, Kurokawa K, Kaito C, Kamura K, Manitra Razanajatovo I, Kusuhara H, Santa T, Sekimizu K. 2004. Quantitative evaluation of the therapeutic effects of antibiotics using silkworms infected with human pathogenic microorganisms. Antimicrob. Agents Chemother. 48:774–779. 10.1128/AAC.48.3.774-779.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ministry of Education, Culture, Sports, Science, and Technology of Japan. 2006. Fundamental guidelines for proper conduct of animal experiments and related activities in academic research institutions. Ministry of Education, Culture, Sports, Science, and Technology of Japan, Tokyo, Japan [Google Scholar]
- 17.Montgomery CP, Boyle-Vavra S, Daum RS. 2010. Importance of the global regulators Agr and SaeRS in the pathogenesis of CA-MRSA USA300 infection. PLoS One 5:e15177. 10.1371/journal.pone.0015177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cooney J, Kienle Z, Foster TJ, O'Toole PW. 1993. The gamma-hemolysin locus of Staphylococcus aureus comprises three linked genes, two of which are identical to the genes for the F and S components of leukocidin. Infect. Immun. 61:768–771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Inoue R, Kaito C, Tanabe M, Kamura K, Akimitsu N, Sekimizu K. 2001. Genetic identification of two distinct DNA polymerases, DnaE and PolC, that are essential for chromosomal DNA replication in Staphylococcus aureus. Mol. Genet. Genomics 266:564–571. 10.1007/s004380100564 [DOI] [PubMed] [Google Scholar]
- 20.Schenk S, Laddaga RA. 1992. Improved method for electroporation of Staphylococcus aureus. FEMS Microbiol. Lett. 73:133–138 [DOI] [PubMed] [Google Scholar]
- 21.Novick RP. 1991. Genetic systems in staphylococci. Methods Enzymol. 204:587–636. 10.1016/0076-6879(91)04029-N [DOI] [PubMed] [Google Scholar]
- 22.Ueda T, Kaito C, Omae Y, Sekimizu K. 2011. Sugar-responsive gene expression and the agr system are required for colony spreading in Staphylococcus aureus. Microb. Pathog. 51:178–185. 10.1016/j.micpath.2011.04.003 [DOI] [PubMed] [Google Scholar]
- 23.Tao L, LeBlanc DJ, Ferretti JJ. 1992. Novel streptococcal-integration shuttle vectors for gene cloning and inactivation. Gene 120:105–110. 10.1016/0378-1119(92)90016-I [DOI] [PubMed] [Google Scholar]
- 24.Kaito C, Hirano T, Omae Y, Sekimizu K. 2011. Digestion of extracellular DNA is required for giant colony formation of Staphylococcus aureus. Microb. Pathog. 51:142–148. 10.1016/j.micpath.2011.04.007 [DOI] [PubMed] [Google Scholar]
- 25.Oogai Y, Matsuo M, Hashimoto M, Kato F, Sugai M, Komatsuzawa H. 2011. Expression of virulence factors by Staphylococcus aureus grown in serum. Appl. Environ. Microbiol. 77:8097–8105. 10.1128/AEM.05316-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Naimi TS, LeDell KH, Boxrud DJ, Groom AV, Steward CD, Johnson SK, Besser JM, O'Boyle C, Danila RN, Cheek JE, Osterholm MT, Moore KA, Smith KE. 2001. Epidemiology and clonality of community-acquired methicillin-resistant Staphylococcus aureus in Minnesota, 1996–1998. Clin. Infect. Dis. 33:990–996. 10.1086/322693 [DOI] [PubMed] [Google Scholar]
- 27.Diep BA, Gill SR, Chang RF, Phan TH, Chen JH, Davidson MG, Lin F, Lin J, Carleton HA, Mongodin EF, Sensabaugh GF, Perdreau-Remington F. 2006. Complete genome sequence of USA300, an epidemic clone of community-acquired meticillin-resistant Staphylococcus aureus. Lancet 367:731–739. 10.1016/S0140-6736(06)68231-7 [DOI] [PubMed] [Google Scholar]
- 28.Yamazaki K, Kato F, Kamio Y, Kaneko J. 2006. Expression of gamma-hemolysin regulated by sae in Staphylococcus aureus strain Smith 5R. FEMS Microbiol. Lett. 259:174–180. 10.1111/j.1574-6968.2006.00236.x [DOI] [PubMed] [Google Scholar]
- 29.Schafer D, Lam TT, Geiger T, Mainiero M, Engelmann S, Hussain M, Bosserhoff A, Frosch M, Bischoff M, Wolz C, Reidl J, Sinha B. 2009. A point mutation in the sensor histidine kinase SaeS of Staphylococcus aureus strain Newman alters the response to biocide exposure. J. Bacteriol. 191:7306–7314. 10.1128/JB.00630-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kelsey JA, Bayles KW, Shafii B, McGuire MA. 2006. Fatty acids and monoacylglycerols inhibit growth of Staphylococcus aureus. Lipids 41:951–961. 10.1007/s11745-006-5048-z [DOI] [PubMed] [Google Scholar]
- 31.Pane-Farre J, Jonas B, Forstner K, Engelmann S, Hecker M. 2006. The sigmaB regulon in Staphylococcus aureus and its regulation. Int. J. Med. Microbiol. 296:237–258. 10.1016/j.ijmm.2005.11.011 [DOI] [PubMed] [Google Scholar]
- 32.Bischoff M, Dunman P, Kormanec J, Macapagal D, Murphy E, Mounts W, Berger-Bachi B, Projan S. 2004. Microarray-based analysis of the Staphylococcus aureus sigmaB regulon. J. Bacteriol. 186:4085–4099. 10.1128/JB.186.13.4085-4099.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Giachino P, Engelmann S, Bischoff M. 2001. Sigma(B) activity depends on RsbU in Staphylococcus aureus. J. Bacteriol. 183:1843–1852. 10.1128/JB.183.6.1843-1852.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gertz S, Engelmann S, Schmid R, Ohlsen K, Hacker J, Hecker M. 1999. Regulation of sigmaB-dependent transcription of sigB and asp23 in two different Staphylococcus aureus strains. Mol. Gen. Genet. 261:558–566. 10.1007/s004380051001 [DOI] [PubMed] [Google Scholar]
- 35.Miyazaki E, Chen JM, Ko C, Bishai WR. 1999. The Staphylococcus aureus rsbW (orf159) gene encodes an anti-sigma factor of SigB. J. Bacteriol. 181:2846–2851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kullik I, Giachino P, Fuchs T. 1998. Deletion of the alternative sigma factor sigmaB in Staphylococcus aureus reveals its function as a global regulator of virulence genes. J. Bacteriol. 180:4814–4820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Homerova D, Bischoff M, Dumolin A, Kormanec J. 2004. Optimization of a two-plasmid system for the identification of promoters recognized by RNA polymerase containing Staphylococcus aureus alternative sigma factor sigmaB. FEMS Microbiol. Lett. 232:173–179. 10.1016/S0378-1097(04)00063-1 [DOI] [PubMed] [Google Scholar]
- 38.Nicholas RO, Li T, McDevitt D, Marra A, Sucoloski S, Demarsh PL, Gentry DR. 1999. Isolation and characterization of a sigB deletion mutant of Staphylococcus aureus. Infect. Immun. 67:3667–3669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gertz S, Engelmann S, Schmid R, Ziebandt AK, Tischer K, Scharf C, Hacker J, Hecker M. 2000. Characterization of the sigma(B) regulon in Staphylococcus aureus. J. Bacteriol. 182:6983–6991. 10.1128/JB.182.24.6983-6991.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Abdelnour A, Arvidson S, Bremell T, Ryden C, Tarkowski A. 1993. The accessory gene regulator (agr) controls Staphylococcus aureus virulence in a murine arthritis model. Infect. Immun. 61:3879–3885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fournier B, Hooper DC. 2000. A new two-component regulatory system involved in adhesion, autolysis, and extracellular proteolytic activity of Staphylococcus aureus. J. Bacteriol. 182:3955–3964. 10.1128/JB.182.14.3955-3964.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Giraudo AT, Cheung AL, Nagel R. 1997. The sae locus of Staphylococcus aureus controls exoprotein synthesis at the transcriptional level. Arch. Microbiol. 168:53–58. 10.1007/s002030050469 [DOI] [PubMed] [Google Scholar]
- 43.Burts ML, DeDent AC, Missiakas DM. 2008. EsaC substrate for the ESAT-6 secretion pathway and its role in persistent infections of Staphylococcus aureus. Mol. Microbiol. 69:736–746. 10.1111/j.1365-2958.2008.06324.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Burts ML, Williams WA, DeBord K, Missiakas DM. 2005. EsxA and EsxB are secreted by an ESAT-6-like system that is required for the pathogenesis of Staphylococcus aureus infections. Proc. Natl. Acad. Sci. U. S. A. 102:1169–1174. 10.1073/pnas.0405620102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Abdallah AM, Gey van Pittius NC, Champion PA, Cox J, Luirink J, Vandenbroucke-Grauls CM, Appelmelk BJ, Bitter W. 2007. Type VII secretion—mycobacteria show the way. Nat. Rev. Microbiol. 5:883–891. 10.1038/nrmicro1773 [DOI] [PubMed] [Google Scholar]
- 46.Green JB, Lower RP, Young JP. 2009. The NfeD protein family and its conserved gene neighbours throughout prokaryotes: functional implications for stomatin-like proteins. J. Mol. Evol. 69:657–667. 10.1007/s00239-009-9304-8 [DOI] [PubMed] [Google Scholar]
- 47.Smith ML, Price SA. 1938. Staphylococcus γ hæmolysin. J. Pathol. Bacteriol. 47:379–393. 10.1002/path.1700470303 [DOI] [Google Scholar]
- 48.Gouaux E, Hobaugh M, Song L. 1997. Alpha-hemolysin, gamma-hemolysin, and leukocidin from Staphylococcus aureus: distant in sequence but similar in structure. Protein Sci. 6:2631–2635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bubeck Wardenburg J, Bae T, Otto M, Deleo FR, Schneewind O. 2007. Poring over pores: alpha-hemolysin and Panton-Valentine leukocidin in Staphylococcus aureus pneumonia. Nat. Med. 13:1405–1406. 10.1038/nm1207-1405 [DOI] [PubMed] [Google Scholar]
- 50.Hayashida A, Bartlett AH, Foster TJ, Park PW. 2009. Staphylococcus aureus beta-toxin induces lung injury through syndecan-1. Am. J. Pathol. 174:509–518. 10.2353/ajpath.2009.080394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Labandeira-Rey M, Couzon F, Boisset S, Brown EL, Bes M, Benito Y, Barbu EM, Vazquez V, Hook M, Etienne J, Vandenesch F, Bowden MG. 2007. Staphylococcus aureus Panton-Valentine leukocidin causes necrotizing pneumonia. Science 315:1130–1133. 10.1126/science.1137165 [DOI] [PubMed] [Google Scholar]
- 52.Voelker U, Dufour A, Haldenwang WG. 1995. The Bacillus subtilis rsbU gene product is necessary for RsbX-dependent regulation of sigma B. J. Bacteriol. 177:114–122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Voelker U, Voelker A, Maul B, Hecker M, Dufour A, Haldenwang WG. 1995. Separate mechanisms activate sigma B of Bacillus subtilis in response to environmental and metabolic stresses. J. Bacteriol. 177:3771–3780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kenny JG, Ward D, Josefsson E, Jonsson IM, Hinds J, Rees HH, Lindsay JA, Tarkowski A, Horsburgh MJ. 2009. The Staphylococcus aureus response to unsaturated long chain free fatty acids: survival mechanisms and virulence implications. PLoS One 4:e4344. 10.1371/journal.pone.0004344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Entenza JM, Vouillamoz J, Glauser MP, Moreillon P. 2004. Efficacy of garenoxacin in treatment of experimental endocarditis due to Staphylococcus aureus or viridans group streptococci. Antimicrob. Agents Chemother. 48:86–92. 10.1128/AAC.48.1.86-92.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nair SP, Bischoff M, Senn MM, Berger-Bachi B. 2003. The sigma B regulon influences internalization of Staphylococcus aureus by osteoblasts. Infect. Immun. 71:4167–4170. 10.1128/IAI.71.7.4167-4170.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kuroda M, Ohta T, Uchiyama I, Baba T, Yuzawa H, Kobayashi I, Cui L, Oguchi A, Aoki K, Nagai Y, Lian J, Ito T, Kanamori M, Matsumaru H, Maruyama A, Murakami H, Hosoyama A, Mizutani-Ui Y, Takahashi NK, Sawano T, Inoue R, Kaito C, Sekimizu K, Hirakawa H, Kuhara S, Goto S, Yabuzaki J, Kanehisa M, Yamashita A, Oshima K, Furuya K, Yoshino C, Shiba T, Hattori M, Ogasawara N, Hayashi H, Hiramatsu K. 2001. Whole genome sequencing of meticillin-resistant Staphylococcus aureus. Lancet 357:1225–1240. 10.1016/S0140-6736(00)04403-2 [DOI] [PubMed] [Google Scholar]
- 58.Baba T, Takeuchi F, Kuroda M, Yuzawa H, Aoki K, Oguchi A, Nagai Y, Iwama N, Asano K, Naimi T, Kuroda H, Cui L, Yamamoto K, Hiramatsu K. 2002. Genome and virulence determinants of high virulence community-acquired MRSA. Lancet 359:1819–1827. 10.1016/S0140-6736(02)08713-5 [DOI] [PubMed] [Google Scholar]
- 59.Senn MM, Giachino P, Homerova D, Steinhuber A, Strassner J, Kormanec J, Fluckiger U, Berger-Bachi B, Bischoff M. 2005. Molecular analysis and organization of the sigmaB operon in Staphylococcus aureus. J. Bacteriol. 187:8006–8019. 10.1128/JB.187.23.8006-8019.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.