Abstract
Prior antibiotic exposure is associated with increased mortality in Gram-negative bacteria-induced sepsis. However, how antibiotic-mediated changes of commensal bacteria promote the spread of enteric pathogenic bacteria in patients remains unclear. In this study, the effects of systemic antibiotic treatment with or without Toll-like receptor (TLR) stimulation on bacterium-killing activity, antibacterial protein expression in the intestinal mucosa, and bacterial translocation were examined in mice receiving antibiotics with or without oral supplementation of dead Escherichia coli or Staphylococcus aureus. We developed a systemic ampicillin, vancomycin, and metronidazole treatment protocol to simulate the clinical use of antibiotics. Antibiotic treatment decreased the total number of bacteria, including aerobic bacteria belonging to the family Enterobacteriaceae and the genus Enterococcus as well as organisms of the anaerobic genera Lactococcus and Bifidobacterium in the intestinal mucosa and lumen. Antibiotic treatment significantly decreased the bacterium-killing activity of the intestinal mucosa and the expression of non-defensin-family proteins, such as RegIIIβ, RegIIIγ, C-reactive protein-ductin, and RELMβ, but not the defensin-family proteins, and increased Klebsiella pneumoniae translocation. TLR stimulation after antibiotic treatment increased NF-κB DNA binding activity, nondefensin protein expression, and bacterium-killing activity in the intestinal mucosa and decreased K. pneumoniae translocation. Moreover, germfree mice showed a significant decrease in nondefensin proteins as well as intestinal defense against pathogen translocation. Since TLR stimulation induced NF-κB DNA binding activity, TLR4 expression, and mucosal bacterium-killing activity in germfree mice, we conclude that the commensal microflora is critical in maintaining intestinal nondefensin protein expression and the intestinal barrier. In turn, we suggest that TLR stimulation induces nondefensin protein expression and reverses antibiotic-induced gut defense impairment.
INTRODUCTION
The human intestines are colonized by trillions of microorganisms that include hundreds of different species of bacteria and viruses (1, 2). These microbes, collectively called the commensal microflora, play an important role in human nutrition and health by promoting nutrient supply, preventing pathogen colonization, and shaping as well as maintaining normal mucosal immunity (3, 4). The most abundant microflora is present in the distal intestine of mammalians and is represented by Gram-negative obligately anaerobe bacteria.
The intestinal tract acts as a major physical barrier between the microflora and internal host tissue and responds to the mucosal innate system through the commensal microflora (5). The intestinal epithelial cells express pattern recognition receptors (PRRs) that protect against microbial invasion while maintaining epithelial barriers in the presence of commensal microflora (6). The best-known PRRs are the Toll-like receptors (TLRs). TLR2 and TLR4 are essential for the recognition of distinct bacterial cell wall components. TLR2 discriminates peptidoglycan, lipoprotein, lipoarabinomannan, and zymosan, whereas TLR4 recognizes lipopolysaccharide (LPS), lipoteichoic acid (LTA), and paclitaxel (originally named taxol). A major downstream effect of TLR signaling is the activation of NF-κB. Paneth cells are important contributors to the limiting of bacterial penetration and to the small-intestinal antimicrobial barrier through synthesis and release of a broad range of antimicrobial peptides and proteins, such as enteric α-defensins (called cryptdins in mice), CRS (cryptdin-related sequence) peptides, lysozyme, RegIIIβ, RegIIIγ, RELMβ (resistin-like molecules β), and C-reactive protein (CRP)-ductin (7). It has been shown that expression of RegIIIγ is dependent on TLR-MyD88-mediated signaling in intestinal epithelial cells and is induced by commensal microbes (8).
The normal microflora acts as a barrier against colonization and invasion of potentially pathogenic microorganisms. Recently, the augmented use of broad-spectrum antibiotics such as ampicillin has led to increased rates of colonization by potentially pathogenic members of the Enterobacteriaceae, such as Klebsiella pneumoniae (9). Moreover, a retrospective cohort study of hospitalized patients demonstrated that prior antibiotic exposure was associated with increased hospital mortality of patients with bacteremia caused by Gram-negative organisms complicated by severe sepsis or septic shock. This may be due to the enhanced antimicrobial resistance of the causative pathogens of patients receiving prior antibiotics (10).
Therefore, we hypothesize that combined antibiotic treatment alters intestinal microbial composition, thus compromising the complex gut defense mechanisms. Through this mechanism, pathogens, such as K. pneumoniae, invade organs, such as the liver and blood. Here, using systemic combined antibiotic treatment in mice, we demonstrated that antibiotic treatment significantly decreased the commensal microflora's NF-κB DNA binding activity as well as nondefensin protein expression in the intestinal mucosa and increased pathogen translocation. Moreover, TLR stimulation induced TLR4 expression and gut defense mechanisms in antibiotic-treated mice and germfree mice. Therefore, oral supplementation of TLR ligands to induce TLR4 expression and NF-κB DNA binding activity in the intestinal mucosa could be a useful strategy for enhancing intestinal defense mechanisms and preventing enteric pathogen translocation in antibiotic-treated patients.
MATERIALS AND METHODS
Animals.
We obtained specific pathogen-free (SPF) and germfree C57BL/6 mice from the National Laboratory Breeding and Research Center (NLBRC, Taipei, Taiwan). Myd88−/− (C57BL/6 background) mice were obtained from Oriental Bioservice, Inc. (Kyoto, Japan). Experiments were in compliance with regulations approved by the National Sun Yat-Sen Animal Experiments Committee.
Experimental design. (i) Experiment 1.
To simulate the use of combined antibiotic treatment in clinical patients with infections, we developed a new broad-spectrum antibiotic treatment regimen in which animals received a combined intramuscular injection of different antibiotics: ampicillin (1,000 mg/liter; Bio Basic Inc.), vancomycin (500 mg/liter; Hospira), and metronidazole (1,000 mg/liter; Sigma) at 100 μl/day for 4 or 6 days. C57BL/6 mice were randomly divided into three groups: group I received an intramuscular injection of normal saline, group II received an intramuscular injection of antibiotics for 4 days, and group III received antibiotics for 6 days. All animals fasted, and sterile water was provided at day 4. The mice were sacrificed at day 6 after antibiotic injection. Bacterial count, intestinal permeability, bacterial translocation (BT) to mesenteric lymph nodes (MLNs), translocation of Escherichia coli or K. pneumoniae to MLNs, liver, and blood, and mucosal bacterium-killing activity were measured in all animals after antibiotic treatment. The bacterial genomic DNA was extracted from the terminal ileum and MLNs for quantitative analysis by PCR and real-time PCR. The terminal ileum was also harvested to analyze NF-κB DNA binding activity and expression of Reg3β, Reg3γ, CRP-ductin, RELMβ, Defcr-rs-10, Crypt1, Crypt4, TLR2, TLR4, and MyD88.
(ii) Experiment 2.
For TLR stimulation in the intestinal mucosa, sterile drinking water supplemented with dead E. coli or S. aureus (2 × 106 CFU/ml) was administered at day 4 to animals receiving antibiotic intramuscular injections for 6 days. C57BL/6 mice were randomly divided into four groups: group I received intramuscular injection of normal saline, group II received intramuscular injection of antibiotics for 6 days, group III received antibiotics for 6 days with water supplemented with dead E. coli at day 4, and group IV received antibiotics for 6 days with water supplemented with dead S. aureus at day 4. The intestines of the animals were collected for assays as described above for experiment 1.
Bacterial content of the intestinal mucosa and lumina.
The collected mucosa and lumen from the terminal ileum were weighed and homogenized in an equal volume of sterile saline. Aliquots of the homogenate from each sample were diluted and 100 μl of the homogenates were plated on different selective agar plates for specific-bacterial strain. The total aerobic bacteria were cultured on tryptic soy broth (TSB) agar plates (Difco); aerobic bacteria belonging to the family Enterobacteriaceae and the genus Enterococcus were cultured on eosin methylene blue and m-Enterococcus agar plates. The anaerobic bacteria Bacteroides, Clostridium perfringens, and Lactobacillus and Bifidobacterium were cultured on Bacteroides bile esculin, tryptose-sulfite-cycloserine, and Bifidobacterium iodoacetate medium-25 agar plates, respectively. The plates were examined for CFU after aerobic incubation at 37°C for 24 h and anaerobic incubation at 37°C for 5 days in an anaerobic chamber.
Measurement of intestinal permeability.
The assay of intestinal permeability was modified from the method described by Otamiri et al. (11). Two ends of a 10-cm segment of small intestine were clipped, 50 μl (25 mg/ml) of fluorescein isothiocyanate (FITC)-dextran (molecular weight [MW], 4,400; Sigma) was injected into the clipped intestinal lumen, and 100 μl of blood was taken from the heart at 30 min later. The sample was analyzed for FITC-dextran concentration at an excitation wavelength of 480 nm and an emission wavelength of 520 nm, and the concentration of FITC-dextran in blood was calculated.
Bacterial translocation to MLNs.
The collected MLNs were weighed and homogenized in an equal volume of sterile saline. Aliquots of the homogenates were plated onto TSB agar plates. The plates were examined after aerobic incubation at 37°C for 24 h.
PCR and quantitative real-time PCR.
Total RNAs were isolated from terminal ileum using total RNA miniprep purification kits (GeneMark). Reverse transcription-generated cDNAs were amplified using PCR. Sets of TLR2 and TLR4 primers were designed according to genes documented in GenBank; sets of Reg3β, Reg3γ, CRP-ductin, RELMβ, Defcr-rs-10, Crypt1, and Crypt4 primers were designed as described in references 12 and 13, and one pair of primers for the GAPDH gene was used as a control.
For the real-time PCRs, 200 ng of the cDNA template was added to 20 μl of mixture containing 12.5 μl of 2× Kapa Sybr fast qPCR master mix (Kapa Biosystems), 2.5 μl of each sense and antisense primer (25 μM), and 5 μl of sterile water. The amplification was performed in a StepOnePlus real-time PCR system (Applied Biosystems 7300).
Bacterial DNA extraction and quantitative real-time PCR.
Bacterial genomic DNA was extracted from terminal ileum using the Qiagen DNA stool kit according to the manufacturer's directions. The number of specific bacterial groups was determined by using a StepOnePlus real-time PCR system (Applied Biosystems 7300). The sequences of specific bacterial primers were as previously published (14).
Western immunoblots.
The RegIIIβ, RELMβ, CRP-ductin, and α-defensin 4 were identified by mouse monoclonal antibodies (R&D Systems); MyD88 and TLR4 were identified by mouse monoclonal and goat polyclonal antibodies, respectively (Santa Cruz Biotechnology Inc.).
K. pneumoniae translocation.
After the animals were anesthetized, the two ends of a 10-cm segment of the small intestine were clipped. Normal saline (500 μl; pH 7.2) containing K. pneumoniae (5 × 107 CFU) was injected into the isolated intestinal segment. After 1 h, MLNs and livers were collected, weighed, and homogenized in an equal volume of sterile saline, and 100 μl of blood was withdrawn from the heart. Blood or aliquots of the homogenates were plated onto TSB agar plates with or without ampicillin (100 μg/ml). The plates were examined after aerobic incubation at 37°C for 24 h.
Determination of the bacterium-killing activity of the mucosa.
The mucosa of the terminal ileum was collected, weighed, and thoroughly suspended in an equal volume of sterile saline by incubation in a shaker (200 rpm) at room temperature for 30 min. Suspended mucosa was centrifuged at 12,000 × g for 5 min, and the supernatant containing antibacterial proteins was collected. A 100-μl bacterial suspension containing E. coli or K. pneumoniae (1 × 103 CFU) were added to 400 μl of supernatant and incubated at room temperature for 30 min. Finally, 100 μl of the supernatant was plated onto LB agar plates with or without ampicillin (100 μg/ml).
Electrophoretic mobility shift assay (EMSA) for NF-κB.
Intestinal mucosae were harvested by centrifugation and used to prepare nuclear extract as described previously (15). The consensus and control oligonucleotides (Santa Cruz Biotechnology Inc.) were labeled by polynucleotide kinase; the NF-κB consensus sequence was 5′AGTTGAGGGGACTTTCCCAGGC3′ (1.75 pmol/liter). The samples were analyzed on a 4% polyacrylamide gel, and the gel was dried and visualized by autoradiography.
Statistical analysis.
All immunoblotting and electrophoretic mobility shift assays were analyzed by densitometric scanning. Data are expressed as means ± standard deviations of the means, and a P value of <0.05 is considered statistically significant. The bacterial counts, intestinal permeability, mucosal bacterium-killing activity, and BT to MLNs between groups were assessed with one-way analysis of variance (ANOVA), followed by Tukey's test.
RESULTS
Antibiotic treatment decreased the intestinal microflora.
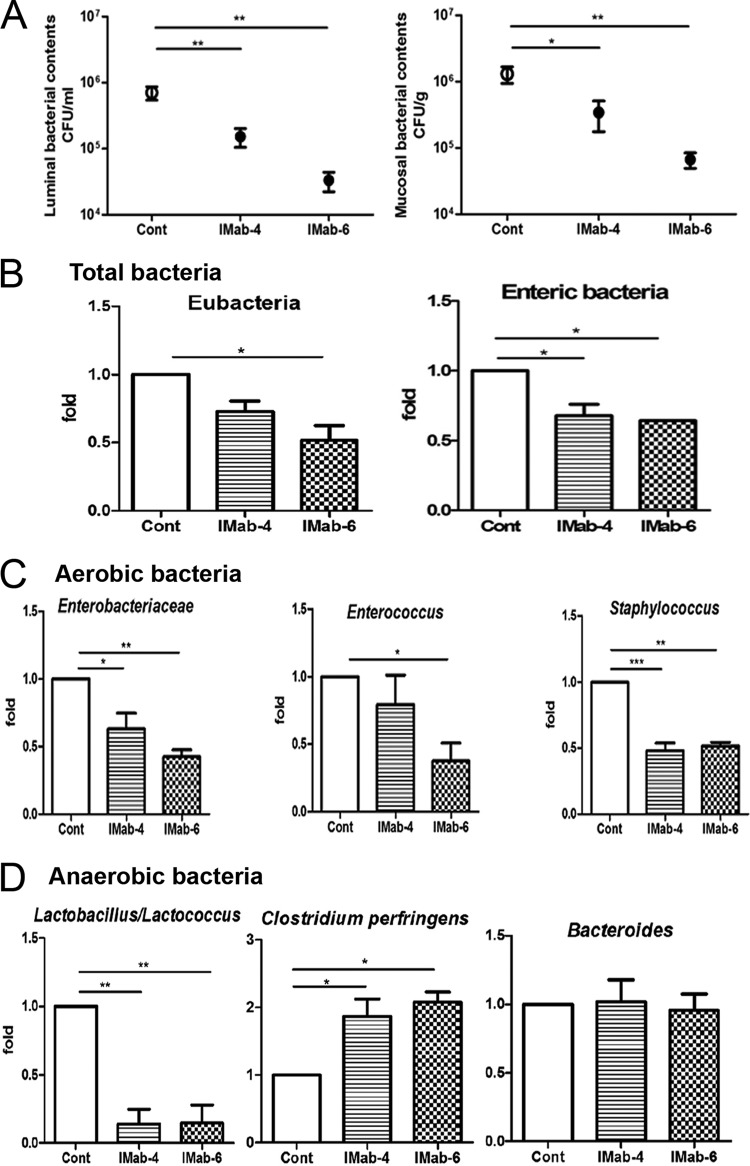
After antibiotic treatment for 4 and 6 days, total aerobic bacterial counts in the lumen significantly decreased by 5.5 × 105 CFU/ml and 6.7 × 105 CFU/ml and in the mucosa by 9.7 × 105 CFU/g and 1.2 × 106 CFU/g, respectively, as demonstrated by plate cultivation (Fig. 1A). Moreover, antibiotic treatment for 6 days significantly decreased counts of the aerobic bacteria of the family Enterobacteriaceae and the genus Enterococcus in the mucosa and lumen (data not shown). Antibiotic treatment for 6 days significantly decreased counts of the anaerobic bacteria Lactobacillus and Bifidobacterium in the mucosa but not in the lumen, whereas those of the anaerobic genus Bacteroides had no difference in the mucosa and in the lumen (data not shown).
FIG 1.
Antibiotic treatment for 4 and 6 days significantly decreased the commensal microflora in the lumen and mucosa in C57BL/6 mice. (A) Total aerobic bacterial counts in the mucosa and lumen of the terminal ileum were significantly decreased after intramuscular antibiotic treatment for 4 or 6 days. (B) Real-time PCR results showed that copy numbers of total Eubacteria organisms and enteric bacteria were significantly decreased after antibiotic treatment for 6 days. (C) Copy numbers of members of the Enterobacteriaceae, Enterococcus, and Staphylococcus were significantly decreased after antibiotic treatment for 6 days, but those of Pseudomonas and Salmonella did not show any difference. (D) Copy numbers of Lactobacillus were significantly decreased after antibiotic treatment for 4 or 6 days, those of Bacteroides did not show any difference, and those of C. perfringens were significantly increased after antibiotic treatment for 6 days. Cont, control; IMab-4, intramuscular antibiotic treatment for 4 days; IMab-6, intramuscular antibiotic treatment for 6 days; *, P < 0.05; **, P < 0.01; ***, P < 0.001. n = 4 to 6/group.
Real-time PCR analysis showed that copy numbers of the total mucosal aerobic bacteria, such as Eubacteria, enteric bacteria (Fig. 1B), and Gram-positive and -negative bacteria (data not shown), all significantly decreased after antibiotic treatment for 6 days. Copy numbers of aerobic bacteria of the family Enterobacteriaceae and members of the genera Enterococcus and Staphylococcus significantly decreased after antibiotic treatment for 6 days (Fig. 1C), but those of Pseudomonas showed no difference (data not shown). Copy numbers of the anaerobic genus Lactobacillus decreased after antibiotic treatment for 4 or 6 days, those of Bacteroides showed no difference, and those of C. perfringens increased after antibiotic treatment for 6 days (Fig. 1D). These results indicate that combined administration of ampicillin, vancomycin, and metronidazole for 4 or 6 days significantly decreases aerobic as well as anaerobic bacteria but increases C. perfringens in the intestine.
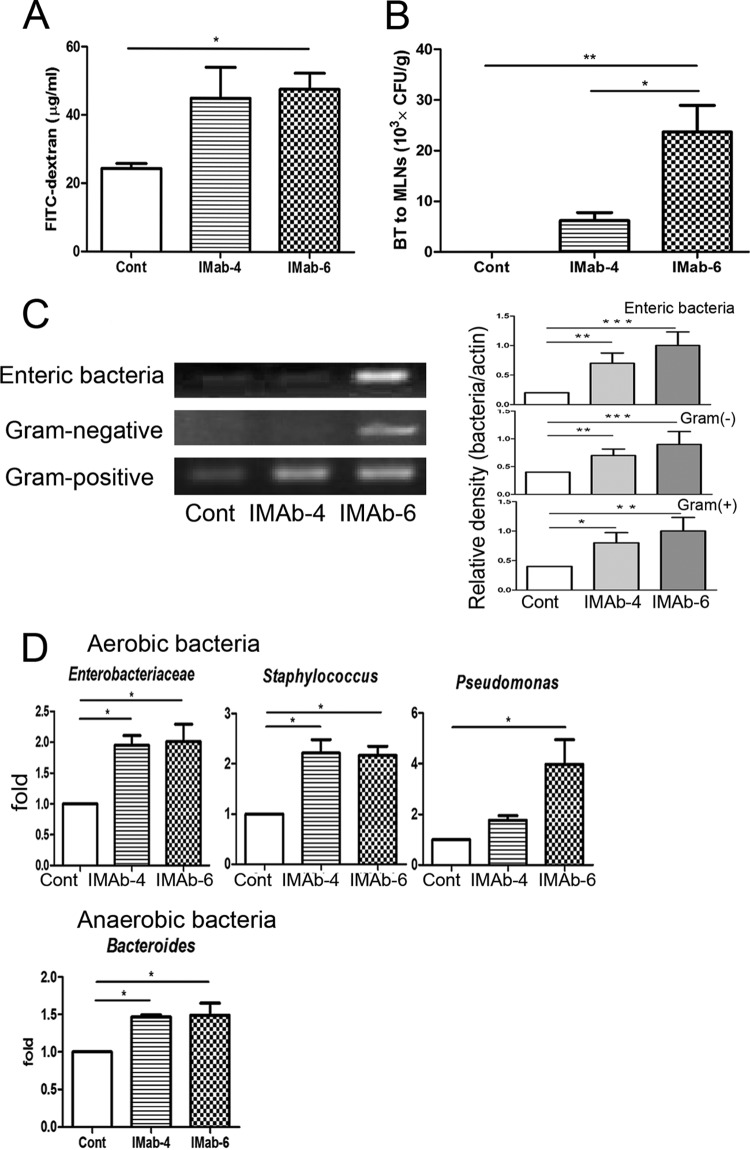
Antibiotic treatment increased intestinal permeability and bacterial translocation.
Antibiotic treatment for 4 days induced a significant 2-fold increase of intestinal permeability compared with the control group (Fig. 2A). Similarly, antibiotic treatment for 6 days induced a significant 24-fold increase of bacterial translocation to MLNs (Fig. 2B). To evaluate specific groups of intestinal microflora that translocated to MLNs, bacterial genomic DNA extracted from MLNs was analyzed by RT-PCR and real-time PCR using specific bacterial primers. Antibiotic treatment for 4 days or 6 days induced a significant increase in translocation of Eubacteria, enteric bacteria, and Gram-positive and -negative to MLNs compared with the control group (Fig. 2C). Antibiotic treatment markedly induced the translocation of aerobic bacteria belonging to the Enterobacteriaceae, Enterococcus, Staphylococcus, and Pseudomonas and the anaerobic genus Bacteroides to MLNs (Fig. 2D), indicating that antibiotic treatment decreases the total amount of bacteria in the intestine and induces intestinal permeability as well as enteric BT to MLNs.
FIG 2.
Antibiotic treatment induced intestinal permeability and bacterial translocation. (A) Antibiotic treatment for 4 days or 6 days significantly increased the intestinal permeability compared with the control group. (B) Antibiotic treatment for 4 or 6 days significantly increased translocation of enteric bacteria to mesenteric lymph nodes (MLNs) compared with the control group. (C) Antibiotic treatment for 6 days induced a significant increase in the translocation of Eubacteria organisms, enteric bacteria, and Gram-positive and -negative bacteria to MLNs. (D) Antibiotic treatment markedly induced the translocation of members of the Enterobacteriaceae, Enterococcus, Staphylococcus, Pseudomonas, and Bacteroides to MLNs. FITC, fluorescein isothiocyanate; Cont, control; IMab-4, intramuscular antibiotic treatment for 4 days; IMab-6, intramuscular antibiotic treatment for 6 days; BT, bacterial translocation; MLNs, mesenteric lymph nodes. *, P < 0.05; **, P < 0.01; ***, P < 0.001. n = 4 to 6/group.
Antibiotic treatment decreased antibacterial protein expression in terminal ileum.
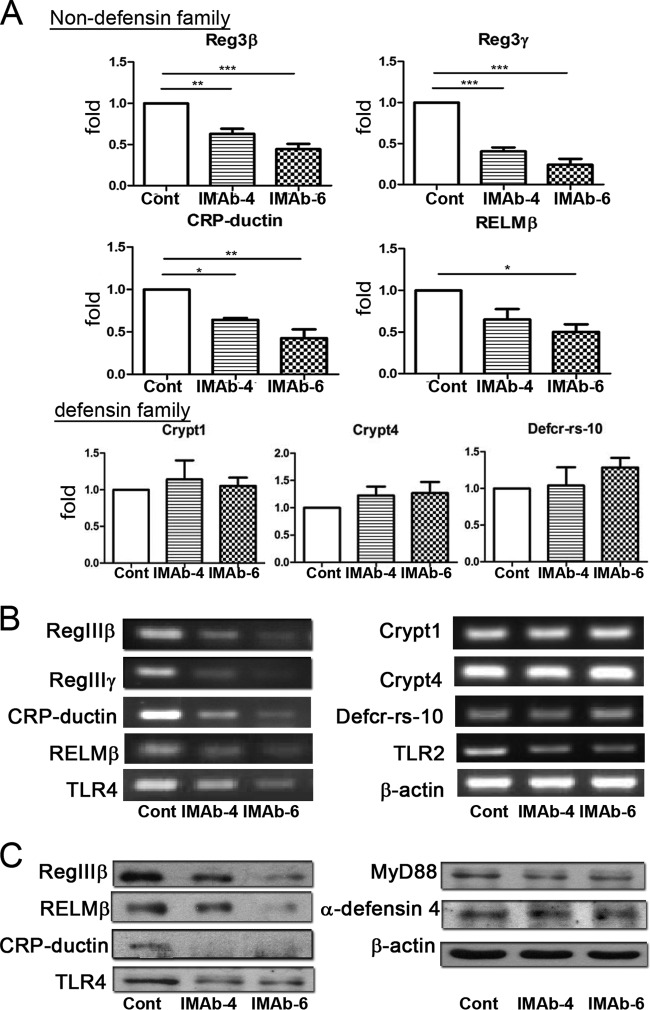
To examine the mechanisms of antibiotic treatment on BT to MLNs, the expression of antibacterial proteins in the terminal ileum was analyzed by RT-PCR, real-time PCR, and immunoblotting. Quantification analysis of real-time PCR results showed that the mRNA levels of non-defensin-family molecules, such as Reg3β, Reg3γ, CRP-ductin, and RELMβ were 37%, 60%, 36%, and 20% decreased, respectively, in mice treated with antibiotics for 4 days compared with the control mice. Furthermore, 56%, 76%, 57%, and 70% decreases in Reg3β, Reg3γ, CRP-ductin, and RELMβ mRNA, respectively, were observed in mice treated with antibiotics for 6 days compared with those of control mice (Fig. 3A). However, no difference in the expression of the defensin-family proteins Defcr-rs-10, Crypt1, and Crypt4 was observed between antibiotic-treated and control mice (Fig. 3A). The results of RT-PCR analysis (Fig. 3B) were consistent with those of real-time PCR analysis. Also, the mRNA levels of TLR4 were significantly decreased but those of TLR2 were only mildly decreased after antibiotic treatment.
FIG 3.
Antibiotic treatment for 4 or 6 days decreased the expression of nondefensin proteins of the intestinal mucosa. Expression of RegIIIβ, RegIIIγ, CRP-ductin, RELMβ, Crypt1, Crypt4, Defcr-rs-10, TLR4, and TLR2 mRNAs in the terminal ileum was analyzed by real-time PCR (A) and RT-PCR (B). (C) Protein expression of RegIIIβ, RELMβ, CRP-ductin, TLR4, MyD88, and α-defensin 4 in the terminal ileum was analyzed by immunoblotting. Cont, control; IMab-4, intramuscular antibiotic treatment for 4 days; IMab-6, intramuscular antibiotic treatment for 6 days; *, P < 0.05; **, P < 0.01; ***, P < 0.001. n = 3/group.
Quantification analysis of immunoblots showed that after antibiotic treatment for 4 and 6 days, there was a significant reduction of RegIIIβ (36% and 74%, respectively) and RELMβ (52% and 81%, respectively) expression (Fig. 3C). However, no significant difference in the expression of α-defensin 4, one of the defensin-family proteins, was observed between antibiotic treated and control mice. In addition, there was a 52% decrease in the expression of TLR4 and a 32% decrease of Myd88 after antibiotic treatment for 6 days compared with the control group (Fig. 3C). These results indicate that antibiotic treatment significantly decreases TLR4 as well as nondefensin protein expression but not α-defensin 4 expression in the intestinal mucosa.
TLR stimulation reversed antibiotic-induced K. pneumoniae translocation and reduced mucosal bacterium-killing activity.
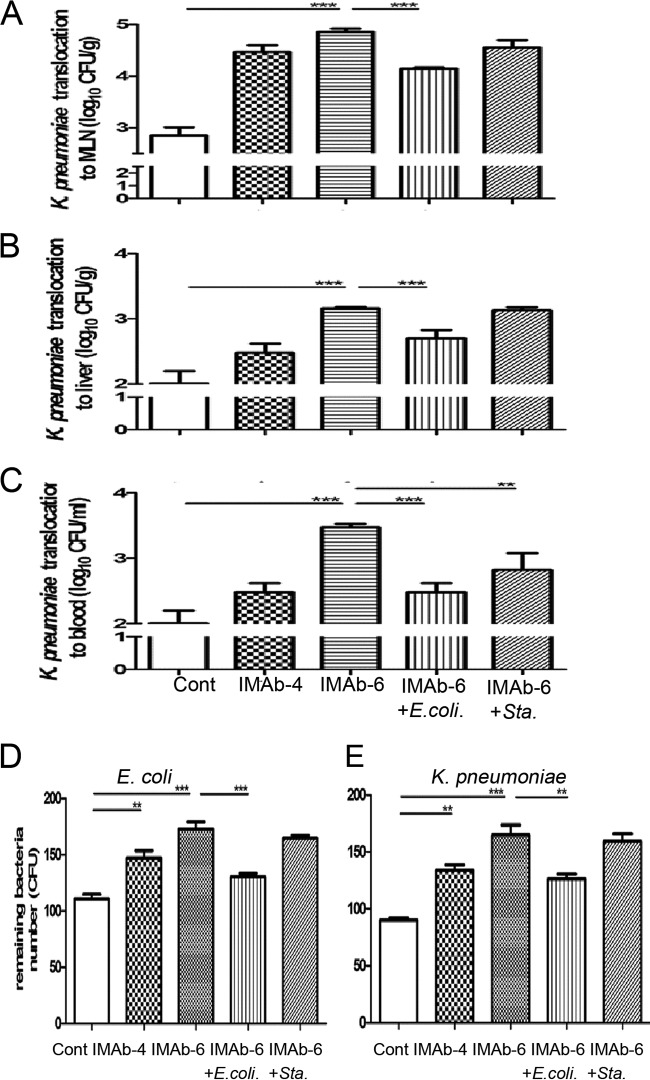
To further examine the effect of antibiotics on gut defense against enteric pathogen invasion, K. pneumoniae (5 × 107 CFU) was injected into the isolated intestinal segment, and MLNs, liver, and blood were collected and cultured 1 h after injection for the evaluation of K. pneumoniae translocation. Antibiotic treatment for 4 and 6 days induced significant 38-fold and 93-fold increases, respectively, of K. pneumoniae translocation to MLNs compared with the control group (Fig. 4A). Moreover, K. pneumoniae translocation to the liver and blood showed significant 21-fold and 41-fold increases, respectively, after antibiotic treatment for 6 days (Fig. 4B and C). Moreover, S. aureus supplementation significantly decreased K. pneumoniae translocation to the blood (Fig. 4C) but mildly decreased the translocation to MLNs as well as the liver compared with the antibiotic treatment group (Fig. 4A and B).
FIG 4.
TLR stimulation of antibiotic-treated mice increased the bacterium-killing activity and decreased the translocation of pathogenic K. pneumoniae. Antibiotic treatment for 6 days significantly increased the translocation of injected K. pneumoniae to mesenteric lymph nodes (MLNs) (A), liver (B), and blood (C). TLR stimulation with dead E. coli supplementation significantly decreased K. pneumoniae translocation to MLNs (A), liver (B), and blood (C), whereas supplementation with dead S. aureus significantly decreased K. pneumoniae translocation only to the blood (C). Antibiotic treatment for 4 and 6 days significantly decreased the mucosal bacterium-killing activity against E. coli (D) and K. pneumoniae (E). TLR stimulation with dead E. coli but not S. aureus significantly increased the mucosal bacterium-killing activity against both E. coli (D) and K. pneumoniae (E). MLNs, mesenteric lymph nodes; Cont, control; IMab-4, intramuscular antibiotic treatment for 4 days; IMab-6, intramuscular antibiotic treatment for 6 days; **, P < 0.01; ***, P < 0.001. n = 4 to 6/group.
To evaluate the effect of antibiotic treatment on the mucosal bacterium-killing activity, the antibacterial proteins were extracted from the mucosa of the terminal ileum and added to the bacterial suspension. Forty-five and fifty-five percent of the inoculated E. coli and K. pneumoniae, respectively, were killed when incubated with the extracted antibacterial proteins from the mucosa of mice without antibiotic treatment (Fig. 4D and E). The mucosal bacterium-killing activity in mice treated with antibiotics for 4 days significantly decreased to 30% for E. coli (Fig. 4D) and 38% for K. pneumoniae (Fig. 4E). The mucosal bacterium-killing activity in mice treated for 6 days further decreased to 17% for E. coli (Fig. 4D) and 24% for K. pneumoniae (Fig. 4E). TLR stimulation with dead E. coli supplementation in mice treated with antibiotics for 6 days significantly increased the mucosal bacterium-killing activity against both E. coli and K. pneumoniae, but supplementation with dead S. aureus had no effect compared with the control group (Fig. 4D and E). These data indicate that TLR stimulation enhances the mucosal bacterium-killing activity, thereby overcoming the loss of antibiotic-induced intestine defense.
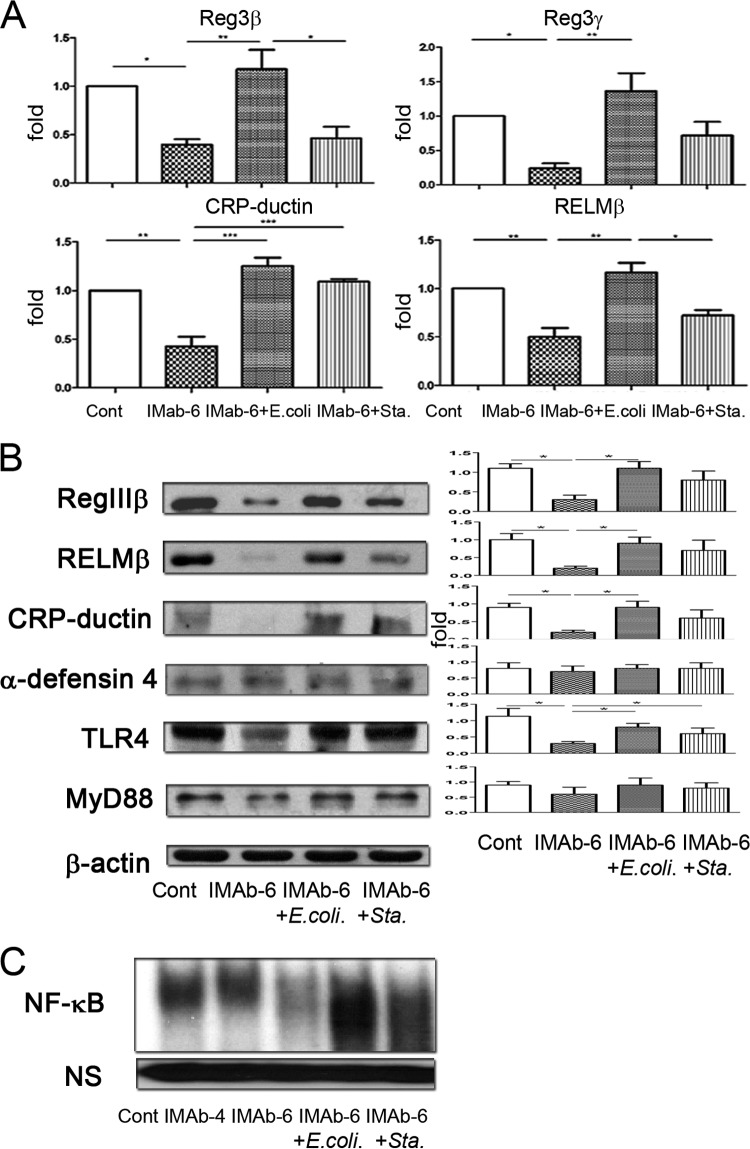
TLR stimulation reversed antibiotic-induced reduction of nondefensin protein expression and NF-κB DNA-binding activity in the intestinal mucosa.
Next, we investigated the mechanism underlying the reversion of antibiotic treatment-induced intestine defense deficit by TLR stimulation. TLR activation in the intestinal mucosa using dead E. coli significantly induced mRNA levels of the non-defensin-family molecules, including Reg3β, Reg3γ, CRP-ductin, and RELMβ, compared with the antibiotic treatment group (Fig. 5A). However, supplementation with dead S. aureus significantly increased the expression of CRP-ductin and mildly increased the expression of Reg3β, Reg3γ, and RELMβ compared with antibiotic treatment for 6 days (Fig. 5A). In contrast, TLR stimulation with dead E. coli or S. aureus had no effect on the mRNA levels of defensin-family molecules Defcr-rs-10, Crypt1, and Crypt4 (data not shown). RT-PCR results were consistent with real-time PCR ones (data not shown).
FIG 5.
TLR stimulation reversed antibiotic treatment-induced reduction of nondefensin proteins and NF-κB DNA binding activity of the intestinal mucosa. (A) TLR stimulation with dead E. coli oral supplementation significantly induced Reg3β, Reg3γ, CRP-ductin, and RELMβ mRNA levels in the intestinal mucosa of antibiotic-treated mice. Dead S. aureus oral supplementation significantly increased the expression of CRP-ductin in the intestinal mucosa. (B) Immunoblotting assays showed that TLR stimulation with dead E. coli induced a significant increase in RegIIIβ, RELMβ, and TLR4 protein expression in antibiotic-treated mice. Dead S. aureus supplementation induced a significant increase in TLR4 protein expression in antibiotic-treated mice. TLR stimulation with dead E. coli or S. aureus had no effect on the expression of α-defensin 4. (C) Antibiotic treatment for 6 days (IMAb-6) significantly decreased NF-κB DNA binding activity in the intestinal mucosa. Dead E. coli or S. aureus supplementation for 2 days reversed the inhibitory effect of antibiotics on NF-κB DNA binding activity in the intestinal mucosa. NS, nonspecific band; Cont, control; IMab-4, intramuscular antibiotic treatment for 4 days; IMab-6, intramuscular antibiotic treatment for 6 days; *, P < 0.05; **, P < 0.01; ***, P < 0.001. n = 3 to 5/group.
Similarly, immunoblotting assays demonstrated that dead E. coli supplementation in antibiotic-treated mice induced a significant increase of RegIIIβ, RELMβ, CRP-ductin, and TLR4 expression and a mild increase of MyD88 expression (Fig. 5B). However, dead S. aureus supplementation induced a significant increase of TLR4 and a moderate increase of RegIIIβ, RELMβ, and CRP-ductin expression in the intestinal mucosa compared with the control group. However, TLR stimulation by supplementation with dead E. coli or S. aureus had no effect on the expression of α-defensin 4 (Fig. 5B).
Antibiotic treatment for 6 days induced a significant decrease of NF-κB DNA binding activity in the terminal ileum, thus indicating that NF-κB activity in the intestinal mucosa is regulated by the commensal microflora. Moreover, the NF-κB DNA binding activity in the terminal ileum was markedly induced when mice received dead E. coli or S. aureus supplementation after antibiotic treatment (Fig. 5C). Taken together, these results indicate that TLR stimulation reverses the inhibitory effect of antibiotic treatment on RegIIIβ, RELMβ, CRP-ductin, and TLR4 expression and NF-κB DNA binding activity in the intestinal mucosa.
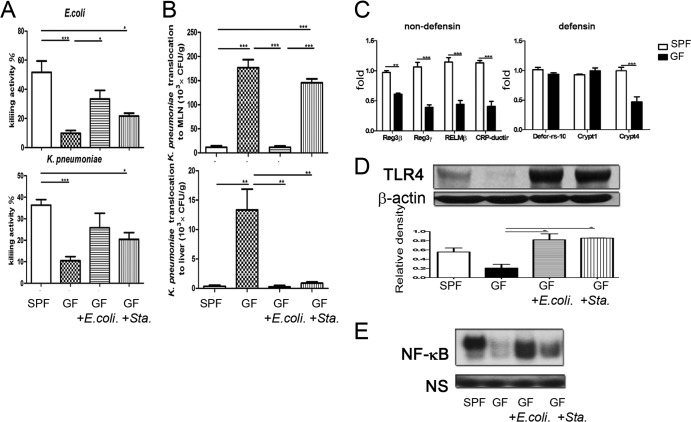
TLR stimulation increased mucosal bacterium-killing activity in germfree mice.
To assess the role of the commensal microflora on gut defense mechanisms, we examined the killing activity and antibacterial protein expression in the intestinal mucosa of germfree mice. The mucosal bacterium-killing activity in germfree mice was significantly decreased to 30% for E. coli and 38% for K. pneumoniae (Fig. 6A) compared with that in specific-pathogen-free (SPF) mice. Next, the effects of TLR stimulation on mucosal bacterium-killing activity in germfree mice were assessed. Supplementation with dead E. coli significantly increased the mucosal bacterium-killing activity in germfree mice against E. coli, whereas supplementation with dead S. aureus mildly increased it (Fig. 6A). Next, after injection of K. pneumoniae (5 × 107 CFU) into the isolated intestinal segment of germfree mice, MLNs and livers were collected for the evaluation of K. pneumoniae translocation. K. pneumoniae translocation to the MLNs and liver showed significant 15-fold and 32-fold increases, respectively, compared with that in SPF mice (Fig. 6B). This suggests that germfree mice decreased the intestinal defense against K. pneumoniae translocation. To evaluate the effect of TLR stimulation on intestinal defense against pathogen translocation, germfree mice were supplemented with dead E. coli or S. aureus for 2 weeks and examined for K. pneumoniae translocation. This treatment significantly decreased K. pneumoniae translocation to MLNs and liver in germfree mice compared with the control group (Fig. 6B). Similarly, oral supplementation of dead S. aureus in germfree mice significantly decreased K. pneumoniae translocation to the liver (Fig. 6B), suggesting that TLR stimulation induces intestinal defense against pathogen translocation in germfree mice.
FIG 6.
TLR stimulation decreased K. pneumoniae translocation and increased mucosal bacterium-killing activity, TLR4 expression, and NF-κB DNA binding activity in the intestinal mucosa in germfree mice. (A) The mucosal bacterium-killing activity against E. coli and K. pneumoniae was significantly decreased in germfree mice compared with SPF mice. TLR stimulation with dead E. coli significantly increased the mucosal bacterium-killing activity against E. coli in germfree mice. (B) K. pneumoniae translocation to MLNs and liver of germfree mice was significantly increased compared with that of SPF mice. TLR stimulation with dead E. coli significantly decreased K. pneumoniae translocation to MLNs and liver. Oral supplementation with dead S. aureus in germfree mice significantly decreased K. pneumoniae translocation to the liver. (C) Reg3β, Reg3γ, RELMβ, CRP-ductin, Defcr-rs-10, Crypt1, and Crypt4 mRNA expression in the intestinal mucosa of germfree and SPF mice. (D) TLR stimulation with dead E. coli or S. aureus (Sta.) significantly induced TLR4 protein expression in the intestinal mucosa in germfree mice. (E) TLR stimulation with dead E. coli or S. aureus significantly induced NF-κB DNA binding activity in the intestinal mucosa in germfree mice. GF, germfree mice; SPF, specific-pathogen-free mice. *, P < 0.05; **, P < 0.01; ***, P < 0.001. n = 4 to 6/group.
TLR stimulation increased TLR4 expression and NF-κB DNA binding activity in the intestinal mucosa of germfree mice.
Next, we studied the molecular mechanisms involved in the mucosal bacterium-killing activity induced by TLR stimulation in germfree mice. PCR results showed that the mRNA levels of Crypt4, Reg3β, Reg3γ, RELMβ, and CRP-ductin were significantly decreased in germfree mice compared with those in SPF mice (Fig. 6C). However, no difference was observed in Defcr-rs-10 and Crypt1 mRNA expression in the intestinal mucosa between the two groups of mice. Similarly, protein levels of RegIIIβ and RELMβ were significantly decreased in germfree mice compared with that in SPF mice (data not shown). However, no difference in protein expression of α-defensin 4 was observed (data not shown). These data indicate that the commensal microflora is important in regulating nondefensin protein expression in the intestinal mucosa.
Next, we examined the effect of TLR stimulation on antibacterial protein expression in the intestinal mucosa of germfree mice. TLR stimulation with dead E. coli or S. aureus significantly induced TLR4 protein expression in germfree mice (Fig. 6D) but had no effect on the mRNA levels of the defensin-family molecules Defcr-rs-10, Crypt1, and Crypt4 (data not shown). Moreover, this treatment significantly induced NF-κB DNA binding activity in the terminal ileum of germfree mice (Fig. 6E). These results indicate that TLR stimulation induces TLR4 expression, NF-κB DNA binding activity in the intestinal mucosa, and intestinal mucosal bacterium-killing activity.
TLR stimulation did not induce RegIIIβ expression in Myd88−/− mice.
Myd88−/− mice were used to examine the role of TLRs in antimicrobial protein expression in the intestinal mucosa induced by supplementation with dead E. coli and S. aureus. PCR results showed that the mRNA levels of Reg3β, Reg3γ, RELMβ, and CRP-ductin were significantly decreased in Myd88−/− mice compared with those in WT mice (Fig. 7A). However, no difference was observed in Defcr-rs-10, Crypt1, and Crypt4 mRNA expression in the intestinal mucosa between the two groups of mice (Fig. 7B). mRNA and protein expression of RegIIIβ was significantly decreased in the intestinal mucosa of Myd88−/− mice compared with that in WT mice (Fig. 7C and D). Moreover, TLR stimulation with dead E. coli or S. aureus had no effect on the mRNA and protein expression of RegIIIβ in the intestinal mucosa of Myd88−/− mice (Fig. 7C and D). These results indicate that Myd88 plays an important role in RegIIIβ expression and dead E. coli or S. aureus supplementation induces antimicrobial protein expression through Myd88.
FIG 7.
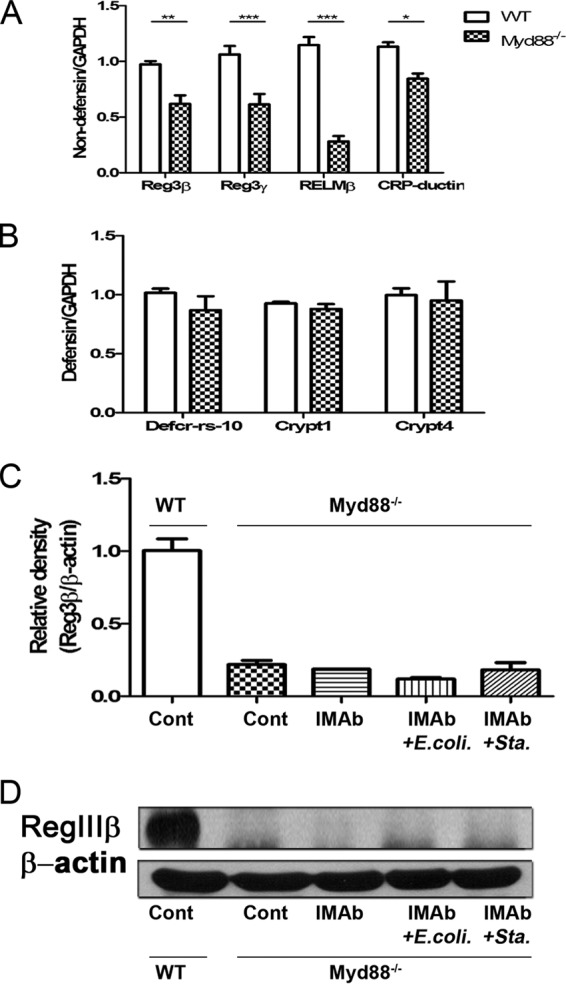
Dead E. coli or S. aureus supplementation did not induce RegIIIβ expression in Myd88−/− mice. (A) Reg3β, Reg3γ, RELMβ, and CRP-ductin mRNA expression in the intestinal mucosa of Myd88−/− and WT mice. (B) Defcr-rs-10, Crypt1, and Crypt4 mRNA expression in the intestinal mucosa of Myd88−/− and WT mice. (C) Dead E. coli or S. aureus supplementation did not stimulate RegIIIβ mRNA expression in the intestinal mucosa of Myd88−/− mice. (D) Dead E. coli or S. aureus supplementation did not stimulate RegIIIβ protein expression in the intestinal mucosa of Myd88−/− mice. Cont, control; IMab, intramuscular antibiotic treatment; *, P < 0.05; **, P < 0.01; ***, P < 0.001. n = 3 to 5/group.
DISCUSSION
The invasion of commensal bacteria into tissues can result in the breakdown of symbiotic host-microorganism interactions, thus contributing to pathologies such as inflammatory bowel diseases (16). In this study, we show that antibiotic treatment for 6 days significantly reduced numbers of aerobic bacteria belonging to the Enterobacteriaceae and Enterococcus in the intestinal mucosa and lumen and numbers of the anaerobic bacteria Lactococcus and Bifidobacterium in the mucosa but did not change numbers of Bacteroides in the mucosa and lumen. It has been reported that antibiotic treatment disrupted the intestinal microflora, leading to increased infection susceptibility by enteric bacteria, such as Listeria monocytogenes (17), C. difficile, Salmonella enterica (18), E. coli, and Enterococcus faecalis (19). Our PCR results demonstrated that the total amount of enteric bacteria was decreased and the amount of major groups of intestinal bacteria, such as the aerobic bacteria of the Enterobacteriaceae and Enterococcus as well as the anaerobic genera Lactobacillus and Lactococcus were significantly decreased in mice receiving antibiotics for 6 days. Our real-time PCR results also showed that the total amounts of enteric bacteria, aerobic bacteria belonging to the Enterobacteriaceae, Enterococcus, and Staphylococcus, and anaerobic bacteria belonging to the genera Lactobacillus and Lactococcus were significantly decreased. However, the anaerobic species C. perfringens was significantly increased. In contrast, the amounts of the aerobic bacteria Pseudomonas and Salmonella as well as the anaerobic genus Bacteroides were unchanged after antibiotic treatment. These results indicate that the antibiotic procedure used in this study can disrupt the intestinal microflora of both the mucosa and the lumen. The commensal microflora plays an important role in maintaining the intestinal defense against colonization by pathogenic bacteria, and antibiotic treatment significantly changes the balance of commensal microflora and decreases the numbers of Lactobacillus and Lactococcus organisms, therefore increasing the mucosal adhesion of potential pathogens, such as C. perfringens. Altogether, these results indicate that systemic combined antibiotic treatment induces pathogen translocation by changing the balance of the commensal microflora and intestinal defense mechanisms.
Recently, bacteria from the family Enterobacteriaceae (e.g., Klebsiella spp. and Enterobacter spp.) have been found to be common causes of a wide range of health care-associated infections and have become resistant to available antimicrobial agents (20). More and more bacteria have been reported to develop antibiotic resistance, e.g., methicillin-resistant S. aureus, vancomycin-resistant Enterococcus spp. (21), and widely antibiotic-resistant bacteria such as Bacteroides (22), Pseudomonas aeruginosa, Salmonella, E. coli, and K. pneumoniae) (23). To investigate specific groups of bacteria that translocated to MLNs after antibiotic treatment, the bacterial genomic DNA extracted from MLNs was analyzed by PCR and real-time PCR. PCR results showed that antibiotic treatment for 6 days induced a marked translocation of enteric bacteria to MLNs, including the aerobic bacteria Enterobacteriaceae and Pseudomonas and the anaerobic bacteria Bacteroides. These results were verified by real-time PCR analysis showing that antibiotic treatment for 6 days significantly increased the translocation of the aerobic bacteria Enterobacteriaceae, Enterococcus, Pseudomonas, and Staphylococcus as well as the anaerobic genus Bacteroides to MLNs. Although antibiotic treatment significantly increased the number of C. perfringens in the mucosa, C. perfringens translocation to MLNs was not increased. Antibiotic treatment surprisingly reduced the number of organisms belonging to the Enterobacteriaceae, Enterococcus, and Staphylococcus in the mucosa but increased the translocation of Enterobacteriaceae, Enterococcus, and Staphylococcus organisms to MLNs. These results indicate that combined antibiotic treatment changes the balance of the intestinal flora and induces the translocation of Bacteroides spp., S. aureus, Enterococcus spp., P. aeruginosa, E. coli, and K. pneumoniae by decreasing the intestinal defense against these bacteria rather than changing the numbers of them in the intestinal mucosa.
It has been shown that broad-spectrum antibiotics such as ampicillin could increase the colonization by pathogenic members of Enterobacteriaceae, such as the ampicillin-resistant K. pneumoniae, and opportunistic pathogens during antibiotic treatment (21, 24). Since our results showed that antibiotic treatment markedly induced translocation of members of the Enterobacteriaceae to MLNs, the translocation of injected pathogenic K. pneumoniae organisms and the nonvirulent E. coli strain DH10B/DsRed was used to investigate the effect of intestinal microflora depletion on intestinal immunity. The E. coli strain DH10B without the virulence plasmid and with a plasmid carrying the dsRED2 gene fragment was used to generate the nonvirulent DH10B/DsRed strain (25). After antibiotic treatment for 6 days, there was a marked increase in the translocation of pathogenic K. pneumoniae to MLNs, liver, and blood (93-fold, 23-fold, and 41-fold, respectively) but not that of DH10B/DsRed. Moreover, in vitro mucosal bacterium-killing assays showed that the killing activities against both K. pneumoniae and DH10B/DsRed were significantly decreased after antibiotic treatment. These results suggest that antibiotic treatment reduces the intestinal defense against bacterial invasion. To further confirm this hypothesis, the expression of antibacterial proteins in the terminal ileum was examined. RT-PCR, real-time PCR, and immunoblotting assays showed that levels of the non-defensin-family molecules RegIIIβ, RegIIIγ, CRP-ductin, and RELMβ were significantly decreased after antibiotic treatment, especially after 6 days of treatment. In contrast, there was no difference in the expression of the defensin-family proteins Defcr-rs-10, Crypt1, and Crypt4. Moreover, the expression of TLR4 was significantly decreased but the expression of TLR2 and MyD88 was slightly decreased after antibiotic treatment. In addition, antibiotic treatment for 6 days induced a profound decrease of NF-κB DNA binding activity in the terminal ileum. Our study demonstrated that in vitro mucosal killing of both K. pneumoniae and E. coli was significantly decreased after antibiotic treatment. Moreover, there was a significant increase in the translocation of injected K. pneumoniae organisms to MLNs, liver, and blood but not in that of E. coli. These results suggest that the expression of nondefensin proteins is triggered by TLR4-MyD88-dependent activation, according to a previous report showing that RegIIIγ expression was regulated by TLR-MyD88-mediated signals (17). It has also been reported that MyD88-deficient mice are susceptible to L. monocytogenes infection (17). In addition, NF-κB plays an important role in triggering the expression of nondefensin proteins through TLR4-MyD88-dependent signaling. Thus, TLR4 expression, NF-κB activation, and subsequent nondefensin protein expression play critical roles in antibiotic-induced reduction of intestinal defense against bacterial translocation and mucosal bacterium-killing activity. Using germfree mice, we further proved that the commensal microflora is critical in maintaining intestinal mucosal immunity and TLR4, Reg3β, Reg3γ, RELMβ, and CRP-ductin expression as well as NF-κB activation in the intestinal mucosa.
TLRs are important pattern recognition molecules in the intestinal epithelium that sense commensal microflora and initiate inflammatory and immune responses (26). However, the relationship between the function of TLRs and intestinal immunity has not been well established. Our data demonstrated that TLR stimulation with dead E. coli but not with dead S. aureus supplementation for 2 days in antibiotic-treated mice markedly decreased the translocation of pathogenic K. pneumoniae to MLNs, liver, and blood (5-fold, 3-fold, and 10-fold, respectively) and significantly increased the mucosal bacterium-killing activity against both K. pneumoniae and E. coli. These results suggest that TLRs play important roles in enhancing intestinal defense mechanisms and preventing invasion by pathogenic bacteria in antibiotic-treated mice and also that dead E. coli is more effective than dead S. aureus in inducing intestinal defense. Furthermore, quantitative analysis of nondefensin protein levels in the distal ileum demonstrated that TLR stimulation in antibiotic-treated mice with dead E. coli significantly increased the expression of TLR4 and of non-defensin-family antibacterial proteins. In contrast, antibiotic-treated mice receiving dead S. aureus for 2 days showed a moderate increase in nondefensin mRNA and protein levels. In addition, TLR stimulation with dead E. coli or S. aureus markedly increased the NF-κB DNA binding activity. These results indicate that TLR stimulation increases NF-κB activation, restores nondefensin protein expression, and increases mucosal bacterium-killing activity, leading to a decreased translocation of pathogenic K. pneumoniae to MLNs, liver, and blood. Moreover, our results show that TLR stimulation enhanced intestinal immune defense to prevent invasion by pathogenic bacteria by increasing expression of nondefensin proteins in antibiotic-treated mice. It is known that NF-κB is a key transcription regulator involved in a variety of inflammatory and innate immune responses. Our results further suggest that NF-κB may be regulated in the intestinal mucosa by the commensal microflora and is associated with the expression of antibacterial proteins and intestinal defense mechanism. Furthermore, germfree mice showed significant decreases in nondefensin proteins as well as in mucosal bacterium-killing activity and an increase in pathogen translocation compared with that in SPF mice. TLR stimulation with dead E. coli or S. aureus in germfree mice significantly increased TLR4 expression, NF-κB DNA binding activity in the intestinal mucosa, and mucosal bacterium-killing activity. Altogether, our data suggest that TLR4 and NF-κB DNA binding activity in the intestinal mucosa play important roles in commensal microflora-induced intestinal defense mechanisms, thereby inducing gut defense in germfree mice through TLR stimulation.
In summary, antibiotic treatment reduced the total numbers of bacteria in the terminal ileum, induced intestinal permeability, and increased the translocation of injected pathogenic K. pneumoniae to MLNs, liver, and blood due to reduction in the mucosal bacterium-killing activity and the expression of nondefensin proteins. TLR stimulation reversed the effect of antibiotic treatment on mucosal bacterium-killing activity, nondefensin proteins, and K. pneumoniae translocation. Moreover, TLR stimulation induced TLR4 expression, NF-κB DNA binding activity, and gut defense mechanisms in germfree mice. Therefore, the commensal microflora is critical in maintaining intestinal nondefensin protein expression as well as in regulating mucosal bacterium-killing activity, and finally, TLR stimulation may induce nondefensin protein expression and reverse antibiotic-induced gut defense deficit.
ACKNOWLEDGMENTS
We gratefully acknowledge Huang Yen Te and Chuang Hsiao Li in Germfree & Gnotobiotic Section, Technical Services Division, National Laboratory Animal Center, National Applied Research Laboratory.
This work was supported by grants from National Science Council (NSC97-2314B-010-030-MY3), Kaohsiung Veterans General Hospital (VGHKS100-055), and VTY Joint Research Program, Tsou's, Foundation (VTY92-P3-19) to L.-W.C.
We declare that no competing financial, professional, or personal interests exist.
Footnotes
Published ahead of print 4 March 2014
REFERENCES
- 1.Antonopoulos DA, Huse SM, Morrison HG, Schmidt TM, Sogin ML, Young VB. 2009. Reproducible community dynamics of the gastrointestinal microbiota following antibiotic perturbation. Infect. Immun. 77:2367–2375. 10.1128/IAI.01520-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ley RE, Lozupone CA, Hamady M, Knight R, Gordon JI. 2008. Worlds within worlds: evolution of the vertebrate gut microbiota. Nat. Rev. Microbiol. 6:776–788. 10.1038/nrmicro1978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen LW, Chang WJ, Chen PH, Liu WC, Hsu CM. 2008. TLR ligand decreases mesenteric ischemia and reperfusion injury-induced gut damage through TNF-alpha signaling. Shock 30:563–570. 10.1097/SHK.0b013e31816a3458 [DOI] [PubMed] [Google Scholar]
- 4.Lotz M, Menard S, Hornef M. 2007. Innate immune recognition on the intestinal mucosa. Int. J. Med. Microbiol. 297:379–392. 10.1016/j.ijmm.2007.03.010 [DOI] [PubMed] [Google Scholar]
- 5.Artis D. 2008. Epithelial-cell recognition of commensal bacteria and maintenance of immune homeostasis in the gut. Nat. Rev. Immunol. 8:411–420. 10.1038/nri2316 [DOI] [PubMed] [Google Scholar]
- 6.Dupaul-Chicoine J, Yeretssian G, Doiron K, Bergstrom KS, McIntire CR, LeBlanc PM, Meunier C, Turbide C, Gros P, Beauchemin N, Vallance BA, Saleh M. 2010. Control of intestinal homeostasis, colitis, and colitis-associated colorectal cancer by the inflammatory caspases. Immunity 32:367-378. 10.1016/j.immuni.2010.02.012 [DOI] [PubMed] [Google Scholar]
- 7.Meyer-Hoffert U, Hornef MW, Henriques-Normark B, Axelsson LG, Midtvedt T, Putsep K, Andersson M. 2008. Secreted enteric antimicrobial activity localises to the mucus surface layer. Gut 57:764–771. 10.1136/gut.2007.141481 [DOI] [PubMed] [Google Scholar]
- 8.Brandl K, Plitas G, Mihu CN, Ubeda C, Jia T, Fleisher M, Schnabl B, DeMatteo RP, Pamer EG. 2008. Vancomycin-resistant enterococci exploit antibiotic-induced innate immune deficits. Nature 455:804–807. 10.1038/nature07250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Parm U, Metsvaht T, Sepp E, Ilmoja ML, Pisarev H, Pauskar M, Lutsar I. 2010. Impact of empiric antibiotic regimen on bowel colonization in neonates with suspected early onset sepsis. Eur. J. Clin. Microbiol. Infect. Dis. 29:807–816. 10.1007/s10096-010-0931-1 [DOI] [PubMed] [Google Scholar]
- 10.Johnson MT, Reichley R, Hoppe-Bauer J, Dunne WM, Micek S, Kollef M. 2011. Impact of previous antibiotic therapy on outcome of Gram-negative severe sepsis. Crit. Care Med. 39:1859–1865. 10.1097/CCM.0b013e31821b85f4 [DOI] [PubMed] [Google Scholar]
- 11.Otamiri T, Sjodahl R, Tagesson C. 1987. An experimental model for studying reversible intestinal ischemia. Acta Chir. Scand. 153:51–56 [PubMed] [Google Scholar]
- 12.Karlsson J, Putsep K, Chu H, Kays RJ, Bevins CL, Andersson M. 2008. Regional variations in Paneth cell antimicrobial peptide expression along the mouse intestinal tract. BMC Immunol. 9:37. 10.1186/1471-2172-9-37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vaishnava S, Behrendt CL, Ismail AS, Eckmann L, Hooper LV. 2008. Paneth cells directly sense gut commensals and maintain homeostasis at the intestinal host-microbial interface. Proc. Natl. Acad. Sci. U. S. A. 105:20858–20863. 10.1073/pnas.0808723105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hunninghake GW, Doerschug KC, Nymon AB, Schmidt GA, Meyerholz DK, Ashare A. 2010. Insulin-like growth factor-1 levels contribute to the development of bacterial translocation in sepsis. Am. J. Respir. Crit. Care Med. 182:517–525. 10.1164/rccm.200911-1757OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li N, Karin M. 1998. Ionizing radiation and short wavelength UV activate NF-kappaB through two distinct mechanisms. Proc. Natl. Acad. Sci. U. S. A. 95:13012–13017. 10.1073/pnas.95.22.13012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kelly D, Conway S, Aminov R. 2005. Commensal gut bacteria: mechanisms of immune modulation. Trends Immunol. 26:326–333. 10.1016/j.it.2005.04.008 [DOI] [PubMed] [Google Scholar]
- 17.Brandl K, Plitas G, Schnabl B, DeMatteo RP, Pamer EG. 2007. MyD88-mediated signals induce the bactericidal lectin RegIII gamma and protect mice against intestinal Listeria monocytogenes infection. J. Exp. Med. 204:1891–1900. 10.1084/jem.20070563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Barman M, Unold D, Shifley K, Amir E, Hung K, Bos N, Salzman N. 2008. Enteric salmonellosis disrupts the microbial ecology of the murine gastrointestinal tract. Infect. Immun. 76:907–915. 10.1128/IAI.01432-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen LW, Wang JS, Hwang B, Chen JS, Hsu CM. 2003. Reversal of the effect of albumin on gut barrier function in burn by the inhibition of inducible isoform of nitric oxide synthase. Arch. Surg. 138:1219–1225. 10.1001/archsurg.138.11.1219 [DOI] [PubMed] [Google Scholar]
- 20.Maragakis LL. 2010. Recognition and prevention of multidrug-resistant Gram-negative bacteria in the intensive care unit. Crit. Care Med. 38:S345–351. 10.1097/CCM.0b013e3181e6cbc5 [DOI] [PubMed] [Google Scholar]
- 21.Kumarasamy KK, Toleman MA, Walsh TR, Bagaria J, Butt F, Balakrishnan R, Chaudhary U, Doumith M, Giske CG, Irfan S, Krishnan P, Kumar AV, Maharjan S, Mushtaq S, Noorie T, Paterson DL, Pearson A, Perry C, Pike R, Rao B, Ray U, Sarma JB, Sharma M, Sheridan E, Thirunarayan MA, Turton J, Upadhyay S, Warner M, Welfare W, Livermore DM, Woodford N. 2010. Emergence of a new antibiotic resistance mechanism in India, Pakistan, and the UK: a molecular, biological, and epidemiological study. Lancet Infect. Dis. 10:597–602. 10.1016/S1473-3099(10)70143-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lofmark S, Jernberg C, Jansson JK, Edlund C. 2006. Clindamycin-induced enrichment and long-term persistence of resistant Bacteroides spp. and resistance genes. J. Antimicrob. Chemother. 58:1160–1167. 10.1093/jac/dkl420 [DOI] [PubMed] [Google Scholar]
- 23.Chadwick P, Niell M. 1973. Transferable antibiotic resistance in E. coli and Klebsiella pneumoniae. Can. Med. Assoc. J. 109:691–696 [PMC free article] [PubMed] [Google Scholar]
- 24.Sullivan A, Edlund C, Nord CE. 2001. Effect of antimicrobial agents on the ecological balance of human microflora. Lancet Infect. Dis. 1:101–114. 10.1016/S1473-3099(01)00066-4 [DOI] [PubMed] [Google Scholar]
- 25.Fratamico PM, Yan X, Liu Y, DebRoy C, Byrne B, Monaghan A, Fanning S, Bolton D. 2010. Escherichia coli serogroup O2 and O28ac O-antigen gene cluster sequences and detection of pathogenic E. coli O2 and O28ac by PCR. Can. J. Microbiol. 56:308–316. 10.1139/W10-010 [DOI] [PubMed] [Google Scholar]
- 26.Takeuchi O, Akira S. 2001. Toll-like receptors; their physiological role and signal transduction system. Int. Immunopharmacol. 1:625–635. 10.1016/S1567-5769(01)00010-8 [DOI] [PubMed] [Google Scholar]