Abstract
Staphylococcus aureus is an important human pathogen that employs a large repertoire of secreted virulence factors to promote disease pathogenesis. Many strains of S. aureus possess a plc gene that encodes a phosphatidylinositol (PI)-specific phospholipase C (PI-PLC) capable of hydrolyzing PI and cleaving glycosyl-PI (GPI)-linked proteins from cell surfaces. Despite being secreted by virulent staphylococci, the contribution of PI-PLC to the capacity of S. aureus to cause disease remains undefined. Our goal in these studies was to understand PI-PLC in the context of S. aureus biology. Among a collection of genetically diverse clinical isolates of S. aureus, community-associated methicillin-resistant S. aureus (CA-MRSA) USA300 secreted the most PI-PLC. Screening a collection of two-component system (TCS) mutants of S. aureus, we identified both the agr quorum-sensing system and the SrrAB TCS to be positive regulators of plc gene expression. Real-time PCR and PI-PLC enzyme assays of the TCS mutants, coupled with SrrA promoter binding studies, demonstrated that SrrAB was the predominant transcriptional activator of plc. Furthermore, plc regulation was linked to oxidative stress both in vitro and in vivo in a SrrAB-dependent manner. A Δplc mutant in a CA-MRSA USA300 background exhibited a survival defect in human whole blood and in isolated neutrophils. However, the same mutant strain displayed no survival defect in murine models of infection or murine whole blood. Overall, these data identify potential links between bacterial responses to the host innate immune system and to oxidative stress and suggest how PI-PLC could contribute to the pathogenesis of S. aureus infections.
INTRODUCTION
Staphylococcus aureus remains a significant human bacterial pathogen worldwide and a leading cause of nosocomial infections in the United States (1). Its capacity to exist as a commensal, colonizing ∼30% of the population at any given time (2), and its tendency to develop resistance to antibiotics are challenges to the control and treatment of S. aureus disease (3, 4). In recent years, hypervirulent strains of methicillin-resistant S. aureus (MRSA) that are capable of infecting otherwise healthy individuals have appeared (5, 6). These community-associated MRSA (CA-MRSA) strains have become a significant public health concern. Although the molecular basis for their greater virulence is not completely understood, recent evidence suggests that CA-MRSA strains are more fit than hospital-acquired MRSA (HA-MRSA) strains and exhibit an increased capacity to secrete virulence determinants (7–9).
S. aureus can cause a wide variety of diseases, from common skin infections, such as cellulitis and abscesses, to more severe and life-threatening processes, such as pneumonia, endovascular disease, and toxic shock. Among its many virulence factors, S. aureus secretes toxins, superantigens, and exoenzymes that promote disease (reviewed in reference 10). Some of these secreted factors have well-defined roles as virulence determinants for specific clinical presentations of staphylococcal infection. For example, toxic shock syndrome toxin 1 is a superantigen that has been associated with toxic shock syndrome (11, 12). However, not all strains of staphylococci secrete the same repertoire of factors, which may explain in part the diverse clinical manifestations of staphylococcal infection. The variability of the exoprotein profile among strains of S. aureus often reflects inherent differences in gene regulation. Expression of many virulence factors is regulated by two-component systems (TCSs), wherein a cell membrane-associated sensor kinase and an intracellular cognate response regulator mediate rapid transduction of extracellular signals to modulate transcriptional responses (13). For example, the agr quorum-sensing system represents an extensively characterized example of a TCS that regulates secreted staphylococcal virulence determinants and differs among strains (14–16).
Although characterizing the regulation and function of secreted virulence factors of S. aureus is important both to understand the pathogenesis of staphylococcal infection and to identify potential new targets for therapeutic intervention, not all proteins secreted by S. aureus have had their biological roles defined. One such staphylococcal exoprotein is phosphatidylinositol (PI)-specific phospholipase C (PI-PLC; SAUSA300_0099), an enzyme that degrades inositol phospholipids and releases glycosyl-PI (GPI)-anchored surface proteins from target membranes (17). Little is known regarding the function of PI-PLC with respect to S. aureus physiology or disease pathogenesis, but several facts suggest that it may serve as a virulence factor for S. aureus. First, PI-PLCs are important virulence factors in the pathogenesis of infection by other Gram-positive bacteria, most notably, Listeria monocytogenes (18) and Bacillus species (19, 20). Second, only S. aureus, the most virulent of the staphylococcal species with respect to human disease, expresses staphylococcal PI-PLC (17). Third, staphylococcal PI-PLC is expressed during infection, as humoral responses to the enzyme are detected in experimental as well as natural disease states (21, 22). Lastly, because PI is not a component of the S. aureus membrane bilayer (23), host cells are the likely targets of PI-PLC enzymatic activity, thus providing a rationale for considering PI-PLC to be a virulence factor. However, despite such features that support PI-PLC as a virulence factor, direct evidence that PI-PLC contributes to the pathogenesis of S. aureus disease is lacking.
We sought to identify the role of PI-PLC in staphylococcal biology and to determine what, if any, contribution that it makes to MRSA virulence. In this study, we report novel observations on the regulation of staphylococcal PI-PLC and suggest a potential role for PI-PLC in the pathogenesis of S. aureus infection.
MATERIALS AND METHODS
Ethics statement.
Written informed consent was obtained for all volunteers according to protocols approved by the Institutional Review Board for human subjects at the University of Iowa.
Bacterial strains, media, and growth conditions.
All bacterial strains and plasmids used in this study are described in Table 1. All Escherichia coli cultures were grown with aeration in Luria-Bertani (LB) broth or on LB agar, and S. aureus strains were grown with aeration in tryptic soy broth (TSB) or on tryptic soy agar (TSA). As necessary for the maintenance of plasmids in E. coli, LB medium was supplemented with the appropriate antibiotic: ampicillin (100 μg/ml), kanamycin (50 μg/ml), or spectinomycin (50 μg/ml). Antibiotics necessary for the maintenance of plasmids in S. aureus included chloramphenicol (10 μg/ml), tetracycline (3 μg/ml), or erythromycin (10 μg/ml). Ampicillin, chloramphenicol, tetracycline, and erythromycin were all obtained from Research Products International (Mount Prospect, IL). Kanamycin was obtained from Thermo Fisher Scientific (Rockford, IL), and spectinomycin was obtained from Sigma-Aldrich (St. Louis, MO).
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Description or function | Source or reference |
|---|---|---|
| Strains | ||
| E. coli strains | ||
| DH5α | Plasmid maintenance | Protein Express |
| TOP10 | Cloning strain | Invitrogen |
| BL21 Star(DE3) | Recombinant protein production | Invitrogen |
| S. aureus strains | ||
| AH1263 | USA300 CA-MRSA Erms (LAC*) | 55 |
| RN4220 | Restriction deficient | 56 |
| MRSA252 | USA200 MRSA | 57 |
| UAMS-1 | Osteomyelitis isolate | 58 |
| Newman | MSSA | 59 |
| COL | HA-MRSA | 60 |
| MW2 | USA400 CA-MRSA | 61 |
| NRS22 | USA600 MRSA | 62 |
| TCH1516 | USA300 CA-MRSA | 63 |
| JMB1145 | LAC* SAUSA300_2337-38Δ | This work |
| JMB1148 | LAC* SAUSA300_0217-18Δ | This work |
| JMB1219 | LAC* SAUSA300_1219-20Δ | This work |
| JMB1223 | LAC* SAUSA300_2035-36Δ | This work |
| JMB1232 | LAC* SAUSA300_2558-59Δ | This work |
| JMB1241 | LAC* SAUSA300_1798-99Δ | This work |
| JMB1357 | LAC* SAUSA300_0254-55Δ | This work |
| JMB1358 | LAC* SAUSA300_1638-39Δ | This work |
| JMB1359 | LAC* SAUSA300_2308-09Δ | This work |
| JMB1377 | LAC* SAUSA300_1865-66Δ | This work |
| JMB1383 | LAC* SAUSA300_1307-08Δ | This work |
| JMB1293 | LAC* SAUSA300_1441-42Δ | This work |
| AH1292 | LAC* Δagr::tetM | 64 |
| AH1558 | LAC* sae::spec | 65 |
| JMB1515 | LAC* SAUSA300_0645-46Δ | This work |
| MJW44 | LAC* SAUSA300_0099Δ(plc) | This work |
| MJW120 | RN4220 Δplc | This work |
| MJW131 | Newman Δplc | This work |
| Plasmids | ||
| pJB38 | Mutation generation vector | 66 |
| pCM28 | Complementation vector | 30 |
| pET28a | Protein expression vector | Invitrogen |
| pSKerm-MCS | Complementation vector | 67 |
| pSK236 | Shuttle vector | 68 |
| pET16b | Protein expression vector | Invitrogen |
| pMW1 | pJB38 SAUSA300_0099Δ | This work |
| pMW4 | pSKerm-MCS SAUSA300_0099 | This work |
| pMW12 | pET28a containing srrA | This work |
| pMW14 | pSK236 containing scn promoter and plc | This work |
| pMW17 | pET16b containing srrB kinase domain | This work |
| pMW20 | pCM28 containing plc | This work |
| pCM29 | pCM28 containing sGFP | 30 |
| pCR2.1 | Cloning vector | Invitrogen |
| pJMB150 | pJB38 SAUSA300_0217-18Δ | This work |
| pJMB155 | pJB38 SAUSA300_2337-38Δ | This work |
| pJMB168 | pJB38 SAUSA300_2035-36Δ | This work |
| pJMB170 | pJB38 SAUSA300_1219-20Δ | This work |
| pJMB181 | pJB38 SAUSA300_2558-59Δ | This work |
| pJMB183 | pJB38 SAUSA300_1798-99Δ | This work |
| pJMB202 | pJB38 SAUSA300_1307-08Δ | This work |
| pJMB204 | pJB38 SAUSA300_1865-66Δ | This work |
| pJMB205 | pJB38 SAUSA300_2308-09Δ | This work |
| pJMB210 | pJB38 SAUSA300_0254-55Δ | This work |
| pJMB211 | pJB38 SAUSA300_1638-39Δ | This work |
| pJMB229 | pJB38 SAUSA300_1441-42Δ | This work |
| pJMB230 | pJB38 SAUSA300_0645-46Δ | This work |
Recombinant DNA and genetic techniques.
All oligonucleotides and primers are listed in Table S1 in the supplemental material and were synthesized by Integrated DNA Technologies (Coralville, IA). Plasmid DNA was electroporated into S. aureus RN4220 as previously described (24). DNA was then moved from RN4220 into other S. aureus strains by the use of bacteriophage 80 alpha (25). S. aureus RNA was obtained by the use of an RNeasy minikit (Qiagen, Valencia, CA) per the manufacturer's protocol. Following purification, the RNA was treated with Turbo DNase (Ambion, Austin, TX) for 2 h at 37°C to remove contaminating DNA.
Expression and purification of recombinant proteins.
A protein expression system consisting of pSK236 containing a heterologous promoter was used to overproduce recombinant PI-PLC containing a C-terminal His6 tag into the culture supernatant. All plc sequences were amplified from S. aureus LAC genomic DNA by PCR using Phusion Hi-Fidelity polymerase, cloned into pCR2.1-TOPO (Invitrogen, Carlsbad, CA), subcloned into pSK236, and sequenced to confirm their identity and the absence of mutations. Full-length plc (amino acids 1 to 329) was amplified using primers PLC-SCNfwd and PLC-SCNrev. The scn promoter was amplified from USA300 genomic DNA using primers SCN-PLCfwd and SCN-PLCrev. The two PCR fragments were combined by overlap PCR using primers SCN-PLCfwd and PLC-SCNrev. This fragment was ligated into the pSK236 vector and electroporated into S. aureus RN4220. S. aureus RN4220 containing the overproduction construct was grown overnight in TSB without antibiotic. The supernatant was collected and passed through a 0.22-μm-pore-size filter (Pall, Ann Arbor, MI), and the protein was precipitated by the addition of ammonium sulfate (85%) for 1 h at room temperature. The precipitated protein was rehydrated in phosphate-buffered saline (PBS) and passed over a nickel-nitrilotriacetic acid-agarose column (Qiagen, Valencia, CA) (see Fig. S1 in the supplemental material). Purified protein fractions were pooled, dialyzed against PBS, concentrated, and stored in 2.5 M glycerol at −80°C.
To purify the kinase domain of SrrB, primers srrBfwd-NdeI and srrBrev-XhoI were used to amplify the sequence from S. aureus LAC genomic DNA that was cloned into pCR2.1-TOPO (Invitrogen, Carlsbad, CA). The srrB fragment was digested from pCR2.1 using NdeI and XhoI and ligated into pET16b (Invitrogen, Carlsbad, CA) using the NdeI and XhoI sites, creating pMW17. The pMW17 vector was transformed into BL21 Star(DE3) (Invitrogen, Carlsbad, CA) cells for protein production. Bacteria were grown to mid-log phase in LB broth, and protein production was stimulated by the addition of 0.1 mM IPTG (isopropyl-β-d-thiogalactopyranoside). Cells were kept at 30°C for 3 h with shaking, and bacteria were pelleted and lysed by sonication. Protein lysates were passed across a nickel-nitrilotriacetic acid-agarose column (Qiagen, Valencia, CA). Purified protein fractions were pooled, dialyzed against PBS, concentrated, and stored in 2.5 M glycerol at −80°C. SrrA was purified in a manner similar to that previously described (26). All protein concentrations were determined using the bicinchoninic acid (BCA) protein assay (Thermo Fisher Scientific, Rockford, IL) per the manufacturer's protocol.
Lipase activity assays.
The lipolytic activity of PI-PLC was determined using the artificial substrate 4-methylumbelliferyl myo-inositol-1-phosphate, N-methyl-morpholine salt (Biosynth International Inc., Itasca, IL) (27). Briefly, reaction mixtures consisted of 10 mM Tris, pH 6.8, 0.8 mM substrate, and 50% culture supernatant. The culture supernatants were obtained by growing various S. aureus strains overnight in TSB at 37°C, spun down to pellet the bacteria, and passed through a 0.45-μm-pore-size low-protein-binding filter (Pall, Ann Arbor, MI). The reaction mixtures were placed in a 96-well plate, and fluorescence was measured using 350-nm excitation/450-nm emission filters on a Spectramax M5 plate reader (Molecular Devices, Sunnyvale, CA). Reactions were allowed to proceed for 30 min at room temperature, with measurements taken every 30 s. The results were either expressed as relative fluorescence units in kinetic data (fluorescence versus time) or normalized to account for the protein concentration at the final endpoint.
Generation of unmarked staphylococcal deletion mutants and complemented strains.
S. aureus chromosomal mutations were created using homologous recombination of a knockout vector introduced by transduction mediated by bacteriophage 80 alpha. Regions of approximately 500 to 1,000 bp upstream and downstream of the plc gene or the genes encoding the TCS response regulator and histidine kinase were generated by PCR. These fragments were fused using extension overlap PCR to generate a 1.0- to 2.0-kb fragment which was restriction enzyme digested and cloned into the polylinker of similarly digested pJB38 (28). The generated vectors were transformed into S. aureus RN4220, and lysates were made with bacteriophage 80 alpha (25). Following transduction into the target S. aureus strains at 30°C, clones exhibiting chloramphenicol resistance were selected on TSA containing chloramphenicol at 45°C. Clones able to grow at the elevated temperature were isolated a second time at 45°C, and bacterial clones were grown in TSB at 30°C. These bacterial cultures were subcultured at 1:1,000 daily for 3 days, the cultures were plated, and bacterial isolates were screened for chloramphenicol sensitivity (29). Antibiotic-sensitive isolates were screened for the chromosomal deletion by PCR and, in the case of plc, by enzymatic analysis.
To complement the Δplc mutant, the plc gene was amplified from USA300 LAC genomic DNA using primers plcCOMPfwd-ClaI and plcCOMPrev-KpnI. The entire plc coding region plus 200 bp upstream and 50 bp downstream was included for complementation. This fragment was cloned into pSKerm-MCS at the ClaI and KpnI restriction sites, resulting in the construction of plasmid pMW4. This plasmid was electroporated into S. aureus derivatives containing the plc deletion for complementation.
To complement the ΔsrrAB mutant, SAUSA300_1441-42 (srrAB) and the native promoter were amplified using USA300 genomic DNA and the primers 1440comp5BamHI and 1440comp3PstI. The PCR fragment was digested with the BamHI and PstI restriction enzymes and gel purified. The digested fragment was ligated into similarly digested pCM28 and verified by PCR. This plasmid was electroporated into RN4220 and subsequently transduced into strain JMB1293 for complementation.
To overexpress PI-PLC in the ΔsrrAB mutant background, the plc gene, including its predicted ribosomal binding site, was amplified from USA300 LAC genomic DNA using primers plcFWDv2-KpnI and plcREV-EcoRI. This fragment was digested with KpnI and EcoRI, gel purified, and ligated into similarly digested pCM28, resulting in the construction of plasmid pMW20. This plasmid was first electroporated into RN4220 for subsequent transduction into strain JMB1293.
Real-time RT-PCR and 5′ RACE.
For reverse transcription-PCR (RT-PCR), cDNA from the S. aureus strains was prepared by incubating RNA with random hexamers in a reverse transcription reaction mixture containing avian myeloblastosis virus (AMV) reverse transcriptase (Roche, Indianapolis, IN). To test for the presence of DNA contamination, identical reaction mixtures lacking AMV reverse transcriptase were also run. All PCRs were performed on either an ABI Prism 7000 real-time PCR detection system (Applied Biosystems, Carlsbad, CA) or an Eppendorf Mastercycler ep realplex2 apparatus (Eppendorf, Hauppauge, NY) with PerfeCTa SYBR green FastMix, ROX (Quanta Biosciences, Gaithersburg, MD), and gene-specific primers. All primers used are described in Table S1 in the supplemental material. Reaction conditions consisted of 40 rounds of denaturation at 95°C for 15 s and annealing/extension at 60°C for 30 s. Single PCR products were confirmed by performing a postamplification melt curve analysis for each reaction. The transcript levels of the target genes were normalized to the level for the gyrB gene (30).
For 5′ rapid amplification of cDNA ends (RACE), a FirstChoice RLM-RACE kit (Ambion, Austin, TX) was used as recommended by the manufacturer. Briefly, 3 to 5 μg of total RNA was ligated to 300 ng of 5′ RACE adaptor using 5 units of T4 RNA ligase (Ambion, Austin, TX) for 1 h at 37°C. RNA was then reverse transcribed with random hexamers and Moloney murine leukemia virus (M-MLV) reverse transcriptase (Ambion, Austin, TX) for 1 h at 42°C. PCR was performed using a primer nested within the gene of interest and a primer specific to the 5′ RACE adaptor (5′ RACE outer primer). The resulting PCR products were cloned into pCR2.1-TOPO and sequenced using the M13rev primer. Ten to 20 clones were sequenced for each primer set.
Oxidative stress assays.
To determine the contribution of select genes in the oxidative stress response of S. aureus, the bacteria were subjected to various concentrations of oxidants in vitro. Briefly, S. aureus cultures were grown overnight in LB broth, subcultured in the morning, and grown to mid-log growth phase (optical density at 550 nm [OD550] = 0.8 to 1.0). The bacteria were washed and resuspended in sterile PBS. Various concentrations of either H2O2 or HOCl were added, and the bacteria were incubated at 37°C for 1 h with shaking. RNA was then collected from the bacteria as previously described.
EMSA.
DNA for electrophoretic mobility shift assay (EMSA) analysis was amplified from the region upstream of plc using primers plcGSsense and plcGSantisense. For nonspecific DNA, the region upstream of rpsC was amplified using primers rpsCGSsense and rpsCGSantisense. PCR products were labeled using a DNA 3′ end biotinylation kit (Thermo Fisher Scientific, Rockford, IL) per the manufacturer's directions before use in a LightShift chemiluminescent EMSA kit (Thermo Fisher Scientific, Rockford, IL). Briefly, labeled DNA was mixed with purified recombinant SrrA for 30 min at room temperature in a reaction buffer supplemented with 50 mM KCl and 5 mM MgCl2. Unlabeled DNA was included in some experiments to test the specificity of the observed shift. All reaction mixtures were electrophoresed into a 5% nondenaturing polyacrylamide gel for 60 to 90 min at 4°C and transferred to an Amersham Hybond-XL membrane (GE Healthcare, Pittsburgh, PA). Membranes were developed and exposed to Biomax (Kodak, Rochester, NY) X-ray film. To phosphorylate SrrA, the purified kinase domain of SrrB (15 μg) was mixed with SrrA (90 μg) in phosphorylation buffer (50 mM Tris, pH 7.6, 50 mM KCl, 20 mM MgCl2, 2 mM ATP) for 30 min at 37°C.
Protein extraction and immunoblot analysis in S. aureus.
Cultures of S. aureus derivatives were grown in TSB to stationary phase (OD550 > 5). To collect proteins from the culture filtrate, spent broth medium was passed through a 0.45-μm-pore-size low-protein-binding filter (Pall, Ann Arbor, MI). Protein concentrations were determined using the BCA protein assay (Thermo Fisher Scientific, Rockford, IL). Protein was added to 5 μl of 5× SDS-PAGE loading buffer containing β-mercaptoethanol, boiled for 5 min, separated on a 12% SDS-polyacrylamide gel, and transferred onto an Immobilon-P membrane (Millipore, Billerica, MA). Membranes were blocked in TBS-T (20 mM Tris-HCl [pH 7.6], 250 mM NaCl, 0.1% Tween 20) containing 5% skim milk for at least 1 h. Membranes were probed with goat polyclonal or mouse monoclonal anti-PI-PLC antibody (1:10,000 or 1:1,000; Elmira Biologicals, Inc., and Iowa State University Hybridoma Facility) in TBS-T overnight at 4°C. After washing, membranes were incubated with goat anti-rabbit or goat anti-mouse IgG antibody conjugated to horseradish peroxidase (Thermo Fisher Scientific, Rockford, IL) for 30 min at room temperature. Blots were visualized using SuperSignal West Pico or Femto chemiluminescent substrate kits (Thermo Fisher Scientific, Rockford, IL). Blots were analyzed by phosphorimaging using a Typhoon 9410 phosphorimager (GE Healthcare, Pittsburgh, PA).
Isolation and infection of neutrophils.
Human neutrophils were isolated from heparinized venous blood from healthy volunteers or from patients with chronic granulomatous disease (CGD) as described previously (31). All three subjects with CGD were unrelated males with abnormal or absent gp91phox (X-CGD) and without a functional NADPH oxidase in their polymorphonuclear neutrophils (PMNs). Isolated neutrophils were maintained at a density equal to or less than 2 × 107/ml in sterile Hanks' balanced salt solution (HBSS; Mediatech, Inc., Manassas, VA). Wild-type and mutant S. aureus strains were opsonized by tumbling in HBSS buffered with 20 mM HEPES, 10% pooled human serum, and 1% human serum albumin (HSA) for 20 min at 37°C. Immediately following opsonization, bacteria were mixed at the appropriate multiplicity of infection (MOI) with isolated neutrophils and tumbled for 10 min at 37°C, as previously described (30). For some experiments, neutrophils were pretreated with diphenyleneiodonium (DPI; 10 μM), a flavoprotein inhibitor that blocks the phagocyte NADPH oxidase (32), prior to S. aureus challenge. After defined periods of phagocytosis of S. aureus, neutrophils were lysed with sterile water (pH 11) and viable bacteria were enumerated by determining the number of CFU after serial dilution.
Flow cytometry.
As a measure of bacterial survival, the fluorescence of bacteria expressing superfolded green fluorescent protein (sGFP) was assessed by flow cytometry, using a previously described method that correlates HOCl-induced bleaching of sGFP to bacterial viability (30, 33). Briefly, S. aureus derivatives that expressed sGFP constitutively were generated through the transduction of pCM29, a plasmid in which sGFP expression is driven by the sarAP1 promoter (30). These bacteria were fed to neutrophils as described previously. At various times after phagocytosis, the fluorescence of S. aureus-containing neutrophils was measured using an Accuri C6 flow cytometry system (Accuri Cytometers, Inc., Ann Arbor, MI). Each sample was analyzed using FCS Express software (De Novo Software, Los Angeles, CA) to determine the geometric mean and the percentage of GFP-positive cells relative to the values for paired DPI-treated controls to determine the mean fluorescence index for each condition tested.
Blood survival assays.
The ability of wild-type and mutant S. aureus to survive in blood was determined under two separate growth conditions, either mid-log phase (OD550 = 0.6 to 1.0) or stationary phase (overnight culture), in TSB. Bacteria were examined after resuspension in conditioned culture medium or after washing and resuspension in fresh medium. For certain experiments, wild-type and mutant strains of USA300 were pelleted in a tabletop centrifuge (8,000 rpm for 5 min), and their supernatants were removed. Cells were then washed 2 times in sterile TSB. Finally, the bacterial pellet was resuspended either in sterile TSB or in filter-sterilized conditioned medium from an overnight culture of USA300. A 100-μl aliquot of these suspensions was added to 1 ml of heparinized human blood obtained from healthy volunteers, or a 50-μl aliquot of bacterial culture was added to 0.5 ml of heparinized mouse blood obtained from 6- to 8-week old female C57BL/6 mice through cardiac puncture. A separate aliquot was used to determine the initial bacterial load by serial dilution and plating on TSA. The infected blood was tumbled at 37°C. After 3 h, serial dilutions in water were performed to determine the endpoint numbers of CFU, which were compared to the initial bacterial load to determine the viable percentage of the initial inoculum.
Mouse infections.
Six- to 8-week old female BALB/c mice were used to study systemic infection of S. aureus. Briefly, S. aureus strains were grown to mid-log phase (OD550 = 0.6 to 1.0) in TSB, centrifuged, and resuspended in PBS. Mice were infected by means of a tail vein injection with a 200-μl aliquot of bacteria (5 × 106 CFU). Mice were housed in microisolation cages following infection and given food and water ad libitum. At day 8 postinfection, the mice were sacrificed and the kidneys were removed aseptically. The tissue samples were homogenized in 1 ml PBS, and the CFU burden was determined by serial dilution and plating on TSA.
To assess the contribution of PI-PLC in a skin infection model, 6- to 8-week old female BALB/c mice were used. The mice were shaved on one side in preparation for the experiment. Bacteria were prepared as described for the systemic infection model and were injected intradermally in a 100-μl aliquot (1 × 107 CFU). Once again, the mice were housed in microisolation cages following infection and given food and water ad libitum. Mice were observed for the presence of abscesses in the injection site for up to 4 days. On day 4, the mice were sacrificed and a 10-mm punch biopsy specimen of the skin surrounding the abscess was collected. Tissue samples were homogenized in 1 ml of PBS, and the CFU burden was determined by serial dilution and plating on TSA. All animal infection experiments were approved by the Institutional Animal Care and Use Committee at the University of Iowa.
Statistical analysis.
All statistical analyses were conducted using Student's t test. P values of <0.05 were determined to be statistically significant.
RESULTS
PI-PLC secretion varies among S. aureus isolates.
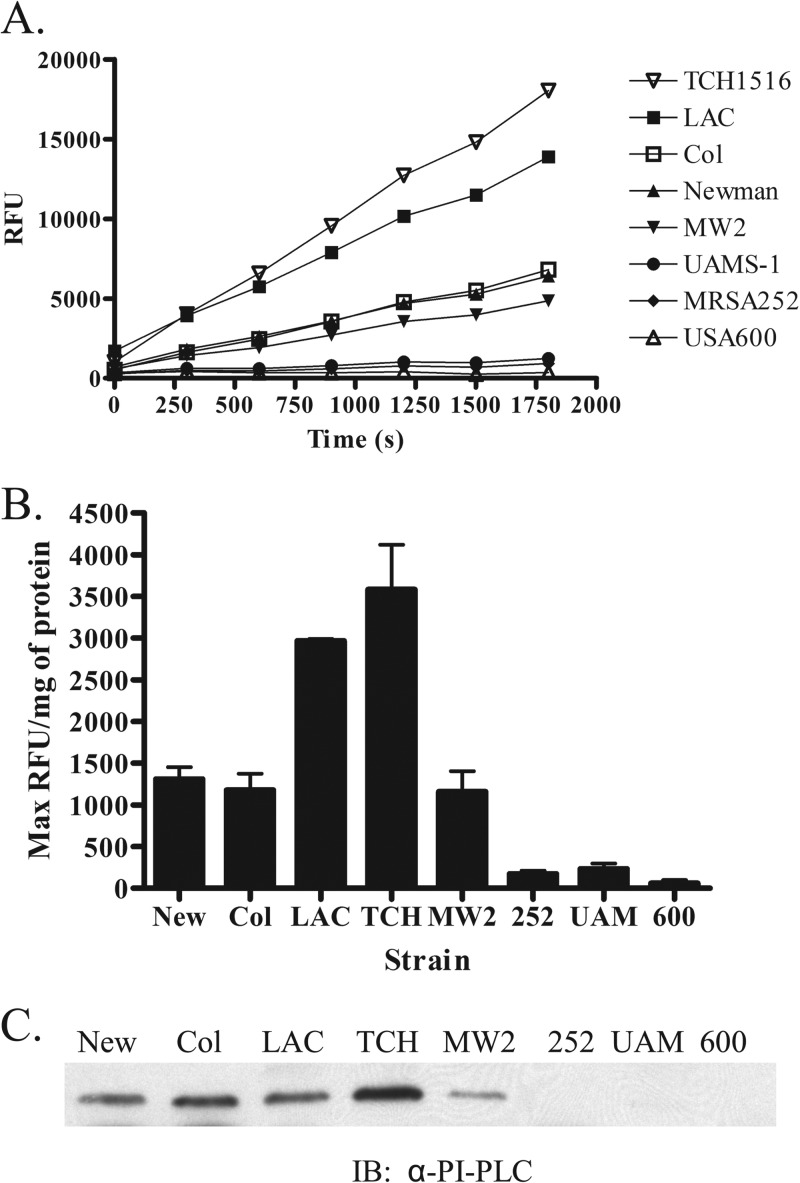
If PI-PLC serves as a virulence factor for S. aureus, the severity of staphylococcal infection might correlate with the presence and extent of PI-PLC expression in clinical isolates. To test this prediction, we assessed PI-PLC secretion across a variety of well-defined strains of S. aureus. We chose for study both methicillin-susceptible (Newman) and -resistant (COL) laboratory strains, two CA-MRSA USA300 isolates (LAC and TCH1516), a CA-MRSA USA400 strain (MW2), two USA200 strains (MRSA252 and UAMS-1), and a USA600 MRSA isolate (NRS22). The extracellular lipolytic activity of each isolate was measured using a spectrofluorometric assay specific for PI-PLC (Fig. 1A). Both USA300 isolates secreted the largest amount of active PI-PLC, whereas the amounts for strains COL, Newman, and MW2 clustered in the middle of the range of values, and strains UAMS-1, MRSA252, and USA600 demonstrated little enzymatic activity (Fig. 1A). Due to variability among strains with respect to the amount of exoproteins secreted, we normalized the maximum fluorescence generated to the protein content. Overall, protein secretion differences were minimal among the strains (data not shown), and the normalized data mirrored the kinetic data, with the USA300 isolates exhibiting the largest amount of activity and strains MRSA252, UAMS-1, and USA600 demonstrating very little activity (Fig. 1B). With one exception, the relative amounts of secreted PI-PLC detected by immunoblotting paralleled the activity data (Fig. 1C); the exception was strain COL, which had less PI-PLC activity than some strains but a similar amount of protein in the immunoblot assay as USA300 LAC. The basis for this slight discordance is not known but could reflect differences in the specific activity of PI-PLC secreted by the two strains, unidentified interactions of PI-PLC with other exoproteins that differ between the two strains, or both. All strains of S. aureus used in this study possess plc, the gene encoding PI-PLC. Although the genome for the USA600 isolate has not been sequenced, we were able to PCR amplify and sequence the plc gene from this strain. The plc gene product from USA600 displayed 98.8% identity to the USA300 (LAC) PI-PLC at the amino acid level, with no mutations in predicted catalytic residues (data not shown). Overall, these data collectively demonstrate that PI-PLC secretion varied among different S. aureus strains, with the highest level exhibited in CA-MRSA USA300 isolates.
FIG 1.
PI-PLC activity and expression vary among S. aureus strains. (A) Kinetic assay demonstrating the PI-PLC activity associated with various S. aureus culture supernatants using a fluorescent artificial substrate over a 30-min time course. The results represent the means for three independent experiments. (B) PI-PLC activity normalized to the protein concentration for culture supernatants. Results are the ratio between the maximum relative fluorescent units (RFU) and protein concentration for a given strain and represent the means ± SEMs for three independent experiments. (C) Representative immunoblot (IB) visualizing the PI-PLC present in the culture supernatants using a monoclonal antibody against PI-PLC. Immunoreactivity was quantitated on a phosphorimager, and the percentages of immunoreactive protein were quantitated for all strains using the TCH1516 signal, which was set to 100%. Abbreviations: New, S. aureus Newman; Col, S. aureus COL; LAC, S. aureus USA300 LAC; TCH, S. aureus USA300 TCH1516; MW2, S. aureus USA400 MW2; 252, S. aureus USA200 MRSA252; UAM, S. aureus USA200 UAMS-1; 600, S. aureus USA600 NRS22.
Regulation of PI-PLC by S. aureus TCSs.
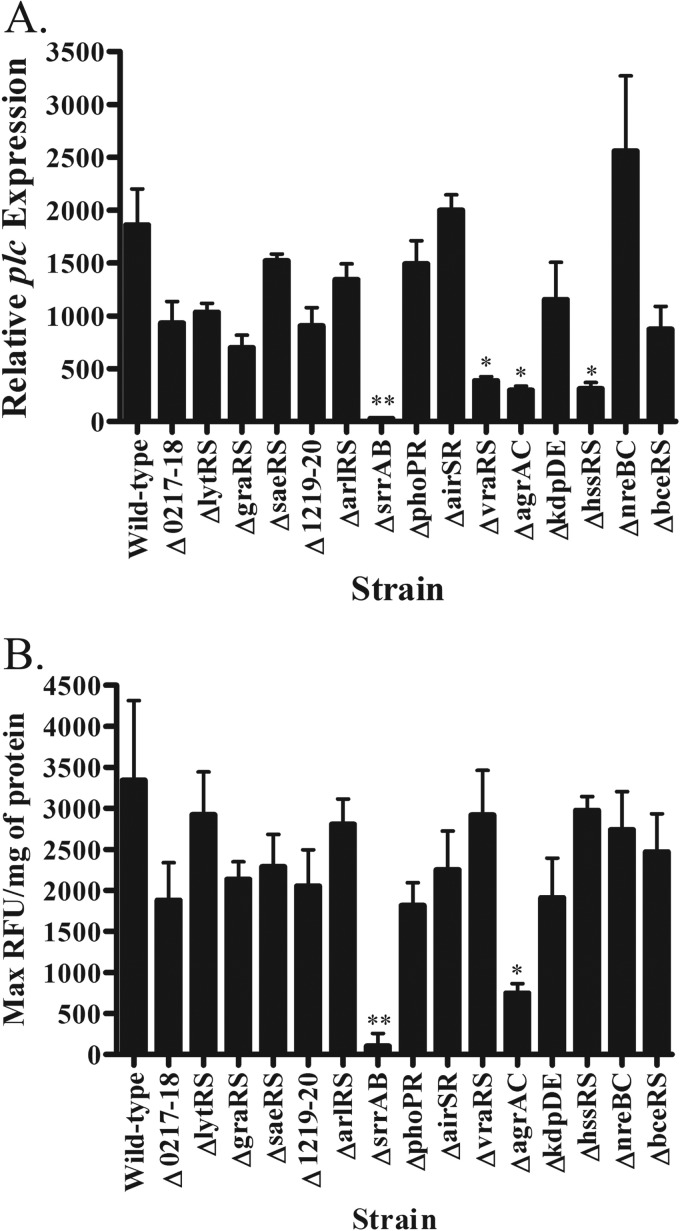
The differences in PI-PLC secretion that we observed (Fig. 1) most likely reflect the differences in the regulation of plc among the strains. Although there is evidence that the accessory gene regulator (agr) influences expression of plc (17), relatively little else is known regarding plc regulation in S. aureus. Using a library of S. aureus two-component system (TCS) mutants, we assessed the contribution of individual S. aureus TCSs to plc transcription. After determining the growth phase for maximal expression of plc (see Fig. S2A in the supplemental material), we screened TCS mutants for relative plc expression during late exponential growth (Fig. 2A). As expected from the previous study, the Δagr mutant displayed approximately 4-fold lower plc transcript levels. A similar 4-fold drop was observed in the ΔvraRS and ΔhssRS mutants. However, the SrrAB TCS had the greatest impact upon plc regulation, with the ΔsrrAB mutant displaying approximately 100-fold lower plc expression. To determine if the levels of secreted PI-PLC corresponded with the transcriptional data, we measured PI-PLC activity in culture supernatants of the TCS mutants. Both the Δagr and ΔsrrAB mutants displayed significantly less PI-PLC activity than the wild-type strain, and the ΔsrrAB strain exhibited the least amount of activity (Fig. 2B). Other observed differences in plc transcript levels, such as in the ΔvraRS and ΔhssRS mutants, were lost at the enzyme level, potentially due to the later time point used for the activity assay. Taken together, these data indicate that SrrAB was the predominant TCS transcriptional activator of plc expression, with other TCSs contributing to a lesser extent.
FIG 2.
Contribution of S. aureus TCSs to the regulation and expression of plc. (A) S. aureus wild-type and TCS mutant strains in USA300 were grown in TSB to late-log growth phase (OD550 = 1.5). The relative expression of plc in each strain was compared. The expression level of plc in each strain was normalized to that of the DNA gyrase subunit B (gyrB). (B) Normalized PI-PLC activity data obtained for each of the TCS mutants from overnight supernatants in relation to those for the wild type. These results represent the means ± SEMs from three independent experiments performed in triplicate. *, P < 0.05; **, P < 0.01.
SrrAB directly regulates plc.
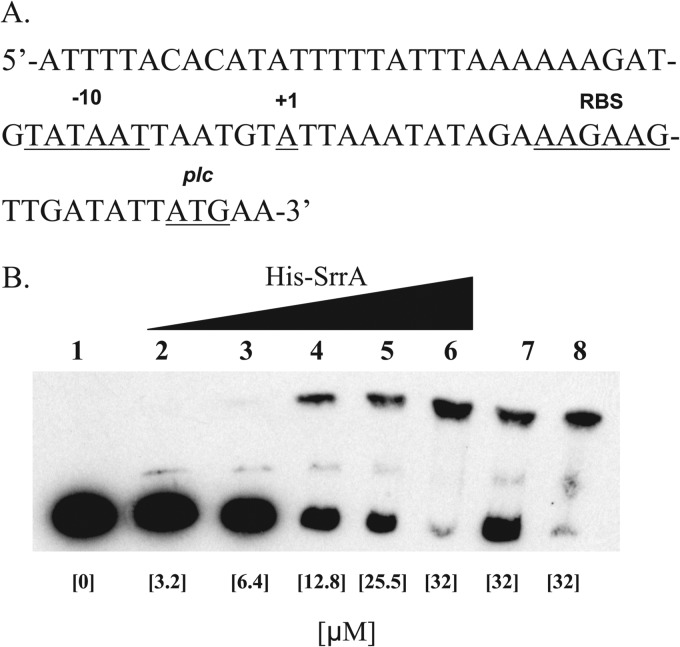
Since the SrrAB TCS had the greatest effect on plc regulation, we reasoned that SrrA would directly bind and induce transcription from the plc promoter. To test this hypothesis, we first examined the kinetics of srrA expression in broth-grown cultures and found that the pattern of expression was similar to what we observed with plc expression (see Fig. S2B in the supplemental material). Next, we mapped the plc promoter region using 5′ RACE analysis to identify a transcriptional start site located 26 bp upstream from the plc translational start site (Fig. 3A). Examination of the sequence revealed identification of a probable ribosomal binding site and a −10 promoter element present in other genes known to be regulated by SrrA (34). However, the consensus sequence for the −35 promoter element and the SrrA binding region were not identified.
FIG 3.
Characterization of the plc promoter region. (A) Depiction of the upstream region of plc. Key features include the annotated translational start site (plc), the identified transcriptional start site (+1), a predicted ribosomal binding site (RBS), and a predicted −10 promoter element (−10). (B) The region directly upstream of plc was bound by phosphorylated His-SrrA. Approximately 5 ng of probe was used for all reactions. Reaction lanes include no His-SrrA (lane 1), various amounts of His-SrrA (lanes 2 to 6), nonfluorescent specific competitor DNA (lane 7), and nonfluorescent nonspecific competitor DNA (lane 8). The concentration of His-SrrA added to each reaction lane is listed. Specific and nonspecific competitor DNA was added at a roughly 50-fold molar excess. Data are representative of those from two independent experiments.
To test for direct regulation of plc, we purified both SrrA and the kinase domain from SrrB. Using the SrrB kinase domain, we phosphorylated SrrA for use in our binding studies. EMSA analysis on the upstream region of plc demonstrated that phosphorylated His-SrrA retarded the mobility of a DNA probe proximal to the plc coding sequence that contained the predicted promoter site (Fig. 3B, lanes 2 to 6). Unphosphorylated His-SrrA failed to generate a similar probe shift (data not shown). This interaction was specific, as unlabeled DNA probe competed off labeled DNA (Fig. 3B, lane 7), whereas unlabeled nonspecific DNA did not (Fig. 3B, lane 8). Overall, these data support our hypothesis that SrrAB directly regulates plc in S. aureus.
Regulation of plc in response to in vitro and in vivo oxidative stress.
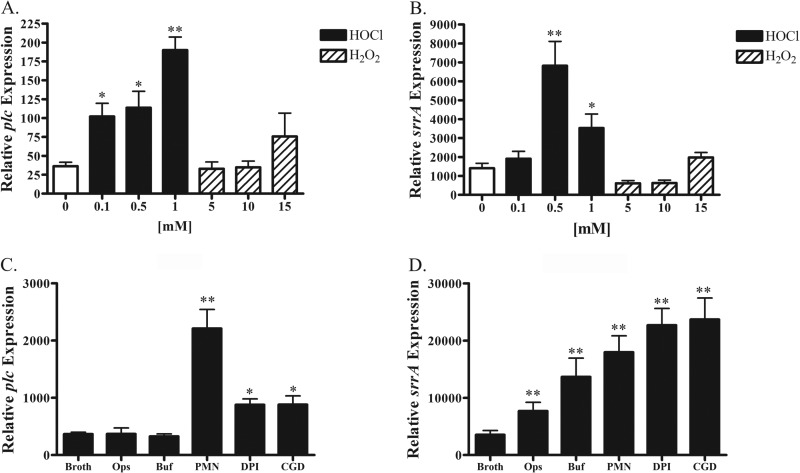
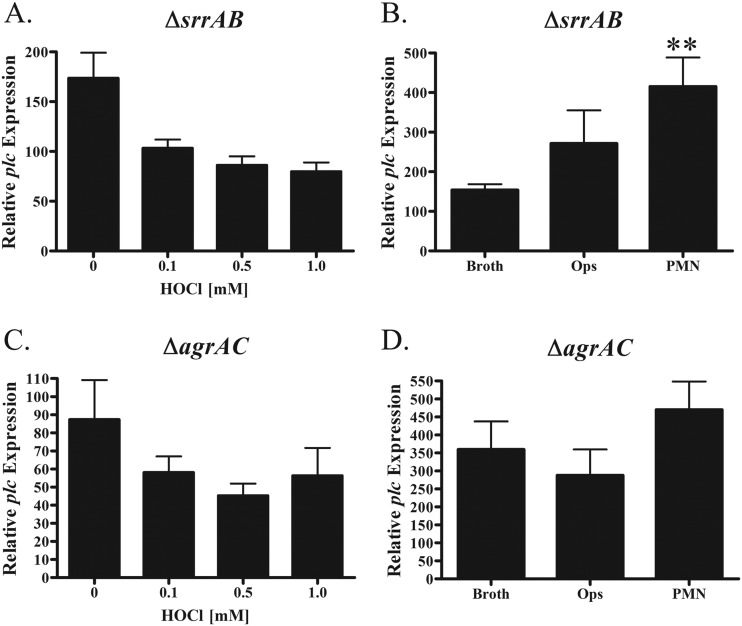
Since published reports have demonstrated that the SrrAB TCS regulates several virulence determinants in response to oxygen levels (35), we tested whether oxidants could influence plc expression in a SrrAB-dependent manner. To test this hypothesis, we used quantitative RT-PCR (qRT-PCR) to measure plc expression by a wild-type S. aureus USA300 (LAC) strain in the presence of various concentrations of either H2O2 or HOCl for 60 min. We found that plc was upregulated in a concentration-dependent manner and that this upregulation was approximately 3-fold in response to HOCl-induced stress at levels as low as 100 μM (Fig. 4A). In contrast to its sensitivity to HOCl, plc expression was not significantly upregulated by H2O2 concentrations as high as 15 mM (Fig. 4A). Similar to plc, regulation of srrA occurred in response to HOCl but not to H2O2 (Fig. 4B). Next we examined the regulation of plc and srrA in a more biologically relevant context by feeding S. aureus to human polymorphonuclear neutrophils (PMNs) and measuring transcriptional expression of intraphagosomal organisms. Whereas S. aureus phagocytosed by normal PMNs upregulated plc approximately 4-fold, organisms ingested by PMNs without a functional NADPH oxidase, as a result of either pharmacologic inhibition with DPI or inherited absence in patients with chronic granulomatous disease, increased plc expression only ∼2-fold compared to expression in broth-grown bacteria (Fig. 4C). In contrast to plc expression, srrA regulation after phagocytosis by PMNs was quite different. Opsonization with human serum alone led to a significant upregulation of srrA (Fig. 4D) by bacteria, whereas exposure to serum did not alter plc expression (Fig. 4C). In addition, srrA was upregulated in normal PMNs as well as those without a functional NADPH oxidase (Fig. 4D), whereas plc expression in DPI-treated PMNs or PMNs from CGD patients was attenuated relative to the responses in normal PMNs (Fig. 4C).
FIG 4.
Regulation of S. aureus plc and srrA in response to in vitro and in vivo oxidative stress. The expression of plc (A) or srrA (B) was compared to that in PBS-treated controls by qRT-PCR in vitro, using various concentrations of either HOCl or H2O2 as sources of oxidative stress for 60 min. In addition, levels of plc (C) or srrA (D) expression were examined in PMNs 10 min after phagocytosis. PMNs treated with DPI or obtained from a patient who suffers from CGD were compared with normal PMNs. Controls included bacteria grown in broth, bacteria that had undergone 20 min of opsonization (Ops), and bacteria that had been resuspended in the PMN buffer for 10 min (Buf). For statistical analysis, all comparisons are against the broth-grown controls. The levels of plc and srrA expression were normalized to the level of expression of the DNA gyrase subunit B (gyrB). All results represent the means ± SEMs from three independent experiments. *, P < 0.05; **, P < 0.01.
We demonstrated that the SrrAB and agr TCSs regulate plc expression (Fig. 2), but the contribution of these regulators to oxidant-dependent induction is not clear. Although plc expression was down 100-fold in the ΔsrrAB background (Fig. 2A), transcript levels were measurable and the effects of HOCl exposure and PMN ingestion could be assessed. At concentrations up to 1 mM, HOCl did not trigger plc upregulation in the ΔsrrAB mutant (Fig. 5A), but unexpectedly, plc was still upregulated following ingestion by normal PMNs (Fig. 5B). The same approach was taken with the Δagr mutant, and again, we found that plc was not upregulated in response to HOCl (Fig. 5C). In addition, the Δagr mutant was unable to upregulate plc following PMN ingestion (Fig. 5D), suggesting that the agr system might be an important factor contributing to SrrAB-independent plc upregulation in PMNs (Fig. 5B). Overall, these data indicate that plc regulation is complex and multifactorial, with the SrrAB and agr systems being significant contributors to the induction mechanism in the presence of specific sources of oxidative stress.
FIG 5.
Contribution of SrrAB and AgrAC TCSs to plc regulation. The relative expression of plc was examined in an S. aureus USA300 ΔsrrAB background (A) or a Δagr background (C) after 60 min of exposure to PBS or various concentrations of HOCl. In addition, the relative expression of plc in PMNs at 10 min postphagocytosis was examined in an S. aureus USA300 ΔsrrAB background (B) or Δagr background (D). Controls included broth-grown bacteria and bacteria that have been opsonized for 20 min (Ops). The level of plc expression was normalized to that of the DNA gyrase subunit B (gyrB), and results represent the means ± SEMs from three independent experiments. **, P < 0.01.
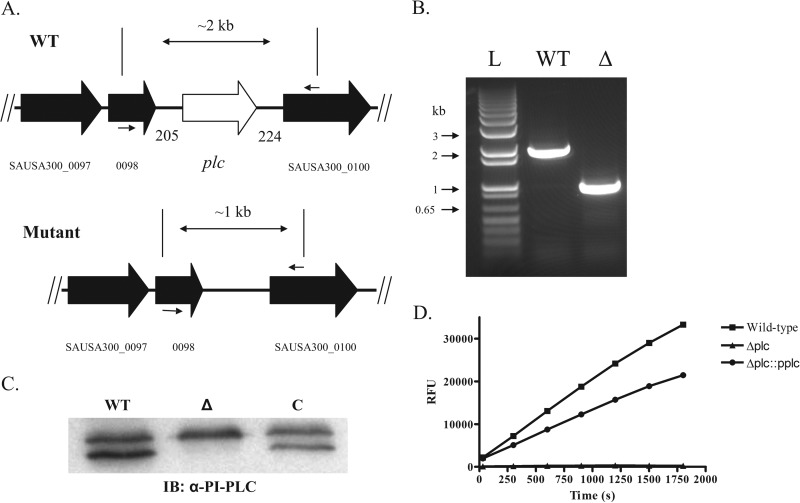
The S. aureus Δplc strain exhibits decreased viability in human blood.
To test the contribution of PI-PLC in the pathogenesis of staphylococcal disease, we constructed a strain with a markerless deletion of the plc gene (Fig. 6A). Work was performed in the S. aureus USA300 (LAC) background, and the deletion of the plc gene was verified by PCR (Fig. 6B), immunoblotting (Fig. 6C), and activity assay (Fig. 6D). The deletion did not produce a general growth defect, as all organisms grew identically in broth culture (see Fig. S3A in the supplemental material) and did not interfere with ingestion by neutrophils, as the phagocytosis of the wild type and mutant was the same (see Fig. S3B in the supplemental material).
FIG 6.
Construction of the Δplc mutant in S. aureus. (A) Chromosomal organization of plc and surrounding genes in an S. aureus USA300 background. The primer sets used for determining genomic deletion are shown. (B) Gel depicting the PCRs run for potential Δplc mutants using the primer set depicted in panel A. Lanes: L, ladder; WT, S. aureus USA300 wild-type genomic DNA; Δ, S. aureus USA300 Δplc genomic DNA. (C) Immunoblot of S. aureus derivative supernatants using goat polyclonal antibody against PI-PLC. Lanes: WT, wild type; Δ, Δplc mutant; C, complemented strain. (D) PI-PLC activity for S. aureus derivatives determined by PI-PLC assay for 30 min. Results for panel D represent the means from three independent experiments performed in triplicate.
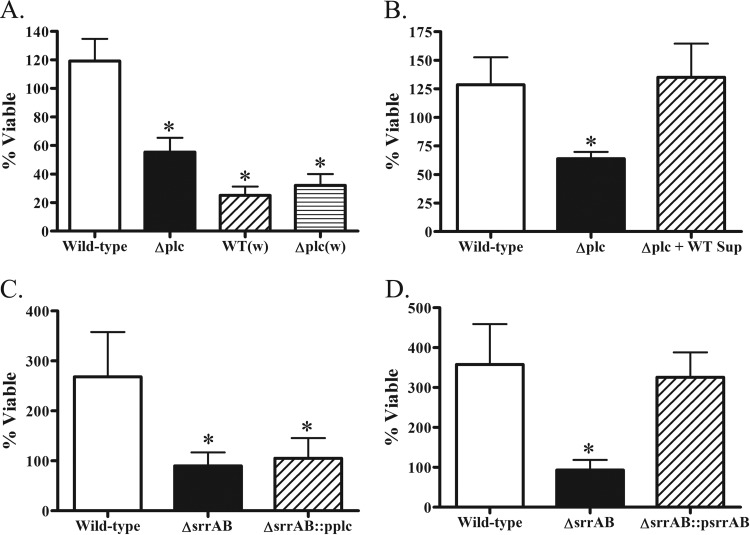
We compared the relative viability of the USA300 (LAC) wild type and Δplc mutant in human blood, using bacteria resuspended in either conditioned culture medium or sterile medium. When suspended in its own conditioned medium, the wild-type strain had moderate growth after 3 h, whereas the Δplc mutant exhibited a significant survival defect, with only about 60% of the initial inoculum being viable after 3 h in human blood (Fig. 7A). However, the difference in viability of the mutant was seen only with stationary-phase bacteria, as exponentially growing bacteria displayed no observable phenotype compared to the wild-type strain (data not shown). Furthermore, there was no difference in viability between the wild-type strain and the Δplc mutant strain when bacteria were washed and resuspended in sterile TSB, though both strains displayed significant defects in viability compared to the wild-type strain suspended in its own conditioned medium (Fig. 7A). Exogenous complementation of plc using supernatant obtained from an overnight culture of wild-type bacteria completely restored the viability phenotype to wild-type levels (Fig. 7B).
FIG 7.
Viability of S. aureus derivatives in whole-blood assays. The viability of S. aureus USA300 Δplc (A and B; stationary phase), ΔsrrAB (C; exponential phase), or ΔsrrAB (D; exponential phase) strains overexpressing PI-PLC exposed to human whole blood was calculated as the number of CFU recovered after 3 h of exposure divided by the number of CFU in the initial bacterial challenge. The numbers of CFU were determined by serial dilution and plating on TSA plates. One set of experiments (A) compared the viability in human blood of the wild-type strain and the Δplc mutant suspended in their own conditioned media or washed (w) and suspended in sterile culture medium. The strains used in experiments involving complementation harbored either control plasmids or the complementing plasmid. The results represent the means ± SEMs for three independent experiments. *, P < 0.05.
Since the SrrAB TCS directly regulated plc (Fig. 2 and 3), we tested the viability of the ΔsrrAB mutant strain in human blood. In agreement with previous studies of SrrAB function in animal models of infection (34, 36), growth of the ΔsrrAB mutant was significantly impaired relative to that of the isogenic parental strain, and the wild-type phenotype was completely restored by complementation (Fig. 7C). However, only exponentially growing bacteria demonstrated differences in viability, as wild-type and ΔsrrAB mutant strains in stationary phase had similar survival (data not shown). As SrrAB regulates many genes, we tested whether or not constitutively secreted PI-PLC could rescue the viability phenotype of the ΔsrrAB mutant. However, a ΔsrrAB strain secreting large amounts of PI-PLC relative to the amount secreted by the wild type (see Fig. S4 in the supplemental material) did not restore the survival defect of the ΔsrrAB mutant (Fig. 7D), suggesting that the ΔsrrAB strain lacked factors in addition to PI-PLC that support survival in human blood.
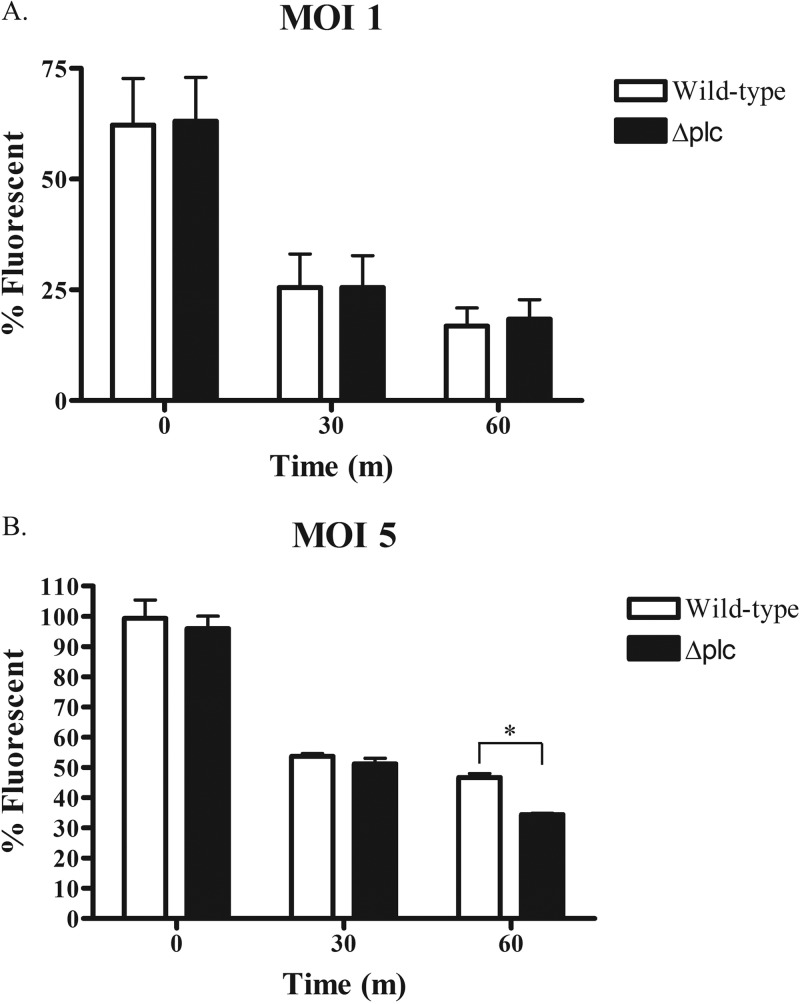
Since whole blood is a complex mixture of cellular and soluble components, we focused our attention on the contributions of PMNs to the phenotype of the Δplc mutant in blood. PMNs are an obvious choice for study in this context, as they are the predominant nucleated cell in human blood and one of the first immune cells encountered in bloodstream infections. We previously demonstrated that oxidant-mediated killing of S. aureus correlates with bleaching of sGFP at late time points after phagocytosis by PMNs (33) and employed this approach to assess relative survival. We observed no differences in the loss of fluorescence between strains ingested by PMNs at an MOI of 1:1 (Fig. 8A). Uptake of the different S. aureus strains was the same for all strains when examined either by flow cytometry (see Fig. S3B in the supplemental material) or by microscopy (data not shown).
FIG 8.
Survival of S. aureus Δplc strains in PMNs. PMNs were challenged with wild-type or Δplc mutant S. aureus USA300 strains grown to mid-log phase at an MOI of either 1:1 (A) or 5:1 (B). Survival was determined by measuring the mean fluorescence index (MFI) of PMNs that had been challenged with GFP-expressing S. aureus derivatives. The results represent the means ± SEMs from at least three independent experiments. Times are in minutes (m). *, P < 0.05.
We previously demonstrated that the oxidant-dependent killing and bleaching of ingested sGFP-expressing S. aureus by human PMNs are less effective as the MOI increases; PMNs kill and bleach organisms less efficiently at an MOI of 5:1 than at one of 1:1 (37). Reasoning that taxing the antimicrobial capacity of PMNs might better uncover phenotypic differences in the fate of wild-type versus Δplc strains, we compared the intracellular fate of wild-type versus mutant S. aureus using an MOI of 5:1. At this MOI, monitoring of bleaching of sGFP as an indicator of loss of viability demonstrated a significant difference in the loss of fluorescence at the 60-min time point, 47% ± 1.3% and 34% ± 0.4% (n = 3) for the wild-type and Δplc mutant strains, respectively (Fig. 8B). These data suggest that PI-PLC is important for the survival of S. aureus in human blood, explained in part by the decreased viability of the Δplc mutant in PMNs.
S. aureus Δplc strains are not attenuated in a mouse model of infection.
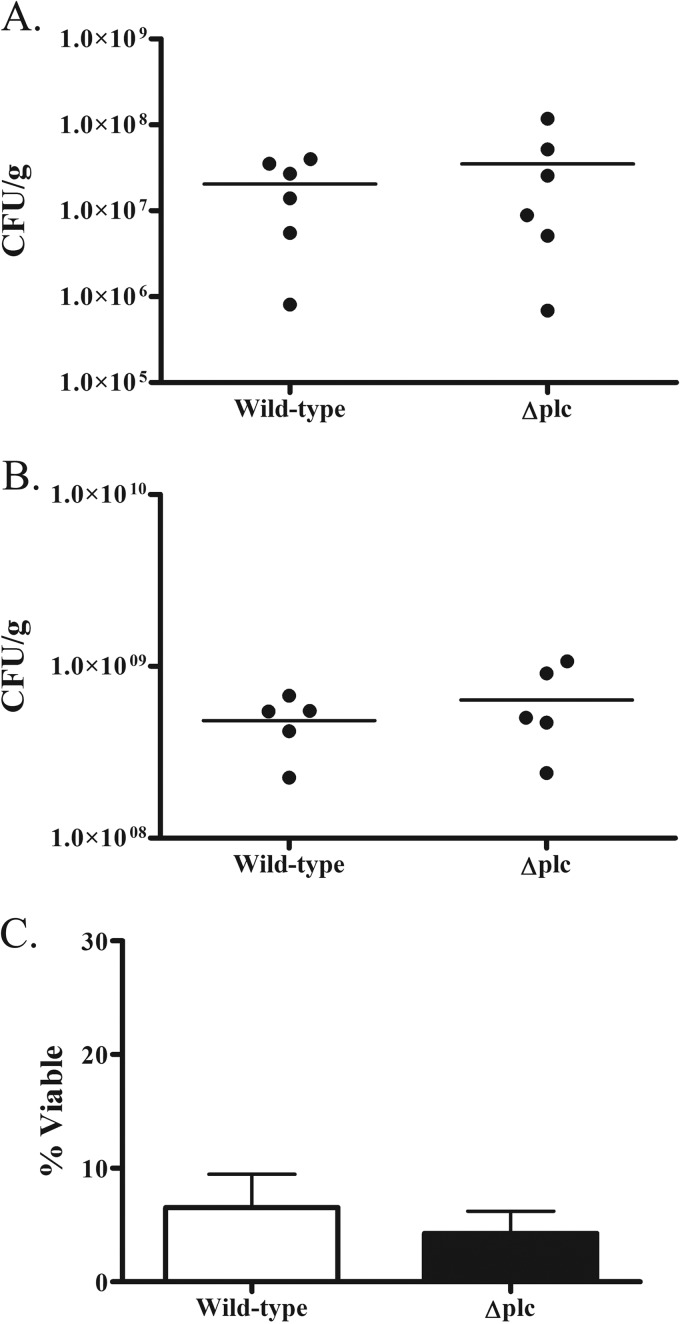
Given that the Δplc strains in their conditioned media exhibited a viability phenotype in human whole blood (Fig. 7A) and ΔsrrAB mutants were attenuated in a murine model of S. aureus infection (36), we tested whether infection in a murine model with the Δplc mutant would be less severe or widespread. However, similar numbers of viable bacteria were recovered from mice challenged with the USA300 or the Δplc mutant strain, whether in a model of systemic infection (Fig. 9A) or a model of skin infection (Fig. 9B). In addition, no differences in lethality were observed between these strains (data not shown), and we were unable to recapitulate the human-whole-blood viability phenotype in the murine system using stationary-phase bacteria, as the Δplc strain grew as well as the wild-type strain in mouse whole blood (Fig. 9C). Also of note, the viability of the wild-type strain in mouse whole blood (Fig. 9C) was strikingly different from that in human whole blood (Fig. 7). Although these data suggest that PI-PLC is not required for full virulence of S. aureus in mice, the murine system did not mirror the interaction of staphylococcal PI-PLC with human blood.
FIG 9.
Challenge of mice with S. aureus USA300 Δplc strains. (A) BALB/c mice were challenged with a bolus of 5 × 106 CFU of wild-type or Δplc mutant S. aureus by tail vein injection. Kidneys were harvested at day 8 postinfection for determination of the number of CFU per gram of tissue. (B) BALB/c mice were shaved and challenged subcutaneously with a bolus of 1 × 107 CFU of wild-type or Δplc mutant S. aureus. A 10-mm punch biopsy specimen was excised from lesions at day 4 postinfection for determination of the number of CFU per gram of tissue. (C) The viability of Δplc strains exposed to mouse whole blood was determined by dividing the number of bacteria recovered after 3 h of exposure to the initial bacterial challenge. The numbers of CFU were determined by serial dilution and plating on TSA plates. The results in panel C represent the means ± SEMs for three independent experiments.
DISCUSSION
Staphylococcal PI-PLC is a secreted enzyme capable of hydrolyzing PI and cleaving GPI-linked proteins from the surface of cells (17) and, like many other exoproteins, is regulated by the agr quorum-sensing system (17). Levels of secreted PI-PLC vary among S. aureus strains (17, 38), and a previous study correlated the presence of secreted PI-PLC and the occurrence of more severe disease, thereby suggesting a role for PI-PLC as a virulence determinant (38). However, evidence that PI-PLC is a virulence factor and elucidation of its impact on human host defenses are lacking. In this study, we provide novel insights into the biology of PI-PLC with respect to its regulation in S. aureus and to its expression in response to oxidants and components of human host defense.
The basis for differences in PI-PLC secretion among isolates of S. aureus is not completely understood. CA-MRSA strains, particularly those of the USA300 lineage (39, 40), constitutively exhibit high levels of activity of the agr quorum-sensing system (14, 16), which modulates expression of many virulence factors that contribute to the severe disease seen with these strains. Consistent with the known relationship between agr and virulence properties, we observed the highest levels of plc expression and PI-PLC activity in USA300 strains (Fig. 1). Conversely, HA-MRSA and USA200 isolates, such as COL and UAMS-1, are known to have weaker agr function (14, 41) and, not surprisingly, exhibited reduced PI-PLC levels (Fig. 1). However, there was residual plc expression and PI-PLC activity in the agr-deficient mutant (Fig. 2), suggesting the presence of additional transcriptional regulators.
By screening a collection of TCS mutants, we identified SrrAB as the main transcriptional activator of plc gene expression (Fig. 2). The SrrAB TCS is a regulatory element conserved among staphylococcal species that controls the expression of virulence factors, including agr, tstH, spa, and ica (26, 34, 35). Ambient oxygen level is one of the environmental signals that modulate SrrAB expression, with microaerobic conditions augmenting SrrAB expression and suppressing RNAIII and other virulence factors (35, 42). We examined plc and srrA expression in response to oxidant stress by challenging USA300 with various concentrations of the reagents H2O2 and HOCl. Although both oxidants triggered upregulation of plc and srrA, HOCl was ∼150-fold more potent on a molar basis than H2O2 (Fig. 4). Because HOCl is the predominant antimicrobial oxidant generated in the phagosomes of human PMNs (43), we anticipated that both plc and srrA would be upregulated in phagocytosed S. aureus. As expected, plc expression increased when staphylococci were ingested by normal PMNs but increased to a lesser extent when they were phagocytosed by PMNs that lacked a functioning oxidase and, consequently, a means to generate HOCl. In contrast, srrA expression increased in PMNs regardless of the presence of oxidants. In fact, expression was greater in DPI-treated neutrophils or neutrophils from CGD patients than in normal PMNs. Furthermore, expression of srrA increased during opsonization of S. aureus and before it was fed to PMNs. These data suggest that stresses on S. aureus in addition to oxidants, such as complement fixation during opsonization, likely contribute to the enhanced levels of SrrAB in the phagosome. Given that binding of SrrA to the plc promoter region needed a relatively large amount of protein to achieve a noticeable shift (Fig. 3), it is possible that SrrA requires a regulatory partner for effective interaction with and transcriptional induction of the plc promoter. Such a prerequisite could explain the selective regulation of plc in response to oxidative stress, whereas additional stimuli influence srrA expression. Taken together, our findings indicate that oxidant stress triggered plc upregulation in a SrrAB-dependent manner and that additional environmental factors contributed to srrA expression in S. aureus.
If PI-PLC is a staphylococcal virulence factor, we reasoned that it might provide a survival advantage to S. aureus in its interactions with elements of host defense. PI-PLC promoted survival of ingested staphylococci in human PMNs at an MOI of 5:1, thereby suggesting that PI-PLC counteracted in part the antimicrobial action of human PMNs. We employed a human-whole-blood model to determine if PI-PLC influenced the outcome of host-microbe interactions that might arise in the circulation during infection. The S. aureus Δplc mutant survived in human whole blood less well than the isogenic wild type, but only for bacteria in stationary phase, when extracellular PI-PLC accumulation was greatest, and only in the presence of its conditioned culture medium. These data suggest that PI-PLC may promote disease during the later stages of infection or in clinical settings where the exoprotein secreted by S. aureus could accumulate. Given the enhanced secretion of PI-PLC by USA300 strains that typically cause skin and soft tissue infections, it may be important for the survival or maintenance of S. aureus in abscesses, an environment with low oxygen tension and microaerobic conditions (reviewed in reference 44) that would promote expression of SrrAB and, consequently, PI-PLC.
Since the SrrAB TCS has been implicated as a virulence factor in both rabbit and murine models of staphylococcal infection (34, 36), its regulation of plc may contribute to disease pathogenesis. However, in contrast to the behavior of the Δplc mutant, the ΔsrrAB mutant exhibited its viability phenotype only during exponential growth and not during stationary phase. Moreover, overexpression of PI-PLC failed to rescue the ΔsrrAB-dependent phenotype of S. aureus in whole blood, suggesting that the ΔsrrAB mutant has a much broader range of defects, as expected, given its role as a global gene regulator.
The Δplc mutant did not demonstrate a survival phenotype in two mouse models of infection, despite the survival defect observed for the Δplc strain in the human-whole-blood infection model and in isolated human PMNs. A mouse-whole-blood infection assay likewise failed to recapitulate a plc-dependent phenotype. Although the basis for the lack of a phenotype in murine blood is unknown, recent evidence demonstrated that the inflammatory response of mice differs greatly from that of humans (45). Furthermore, murine neutrophils differ from human neutrophils with respect to several attributes important for antimicrobial action (46–48), including intracellular signaling pathways mediated through phosphoinositide 3-kinase (PI3K) (49). The phosphoinositide-dependent signaling pathway would be especially relevant to links between PI-PLC and human disease, given the role of PI3K in the oxidase-dependent production of reactive oxygen species by PMNs and the potential for disruption by staphylococcal PI-PLC. The failure of the murine system to mirror the pathogenic effects of staphylococcal PI-PLC seen in human blood or isolated PMNs reinforces the recent recognition that the mouse is not an informative model system to study some human diseases and pathogenic processes.
In light of our data implicating PI-PLC as a staphylococcal virulence factor, the molecular targets for PI-PLC in PMNs now need to be identified. Since staphylococcal PI-PLC is approximately 27 to 39% identical to the PI-PLC found in Listeria and Bacillus species (17), studies from these organisms might shed some insight into the function of the enzyme in S. aureus. The PI-PLC of L. monocytogenes promotes disease by modulating activation of the phagocyte NADPH oxidase (50, 51) and by disrupting membrane-bound compartments (52), thereby releasing viable listeria into the host cell cytoplasm. In contrast to the enzyme in L. monocytogenes, staphylococcal PI-PLC lacks cytolytic activity (53), and S. aureus does not escape into the neutrophil cytoplasm after phagocytosis (54). In addition, unlike the PI-PLC in L. monocytogenes, staphylococcal PI-PLC can cleave GPI-linked proteins that may influence signaling cascades in immune cells. Immunomodulation of signal transduction pathways has been implicated in the observed suppression of the immune response of dendritic cells and T cells by PI-PLC secreted from Bacillus anthracis (20). Additional studies of staphylococcal PI-PLC are needed to define the full range of its enzymatic activity and to identify the targets on cells that alter subsequent host responses.
Overall, the data presented here elucidate some of the regulatory factors that influence PI-PLC expression, demonstrate that PI-PLC promoted survival of S. aureus in human whole blood and PMNs, and provide evidence that PI-PLC may contribute to the virulence of S. aureus. The relative prominence of PI-PLC among the virulent exoproteins of S. aureus, the full repertoire of its regulatory factors, and its relationship to SrrAB remain to be defined.
Supplementary Material
ACKNOWLEDGMENTS
We express special thanks to the members of the W. M. Nauseef and A. R. Horswill labs for their help in these studies and members of Fayyaz Sutterwala's lab for help in performing the animal studies. We thank Barry Kreiswirth for providing the USA600 strain.
We acknowledge funding from National Institute of Allergy and Infectious Diseases (NIAID) grant AI078921 (to A.R.H.), grants AI070958 and AI044642 (to W.M.N.), and T32 training grant AI007343 (to M.J.W.). The W. M. Nauseef lab is also supported by a merit review award and use of facilities at the Iowa City Department of Veterans Affairs (VA) Medical Center, Iowa City, IA.
Footnotes
Published ahead of print 22 January 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/IAI.01168-13.
REFERENCES
- 1.Hidron AI, Edwards JR, Patel J, Horan TC, Sievert DM, Pollock DA, Fridkin SK, National Healthcare Safety Network Team, Participating National Healthcare Safety Network Facilities 2008. NHSN annual update: antimicrobial-resistant pathogens associated with healthcare-associated infections: annual summary of data reported to the National Healthcare Safety Network at the Centers for Disease Control and Prevention, 2006-2007. Infect. Control Hosp. Epidemiol. 29:996–1011. 10.1086/591861 [DOI] [PubMed] [Google Scholar]
- 2.Wertheim HF, Melles DC, Vos MC, van Leeuwen W, van Belkum A, Verbrugh HA, Nouwen JL. 2005. The role of nasal carriage in Staphylococcus aureus infections. Lancet Infect. Dis. 5:751–762. 10.1016/S1473-3099(05)70295-4 [DOI] [PubMed] [Google Scholar]
- 3.Drago L, De Vecchi E, Nicola L, Gismondo MR. 2007. In vitro evaluation of antibiotics' combinations for empirical therapy of suspected methicillin resistant Staphylococcus aureus severe respiratory infections. BMC Infect. Dis. 7:111. 10.1186/1471-2334-7-111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dulon M, Haamann F, Peters C, Schablon A, Nienhaus A. 2011. MRSA prevalence in European healthcare settings: a review. BMC Infect. Dis. 11:138. 10.1186/1471-2334-11-138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.King MD, Humphrey BJ, Wang YF, Kourbatova EV, Ray SM, Blumberg HM. 2006. Emergence of community-acquired methicillin-resistant Staphylococcus aureus USA 300 clone as the predominant cause of skin and soft-tissue infections. Ann. Intern. Med. 144:309–317. 10.7326/0003-4819-144-5-200603070-00005 [DOI] [PubMed] [Google Scholar]
- 6.Francis JS, Doherty MC, Lopatin U, Johnston CP, Sinha G, Ross T, Cai M, Hansel NN, Perl T, Ticehurst JR, Carroll K, Thomas DL, Nuermberger E, Bartlett JG. 2005. Severe community-onset pneumonia in healthy adults caused by methicillin-resistant Staphylococcus aureus carrying the Panton-Valentine leukocidin genes. Clin. Infect. Dis. 40:100–107. 10.1086/427148 [DOI] [PubMed] [Google Scholar]
- 7.Collins J, Rudkin J, Recker M, Pozzi C, O'Gara JP, Massey RC. 2010. Offsetting virulence and antibiotic resistance costs by MRSA. ISME J. 4:577–584. 10.1038/ismej.2009.151 [DOI] [PubMed] [Google Scholar]
- 8.D'Agata EM, Webb GF, Horn MA, Moellering RC, Jr, Ruan S. 2009. Modeling the invasion of community-acquired methicillin-resistant Staphylococcus aureus into hospitals. Clin. Infect. Dis. 48:274–284. 10.1086/595844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang R, Braughton KR, Kretschmer D, Bach TH, Queck SY, Li M, Kennedy AD, Dorward DW, Klebanoff SJ, Peschel A, DeLeo FR, Otto M. 2007. Identification of novel cytolytic peptides as key virulence determinants for community-associated MRSA. Nat. Med. 13:1510–1514. 10.1038/nm1656 [DOI] [PubMed] [Google Scholar]
- 10.Larkin EA, Carman RJ, Krakauer T, Stiles BG. 2009. Staphylococcus aureus: the toxic presence of a pathogen extraordinaire. Curr. Med. Chem. 16:4003–4019. 10.2174/092986709789352321 [DOI] [PubMed] [Google Scholar]
- 11.Bergdoll MS, Crass BA, Reiser RF, Robbins RN, Davis JP. 1981. A new staphylococcal enterotoxin, enterotoxin F, associated with toxic-shock-syndrome Staphylococcus aureus isolates. Lancet i:1017–1021 [DOI] [PubMed] [Google Scholar]
- 12.Schlievert PM, Shands KN, Dan BB, Schmid GP, Nishimura RD. 1981. Identification and characterization of an exotoxin from Staphylococcus aureus associated with toxic-shock syndrome. J. Infect. Dis. 143:509–516. 10.1093/infdis/143.4.509 [DOI] [PubMed] [Google Scholar]
- 13.Krell T, Lacal J, Busch A, Silva-Jimenez H, Guazzaroni ME, Ramos JL. 2010. Bacterial sensor kinases: diversity in the recognition of environmental signals. Annu. Rev. Microbiol. 64:539–559. 10.1146/annurev.micro.112408.134054 [DOI] [PubMed] [Google Scholar]
- 14.Li M, Diep BA, Villaruz AE, Braughton KR, Jiang X, DeLeo FR, Chambers HF, Lu Y, Otto M. 2009. Evolution of virulence in epidemic community-associated methicillin-resistant Staphylococcus aureus. Proc. Natl. Acad. Sci. U. S. A. 106:5883–5888. 10.1073/pnas.0900743106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Peng HL, Novick RP, Kreiswirth B, Kornblum J, Schlievert P. 1988. Cloning, characterization, and sequencing of an accessory gene regulator (agr) in Staphylococcus aureus. J. Bacteriol. 170:4365–4372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tsuji BT, Rybak MJ, Cheung CM, Amjad M, Kaatz GW. 2007. Community- and health care-associated methicillin-resistant Staphylococcus aureus: a comparison of molecular epidemiology and antimicrobial activities of various agents. Diagn. Microbiol. Infect. Dis. 58:41–47. 10.1016/j.diagmicrobio.2006.10.021 [DOI] [PubMed] [Google Scholar]
- 17.Daugherty S, Low MG. 1993. Cloning, expression, and mutagenesis of phosphatidylinositol-specific phospholipase C from Staphylococcus aureus: a potential staphylococcal virulence factor. Infect. Immun. 61:5078–5089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Camilli A, Goldfine H, Portnoy DA. 1991. Listeria monocytogenes mutants lacking phosphatidylinositol-specific phospholipase C are avirulent. J. Exp. Med. 173:751–754. 10.1084/jem.173.3.751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Callegan MC, Cochran DC, Kane ST, Gilmore MS, Gominet M, Lereclus D. 2002. Contribution of membrane-damaging toxins to Bacillus endophthalmitis pathogenesis. Infect. Immun. 70:5381–5389. 10.1128/IAI.70.10.5381-5389.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zenewicz LA, Wei Z, Goldfine H, Shen H. 2005. Phosphatidylinositol-specific phospholipase C of Bacillus anthracis down-modulates the immune response. J. Immunol. 174:8011–8016 [DOI] [PubMed] [Google Scholar]
- 21.Burlak C, Hammer CH, Robinson MA, Whitney AR, McGavin MJ, Kreiswirth BN, Deleo FR. 2007. Global analysis of community-associated methicillin-resistant Staphylococcus aureus exoproteins reveals molecules produced in vitro and during infection. Cell. Microbiol. 9:1172–1190. 10.1111/j.1462-5822.2006.00858.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Holtfreter S, Nguyen TT, Wertheim H, Steil L, Kusch H, Truong QP, Engelmann S, Hecker M, Volker U, van Belkum A, Broker BM. 2009. Human immune proteome in experimental colonization with Staphylococcus aureus. Clin. Vaccine Immunol. 16:1607–1614. 10.1128/CVI.00263-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Beining PR, Huff E, Prescott B, Theodore TS. 1975. Characterization of the lipids of mesosomal vesicles and plasma membranes from Staphylococcus aureus. J. Bacteriol. 121:137–143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schenk S, Laddaga RA. 1992. Improved method for electroporation of Staphylococcus aureus. FEMS Microbiol. Lett. 73:133–138 [DOI] [PubMed] [Google Scholar]
- 25.Novick RP. 1991. Genetic systems in staphylococci. Methods Enzymol. 204:587–636. 10.1016/0076-6879(91)04029-N [DOI] [PubMed] [Google Scholar]
- 26.Ulrich M, Bastian M, Cramton SE, Ziegler K, Pragman AA, Bragonzi A, Memmi G, Wolz C, Schlievert PM, Cheung A, Doring G. 2007. The staphylococcal respiratory response regulator SrrAB induces ica gene transcription and polysaccharide intercellular adhesin expression, protecting Staphylococcus aureus from neutrophil killing under anaerobic growth conditions. Mol. Microbiol. 65:1276–1287. 10.1111/j.1365-2958.2007.05863.x [DOI] [PubMed] [Google Scholar]
- 27.Anderson D, Fodge D, Hsiao HY, Liu L. November 2004. Enzyme treatment. US patent 20,040,223,961 A1
- 28.Bose JL, Fey PD, Bayles KW. 2013. Genetic tools to enhance the study of gene function and regulation in Staphylococcus aureus. Appl. Environ. Microbiol. 79:2218–2224. 10.1128/AEM.00136-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bae T, Schneewind O. 2006. Allelic replacement in Staphylococcus aureus with inducible counter-selection. Plasmid 55:58–63. 10.1016/j.plasmid.2005.05.005 [DOI] [PubMed] [Google Scholar]
- 30.Pang YY, Schwartz J, Thoendel M, Ackermann LW, Horswill AR, Nauseef WM. 2010. agr-dependent interactions of Staphylococcus aureus USA300 with human polymorphonuclear neutrophils. J. Innate Immun. 2:546–559. 10.1159/000319855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nauseef WM. 2007. Isolation of human neutrophils from venous blood. Methods Mol. Biol. 412:15–20. 10.1007/978-1-59745-467-4_2 [DOI] [PubMed] [Google Scholar]
- 32.Ellis JA, Mayer SJ, Jones OT. 1988. The effect of the NADPH oxidase inhibitor diphenyleneiodonium on aerobic and anaerobic microbicidal activities of human neutrophils. Biochem. J. 251:887–891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schwartz J, Leidal KG, Femling JK, Weiss JP, Nauseef WM. 2009. Neutrophil bleaching of GFP-expressing staphylococci: probing the intraphagosomal fate of individual bacteria. J. Immunol. 183:2632–2641. 10.4049/jimmunol.0804110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pragman AA, Yarwood JM, Tripp TJ, Schlievert PM. 2004. Characterization of virulence factor regulation by SrrAB, a two-component system in Staphylococcus aureus. J. Bacteriol. 186:2430–2438. 10.1128/JB.186.8.2430-2438.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yarwood JM, McCormick JK, Schlievert PM. 2001. Identification of a novel two-component regulatory system that acts in global regulation of virulence factors of Staphylococcus aureus. J. Bacteriol. 183:1113–1123. 10.1128/JB.183.4.1113-1123.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Richardson AR, Dunman PM, Fang FC. 2006. The nitrosative stress response of Staphylococcus aureus is required for resistance to innate immunity. Mol. Microbiol. 61:927–939. 10.1111/j.1365-2958.2006.05290.x [DOI] [PubMed] [Google Scholar]
- 37.Femling JK, Nauseef WM, Weiss JP. 2005. Synergy between extracellular group IIA phospholipase A2 and phagocyte NADPH oxidase in digestion of phospholipids of Staphylococcus aureus ingested by human neutrophils. J. Immunol. 175:4653–4661 [DOI] [PubMed] [Google Scholar]
- 38.Marques MB, Weller PF, Parsonnet J, Ransil BJ, Nicholson-Weller A. 1989. Phosphatidylinositol-specific phospholipase C, a possible virulence factor of Staphylococcus aureus. J. Clin. Microbiol. 27:2451–2454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Loughman JA, Fritz SA, Storch GA, Hunstad DA. 2009. Virulence gene expression in human community-acquired Staphylococcus aureus infection. J. Infect. Dis. 199:294–301. 10.1086/595982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Montgomery CP, Boyle-Vavra S, Adem PV, Lee JC, Husain AN, Clasen J, Daum RS. 2008. Comparison of virulence in community-associated methicillin-resistant Staphylococcus aureus pulsotypes USA300 and USA400 in a rat model of pneumonia. J. Infect. Dis. 198:561–570. 10.1086/590157 [DOI] [PubMed] [Google Scholar]
- 41.Cassat J, Dunman PM, Murphy E, Projan SJ, Beenken KE, Palm KJ, Yang SJ, Rice KC, Bayles KW, Smeltzer MS. 2006. Transcriptional profiling of a Staphylococcus aureus clinical isolate and its isogenic agr and sarA mutants reveals global differences in comparison to the laboratory strain RN6390. Microbiology 152:3075–3090. 10.1099/mic.0.29033-0 [DOI] [PubMed] [Google Scholar]
- 42.Throup JP, Zappacosta F, Lunsford RD, Annan RS, Carr SA, Lonsdale JT, Bryant AP, McDevitt D, Rosenberg M, Burnham MK. 2001. The srhSR gene pair from Staphylococcus aureus: genomic and proteomic approaches to the identification and characterization of gene function. Biochemistry 40:10392–10401. 10.1021/bi0102959 [DOI] [PubMed] [Google Scholar]
- 43.Klebanoff SJ, Kettle AJ, Rosen H, Winterbourn CC, Nauseef WM. 2013. Myeloperoxidase: a front-line defender against phagocytosed microorganisms. J. Leukoc. Biol. 93:185–198. 10.1189/jlb.0712349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cheng AG, DeDent AC, Schneewind O, Missiakas D. 2011. A play in four acts: Staphylococcus aureus abscess formation. Trends Microbiol. 19:225–232. 10.1016/j.tim.2011.01.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Seok J, Warren HS, Cuenca AG, Mindrinos MN, Baker HV, Xu W, Richards DR, McDonald-Smith GP, Gao H, Hennessy L, Finnerty CC, Lopez CM, Honari S, Moore EE, Minei JP, Cuschieri J, Bankey PE, Johnson JL, Sperry J, Nathens AB, Billiar TR, West MA, Jeschke MG, Klein MB, Gamelli RL, Gibran NS, Brownstein BH, Miller-Graziano C, Calvano SE, Mason PH, Cobb JP, Rahme LG, Lowry SF, Maier RV, Moldawer LL, Herndon DN, Davis RW, Xiao W, Tompkins RG, Inflammation Host Response to Injury, Large Scale Collaborative Research Program 2013. Genomic responses in mouse models poorly mimic human inflammatory diseases. Proc. Natl. Acad. Sci. U. S. A. 110:3507–3512. 10.1073/pnas.1222878110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Noguchi N, Nakano K, Aratani Y, Koyama H, Kodama T, Niki E. 2000. Role of myeloperoxidase in the neutrophil-induced oxidation of low density lipoprotein as studied by myeloperoxidase-knockout mouse. J. Biochem. 127:971–976. 10.1093/oxfordjournals.jbchem.a022713 [DOI] [PubMed] [Google Scholar]
- 47.Rausch PG, Moore TG. 1975. Granule enzymes of polymorphonuclear neutrophils: a phylogenetic comparison. Blood 46:913–919 [PubMed] [Google Scholar]
- 48.Eisenhauer PB, Lehrer RI. 1992. Mouse neutrophils lack defensins. Infect. Immun. 60:3446–3447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Condliffe AM, Davidson K, Anderson KE, Ellson CD, Crabbe T, Okkenhaug K, Vanhaesebroeck B, Turner M, Webb L, Wymann MP, Hirsch E, Ruckle T, Camps M, Rommel C, Jackson SP, Chilvers ER, Stephens LR, Hawkins PT. 2005. Sequential activation of class IB and class IA PI3K is important for the primed respiratory burst of human but not murine neutrophils. Blood 106:1432–1440. 10.1182/blood-2005-03-0944 [DOI] [PubMed] [Google Scholar]
- 50.Lam GY, Fattouh R, Muise AM, Grinstein S, Higgins DE, Brumell JH. 2011. Listeriolysin O suppresses phospholipase C-mediated activation of the microbicidal NADPH oxidase to promote Listeria monocytogenes infection. Cell Host Microbe 10:627–634. 10.1016/j.chom.2011.11.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sibelius U, Schulz EC, Rose F, Hattar K, Jacobs T, Weiss S, Chakraborty T, Seeger W, Grimminger F. 1999. Role of Listeria monocytogenes exotoxins listeriolysin and phosphatidylinositol-specific phospholipase C in activation of human neutrophils. Infect. Immun. 67:1125–1130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bannam T, Goldfine H. 1999. Mutagenesis of active-site histidines of Listeria monocytogenes phosphatidylinositol-specific phospholipase C: effects on enzyme activity and biological function. Infect. Immun. 67:182–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Low MG, Finean JB. 1977. Modification of erythrocyte membranes by a purified phosphatidylinositol-specific phospholipase C (Staphylococcus aureus). Biochem. J. 162:235–240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kobayashi SD, Braughton KR, Palazzolo-Ballance AM, Kennedy AD, Sampaio E, Kristosturyan E, Whitney AR, Sturdevant DE, Dorward DW, Holland SM, Kreiswirth BN, Musser JM, DeLeo FR. 2010. Rapid neutrophil destruction following phagocytosis of Staphylococcus aureus. J. Innate Immun. 2:560–575. 10.1159/000317134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Boles BR, Thoendel M, Roth AJ, Horswill AR. 2010. Identification of genes involved in polysaccharide-independent Staphylococcus aureus biofilm formation. PLoS One 5:e10146. 10.1371/journal.pone.0010146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kreiswirth BN, Lofdahl S, Betley MJ, O'Reilly M, Schlievert PM, Bergdoll MS, Novick RP. 1983. The toxic shock syndrome exotoxin structural gene is not detectably transmitted by a prophage. Nature 305:709–712. 10.1038/305709a0 [DOI] [PubMed] [Google Scholar]
- 57.Holden MT, Feil EJ, Lindsay JA, Peacock SJ, Day NP, Enright MC, Foster TJ, Moore CE, Hurst L, Atkin R, Barron A, Bason N, Bentley SD, Chillingworth C, Chillingworth T, Churcher C, Clark L, Corton C, Cronin A, Doggett J, Dowd L, Feltwell T, Hance Z, Harris B, Hauser H, Holroyd S, Jagels K, James KD, Lennard N, Line A, Mayes R, Moule S, Mungall K, Ormond D, Quail MA, Rabbinowitsch E, Rutherford K, Sanders M, Sharp S, Simmonds M, Stevens K, Whitehead S, Barrell BG, Spratt BG, Parkhill J. 2004. Complete genomes of two clinical Staphylococcus aureus strains: evidence for the rapid evolution of virulence and drug resistance. Proc. Natl. Acad. Sci. U. S. A. 101:9786–9791. 10.1073/pnas.0402521101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gillaspy AF, Hickmon SG, Skinner RA, Thomas JR, Nelson CL, Smeltzer MS. 1995. Role of the accessory gene regulator (agr) in pathogenesis of staphylococcal osteomyelitis. Infect. Immun. 63:3373–3380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Baba T, Bae T, Schneewind O, Takeuchi F, Hiramatsu K. 2008. Genome sequence of Staphylococcus aureus strain Newman and comparative analysis of staphylococcal genomes: polymorphism and evolution of two major pathogenicity islands. J. Bacteriol. 190:300–310. 10.1128/JB.01000-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dyke KG, Jevons MP, Parker MT. 1966. Penicillinase production and intrinsic resistance to penicillins in Staphylococcus aures. Lancet i:835–838 [DOI] [PubMed] [Google Scholar]
- 61.Baba T, Takeuchi F, Kuroda M, Yuzawa H, Aoki K, Oguchi A, Nagai Y, Iwama N, Asano K, Naimi T, Kuroda H, Cui L, Yamamoto K, Hiramatsu K. 2002. Genome and virulence determinants of high virulence community-acquired MRSA. Lancet 359:1819–1827. 10.1016/S0140-6736(02)08713-5 [DOI] [PubMed] [Google Scholar]
- 62.McDougal LK, Steward CD, Killgore GE, Chaitram JM, McAllister SK, Tenover FC. 2003. Pulsed-field gel electrophoresis typing of oxacillin-resistant Staphylococcus aureus isolates from the United States: establishing a national database. J. Clin. Microbiol. 41:5113–5120. 10.1128/JCM.41.11.5113-5120.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Highlander SK, Hulten KG, Qin X, Jiang H, Yerrapragada S, Mason EO, Jr, Shang Y, Williams TM, Fortunov RM, Liu Y, Igboeli O, Petrosino J, Tirumalai M, Uzman A, Fox GE, Cardenas AM, Muzny DM, Hemphill L, Ding Y, Dugan S, Blyth PR, Buhay CJ, Dinh HH, Hawes AC, Holder M, Kovar CL, Lee SL, Liu W, Nazareth LV, Wang Q, Zhou J, Kaplan SL, Weinstock GM. 2007. Subtle genetic changes enhance virulence of methicillin resistant and sensitive Staphylococcus aureus. BMC Microbiol. 7:99. 10.1186/1471-2180-7-99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kiedrowski MR, Kavanaugh JS, Malone CL, Mootz JM, Voyich JM, Smeltzer MS, Bayles KW, Horswill AR. 2011. Nuclease modulates biofilm formation in community-associated methicillin-resistant Staphylococcus aureus. PLoS One 6:e26714. 10.1371/journal.pone.0026714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Nygaard TK, Pallister KB, Ruzevich P, Griffith S, Vuong C, Voyich JM. 2010. SaeR binds a consensus sequence within virulence gene promoters to advance USA300 pathogenesis. J. Infect. Dis. 201:241–254. 10.1086/649570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wormann ME, Reichmann NT, Malone CL, Horswill AR, Grundling A. 2011. Proteolytic cleavage inactivates the Staphylococcus aureus lipoteichoic acid synthase. J. Bacteriol. 193:5279–5291. 10.1128/JB.00369-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Olson ME, King JM, Yahr TL, Horswill AR. 2013. Sialic acid catabolism in Staphylococcus aureus. J. Bacteriol. 195:1779–1788. 10.1128/JB.02294-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gaskill ME, Khan SA. 1988. Regulation of the enterotoxin B gene in Staphylococcus aureus. J. Biol. Chem. 263:6276–6280 [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.