Abstract
Lyme disease, caused by Borrelia burgdorferi, is the most commonly reported vector-borne disease in the United States. Many patients treated for early Lyme disease incur another infection in subsequent years, suggesting that previous exposure to B. burgdorferi may not elicit a protective immune response. However, identical strains are almost never detected from patients who have been infected multiple times, suggesting that B. burgdorferi exposure may elicit strain-specific immunity. Probabilistic and simulation models assuming biologically realistic data derived from patients in the northeastern United States suggest that patients treated for early Lyme disease develop protective immunity that is strain specific and lasts for at least 6 years.
INTRODUCTION
Lyme disease, caused by Borrelia burgdorferi, is the most commonly reported vector-borne infection in the United States, with over 30,000 reported cases in 2012 (1). Erythema migrans, the characteristic skin lesion that arises at the site of B. burgdorferi inoculation by an infected Ixodes sp. tick, is the earliest objective clinical manifestation. As many as 15% of patients with an erythema migrans skin lesion who are treated with antibiotics will develop another erythema migrans lesion within 5 years due to a subsequent tick bite (2). The large proportion of patients experiencing repeated B. burgdorferi infections suggests that previous exposure does not elicit a protective immune response. However, whether patients might develop protective immunity against specific strains of B. burgdorferi has not been investigated. In a recent report of 17 patients who had each experienced two or more episodes of B. burgdorferi infection, in only 1 patient was the same strain observed on more than one occasion (3). In this patient, who had four episodes of erythema migrans in less than 20 years, the same borrelial strain was isolated only during the first and third episodes, which were 5 years apart. The present study investigated whether the findings in the previous study of 17 patients, each of whom had one or more recurrences of culture-confirmed Lyme disease, are consistent with development of protective immunity against specific strains of B. burgdorferi.
Maintenance of natural variation among strains within several pathogenic species has been driven by strain-specific protective immunity in hosts (4). It has been proposed that polymorphisms in B. burgdorferi populations, such as that observed at the outer surface protein C (ospC) locus (3, 5–8), are maintained selectively by strain-specific immunity in natural hosts (6, 9). However, the evidence to support this hypothesis is indirect and limited to a data set showing that laboratory mice immunized with one OspC protein were selectively protected only against infection with B. burgdorferi expressing the same OspC protein, not against strains expressing a different OspC protein (10). In this study, we provide empirical evidence and statistical support for the presence of strain-specific immunity in patients treated for early Lyme disease. Additionally, the results of this study provide a quantitative estimate of the duration of strain-specific immunity.
MATERIALS AND METHODS
Prevalence of each B. burgdorferi strain in patients.
For this study, we analyzed the occurrence of identical strains of B. burgdorferi in a cohort of 17 patients with multiple episodes of culture-confirmed erythema migrans (3). We used both multinomial probability analyses and a stochastic simulation model to evaluate whether only 1 or fewer of 17 patients could have identical strains of B. burgdorferi in recurrent infections due to chance alone. All analyses estimated the probability of recovering a particular strain of B. burgdorferi from a patient due to chance alone, based on empirical patient data (3, 11). Since initial infections in patients cannot be influenced by immune responses to prior B. burgdorferi infections, the probability of observing each strain of B. burgdorferi by chance alone can be estimated as the frequency with which each strain is observed in patients with no history of infection. These data were obtained from the work of Nadelman et al. (3) (data set 1) and Wormser et al. (11) (data set 2) (see the supplemental material for further details). Data set 1 comprised the strains of B. burgdorferi recovered from the initial episode of erythema migrans in the 17 patients reported by Nadelman et al. (3). Data set 2 consisted of the subset of strains cultured from the erythema migrans lesions of patients reported by Wormser et al. (11) who had no history of B. burgdorferi infection and who were not included in the report by Nadelman et al. (3). Data set 2 provided a more robust frequency distribution of the B. burgdorferi strains causing primary infections due to the larger number of borrelial isolates (200 versus 17). Both data sets are based on patients with early Lyme disease from the same geographic area of New York State. In addition to the distributions described above, an even probability distribution (data set 3), which assumed that patients were equally likely to be infected with each of the B. burgdorferi strains, was used in the analyses described below.
Multinomial probability analyses.
The probability that the same strain would be recovered multiple times in only 1 or fewer of 17 patients who experienced repeated infections by chance alone was calculated using a multinomial probability distribution involving the sum of (i) the probability that the same strain would never be cultured multiple times from any patient (equation 1) and (ii) the probability that the same strain would be cultured exactly twice from exactly 1 of the 17 patients (equation 2), as follows:
| (1) |
| (2) |
where n is the number of B. burgdorferi strains, a is the number of patients from which B. burgdorferi was cultured on 2 occasions (n = 14), b is the number of patients from which B. burgdorferi was cultured on 3 occasions (n = 2), and c is the number of patients from which B. burgdorferi was cultured on 4 occasions (n = 1). Pi—the probability of strain type i being cultured from a patient due to chance alone—was derived from the frequency of each strain found in primary infections as described above (data sets 1 to 3).
Stochastic simulation model of the presence and duration of strain-specific immunity.
The presence and duration of strain-specific immunity necessary to account for the observed pattern of strains of B. burgdorferi recovered from patients with consecutive, repeated infections (3) were evaluated using a stochastic simulation (see the supplemental material). The model simulated the strains of infecting B. burgdorferi that each of 17 patients incurred either with or without strain-specific immunity. The parameters of the model included the number of years in which each patient could have contracted a B. burgdorferi infection, the probability that a patient was challenged with a particular B. burgdorferi strain following a tick bite, and the duration of strain-specific immunity (Table 1).
TABLE 1.
Model parameters and ranges of values
| Parameter class | Parameter values in model |
|---|---|
| Yrs included in the study | Time from primary infection of each patient until 2011 |
| Time from primary infection to final infection of each patient | |
| Probability of each strain in primary human infections | Data set 1a (3) |
| Data set 2a (11) | |
| Data set 3 (even distribution) | |
| Duration of strain-specific immunity | 0–15 yr |
| Probability of challenge with an infectious tick | 0–100% |
| Proportion of patients with strain-specific immunity | 0–100% |
See the supplemental material and the text for a detailed explanation.
Model parameters.
The number of years during which each of the 17 patients in the model could have contracted a B. burgdorferi infection was calculated as either (i) the number of years between the initial infection for that patient and 2011, the last year that a positive B. burgdorferi culture was obtained from any patient (“extended availability”), or (ii) the number of years between the initial and final infections for each patient (“limited availability”). The former set of parameters ranged from 3 to 18 years (mean = 11.12 years; median = 12 years), while the latter set of parameters ranged from 2 to 15 years (mean = 5.65 years; median = 5 years).
The probability of infection with each strain, assuming no strain-specific immunity, can be derived from data set 1, 2, or 3 by assuming that the patient was bitten by an infected tick. The probability that a patient was bitten by an infected tick cannot be calculated from empirical data. Thus, the value taken for the infectious tick bite parameter was selected such that there were at least 34 total infections, i.e., each patient had 2 separate infections, on average, and at most 39 total infections, i.e., the total number of infections observed in the 17 patients reported by Nadelman et al. (3). All iterations that had fewer than 34 or more than 39 total infections were excluded from the analyses. The unlikely assumption that the probability of a tick bite was equivalent across all patients and all years was implicit in the model. However, this assumption did not affect the qualitative conclusions and is conservative with respect to the quantitative estimates of the duration of strain-specific immunity. That is, higher or more variable values for the tick bite probability parameter require greater durations of strain-specific immunity to arrive at an outcome with only one patient experiencing two episodes of erythema migrans caused by the same strain of B. burgdorferi (3).
The parameter describing the duration of strain-specific immunity ranged from 0 to 15 years in the stochastic models. It was assumed that the duration of strain-specific immunity was constant across all patients and across all strains. The former assumption was relaxed in subsequent iterations of the model. The latter assumption is conservative and reflects the minimum duration of strain-specific immunity for each B. burgdorferi strain.
Model implementation.
The model simulated infections in 17 patients by choosing the strain type causing the infection each year that the patient was exposed to an infected tick. We simulated 10,000 sets of 17 patients for every combination of parameter values used in the stochastic model. Parameter combinations included (i) 2 parameter sets describing the number of years a patient could have contracted a B. burgdorferi infection (either extended or limited availability), (ii) 3 parameter sets describing the probability that patients were challenged with a particular B. burgdorferi strain (either data set 1, 2, or 3), and (iii) 15 parameter values describing the duration of strain-specific immunity, resulting in 90 models evaluated (Table 1).
The strain of B. burgdorferi infecting each patient in the initial year he or she was available was determined in each iteration of the simulation by drawing from the appropriate probability distribution (data set 1, 2, or 3) (Fig. 1). In each subsequent year that the patient could have contracted a B. burgdorferi infection (extended or limited availability), the model determined if the patient was fed upon by a potentially infectious tick (Table 1). The probability that the patient was challenged by an infected tick—which incorporates the chance of being bitten by a tick each year, the probability that the tick is infected, and the probability that the bacterium is transmitted to the host—was chosen at random in each iteration of the model. If the patient was exposed to an infected tick, the strain carried by that tick was inferred from the same probability distribution used to choose the strain in the initial infection (data set 1, 2, or 3). If a previous infection with the same strain as that carried by the feeding tick occurred within the duration of the strain-specific immunity time set in that iteration of the model (Table 1), the current tick bite did not result in a new infection. Otherwise, the current tick bite resulted in a new infection with the strain carried by the feeding tick. The total number of infections in each patient, as well as the number of repeated infections with the same strain, was tabulated for each iteration of the model. All iterations of the model resulting in fewer than 34 or more than 39 total infections across all patients were excluded from further analyses. An additional set of simulations was performed to evaluate the effect of variation among patients in the development of protective immunity. In these simulations, the fraction of patients who developed strain-specific immunity varied between 0 and 100% (10% intervals), with the duration of immunity set to 5, 10, or 15 years.
FIG 1.
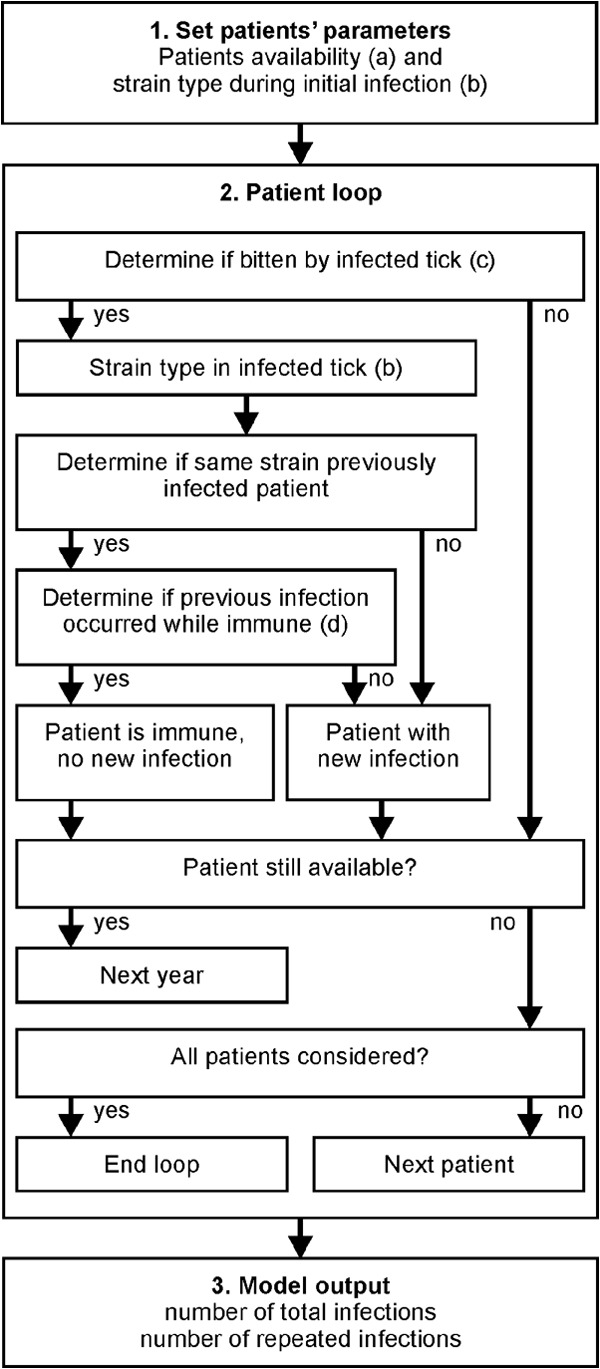
Schematic representation of the stochastic simulation model. (1) Set patients' parameters. The model uses the patient availability from the appropriate data set (limited or extended availability) (a) and determines the strain in the initial infection of each patient from the appropriated strain probability distribution (data set 1, 2, or 3) (b). (2) Patient loop. For each patient, the model first determines, for each year that the patient is available, if the patient is bitten by an infected tick (c). If the patient is exposed to an infected tick, the model determines the B. burgdorferi strain carried by the infecting tick (b). If the patient has previously been infected by the strain in the infecting tick within the duration of strain-specific immunity (d), that tick bite does not result in a new infection. If the patient is not immune, a new infection is recorded. The process is repeated until the patient is no longer available, at which point the model simulates the next patient until the infections in all patients have been simulated. (3) Model output. The model saves the total number of infections and the number of patients that have been infected by the same strain multiple times for each iteration of the model. For each combination of parameters (Table 1), data from 10,000 iterations are recorded.
RESULTS
We used both multinomial probability analyses and a stochastic simulation model to determine whether the empirical data from 17 patients, each of whom had one or more recurrences of culture-confirmed Lyme disease, are consistent with development of protective immunity against specific strains of B. burgdorferi. The probability that the same strain of B. burgdorferi would be recovered only once due to random chance in 17 patients treated for early Lyme disease who experienced at least one recurrence of erythema migrans ranged from 0.011 to 0.054 (Table 2). Assuming that the probability that a particular borrelial strain infects humans due to chance alone is equivalent to the frequency with which each strain has been observed in primary infections of patients (data set 1 [3] or 2 [11]), it is highly improbable that only one or fewer patients would have developed a recurrent infection with the same borrelial strain (P < 0.035). Furthermore, making the very conservative assumption that all strains are equally likely to cause human infections (data set 3), the probability of 1 or fewer of 17 patients being infected with the same strain on multiple occasions by chance alone remains unlikely (P < 0.054).
TABLE 2.
Probability that only 1 of 17 patients would have been infected with an identical strain of B. burgdorferi in a recurrent episode of Lyme disease due to chance alone
| Data source for frequency distribution of strain types | Probability of 1 or fewer patients becoming infected with the same strain of B. burgdorferi |
|---|---|
| Data set 1a (3) | 0.035 |
| Data set 2a (11) | 0.011 |
| Data set 3 (even distribution) | 0.054 |
See text for a detailed explanation.
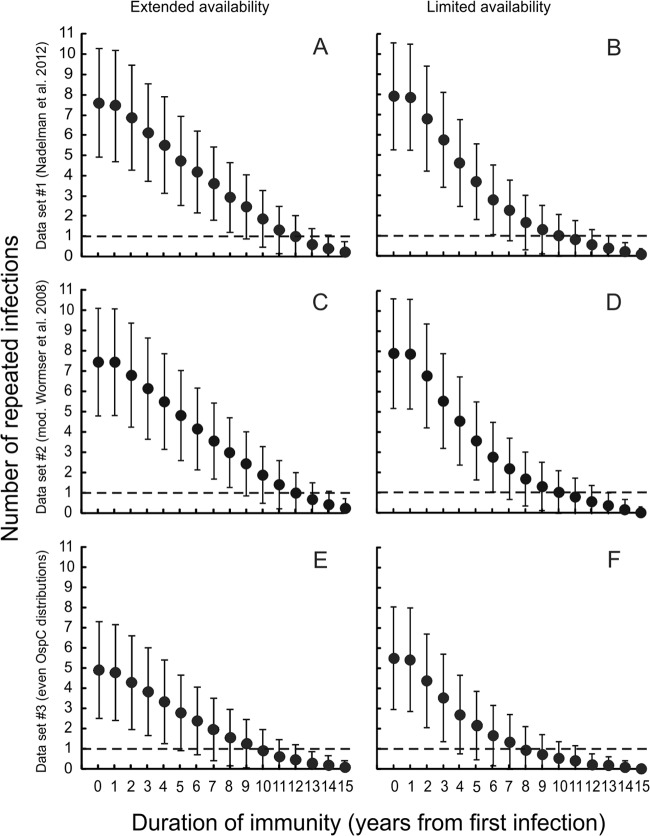
All stochastic simulation models demonstrated that a far larger proportion of patients would be expected to have experienced multiple infections with the same B. burgdorferi strain than was actually observed by Nadelman et al. (3), assuming that patients did not acquire strain-specific immunity (Fig. 2). All simulations, regardless of the parameter sets used, always resulted in more than 1 of the 17 patients developing an infection caused by the same B. burgdorferi strain if no strain-specific immunity was assumed in the model. Furthermore, the duration of strain-specific immunity would have had to last for at least 4 years for only one or fewer patients to present with the same B. burgdorferi strain on multiple occasions, regardless of the parameters used in the model.
FIG 2.
The number of patients experiencing multiple infections caused by the same B. burgdorferi strain is a function of the duration of strain-specific immunity. The duration of strain-specific immunity must be at least 4 years for one or fewer patients to present with the same B. burgdorferi strain on multiple occasions, regardless of the parameter sets investigated (dashed line). Data in the left column (A, C, and E) represent models that assume that patients could have contracted a B. burgdorferi infection from the date of the primary infection through 2011 (extended availability); data in the right column (B, D, and F) represent models that assume that patients could have contracted a B. burgdorferi infection from the date of the primary infection through the final infection of that patient (limited availability). Panels A and B assume the strain probability distribution derived from data set 1 (Nadelman et al. 2012 [3]) as described in the text; panels C and D assume the strain probability derived from data set 2 (mod. Wormser et al. 2008 [11]). Panels E and F assume equal probabilities per strain (data set 3). Error bars represent the ranges of results for 10,000 simulations.
Assuming the empirically determined stochastic expectation that patients were challenged with a particular borrelial strain (data set 2), nearly 8 of the 17 patients evaluated would have been expected to have multiple infections with the same strain if patients did not develop strain-specific immunity (Fig. 2). Assuming these model parameters, the duration of strain-specific immunity must exceed 6 years before the same strain would be expected to be recovered on multiple occasions from one or fewer patients.
Although all model parameters affected the quantitative estimate of the duration of strain-specific immunity, no combination of parameters resulted in observations similar to those reported by Nadelman et al. (3) without invoking strain-specific immunity. Furthermore, the probability distribution for acquiring strains by chance alone, using any empirically determined distribution (data set 1 or 2), had little effect on the quantitative estimate of the duration of strain-specific immunity (Fig. 1). All models using empirically determined parameters required between 6 and 9 years of strain-specific immunity for multiple infections with the same strain to occur in only one or fewer of the simulated patients. Even using the very conservative assumption that all B. burgdorferi strains are equally likely to infect patients required a duration of at least 4 years of strain-specific immunity. The duration that each patient was available to become infected had a small but consistent effect on the quantitative estimate of the duration of strain-specific immunity: simulations where patients were available to become infected for shorter times had lower estimates of the minimum duration of strain-specific immunity (Fig. 2).
The simulation model was extremely sensitive to the total number of infections incurred by each patient. Simulations in which the total number of infections did not exceed 34 (twice the number of patients) predicted that many patients would not incur multiple infections with the same strain, independent of the duration of strain-specific immunity. In contrast, simulations in which the total number of infections exceeded 39 (the total number experienced by the patients reported by Nadelman et al. [3]) inflated the number of patients infected with the same strain on multiple occasions when the duration of strain-specific immunity was low.
The simulation model does not require that all patients develop strain-specific immunity in order to explain the previously observed data (3). However, the duration of strain-specific immunity needed is inversely correlated with the proportion of patients experiencing strain-specific immunity (Fig. 3); as the duration of strain-specific immunity in the model decreases, a larger proportion of patients would need to develop strain-specific immunity to account for the data previously reported (3).
FIG 3.
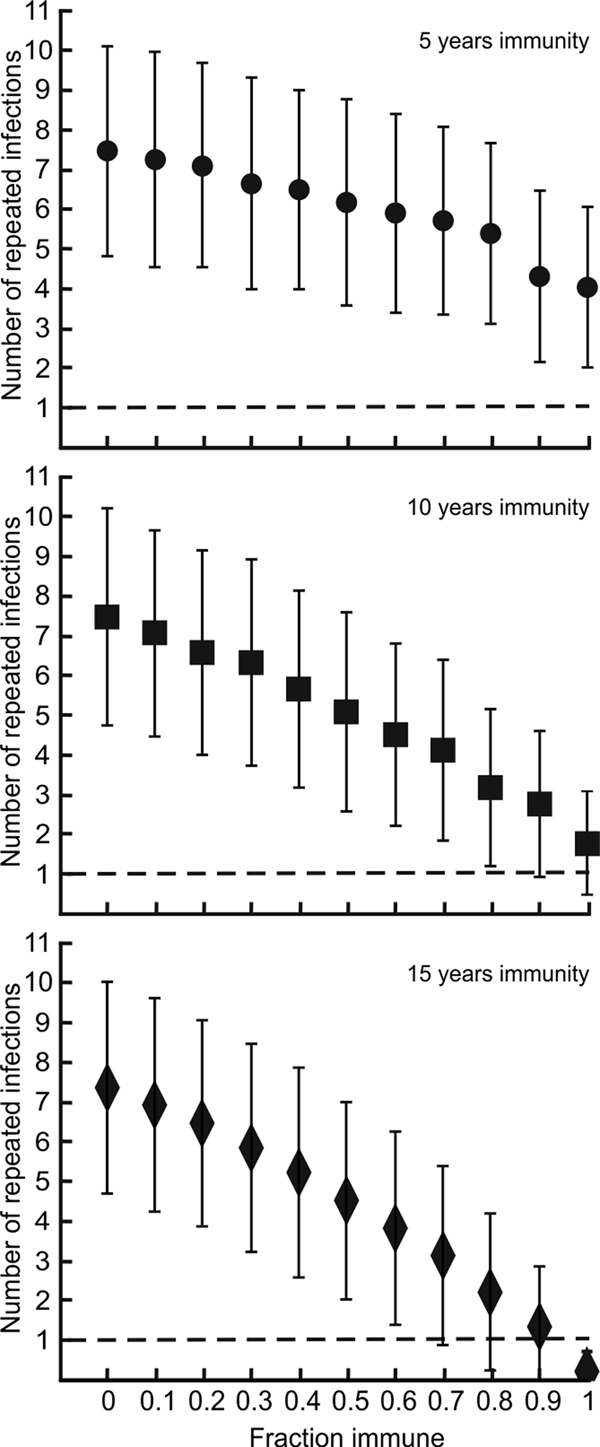
The number of patients experiencing multiple infections caused by the same B. burgdorferi strain is a function of the fraction of patients who develop strain-specific immunity. At least 70% of the patients must have developed strain-specific immunity for one or fewer patients to present with the same B. burgdorferi strain on multiple occasions (dashed line), regardless of the parameter sets investigated. Data shown were obtained by assuming strain-specific immunity of 5 years, 10 years, and 15 years. The analyses shown assume the probability distribution of B. burgdorferi strains derived from the work of Nadelman et al. (3). Error bars represent ranges of results for 10,000 simulations.
DISCUSSION
The present analyses suggest that it is highly unlikely that only 1 of 17 patients would have been infected with an identical strain of B. burgdorferi in a recurrent episode of Lyme disease in the absence of strain-specific immunity. Furthermore, the duration of strain-specific immunity needed to be at least 4 years to explain the data actually observed (3). The presence and long duration of strain-specific immunity that the models suggest imply that humans, once infected, are highly unlikely to acquire a subsequent infection caused by the same strain of B. burgdorferi. Statistical models assuming a biologically realistic distribution of the expectation of a particular strain of B. burgdorferi strain causing infection in human patients (derived from multiple data sets from patients in the same geographical region) demonstrated that the probability that so few patients would be infected by the same borrelial strain by chance alone is between 0.011 and 0.035. Stochastic simulation models confirmed that strain-specific immunity was necessary to explain the reported patient data and further estimated that strain-specific immunity must exceed 6 years if biologically realistic parameters are assumed.
Although transient immunity to reinfection with the same strain of B. burgdorferi has been demonstrated in laboratory animals, no attempt has been made to determine if challenge with different strains of B. burgdorferi might be able to cause infection (12–15). It was also demonstrated experimentally in mice that immunization with outer surface protein C (OspC) of one strain of B. burgdorferi was protective against infection by that strain but not two different strains (10). However, strain-specific immunity is not likely to be restricted to OspC and may also develop against other B. burgdorferi strain-specific surface proteins. Future clinical studies should aim to expand the recognition and characterization of the antigens that could be involved in strain-specific immunity in humans. Furthermore, the above-described study employed an immunization protocol by needle inoculation rather than natural infection and challenge by tick feeding. Our investigation is thus unique in that it is the first study to provide evidence for strain-specific immunity in either humans or animals due to natural infection.
The qualitative results as well as the quantitative estimates of the duration of strain-specific immunity from the stochastic simulation model were robust to the assumptions inherent in the choice of parameters. Altering the parameter set describing the expected probability of individuals encountering a tick infected with a particular strain of B. burgdorferi due to chance alone had little effect on the quantitative estimate of the duration of strain-specific immunity. All simulations, even those using the very conservative assumption that all strains are equally likely to infect patients, resulted in an estimated minimum duration of strain-specific immunity of 4 years (Fig. 2). For all of the parameter sets using empirical patient data in the analyses, the minimum duration of strain-specific immunity was estimated to be 6 to 9 years.
The simulations make the unlikely assumption that all patients experience the same duration of strain-specific immunity. However, relaxing this assumption did not change the qualitative result that strain-specific immunity is necessary to observe so few patients infected with the same strain on repeated occasions. The duration of strain-specific immunity is, however, inversely correlated with the proportion of patients experiencing strain-specific immunity (Fig. 3). That is, as the proportion of patients acquiring strain-specific immunity decreases, the estimate of the duration of strain-specific immunity in the other patients increases. While we expect that there is variation among patients in both the extent and duration of strain-specific immunity, the presented model is conservative with respect to this variation. It is also likely that the presence and duration of strain-specific immunity elicited vary among infecting strains. While this phenomenon needs to be investigated empirically, the model is also conservative with respect to this variation.
The models assumed that each strain elicited a protective response against future infections with the same strain but did not affect the probability of infection caused by other strains. It is possible that partial or full protective immunity also develops against additional strains, although this could not be established with the current data set. Data sets with larger numbers of patients with recurrent infections or controlled experiments in laboratory animals are necessary to determine the extent of cross immunity among strains.
Strain-specific immunity is thought to exist with other bacterial pathogens, such as Streptococcus pneumoniae (16). However, the data set on which our analyses and conclusions were drawn regarding strain-specific immunity in Lyme disease is rather unique. The Lyme disease data set represented 17 patients who were treated with antibiotics for two or more episodes of early Lyme disease that were not only confirmed by culture but also typed in such a way that each cultured strain of B. burgdorferi could readily be distinguished from another. The analyses presented indicate that patients treated for early Lyme disease develop type-specific immunity that will last for a minimum of 4 years. However, since at least 16 strains of B. burgdorferi, defined by the allele at the ospC locus, have been shown to infect humans in the United States (17), it is essential that people in areas where Lyme disease is endemic continue to utilize effective prophylaxis measures to prevent future tick bites, even if they have protective immunity to certain strains due to a previous infection.
Supplementary Material
ACKNOWLEDGMENTS
We thank Diane Holmgren and Susan Bittker for helpful comments and/or assistance in the laboratory.
This work was supported in part by grants from the NIH (grants AR41511, AI45801, AI076342, and AI097137) and the Burroughs Wellcome Fund.
R. B. Nadelman reports receiving consulting fees from Guidepoint Global and Decision Resources and serving as an expert witness in medical malpractice cases regarding Lyme disease. G. P. Wormser reports receiving research grants from Immunetics, Inc., Bio-Rad, DiaSorin, Inc., and bioMérieux SA. He owns equity in Abbott, has been an expert witness in malpractice cases involving Lyme disease, is an unpaid board member of the American Lyme Disease Foundation, has been an expert witness regarding Lyme disease in a disciplinary action for the Missouri Board of Registration for the Healing Arts, and is a consultant to Baxter for Lyme vaccine development.
Footnotes
Published ahead of print 13 January 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/IAI.01451-13.
REFERENCES
- 1.CDC. 2013. Lyme disease—United States. MMWR Morb. Mortal. Wkly. Rep. 62:ND-579–ND-592 [Google Scholar]
- 2.Nowakowski J, Nadelman RB, Sell R, McKenna D, Cavaliere LF, Holmgren D, Gaidici A, Wormser GP. 2003. Long-term follow-up of patients with culture-confirmed Lyme disease. Am. J. Med. 115:91–96. 10.1016/S0002-9343(03)00308-5 [DOI] [PubMed] [Google Scholar]
- 3.Nadelman RB, Hanincová K, Mukherjee P, Liveris D, Nowakowski J, McKenna D, Brisson D, Cooper D, Bittker S, Madison G, Holmgren D, Schwartz I, Wormser GP. 2012. Differentiation of reinfection from relapse in recurrent Lyme disease. N. Engl. J. Med. 367:1883–1890. 10.1056/NEJMoa1114362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cheesman S, Tanabe K, Sawai H, O'Mahony E, Carter R. 2009. Strain-specific immunity may drive adaptive polymorphism in the merozoite surface protein 1 of the rodent malaria parasite Plasmodium chabaudi. Infect. Genet. Evol. 9:248–255. 10.1016/j.meegid.2008.12.002 [DOI] [PubMed] [Google Scholar]
- 5.Brisson D, Dykhuizen DE. 2004. ospC diversity in Borrelia burgdorferi: different hosts are different niches. Genetics 168:713–722. 10.1534/genetics.104.028738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Qiu WG, Dykhuizen DE, Acosta MS, Luft BJ. 2002. Geographic uniformity of the Lyme disease spirochete (Borrelia burgdorferi) and its shared history with tick vector (Ixodes scapularis) in the northeastern United States. Genetics 160:833–849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Haven J, Vargas LC, Mongodin EF, Xue V, Hernandez Y, Pagan P, Fraser-Liggett CM, Schutzer SE, Luft BJ, Casjens SR, Qiu WG. 2011. Pervasive recombination and sympatric genome diversification driven by frequency-dependent selection in Borrelia burgdorferi, the Lyme disease bacterium. Genetics 189:951–966. 10.1534/genetics.111.130773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barbour AG, Travinsky B. 2010. Evolution and distribution of the ospC gene, a transferable serotype determinant of Borrelia burgdorferi. mBio 1:e00153–10. 10.1128/mBio.00153-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang IN, Dykhuizen DE, Qiu W, Dunn JJ, Bosler EM, Luft BJ. 1999. Genetic diversity of ospC in a local population of Borrelia burgdorferi sensu stricto. Genetics 151:15–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Probert WS, Crawford M, Cadiz RB, LeFebvre RB. 1997. Immunization with outer surface protein (Osp) A, but not OspC, provides cross-protection of mice challenged with North American isolates of Borrelia burgdorferi. J. Infect. Dis. 175:400–405. 10.1093/infdis/175.2.400 [DOI] [PubMed] [Google Scholar]
- 11.Wormser G, Brisson D, Liveris D, Hanincova K, Sandigursky S, Nowakowski J, Nadelman R, Ludin S, Schwartz I. 2008. Borrelia burgdorferi genotype predicts the capacity for hematogenous dissemination during early Lyme disease. J. Infect. Dis. 198:1358–1364. 10.1086/592279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Piesman J, Dolan MC, Happ CM, Luft BJ, Rooney SE, Mather TN, Golde WT. 1997. Duration of immunity to reinfection with tick-transmitted Borrelia burgdorferi in naturally infected mice. Infect. Immun. 65:4043–4047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Foley DM, Gayek RJ, Skare JT, Wagar EA, Champion CI, Blanco DR, Lovett MA, Miller JN. 1995. Rabbit model of Lyme borreliosis: erythema migrans, infection-derived immunity, and identification of Borrelia burgdorferi proteins associated with virulence and protective immunity. J. Clin. Invest. 96:965–975. 10.1172/JCI118144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Foley DM, Wang YP, Wu XY, Blanco DR, Lovett MA, Miller JN. 1997. Acquired resistance to Borrelia burgdorferi infection in the rabbit. Comparison between outer surface protein A vaccine- and infection-derived immunity. J. Clin. Invest. 99:2030–2035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Barthold SW. 1993. Antigenic stability of Borrelia burgdorferi during chronic infections of immunocompetent mice. Infect. Immun. 61:4955–4961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Musher DM. 1992. Infections caused by Streptococcus pneumoniae: clinical spectrum, pathogenesis, immunity, and treatment. Clin. Infect. Dis. 14:801–807. 10.1093/clinids/14.4.801 [DOI] [PubMed] [Google Scholar]
- 17.Dykhuizen DE, Brisson D, Sandigursky S, Wormser GP, Nowakowski J, Nadelman RB, Schwartz I. 2008. The propensity of different Borrelia burgdorferi sensu stricto genotypes to cause disseminated infections in humans. Am. J. Trop. Med. Hyg. 78:806–810 [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.