Abstract
Ureaplasma species commonly colonize the adult urogenital tract and are implicated in invasive diseases of adults and neonates. Factors that permit the organisms to cause chronic colonization or infection are poorly understood. We sought to investigate whether host innate immune responses, specifically, antimicrobial peptides (AMPs), are involved in determining the outcome of Ureaplasma infections. THP-1 cells, a human monocytoid tumor line, were cocultured with Ureaplasma parvum and U. urealyticum. Gene expression levels of a variety of host defense genes were quantified by real-time PCR. In vitro antimicrobial activities of synthetic AMPs against Ureaplasma spp. were determined using a flow cytometry-based assay. Chromosomal histone modifications in host defense gene promoters were tested by chromatin immunoprecipitation (ChIP). DNA methylation status in the AMP promoter regions was also investigated. After stimulation with U. parvum and U. urealyticum, the expression of cell defense genes, including the AMP genes (DEFB1, DEFA5, DEFA6, and CAMP), was significantly downregulated compared to that of TNFA and IL-8, which were upregulated. In vitro flow cytometry-based antimicrobial assay revealed that synthetic peptides LL-37, hBD-3, and hBD-1 had activity against Ureaplasma spp. Downregulation of the AMP genes was associated with chromatin modification alterations, including the significantly decreased histone H3K9 acetylation with U. parvum infection. No DNA methylation status changes were detected upon Ureaplasma infection. In conclusion, AMPs have in vitro activity against Ureaplasma spp., and suppression of AMP expression might be important for the organisms to avoid this aspect of the host innate immune response and to establish chronic infection and colonization.
INTRODUCTION
Ureaplasmas belong to the class Mollicutes, bacteria that lack a cell wall, and have been known for decades to colonize the human urogenital tract. Ureaplasmas are adapted to life as obligate parasites in eukaryotic hosts and use urea as their sole source of energy. There are two species that occur in humans, Ureaplasma parvum and Ureaplasma urealyticum. Ureaplasmas are frequently isolated from the adult lower urogenital tract, with a prevalence reaching 70% in some populations, and they also are commonly found in the respiratory tracts of preterm neonates, in whom they occasionally cause invasive disease (1). Although they are often considered commensals, numerous studies have implicated ureaplasmas in a wide variety of disorders, including chorioamnionitis, postpartum endometritis, nongonococcal urethritis (NGU), infertility, urinary calculi, arthritis, and preterm birth, as well as pneumonia, meningitis, and chronic lung disease in neonates (1). The host immune response to Ureaplasma infections involves elevated proinflammatory cytokines, such as interleukin 1α (IL-1α), IL-1β, IL-6, IL-8, monocyte chemoattractant protein 1 (MCP-1), and tumor necrosis factor alpha (TNF-α), as well as infiltration of neutrophils and macrophages at infection sites (2, 3). Very little is understood regarding how ureaplasmas cause human disease and, in particular, how they manage to establish chronic colonization and infection in the human urogenital tract.
Concerted attempts have been made to try to identify virulent strains of Ureaplasma as well as specific virulence factors. However, our previous studies and other reports have indicated that the virulence of Ureaplasma spp. is not likely associated with specific serovars (4, 5). Other studies have suggested that the various clinical outcomes, ranging from asymptomatic colonization to severe inflammation and invasive disease, might be primarily dependent on host-specific factors (6). Despite increasing recognition of the importance of these common organisms in human disease, little work has been done to characterize the interactions between Ureaplasma and the host that lead to diverse outcomes. The key question of how the organism manages to evade immune surveillance during the establishment of chronic infections is unanswered.
Ureaplasmas reside primarily in the mucosa in close association with epithelial cells. Innate immunity provides the first line of defense against infecting organisms in the mucosa. Antimicrobial peptides (AMPs) play an important role in the defense against a wide range of Gram-negative and Gram-positive bacteria, fungi, and viruses (7–9). AMPs act as endogenous antibiotics in the direct inhibition of pathogens and also as modulators of innate and adaptive immune responses. The most extensively studied human AMPs are defensins and cathelicidin, which are cationic, amphipathic peptides capable of disrupting microbial membranes (8). Human defensins include six α-defensins (HNP-1 to -4 and HD5 and -6, encoded by genes DEFA1 to -6), four major β-defensins (hBD-1 to -4, encoded by genes DEFB1, DEFB4, DEFB103, and DEFB104, respectively) and at least two other forms of β-defensins, and θ-defensins. The α-defensins HNP-1 to -4 are expressed mainly by neutrophils, while HD5, HD6, and hBDs are expressed predominantly by mucosal epithelial cells. Human cathelicidin (hCAP18/LL-37) is encoded by the single gene CAMP and is expressed mainly in granulocytes and epithelial cells of the skin, lungs, and gut. Extensive studies have characterized the activities of human AMPs against a variety of microorganisms (8). However, only a limited number of studies involved the Mollicutes (10–12), and there have been almost none that included Ureaplasma. Published reports suggest that canine β-defensins and cathelicidin display low activity against U. urealyticum (13, 14), and two AMPs from frogs show activity against Ureaplasma clinical isolates from humans (15, 16). A recent study indicated that a mouse cystatin-related epididymal spermatogenetic (CRES) protein had anti-Ureaplasma activity (17).
The coexistence of bacterial pathogens with their eukaryotic hosts over long periods has permitted the evolution of selective mechanisms by which pathogens manipulate host cell functions for survival, replication, and escape from early innate as well as adaptive immune responses. Recent reports demonstrate that some bacterial pathogens are able to induce host cell chromatin remodeling and thus impose their own transcriptional signature onto host cells, thereby suppressing host defense responses and avoiding immune surveillance (18). For example, a family of bacterial toxins from Listeria monocytogenes, the cholesterol-dependent cytolysins (CDC), were reported to induce a global dephosphorylation of H3S10, correlating with repression of host immunity genes (19). Garcia-Garcia et al. reported that infection of human monocytoid THP-1 cells by the intracellular pathogen Anaplasma phagocytophilum led to silencing of host defense genes, a process dependent on the upregulation of histone deacetylase 1 (HDAC1) (20).
We hypothesize that Ureaplasma spp. are able to manipulate host AMP gene expression by epigenetic mechanisms and thus avoid killing by this important arm of the innate immune system in order to create a persistent infection. We carried out coinfection studies with the human THP-1 monocytoid tumor cell line and Ureaplasma spp. and examined the effect on expression of AMP genes. The antimicrobial activity of AMPs against Ureaplasma spp. and possible epigenetic mechanisms involved in the Ureaplasma-host cell interactions were also explored.
MATERIALS AND METHODS
Cell line.
Human THP-1 cells (ATCC CCL-240) were grown in RPMI 1640 medium (Life Technologies, Carlsbad, CA) containing 10% fetal bovine serum (FBS).
Bacterial culture.
U. parvum (serovar 3, ATCC 27815) and U. urealyticum (serovar 10, ATCC 33699) were grown in regular Shepard's 10B urea broth overnight at 37°C until color changed. One-milliliter aliquots were made and stored at −80°C as a regular stock for the antimicrobial activity assay. The numbers of CFU of the frozen stock were determined.
For the cell stimulation study, to eliminate the background effects from the regular Shepard's 10B urea broth, overnight cultures of U. parvum and U. urealyticum were harvested by centrifugation at 10,000 × g for 90 min at 4°C and washed three times in 30 ml of phosphate-buffered saline (PBS) by centrifugation at 10,000 × g for 20 min at 4°C. Bacteria were resuspended in 1 ml of PBS containing 20% FBS, and 0.1-ml aliquots were made. The aliquots were stored at −80°C, and one tube was thawed for CFU determination.
THP-1 cell stimulation.
Freshly grown THP-1 cells at a concentration of 5 × 105 cells/ml were distributed into a 24-well plate (0.5 ml/well). Washed U. parvum or U. urealyticum stocks were thawed and added to the wells at a ratio of 5 CFU/cell. Phorbol myristate acetate (12-O-tetradecanoyl-phorbol-13-acetate [PMA]; Sigma, St. Louis, MO) was used (200 nM final concentration) as a stimulation control. After addition of 0.5 ml of fresh RPMI 1640 medium to each well, the plate was incubated at 37°C plus 5% CO2 for various periods.
RNA isolation and quantitative RT-PCR.
THP-1 cells cocultured with ureaplasmas were harvested at 4, 8, 24, and 48 h. Total RNA was purified using TRIzol reagent (Life Technologies). cDNA was synthesized using the SuperScript First-Strand Synthesis System for real-time PCR (RT-PCR) (Life Technologies). Real-time PCR was performed using a LightCycler 2.0 (Roche Diagnostics, Indianapolis, IN) to quantify gene expression levels of a variety of host defense genes, including DEFB1 and DEFA5, DEFA6, and CAMP, as well as IL-8 and TNFA. Primer sequences were from published resources or the qPrimerDepot database (21) (Table 1). The resulting amplicons were examined by melting peaks and agarose gel, and only those primers that generated a single amplicon were chosen. The relative gene expression level of each defense gene was calculated using the threshold cycle (2−ΔΔCT) method (22). RPLP0 was used as a housekeeping gene for normalization, and uninfected cells were used as a reference.
TABLE 1.
Primers for gene expression quantification, ChIP DNA quantification, and DNA methylation analysis
| Gene | Primer name | Primer sequence (5′–3′) | Application | Source |
|---|---|---|---|---|
| ACP6 | ACP6-eF | ATAGCTGGGGGTTCCACTCT | Gene expression quantification | qPrimerDepot |
| ACP6-eR | AAAATGGTGCAGGTCGTGTT | |||
| CAMP | CAMP-eF | GGGCACACTGTCTCCTTCAC | Gene expression quantification | qPrimerDepot |
| CAMP-eR | TCGGATGCTAACCTCTACCG | |||
| CAT | CAT-eF | ACGGGGCCCTACTGTAATAA | Gene expression quantification | qPrimerDepot |
| CAT-eR | AGATGCAGCACTGGAAGGAG | |||
| DEFA5 | DEFA5-eF | GGACTCACGGGTAGCACAAC | Gene expression quantification | qPrimerDepot |
| DEFA5-eR | CCTTTGCAGGAAATGGACTC | |||
| DEFA6 | DEFA6-eF | GACCTTCTGCAATGGCAAGT | Gene expression quantification | qPrimerDepot |
| DEFA6-eR | AGGACTTTGCCGTCTCCTTT | |||
| DEFB1 | DEFB1-eF | GGGCAGGCAGAATAGAGACA | Gene expression quantification | qPrimerDepot |
| DEFB1-eR | TTTTGTCTGAGATGGCCTCA | |||
| IL-8 | IL-8 F | CTAGGACAAGAGCCAGGAAGA | Gene expression quantification | 49 |
| IL-8 R | AACCCTCTGCACCCAGTTTTC | |||
| MPO | MPO-eF | TCCCGAAGTAAGAGGGTGTG | Gene expression quantification | qPrimerDepot |
| MPO-eR | CCCTGTCTCCTCACCAACC | |||
| RPLP0 | RPLP0-F | ATCTGCTTGGAGCCCACAT | Gene expression quantification | qPrimerDepot |
| RPLP0-R | GCGACCTGGAAGTCCAACTA | |||
| TNFA | TNFa1-F | CCCAGGCAGTCAGATCATCTTC | Gene expression quantification | 50 |
| TNFa1-R | GGTTTGCTACAACATGGGCTACA | |||
| CAMP | CAMP-F1 | GGCTTGGGAACATTTTGAGA | ChIP PCR | This study |
| CAMP-R1 | ATCCCCTTCTGCATCCTTCT | |||
| DEFA5 | DEFA5-F1 | GGAGCATCAAAGGGATCTTG | ChIP PCR | This study |
| DEFA5-R1 | TGAGGAGTCAGCCTGGATTT | |||
| DEFA6 | DEFA6-F1 | AGCATCAAAGGGACATGGAG | ChIP PCR | This study |
| DEFA6-R1 | AGGAGCCAGCCTGGATTTAT | |||
| DEFB103 | DEFB103-F1 | CAGTCTGGGCAGCATAGTGA | ChIP PCR | This study |
| DEFB103-R1 | GCCTCCACACATGGCTAAAT | |||
| DEFB104 | DEFB104-F1 | AGAAACGATTCAGGGAAGCA | ChIP PCR | This study |
| DEFB104-R1 | GAGGTGTTGGGGCTACAGAA | |||
| DEFB1 | DEFB1-F1 | TCCAGAAACCCCATCAGAAC | ChIP PCR | This study |
| DEFB1-R1 | CCGCTGGATTTAGCTTTCAG | |||
| TNFA | TNFA-F1 | ACCACAGCAATGGGTAGGAG | ChIP PCR | This study |
| TNFA-R1 | GTCCCCATACTCGACTTCCA | |||
| DEFA6 | defa6_msF | GGTTTTTATTAGGGTTTTTATGTGG | DNA methylation | This study |
| defa6_msR | ACCCATTTACTCTAAAATCCAACTTT | |||
| DEFB1 | defb1_msF1 | TTAAGGAAAATTTGAGGGATATTTG | DNA methylation | This study |
| defb1_msR1 | CTTCCTCTAACCAAAACTACCTTCTC | |||
| DEFB104A | defb104a_msF2 | GTGTTTTTTTTAAGGGTAAATGTTAGG | DNA methylation | This study |
| defb104a_msR2 | AATAACAACAAAAAATACTTAATAAC | |||
| defb4_msF1 | TGAGGATGGGGTTTATTATGTTTT | |||
| DEFB4 | defb4_msR1 | CCACTCACCTACTACCCACTCTATAC | DNA methylation | This study |
| defb4_msF2 | TTTTGAAAATATGGGGTTTTTTTATAA | |||
| defb4_msR2 | CACTCAAAAAATATCAACTCCACAA | |||
| TNFA | TNFa_msF | TTTATTTTTTTATTTAAGGGGAAATG | DNA methylation | This study |
| TNFa_msR | CCATATACCAAACATCCTATCTCTC |
In vitro antimicrobial activity assay.
Synthetic hBD1-, hBD-2, hBD-3, hBD-4, HNP-1, HNP-2, and LL-37 peptides (all from Ana Spec, Inc., Fremont, CA, except for hBD-2, which was from Phoenix Pharmaceuticals, Burlingame, CA) were dissolved in water or dimethyl sulfoxide (DMSO) according to the manufacturer's instructions to make the stock solutions of 1 mg/ml. A flow cytometric method of viability determination using the membrane potential-sensitive dye bis-(1,3-dibutylbarbituric acid) trimethine axonal [DiBAC4(3)] was adapted from the work of Nuding et al. (23). Preliminary experiments indicated that the activity of AMPs was dependent on both the bacterial and AMP concentrations and exhibited no differences with incubation times between 30 and 90 min (data not shown). Thus, a 30-min incubation time and a bacterial concentration of about 1 × 105 CFU/ml was chosen for all subsequent experiments.
Frozen stocks of U. parvum (serovar 3, ATCC 27815) and U. urealyticum (serovar 10, ATCC 33699) were quickly thawed at 37°C and diluted with 10B broth to reach the concentration of 1 × 105 CFU/ml. Ureaplasmas were incubated with AMPs at concentrations ranging from 6.25 to 100 μg/ml (serial 1:1 dilutions) in a final volume of 100 μl for 30 min at 37°C. Bacteria incubated with the same volume of corresponding AMP solvents (water or DMSO) served as negative controls. After incubation, the anionic dye DiBAC4(3) (AnaSpec, Inc.) was added to the cultures to a final concentration of 1 μg/ml to label the depolarized bacteria. After incubation for 10 min at 37°C, the bacteria were centrifuged for 10 min at 6,000 × g at room temperature. Pellets were resuspended in 100 μl of PBS, and the depolarized fluorescent bacteria were detected with an Accuri C6 flow cytometer (BD Biosciences, San Diego, CA).
A total of 5,000 to 10,000 events were collected at a low flow rate (14 μl/min) from each sample. The FL1 channel was used to detect the emission of DiBAC4(3) (maximum wavelength = 516 nm), and a dot plot with FL1 side scatter was generated. The gates for differentiation of viable, nondepolarized and depolarized bacteria were determined according to the untreated negative control and heat-killed positive control. The survival rates were calculated by normalizing the percentage of viable, nondepolarized cells in the experimental groups to the values for their negative controls.
ChIP.
Histone modification patterns in the 1,000-bp proximal promoter regions of defense genes were analyzed by chromatin immunoprecipitation (ChIP) and quantitative PCR (qPCR). THP-1 cells (1 × 106) were stimulated with U. parvum, U. urealyticum, or 200 nM PMA as described above for 16 h and cross-linked with 1% formaldehyde. Cells were lysed with lysing buffer (50 mM Tris-HCl [pH 8.0], 10 mM EDTA, 1% [wt/vol] SDS, Halt protease inhibitor cocktail [1:100 dilution from 100× stock; Thermo Scientific, IL], 1 mM phenylmethylsulfonyl fluoride [PMSF], 20 mM sodium butyrate), and nuclei were harvested by centrifugation at 4,000 × g for 5 min at 4°C. Chromatin was sheared on ice with a Microson XL ultrasonic cell disruptor (Heat Systems, Farmingdale, NY) at 30% output power three times for 30 s each time, with 30-s pauses between. Sheared chromatin was incubated at 4°C for 2 h with protein A/G Dynabeads (Life Technologies) that conjugated with anti-acetyl-histone H3 (Lys9) (catalog no. 07-352), anti-trimethyl-histone H3 (Lys9) (catalog no. 07-442), or anti-trimethyl-histone H3 (Lys4) (catalog no. 07-473) (All from Millipore, CA). Isotype-matched antibody was included as a negative ChIP control. Immunoprecipitated protein DNA was digested with proteinase K (50 μg/ml) for 30 min at 56°C and purified with QIAquick nucleotide removal columns (Qiagen, Germantown, MD). The resulted DNA was quantified by qPCR using primers specific for the gene promoter region. Primers were designed by Primer 3 (Table 1). The relative target DNA changes were calculated by comparing the percentage of precipitated DNA (percent input) in stimulated cells to that in the uninfected control.
DNA methylation analysis.
Genomic DNA of infected and uninfected cells was purified with the Qiagen Blood and Tissue kit (Qiagen). Bisulfite conversion was carried out using the Epitect bisulfite kit (Qiagen) according to the manufacturer's instructions. Primers specific for the CpG-rich promoter regions were designed using the online software MethPrimer (http://www.urogene.org/methprimer/) (Table 1). PCR products were sequenced and analyzed using CLC DNA workbench 5.
Statistical analysis.
The Q-Q plot was first conducted to examine data distribution and did not show obvious deviations from the normality assumption. To account for potential dependence of measurements obtained on the same experiment occasion, the paired t test was used to assess the statistical significance of the difference in relative gene expression levels between the tested genes (DEFB1, DEFA5, DEFA6, CAMP, ACP6, CAT, and MPO) and the reference genes (IL-8 and TNFA) for THP-1 cells stimulated with U. parvum and U. urealyticum at 4, 8, 24, and 48 h. The paired t test was also conducted to compare the histone modification alterations in the defense gene promoters with the reference gene (TNFA) in the THP-1 cells infected by U. parvum and U. urealyticum, stratified by the types of antibodies (H3K9ac, H3K4me3, and H3K9me3). The histone modification patterns for U. parvum- and U. urealyticum-infected THP-1 cells were further compared with PMA, stratified by the defense genes. A P value of <0.05 was considered statistically significant.
RESULTS
Host defense gene expression is downregulated in Ureaplasma-infected cells.
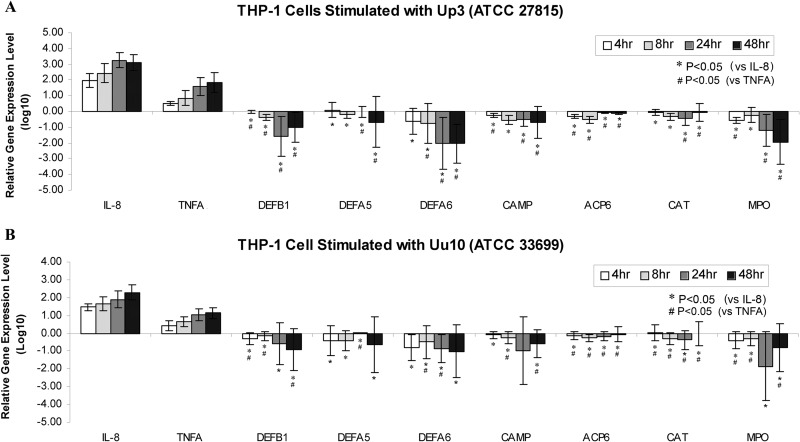
The expression of 7 genes (DEFB1, DEFA5, DEFA6, CAMP, ACP6, CAT, and MPO) encoding AMPs or otherwise involved in oxidative and enzymatic defense responses was quantified by RT-PCR. Results showed that upon infection with U. parvum or U. urealyticum, the expression of IL-8 and TNF-α was upregulated (Fig. 1), which is consistent with previous reports (24, 25). Interestingly, on the other hand, the expression of four AMP genes, DEFB1, DEFA5, DEFA6, and CAMP, was significantly downregulated (P < 0.05) compared to that of IL-8 or TNFA in THP-1 cells at most time points of infection. Notably, the expression of ACP6, CAT, and MPO was also significantly (P < 0.05) downregulated compared to that of IL-8 and TNFA. It is known that cellular defense genes are organized as clusters in the chromosomes; e.g., most of the defensins are localized on chromosome 8p23 (26). Thus, it is possible that the host defense gene expression is globally suppressed by infection with Ureaplasma spp.
FIG 1.
Host defense gene expression in THP-1 cells upon Ureaplasma stimulation. A total of 5 × 105 THP-1 cells were cocultured with U. parvum (serovar 3, ATCC 27815, 5 CFU/cell) (A) or U. urealyticum (serovar 10, ATCC 33699, 5 CFU/cell) (B) for 4, 8, 24, and 48 h. Gene expression levels of DEFB1, DEFA5, DEFA6, CAMP, ACP6, CAT, and MPO were quantified by real-time PCR using cDNA synthesized from RNA harvested from each well. RPLP0 was used as a housekeeping gene for normalization. Untreated cells were used as a reference. The relative gene expression level of each tested gene was calculated using the 2−ΔΔCT method and is presented in log10 form. Data represent mean relative gene expression levels ± standard deviations (n = 4 independent experiments). Numbers that are <0 denote downregulation, and those that are >0 indicate upregulation.
In vitro activities of AMPs against Ureaplasma spp.
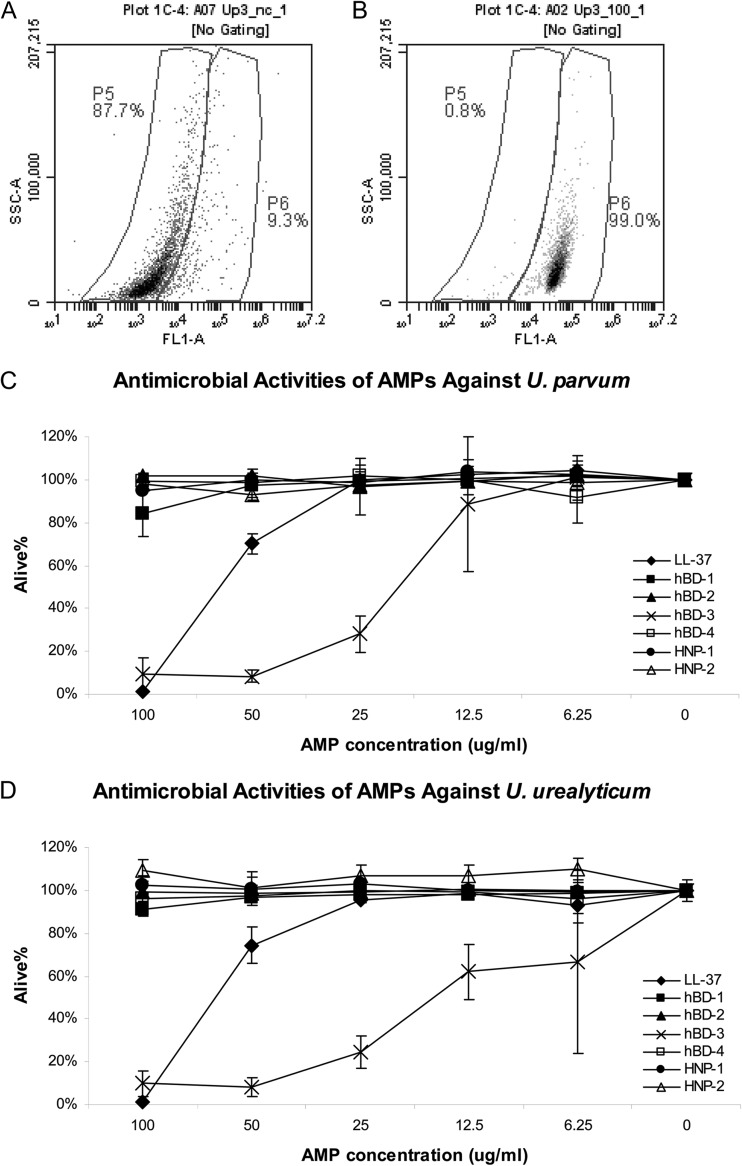
We next explored whether the downregulated AMPs have any adverse effect on the survival of Ureaplasma spp. by using an in vitro antimicrobial assay in which the viability of the ureaplasmas was monitored by flow cytometry (Fig. 2). The viable, nondepolarized cells were not labeled by DiBAC4(3) and appeared in the low-fluorescence gate in the flow cytometry plot, while the depolarized cells were fluorescently labeled and were located in the high-fluorescence gate. In the representative examples shown, about 88% of the U. parvum cells were viable in the untreated culture from thawed stock and were plotted in the lower-fluorescence gate P5 (Fig. 2A). Following incubation with 100 μg/ml of LL-37, the majority of the bacteria were depolarized and were plotted in the higher-fluorescence gate P6 (Fig. 2B). LL-37 and hBD-3 directly impaired the viability of U. parvum and U. urealyticum in a dose-dependent manner in the range of 6.25 to 100 μg/ml (Fig. 2B, C, and D). The activities of LL-37 against U. parvum and U. urealyticum were similar, with 50% inhibitory concentrations (IC50s) of about 65 μg/ml. At a concentration of 100 μg/ml, more than 98% of the bacteria were depolarized. hBD-3 showed higher activities than did LL-37 against the two Ureaplasma spp., with IC50s of about 19 and 17 μg/ml for U. parvum and U. urealyticum, respectively. The observed activity of hBD-1 against ureaplasmas was much lower than those of LL-37 and hBD-3 under the tested condition. No antimicrobial activity was observed for hBD-2, hBD-4, HNP-1, or HNP-2 under the tested condition.
FIG 2.
In vitro antimicrobial activity assay. Diluted stocks of U. parvum or U. urealyticum were incubated with synthetic AMP or AMP solvents at the indicated concentrations for 30 min at 37°C. After treatment, bacteria were incubated with DiBAC4, to label depolarized cells. Control viable nondepolarized bacteria from freshly thawed stock (low-fluorescence gate P5 [A]) and depolarized cells after treatment with LL-37 (depolarized high-fluorescence gate P6 [B]) are shown as representative examples. A total of 5,000 to 10,000 events were collected from each experimental treatment. The survival rates were calculated by normalizing the percentage of viable, nondepolarized cells in the experimental group to the value for the negative controls (C and D). Data represent average survival rates ± standard deviations from 3 experiments.
These data demonstrate that selected human AMPs, at least LL-37 and hBD-3, can directly damage Ureaplasma species, and thus suppression of the AMPs by Ureaplasma infection would compromise host antimicrobial defense and potentially facilitate the survival of Ureaplasma spp. on the mucosal surface and the establishment of chronic infection.
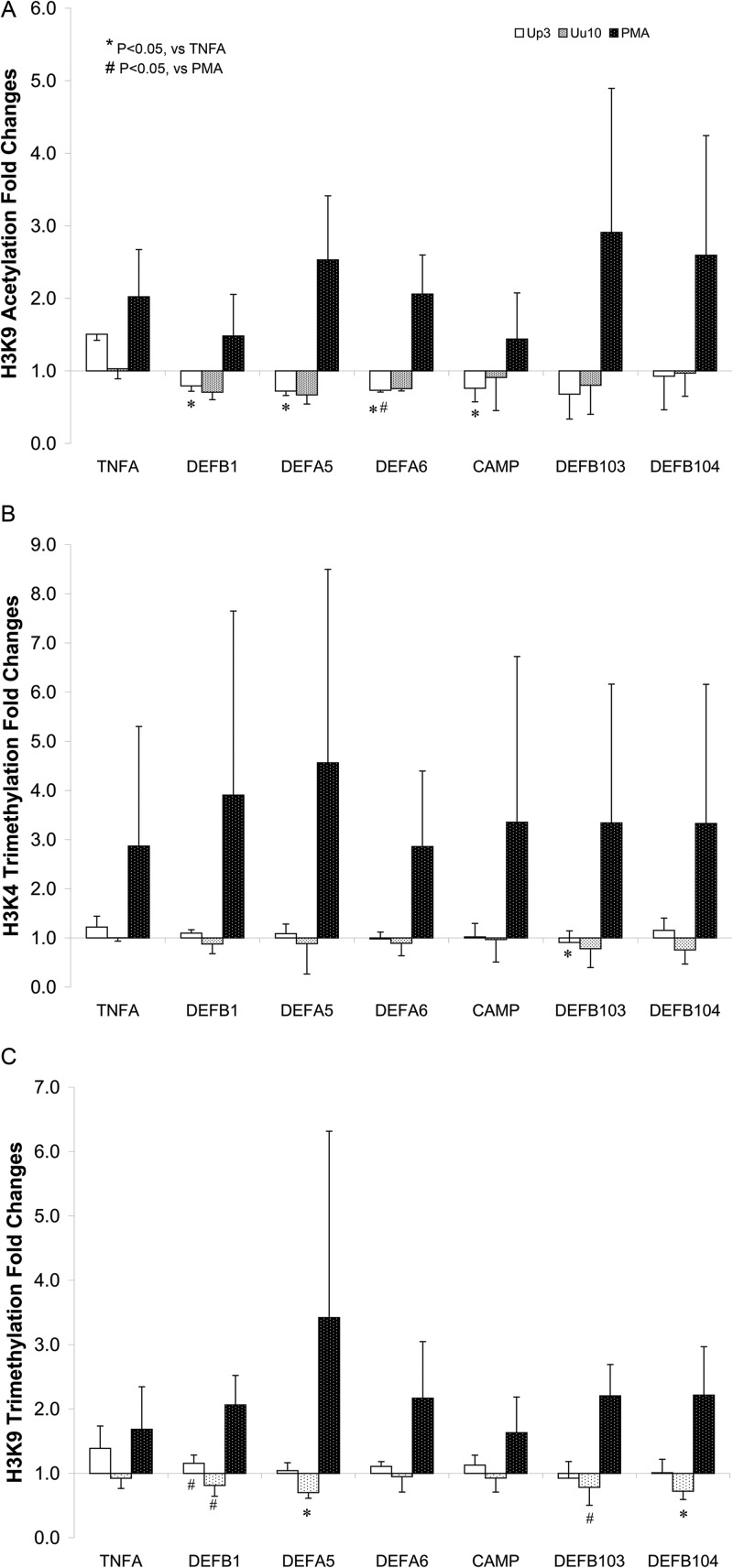
Ureaplasma infection induces host defense gene chromatin modifications.
It is known that silencing of the genes is often associated with epigenetic modifications in the chromatin (5). To test for possible host chromatin alterations upon Ureaplasma infection, we employed chromatin immunoprecipitation (ChIP) to look for histone modifications in the AMP gene promoters in the presence or absence of Ureaplasma infection (Fig. 3). The results indicate that U. parvum or U. urealyticum infection is associated with a decrease in histone H3 Lys9 (H3K9) acetylation, a modification related to gene activation, in the promoters of DEFB1, DEFA5, DEFA6, CAMP, DEFB103, and DEFB104 (Fig. 3A). Compared to TNFA under the same infection condition, the decrease of H3K9 acetylation in DEFB1, DEFA5, DEFA6, and CAMP promoters was significant (P < 0.05) in cells infected with U. parvum but not U. urealyticum. On the other hand, in cells stimulated with PMA, H3K9 histone acetylation in the six genes was uniformly increased. Except for DEFB103, there were no significant changes in histone H3 Lys4 (H3K4) trimethylation, another gene activation modification, in cells infected with U. parvum or U. urealyticum (Fig. 3B). Infection with U. urealyticum caused a slight but significant (P < 0.05) decrease in H3K9 trimethylation (a gene repression modification) of DEFA5 and DEFB104 compared to that of TNFA (Fig. 3C). The H3K9 trimethylation alteration in genes DEFB1 and DEFB103 caused by U. parvum or U. urealyticum infection was also significant (P < 0.05) compared to the alteration in cells stimulated with PMA. Thus, the downregulation of AMP genes by the infection of Ureaplasma spp. correlated with host cell chromatin modifications; however, the two species exhibit differences in the ability to carry out histone modification.
FIG 3.
Histone modification alterations in the defense gene promoters in Ureaplasma-infected THP-1 cells. THP-1 cells (1 × 106) were cocultured with U. parvum or U. urealyticum (5 CFU/cell) or PMA (200 nM) for 16 h. Formaldehyde-cross-linked, sheared chromatin was immunoprecipitated using anti-acetyl-histone H3 (Lys9 [A]) or anti-trimethyl-histone H3 (Lys9 [B] or Lys4 [C]). Immunoprecipitated DNA fragments were quantified by qPCR using primers specific for the promoter regions of the genes tested. Histone modification alterations were expressed as the ratio of immunoprecipitated chromatin target (% input) from stimulated to unstimulated cells. Data are the average fold changes of histone modifications ± standard deviations of three independent experiments.
No DNA methylation changes were detected in the THP-1 cells after coculture with ureaplasmas (data not shown). Thus, this mechanism does not seem to play an essential role in regulating AMP gene expression during ureaplasma infection.
DISCUSSION
Data in these experiments demonstrate for the first time in a human cell line that in vitro infection with U. parvum or U. urealyticum causes downregulation of host defense gene expression and that synthetic AMP peptides have bactericidal activities against Ureaplasma spp. in vitro. An epigenetic process by which Ureaplasma spp. suppress the host defense genes has been identified, although the molecular mechanism(s) remain to be characterized.
Our data indicate that human AMPs, LL-37 and BD-3, are active in damaging Ureaplasma spp. that infect humans. Previous studies in other laboratories have demonstrated that three canine β-defensins have low bactericidal activity against U. urealyticum (13). Canine cathelicidin also had a low activity against U. urealyticum, while the activity against Ureaplasma canigenitalium, a species that infects dogs, was higher (14), indicating possible species specificities of the AMP targets. Among the seven synthetic human AMPs tested in this study, LL-37 and BD-3 showed promising activity against both U. parvum and U. urealyticum, while the activity of BD-1 was low and no activity was observed for BD-2, BD-4, HNP-1, or HNP-2. Looking at other Mollicutes, it has been found that BD-2 and BD-3, but not BD-1, possess activity against Mycoplasma pneumoniae (11). It is also noteworthy that Mycoplasma fermentans and Mycoplasma hyorhinis were susceptible to four membrane-active peptides: alamethicin, dernaseptin B2, gramicidin S, and surfactin (27). We noticed that the in vitro antimicrobial activity of synthetic LL-37 and BD-3 under the tested concentrations was mainly to damage the membrane and depolarize the ureaplasmas. Some depolarized ureaplasmas were still viable, probably because the membrane damage was not severe enough under the tested AMP concentrations and could be repaired once the AMPs were diluted (tested by a parallel CFU dilution assay; data not shown).
Because of their lack of a cell wall, ureaplasmas are naturally resistant to cell wall-active antibiotics, leaving the treatment dependent on the classes that affect protein or DNA synthesis, primarily macrolides, tetracyclines, and fluoroquinolones. However, resistance to all these classes of antibiotics has been identified in clinical isolates (28, 51). Hence, there is an increasing interest in identifying new antibiotics and treatment strategies against ureaplasmas. The AMPs are among the compounds under investigation for their general antimicrobial potency, and several of them are undergoing clinical evaluation (29, 30). Regarding the Mollicutes, a recombinant plasmid expressing a gene for an AMP, melittin, was introduced into cell line and animal models, and the results suggested efficacy in the treatment of infections caused by Mycoplasma hominis and Mycoplasma gallisepticum (31–34). Our study may provide another promising new strategy for treating ureaplasma infections. Two individual peptides, LL-37 and BD-3, were found to be active against ureaplasmas in vitro. It is known that the in vivo conditions are much different, i.e., that the bacteria could be attacked by multiple AMPs of different structural classes, and that the local concentrations of AMPs could be much higher (35). Because AMPs are multifunctional molecules that not only serve as gene-coded antibiotics but also influence diverse cellular processes involved in resolution of infection and repair of damaged epithelia (36), subsequent studies are needed to clarify a possible role for AMPs in treating ureaplasmal infections in vivo.
Although it is clear from the epidemiological studies that ureaplasmas are associated with many invasive infections, the mechanism(s) of pathogenicity of this organism is still under debate. We speculate that the persistent colonization might be associated with low virulence and an ability to suppress host innate immune responses. Data from a sheep model confirmed that chronic fetal exposure to U. parvum suppressed the lipopolysaccharide (LPS)-induced innate immune response with decreased lung proinflammatory and anti-inflammatory cytokine expression, fewer CD3+ T lymphocytes, and decreased in vitro monocyte responsiveness (37). In this study, we showed that U. parvum and U. urealyticum infection of THP-1 cells caused selective downregulation of the AMP genes and other cell defense genes, providing further proof of the ability of Ureaplasma spp. to manipulate the host defense response. This finding may have an important clinical implication. In many cases, there are coinfections of Ureaplasma spp. together with other microorganisms (38). It is possible that the impaired innate immune response caused by Ureaplasma spp. could increase the susceptibility of the host to the adverse effects of other pathogens. In studies of other mycoplasmas involved in human and animal diseases, suppression of host immune responses has also been observed. Reports have documented that both humoral and cellular immunity was suppressed in patients with M. pneumoniae infection (39, 40), although a mechanism has not been identified. A global suppression of ovine peripheral blood mononuclear cells by M. ovipneumoniae has also been reported, again without a clear description of the pathogenic process employed by the organism (41).
Pathogen-induced epigenetic alterations in host cells have been observed in many other microbes. The protein effector OspF in Shigella flexneri blocks the access of transcription factor NF-κB to chromatin, resulting in attenuated expression of host genes involved in immune responses (42). An epigenetic regulatory mechanism has been described in which the histone deacetylase inhibitors butyrate and trichostatin A enhance the expression of cathelicidin in lung epithelial cells by increasing histone acetylation (43). Our results indicate that upon Ureaplasma infection, the decrease of histone 3 acetylation in the promoter of AMP genes, including CAMP (encoding cathelicidin LL-37), is associated with downregulated gene expression, similar to the reported effects of butyrate and trichostatin A. Our results also suggest that while U. parvum and U. urealyticum may regulate the host AMP expression similarly in the site of H3K9 acetylation, the histone codes, a combination of multiple histone modifications, employed by the two organisms might be different. This might influence the virulence and host adaptation of the two organisms.
In this work, we also reported a new approach in monitoring the antimicrobial activity of AMPs against Ureaplasma by flow cytometry. This is a fast and accurate method for counting the viability of the microorganisms compared to the traditional CFU counting method (23). The use of flow cytometry with DNA dyes and membrane-impermeant probes for determining the viability of various Mycoplasma species has been developed recently (44–48). We have for the first time applied this method to Ureaplasma spp. to identify and enumerate viable cells in the antimicrobial assay. Up to 10,000 cells can be analyzed within minutes using flow cytometry, in contrast to the traditional CFU plating method, in which several hundred colonies are counted by visual observation. However, one drawback is that this method may not differentiate membrane-damaged but potentially viable and dead bacteria.
There are limitations to this study. First, the reported results were based only on the ATCC type strains. Future studies should consider clinical strains with different patient origins for their ability to suppress the host defense response. Second, due to the availability of high-quality RT-PCR primers for quantifying the gene expression level and the ChIP DNA amount, only a limited number of AMP genes were examined in this study. To obtain a whole picture of global gene expression changes upon ureaplasma infection, other methods, such as transcriptome sequencing (RNA-seq), should be considered.
In summary, our in vitro study with human cells demonstrated the novel finding that Ureaplasma infection induces the suppression of AMP gene transcription likely involving the modulation of histone acetylation by an unknown mechanism. This impairment of cellular AMP gene activation may play a role in the ability of Ureaplasma spp. to avoid the host immune system and facilitate the establishment of chronic infection. Future studies to better characterize the mechanism of Ureaplasma-mediated AMP gene suppression are warranted.
ACKNOWLEDGMENTS
This study was supported by a grant from the Kaul Pediatric Research Institute, Children's Hospital of Alabama.
Technical support from Lynn B. Duffy and Amy E. Ratliff is greatly appreciated.
Footnotes
Published ahead of print 3 February 2014
REFERENCES
- 1.Waites KB, Schelonka RL, Xiao L, Grigsby PL, Novy MJ. 2009. Congenital and opportunistic infections: Ureaplasma species and Mycoplasma hominis. Semin. Fetal Neonatal Med. 14:190–199. 10.1016/j.siny.2008.11.009 [DOI] [PubMed] [Google Scholar]
- 2.Viscardi RM, Hasday JD. 2009. Role of Ureaplasma species in neonatal chronic lung disease: epidemiologic and experimental evidence. Pediatr. Res. 65:84R–90R. 10.1203/PDR.0b013e31819dc2f9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schelonka RL, Waites KB. 2007. Ureaplasma infection and neonatal lung disease. Semin. Perinatol. 31:2–9. 10.1053/j.semperi.2007.01.001 [DOI] [PubMed] [Google Scholar]
- 4.Xiao L, Paralanov V, Glass JI, Duffy LB, Robertson JA, Cassell GH, Chen Y, Waites KB. 2011. Extensive horizontal gene transfer in ureaplasmas from humans questions the utility of serotyping for diagnostic purposes. J. Clin. Microbiol. 49:2818–2826. 10.1128/JCM.00637-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Paralanov V, Lu J, Duffy LB, Crabb DM, Shrivastava S, Methe BA, Inman J, Yooseph S, Xiao L, Cassell GH, Waites KB, Glass JI. 2012. Comparative genome analysis of 19 Ureaplasma urealyticum and Ureaplasma parvum strains. BMC Microbiol. 12:88. 10.1186/1471-2180-12-88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Reyes L, Reinhard M, Brown MB. 2009. Different inflammatory responses are associated with Ureaplasma parvum-induced UTI and urolith formation. BMC Infect. Dis. 9:9. 10.1186/1471-2334-9-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rogan MP, Geraghty P, Greene CM, O'Neill SJ, Taggart CC, McElvaney NG. 2006. Antimicrobial proteins and polypeptides in pulmonary innate defence. Respir. Res. 7:29. 10.1186/1465-9921-7-29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Doss M, White MR, Tecle T, Hartshorn KL. 2010. Human defensins and LL-37 in mucosal immunity. J. Leukoc. Biol. 87:79–92. 10.1189/jlb.0609382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zasloff M. 2002. Antimicrobial peptides of multicellular organisms. Nature 415:389–395. 10.1038/415389a [DOI] [PubMed] [Google Scholar]
- 10.Béven L, Wroblewski H. 1997. Effect of natural amphipathic peptides on viability, membrane potential, cell shape and motility of mollicutes. Res. Microbiol. 148:163–175. 10.1016/S0923-2508(97)87647-4 [DOI] [PubMed] [Google Scholar]
- 11.Kuwano K, Tanaka N, Shimizu T, Kida Y. 2006. Antimicrobial activity of inducible human beta defensin-2 against Mycoplasma pneumoniae. Curr. Microbiol. 52:435–438. 10.1007/s00284-005-0215-7 [DOI] [PubMed] [Google Scholar]
- 12.Fassi Fehri L, Wroblewski H, Blanchard A. 2007. Activities of antimicrobial peptides and synergy with enrofloxacin against Mycoplasma pulmonis. Antimicrob. Agents Chemother. 51:468–474. 10.1128/AAC.01030-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sang Y, Ortega MT, Blecha F, Prakash O, Melgarejo T. 2005. Molecular cloning and characterization of three beta-defensins from canine testes. Infect. Immun. 73:2611–2620. 10.1128/IAI.73.5.2611-2620.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sang Y, Teresa Ortega M, Rune K, Xiau W, Zhang G, Soulages JL, Lushington GH, Fang J, Williams TD, Blecha F, Melgarejo T. 2007. Canine cathelicidin (K9CATH): gene cloning, expression, and biochemical activity of a novel pro-myeloid antimicrobial peptide. Dev. Comp. Immunol. 31:1278–1296. 10.1016/j.dci.2007.03.007 [DOI] [PubMed] [Google Scholar]
- 15.Lee WH, Zhang J, Zhang YX, Jin Y, Lai R, Zhang Y. 2005. Maximin 9, a novel free thiol containing antimicrobial peptide with antimycoplasma activity from frog Bombina maxima. FEBS Lett. 579:4443–4448. 10.1016/j.febslet.2005.07.010 [DOI] [PubMed] [Google Scholar]
- 16.Wang T, Zhang J, Shen JH, Jin Y, Lee WH, Zhang Y. 2005. Maximins S, a novel group of antimicrobial peptides from toad Bombina maxima. Biochem. Biophys. Res. Commun. 327:945–951. 10.1016/j.bbrc.2004.12.094 [DOI] [PubMed] [Google Scholar]
- 17.Wang L, Yuan Q, Chen S, Cai H, Lu M, Liu Y, Xu C. 2012. Antimicrobial activity and molecular mechanism of the CRES protein. PLoS One 7:e48368. 10.1371/journal.pone.0048368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hamon MA, Cossart P. 2008. Histone modifications and chromatin remodeling during bacterial infections. Cell Host Microbe 4:100–109. 10.1016/j.chom.2008.07.009 [DOI] [PubMed] [Google Scholar]
- 19.Hamon MA, Batsche E, Regnault B, Tham TN, Seveau S, Muchardt C, Cossart P. 2007. Histone modifications induced by a family of bacterial toxins. Proc. Natl. Acad. Sci. U. S. A. 104:13467–13472. 10.1073/pnas.0702729104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Garcia-Garcia JC, Barat NC, Trembley SJ, Dumler JS. 2009. Epigenetic silencing of host cell defense genes enhances intracellular survival of the rickettsial pathogen Anaplasma phagocytophilum. PLoS Pathog. 5:e1000488. 10.1371/journal.ppat.1000488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cui W, Taub DD, Gardner K. 2007. qPrimerDepot: a primer database for quantitative real time PCR. Nucleic Acids Res. 35:D805–D809. 10.1093/nar/gkl767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) method. Methods 25:402–408. 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- 23.Nuding S, Fellermann K, Wehkamp J, Mueller HA, Stange EF. 2006. A flow cytometric assay to monitor antimicrobial activity of defensins and cationic tissue extracts. J. Microbiol. Methods 65:335–345. 10.1016/j.mimet.2005.08.004 [DOI] [PubMed] [Google Scholar]
- 24.Manimtim WM, Hasday JD, Hester L, Fairchild KD, Lovchik JC, Viscardi RM. 2001. Ureaplasma urealyticum modulates endotoxin-induced cytokine release by human monocytes derived from preterm and term newborns and adults. Infect. Immun. 69:3906–3915. 10.1128/IAI.69.6.3906-3915.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Peltier MR, Tee SC, Smulian JC. 2008. Effect of progesterone on proinflammatory cytokine production by monocytes stimulated with pathogens associated with preterm birth. Am. J. Reprod. Immunol. 60:346–353. 10.1111/j.1600-0897.2008.00633.x [DOI] [PubMed] [Google Scholar]
- 26.Taudien S, Groth M, Huse K, Petzold A, Szafranski K, Hampe J, Rosenstiel P, Schreiber S, Platzer M. 2010. Haplotyping and copy number estimation of the highly polymorphic human beta-defensin locus on 8p23 by 454 amplicon sequencing. BMC Genomics 11:252. 10.1186/1471-2164-11-252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nir-Paz R, Prevost MC, Nicolas P, Blanchard A, Wroblewski H. 2002. Susceptibilities of Mycoplasma fermentans and Mycoplasma hyorhinis to membrane-active peptides and enrofloxacin in human tissue cell cultures. Antimicrob. Agents Chemother. 46:1218–1225. 10.1128/AAC.46.5.1218-1225.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xiao L, Crabb DM, Duffy LB, Paralanov V, Glass JI, Hamilos DL, Waites KB. 2011. Mutations in ribosomal proteins and ribosomal RNA confer macrolide resistance in human Ureaplasma spp. Int. J. Antimicrob. Agents 37:377–379. 10.1016/j.ijantimicag.2010.12.012 [DOI] [PubMed] [Google Scholar]
- 29.Marr AK, Gooderham WJ, Hancock RE. 2006. Antibacterial peptides for therapeutic use: obstacles and realistic outlook. Curr. Opin. Pharmacol. 6:468–472. 10.1016/j.coph.2006.04.006 [DOI] [PubMed] [Google Scholar]
- 30.Alba A, Lopez-Abarrategui C, Otero-Gonzalez AJ. 2012. Host defense peptides: an alternative as antiinfective and immunomodulatory therapeutics. Biopolymers 98:251–267. 10.1002/bip.22076 [DOI] [PubMed] [Google Scholar]
- 31.Lazarev VN, Parfenova TM, Gularyan SK, Misyurina OY, Akopian TA, Govorun VM. 2002. Induced expression of melittin, an antimicrobial peptide, inhibits infection by Chlamydia trachomatis and Mycoplasma hominis in a HeLa cell line. Int. J. Antimicrob. Agents 19:133–137. 10.1016/S0924-8579(01)00479-4 [DOI] [PubMed] [Google Scholar]
- 32.Lazarev VN, Shkarupeta MM, Kostryukova ES, Levitskii SA, Titova GA, Akopian TA, Govorun VM. 2007. Recombinant plasmid constructs expressing gene for antimicrobial peptide melittin for the therapy of mycoplasma and chlamydia infections. Bull. Exp. Biol. Med. 144:452–456. 10.1007/s10517-007-0350-1 [DOI] [PubMed] [Google Scholar]
- 33.Lazarev VN, Shkarupeta MM, Titova GA, Kostrjukova ES, Akopian TA, Govorun VM. 2005. Effect of induced expression of an antimicrobial peptide melittin on Chlamydia trachomatis and Mycoplasma hominis infections in vivo. Biochem. Biophys. Res. Commun. 338:946–950. 10.1016/j.bbrc.2005.10.028 [DOI] [PubMed] [Google Scholar]
- 34.Lazarev VN, Stipkovits L, Biro J, Miklodi D, Shkarupeta MM, Titova GA, Akopian TA, Govorun VM. 2004. Induced expression of the antimicrobial peptide melittin inhibits experimental infection by Mycoplasma gallisepticum in chickens. Microbes Infect. 6:536–541. 10.1016/j.micinf.2004.02.006 [DOI] [PubMed] [Google Scholar]
- 35.Diamond G, Beckloff N, Weinberg A, Kisich KO. 2009. The roles of antimicrobial peptides in innate host defense. Curr. Pharm. Des. 15:2377–2392. 10.2174/138161209788682325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lai Y, Gallo RL. 2009. AMPed up immunity: how antimicrobial peptides have multiple roles in immune defense. Trends Immunol. 30:131–141. 10.1016/j.it.2008.12.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kallapur SG, Kramer BW, Knox CL, Berry CA, Collins JJ, Kemp MW, Nitsos I, Polglase GR, Robinson J, Hillman NH, Newnham JP, Chougnet C, Jobe AH. 2011. Chronic fetal exposure to Ureaplasma parvum suppresses innate immune responses in sheep. J. Immunol. 187:2688–2695. 10.4049/jimmunol.1100779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Onderdonk AB, Delaney ML, DuBois AM, Allred EN, Leviton A, Extremely Low Gestational Age Newborns (ELGAN) Study Investigators 2008. Detection of bacteria in placental tissues obtained from extremely low gestational age neonates. Am. J. Obstet. Gynecol. 198:110.e1–110.e7. 10.1016/j.ajog.2007.05.044 [DOI] [PubMed] [Google Scholar]
- 39.Tsunekawa H, Takagi E, Kishimoto H, Shimokata K. 1987. Depressed cellular immunity in Mycoplasma pneumoniae pneumonia. Eur. J. Respir. Dis. 70:293–299 [PubMed] [Google Scholar]
- 40.Stelmach I, Podsiadlowicz-Borzecka M, Grzelewski T, Majak P, Stelmach W, Jerzynska J, Poplawska M, Dziadek J. 2005. Humoral and cellular immunity in children with Mycoplasma pneumoniae infection: a 1-year prospective study. Clin. Diagn. Lab. Immunol. 12:1246–1250. 10.1128/CDLI.12.10.1246-1250.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shahzad W, Ajuwape AT, Rosenbusch RF. 2010. Global suppression of mitogen-activated ovine peripheral blood mononuclear cells by surface protein activity from Mycoplasma ovipneumoniae. Vet. Immunol. Immunopathol. 136:116–121. 10.1016/j.vetimm.2010.02.001 [DOI] [PubMed] [Google Scholar]
- 42.Arbibe L, Kim DW, Batsche E, Pedron T, Mateescu B, Muchardt C, Parsot C, Sansonetti PJ. 2007. An injected bacterial effector targets chromatin access for transcription factor NF-kappaB to alter transcription of host genes involved in immune responses. Nat. Immunol. 8:47–56. 10.1038/ni1423 [DOI] [PubMed] [Google Scholar]
- 43.Kida Y, Shimizu T, Kuwano K. 2006. Sodium butyrate up-regulates cathelicidin gene expression via activator protein-1 and histone acetylation at the promoter region in a human lung epithelial cell line, EBC-1. Mol. Immunol. 43:1972–1981. 10.1016/j.molimm.2005.11.014 [DOI] [PubMed] [Google Scholar]
- 44.Assunção P, Antunes NT, Rosales RS, Poveda C, de la Fe C, Poveda JB, Davey HM. 2007. Application of flow cytometry for the determination of minimal inhibitory concentration of several antibacterial agents on Mycoplasma hyopneumoniae. J. Appl. Microbiol. 102:1132–1137. 10.1111/j.1365-2672.2006.03170.x [DOI] [PubMed] [Google Scholar]
- 45.Assunção P, de la Fe C, Antunes NT, Rosales RS, Ruiz de Galarreta CM, Poveda JB. 2006. Use of flow cytometry for enumeration of Mycoplasma mycoides subsp. mycoides large-colony type in broth medium. J. Appl. Microbiol. 100:878–884. 10.1111/j.1365-2672.2005.02858.x [DOI] [PubMed] [Google Scholar]
- 46.Assunção P, Diaz R, Comas J, de Galarreta CM, Gonzalez-Llamazares OR, Poveda JB. 2005. Evaluation of Mycoplasma hyopneumoniae growth by flow cytometry. J. Appl. Microbiol. 98:1047–1054. 10.1111/j.1365-2672.2005.02536.x [DOI] [PubMed] [Google Scholar]
- 47.Mohammadpour HA, Tracy CR, Redelman D, duPre SA, Hunter KW. 2010. Flow cytometric method for quantifying viable Mycoplasma agassizii, an agent of upper respiratory tract disease in the desert tortoise (Gopherus agassizii). Lett. Appl. Microbiol. 50:347–351. 10.1111/j.1472-765X.2010.02800.x [DOI] [PubMed] [Google Scholar]
- 48.Soehnlen MK, Kunze ME, Karunathilake KE, Henwood BM, Kariyawasam S, Wolfgang DR, Jayarao BM. 2011. In vitro antimicrobial inhibition of Mycoplasma bovis isolates submitted to the Pennsylvania Animal Diagnostic Laboratory using flow cytometry and a broth microdilution method. J. Vet. Diagn. Invest. 23:547–551. 10.1177/1040638711404155 [DOI] [PubMed] [Google Scholar]
- 49.Schmeck B, Lorenz J, N′Guessan PD, Opitz B, van Laak V, Zahlten J, Slevogt H, Witzenrath M, Flieger A, Suttorp N, Hippenstiel S. 2008. Histone acetylation and flagellin are essential for Legionella pneumophila-induced cytokine expression. J. Immunol. 181:940–947 [DOI] [PubMed] [Google Scholar]
- 50.Boeuf P, Vigan-Womas I, Jublot D, Loizon S, Barale JC, Akanmori BD, Mercereau-Puijalon O, Behr C. 2005. CyProQuant-PCR: a real time RT-PCR technique for profiling human cytokines, based on external RNA standards, readily automatable for clinical use. BMC Immunol. 6:5. 10.1186/1471-2172-6-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Xiao L, Crabb DM, Duffy LB, Paralanov V, Glass JI, Waites KB. 2012. Chromosomal mutations responsible for fluoroquinolone resistance in Ureaplasma species in the United States. Antimicrob. Agents Chemother. 56:2780–2783. 10.1128/AAC.06342-11 [DOI] [PMC free article] [PubMed] [Google Scholar]