Abstract
Transmissible strains of Pseudomonas aeruginosa have been described for cystic fibrosis (CF) and may be associated with a worse prognosis. Using a comprehensive strain biobank spanning 3 decades, we sought to determine the prevalence and stability of chronic P. aeruginosa infection in an adult population. P. aeruginosa isolates from sputum samples collected at initial enrollment in our adult clinic and at the most recent clinic visit were examined by a combination of pulsed-field gel electrophoresis and multilocus sequence typing and compared against a collection of established transmissible and local non-CF bronchiectasis (nCFB) isolates. A total of 372 isolates from 107 patients, spanning 674 patient-years, including 66 patients with matched isolates from initial and final encounters, were screened. A novel clone with increased antibacterial resistance, termed the prairie epidemic strain (PES), was found in 29% (31/107 patients) of chronically infected patients referred from multiple prairie-based CF centers. This isolate was not found in those diagnosed with CF as adults or in a control population with nCFB. While 90% (60/66 patients) of patients had stable infection over a mean of 10.8 years, five patients experienced strain displacement of unique isolates, with PES occurring within 2 years of transitioning to adult care. PES has been present in our cohort since at least 1987, is unique to CF, generally establishes chronic infection during childhood, and has been found in patients at the time of transition of patients from multiple prairie-based CF clinics, suggesting broad endemicity. Studies are under way to evaluate the clinical implications of PES infection.
INTRODUCTION
Chronic airway infection with cystic fibrosis (CF), punctuated by repeated bouts of acute perturbations, results in progressive airway destruction, leading to bronchiectasis and respiratory failure. Pseudomonas aeruginosa, the archetypical CF pathogen, infects 70% of patients (1–3). The acquisition of this organism and its conversion to a hyper-alginate-producing mucoid phenotype are associated with deteriorating clinical status, increased treatment requirements, and progression to end-stage disease (4–8). P. aeruginosa, which is ubiquitous in the environment (9–11), was initially presumed to be acquired from each patient's local environment and not transmitted among patients (12). However, multiple local, national, and transcontinental epidemic strains of P. aeruginosa have now been described. Several of these strains are associated with an accelerated clinical deterioration notable for increased treatment burden (13, 14), increased exacerbation frequency (14), and progression to respiratory failure (9). For further information on transmissible strains of P. aeruginosa in CF, the reader is referred to a recent review by Fothergill et al. (15).
Despite extensive study of epidemic strains, little is known regarding the stability of infection or the circumstances surrounding acquisition (16). Most studies have been prospective and are thus limited by their short duration of follow-up and lack of historical context. These studies are therefore able to report point prevalences of infection but provide little information on incidence or when infections first occurred (13, 17). Accordingly, the development of rational, evidence-based infection control protocols to limit the spread of transmissible organisms has been limited. Making use of a strain biobank spanning 25 years, we were able to examine the incidence, prevalence, and stability of strains of P. aeruginosa within an adult CF cohort managed with traditional infection control strategies. A novel clonal isolate causing chronic infections in several generations of patients transitioning to adult CF care over 25 years was found in 1/3 of our population. This clone was found in patients referred from multiple Prairie Provinces, suggesting a broad distribution across western Canada. Despite its high prevalence in adult populations, incident infections were rarely observed, suggesting that its risk for acquisition is highest during the developing years.
MATERIALS AND METHODS
Clinic population and structure.
The Calgary Adult Cystic Fibrosis Clinic (CACFC) serves the needs of all CF patients living in southern Alberta, a province of 3.65 million individuals living in an area spanning 661,848 km2. Patients attend the clinic at least quarterly and are hospitalized exclusively at the Foothills Medical Centre. Prior to 2009, traditional infection control standards were limited to a common waiting room where personal contact was excluded to a 1-m “bubble” and good cough and hand hygiene was strictly enforced. Since then, universal segregation has been adopted, with patients immediately escorted to private rooms and seen by circulating health care workers. Inpatients are excluded from sharing rooms with other individuals with CF. Upon enrollment at the CACFC, all patients consent to the regular submission and banking of sputum and bacterial isolates from each and every encounter. All pathogens recovered by culture are stored at −80°C in the CACFC strain biobank. Ethical approval for this study was granted by the Calgary Health Region Ethics Board (CHREB) (approval no. E23087).
Patient and strain selection.
CACFC attendees had a diagnosis of CF according to established criteria (18). Patients were randomly selected and categorized as follows: (i) active patients, i.e., patients who were being monitored and managed for chronic P. aeruginosa infection at the time of study; (ii) transferred patients, i.e., those who had left the care of our clinic and moved to an area outside southern Alberta; and (iii) patients who experienced end-stage CF lung disease and either succumbed to CF or received lung transplantation. Chronic infection was defined per the Leeds criteria (19). A distinct morphotype of P. aeruginosa was defined a priori as a macroscopically distinct colony type on MacConkey agar. To assess the stability of infection, sputum isolates were assessed at two time points: the first encounter (FE; enrollment in clinic) and the most recent encounter (RE; last clinic visit or visit prior to transplant/death/leaving the catchment area). Every distinct morphotype from each sputum sample (up to a maximum of five morphotypes/sample) was evaluated. Extracted clinical data were limited to age of diagnosis, residence at the time of enrollment, and referral source. Patients with confirmed non-CF bronchiectasis (nCFB) were used as local controls. Multiple established transmissible strains of P. aeruginosa were used as controls.
Molecular typing.
Pulsed-field gel electrophoresis (PFGE) was used as the primary screening procedure for all isolates, using a protocol modified from the work of Aaron et al. (9). Gels were electrophoresed using a Chef Mapper XA system (Bio-Rad, Hercules, CA) under the following conditions: buffer temperature, 10°C; voltage, 6.0 V/cm; run time, 20 h; included angle, 120°; initial switch time, 5 s; and final switch time, 45 s. Strains that had banding patterns that were ≥80% identical were considered related, conforming to the Tenover criteria, in which isolates with 1 to 3 band differences are still considered related (20, 21). PFGE profiles were compared using BioNumerics, version 7.0 (Applied Maths, Austin, TX). Dendrograms were generated with a 2.0% position tolerance by using the unweighted-pair group method using average linkages (UPGMA) and the Sørensen-Dice similarity coefficient.
Clusters involving isolates from three or more patients were considered clonal a priori. Patients who had isolates with discrepant PFGE profiles between FE and RE samples underwent multilocus sequence typing (MLST) to determine if relatedness existed despite changes in PFGE profiles, using a method developed by Curran et al. (22). The procedure followed methods from previously published works (23–25). Sequences were compared to those publically available at the P. aeruginosa MLST online database (http://pubmlst.org/paeruginosa/) to identify allele and sequence types. Clonality was defined when MLST results for isolates matched for each of the seven sequenced housekeeping loci. Clonally related isolates were defined as those isolates from the same patient at different points in time that had ≤2 loci altered. Patients with discordant isolates identified at FE and RE were a priori considered to have undergone a “superinfection” if a new strain had displaced a prior colonizing P. aeruginosa strain, as demonstrated by discrepancies by both PFGE and MLST.
Antibiotic susceptibility testing.
Antibiotic susceptibility to tobramycin, ceftazidime, ciprofloxacin, and meropenem (Oxoid, Basingstoke, Hampshire, England) was determined using the Kirby-Bauer disc diffusion method and interpreted according to current Clinical and Laboratory Standards Institute guidelines (26). Nonsusceptibility refers to isolates with both intermediate and resistant MIC values. Multidrug resistance (MDR) and pan-drug resistance (PDR) were defined as nonsusceptibility to ≥3 and 4 classes of antibacterials, respectively (27).
Statistics.
Statistical analysis was performed using Stata, version 11.0 (Stata-Corp, TX). Asymmetrically distributed variables were reported as medians with interquartile ranges (IQRs) and were compared using the rank sum test for pairs and the median test for multiples. Differences in proportions among categorical data were assessed using Fisher's exact test for pairwise comparisons and the chi-square test for multiple groups.
RESULTS
Patient population.
A cohort of 113 patients was included in the study (Fig. 1). This population included 107 patients with chronic P. aeruginosa infection and 6 patients with transient infection. Of those included in the cohort, 72 (63%) patients are currently active within the CACFC cohort, 12 (11%) patients were transferred to another CF clinic, and 29 (26%) patients experienced respiratory failure and either received a life-saving lung transplant (24) or succumbed to CF (5). Among the members of the cohort, 101 (89%) patients had received prior care at a pediatric CF care center, and 12 were adults diagnosed with CF late in life, among which 9 received care only at CACFC.
FIG 1.
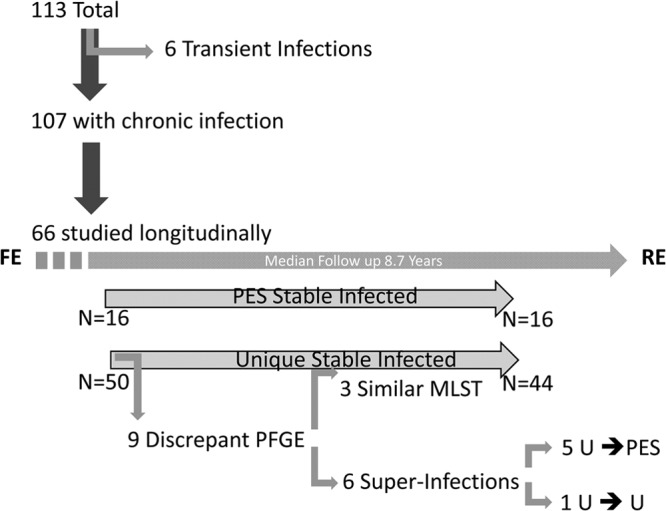
Patients included in the study. Of the 106 patients with chronic infection, 66 had matched first encounter (FE) and recent encounter (RE) isolates screened. Whereas PES isolates demonstrated stability over time, 12% of patients with unique (U) isolates experienced superinfections, predominately with PES. Genomic changes resulting in discrepant PFGE profiles of clonally related unique isolates were identified to have occurred in three instances, owing to apparent clonal relatedness by MLST.
Strains.
P. aeruginosa morphotypes were screened in 93 FE and 92 RE sputum samples. Sixty-six patients had matched FE and RE isolates screened. A mean of 1.89 (standard deviation [SD], ±0.3) morphotypes were screened from each sputum sample. A total of 342 isolates (156 FE and 186 RE isolates) were included in the initial portion of the study. Seven isolates could not be typed successfully by PFGE. Ten strains causing transient infections (having failed to establish chronic infection), obtained from six patients at eight time points, were also included. In those patients with discrepant FE and RE isolates, 54 serial isolates, representing one randomly selected morphotype per available year, were screened to determine when “superinfection” occurred. Thirty-five isolates from nine patients who had discrepant PFGE profiles over time underwent MLST to clarify if changes in PFGE profiles were due to strain displacement or, rather, to mutation of a chronic colonizing strain.
Forty representatives of epidemic strains were included, including the Liverpool epidemic strain (LES)/strain A (6/10), strain B (10), the Midlands strain (Md1) (2), the Manchester epidemic strain (1), Australian epidemic strains AUST-01 (3), AUST-02 (2), AUST-03 (1), AUST-04 (1), and AUST-6 (2), and isolates 0207 (1) and 0227 (1). Two clone C isolates, representing a P. aeruginosa strain with global distribution in the environment but found within CF patients, were also included (29, 30). Seventeen strains of P. aeruginosa isolated from 16 patients with nCFB were used as controls.
Prevalence of clonal infection within the CACFC.
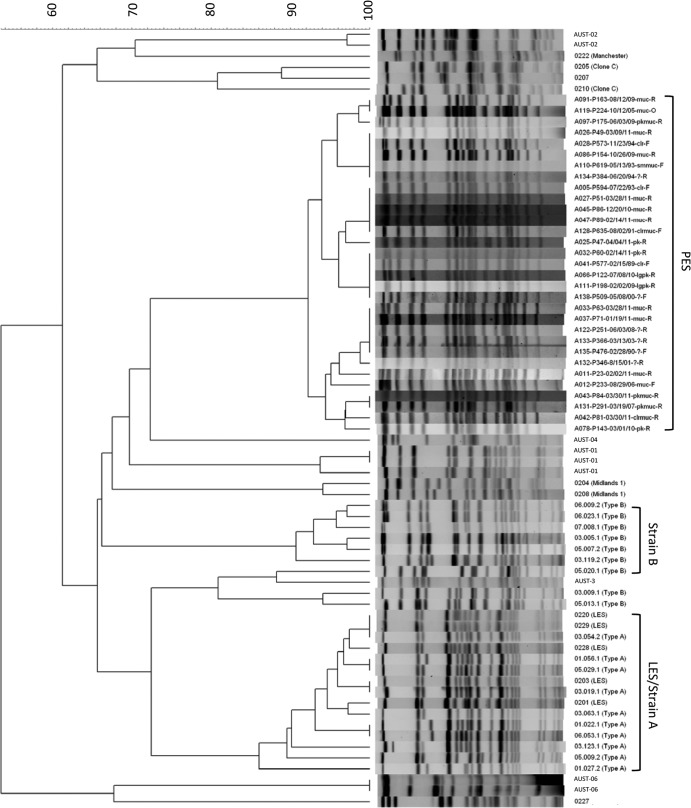
The majority of patients in the cohort (81/113 patients [71%]) were infected with unique/nonclonal strains of P. aeruginosa. Only one patient was identified as having a previously described transmissible strain (data not shown). This patient had been chronically infected with LES since her transition from a CF clinic in Ontario 2 years prior. Ninety-seven strains formed one dominant shared clonal complex, which was identified in 29% (31/107 patients) of patients with chronic infection. The identified clonal strain was not similar to described epidemic strains (Fig. 2) and corresponded to MLST sequence type 192.
FIG 2.
PES represents a novel transmissible P. aeruginosa strain distinct from previously reported epidemic strains included in the strain collection. Only one isolate from each patient with PES is displayed, for the sake of simplicity. PES isolates share a 92% degree of similarity, whereas LES/strain A isolates share 86% similarity.
Chronic infection with the clonal isolate was strongly associated with attendance at a pediatric CF clinic (33% [31/95 patients] versus 0% [0/12 patients] for those diagnosed as adults; P = 0.02). Furthermore, colonization with this isolate was related to the referral source: the local pediatric CF clinic (25/61 patients [41%]), other Prairie Province clinics (3/9 patients [33%]), British Columbia clinics (0/3 patients [0%]), eastern province clinics (0/6 patients [0%]), clinics outside Canada (0/1 patient [0%]), or newly diagnosed patients first assessed at the CACFC (0/9 patients [0%]) (P = 0.06). No difference in rates of prairie epidemic strain (PES) infection were observed through three separate decades of patients enrolling in the CACFC, with rates of 27% (3/11 patients) for 1980 to 1990, 37% (14/38 patients) for 1990 to 2000, and 22% (14/65 patients) for 2000 to 2010 (P = 0.199). The strain was observed exclusively in chronic infections and was not seen in transient infections (29% [97/332 isolates] of chronic infections versus 0% [0/10 isolates] of transient infections; P = 0.06). Patients infected with this strain were not observed to be coinfected at the same time point with other P. aeruginosa strains.
Common environmental sources of acquisition were deemed exceedingly unlikely, as infected patients lived in 13 different communities in Alberta (spanning an area of 196,350 km2) and two other prairie-based provinces upon enrolling in the CACFC. Accordingly, we have putatively termed this clonal complex the prairie epidemic strain (PES). PES was uniquely associated with chronic infections in CF patients and was not found in any patient with nCFB (29% [31/107 patients] of CF patients versus 0% [0/16 patients] of nCFB patients) (P = 0.01).
Stability of infection.
Sixty-two percent of patients (66/107 patients) with both FE and RE isolates were screened, enabling an assessment of strain stability. These patients were monitored for a median of 8.7 years (IQR, 2.8 to 15.8 years), with a total microbiologic follow-up of 651.4 patient-years. Of those patients with chronic infection assessed at only one point, 23 (21%) had only FE isolates screened, and 18 (17%) had only RE isolates screened.
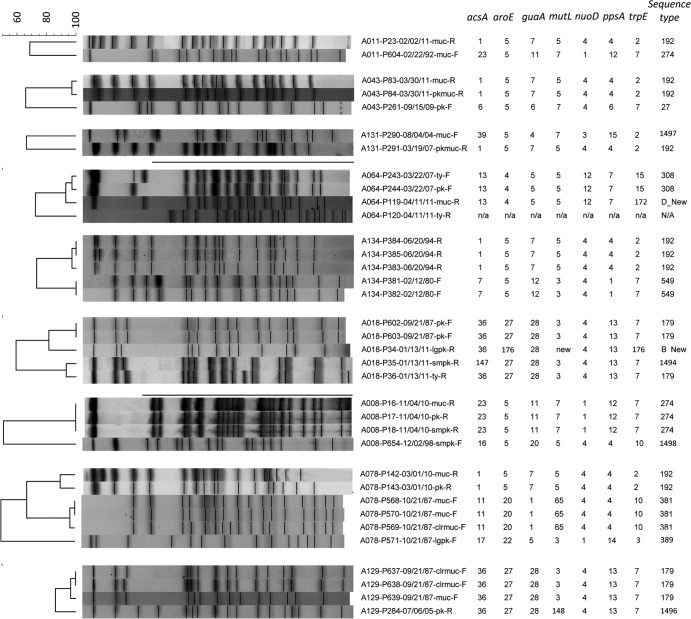
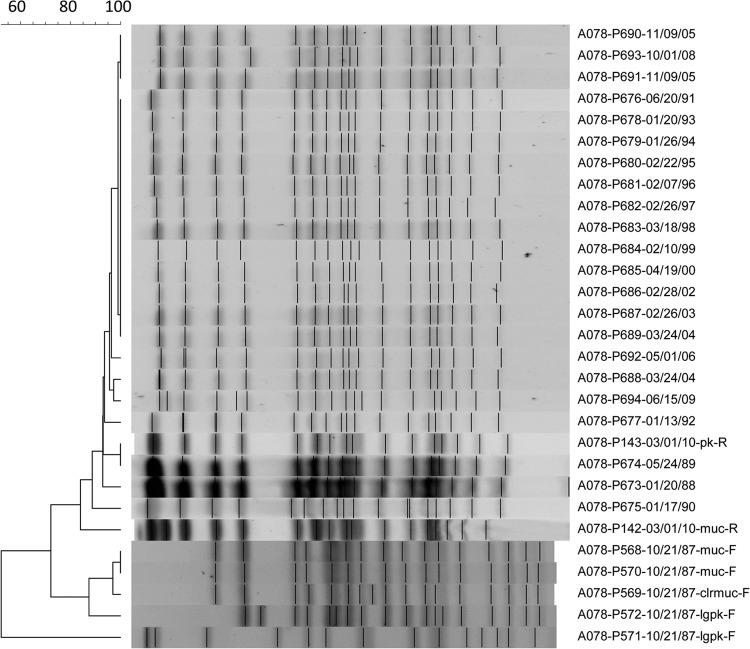
PFGE profiles were concordant for 87% (57/66 patients) of patients with matched FE and RE sets, even when samples were separated in time by as much as 25 years. Thirty-five isolates from 9 patients with discrepant PFGE patterns between FE and RE samples were subjected to MLST (Fig. 3). In three cases, while clonality could be inferred, at least one locus was altered, suggesting clonally related isolates in each instance, but conferring different MLST types. A new strain could be observed to displace existing colonizers in six instances of discrepant profiles. Of these “superinfections,” five represented incidences of PES infection. From these five patients, 54 additional P. aeruginosa serial yearly isolates obtained over 58 patient-years between FE and RE were screened. Acquisition of PES occurred within 2 years of transition to the CACFC and was then stable for the total length of follow-up (Fig. 4). Notably, each of these patients was chronically infected with a unique strain that was no longer observed after PES superinfection.
FIG 3.
Nine patients had discrepant PFGE profiles between FE and RE samples. Superinfections can be seen to have occurred in six cases: five with PES and one with a new, unique strain. Three patients with significant differences in PFGE profiles still showed evidence of clonal relatedness by MLST. Samples are named as follows: patient ID (A number)/strain ID (P number)/date collected (month/day/year)-morphotype. n/a or N/A, samples that were unable to be amplified despite repeated attempts.
FIG 4.
Longitudinal follow-up of a patient with P. aeruginosa CF isolates through 23 years. A single example of a PFGE profile is shown for a patient who acquired PES superinfection during follow-up at the CACFC, including all morphotypes from FE and RE samples and at least one isolate collected yearly in between. Samples are named as follows: patient ID-strain ID-date collected-morphotype.
The risk of PES superinfection occurring throughout the study was 1.2% per patient-year. Once chronic infection was established, PES was never observed to be displaced by other colonizing P. aeruginosa strains, despite a median length of culture follow-up of 10.9 years (IQR, 5.7 to 16.2 years), for a total of 261.4 patient-years of follow-up.
Antimicrobial resistance.
Four hundred isolates were screened for antimicrobial susceptibility, including 249 unique isolates, 89 PES isolates, 16 nCFB isolates, 18 LES isolates, 10 strain B isolates, and 18 other epidemic/global isolates. Antimicrobial nonsusceptibility was found to be more common among the epidemic strains, particularly LES (Fig. 5). Within the CACFC, PES was found to have higher rates of nonsusceptibility to ≥1 class (51% versus 31%; odds ratio [OR] = 1.63; 95% confidence interval [95% CI] = 1.24 to 2.17; P = 0.001) and ≥2 classes (20% versus 12%; OR = 1.73; 95% CI = 1.02 to 2.97; P = 0.04) of antimicrobials than the case for unique isolates. In particular, drug nonsusceptibility rates were increased in PES relative to unique isolates for ceftazidime (12% versus 5%; OR = 2.35; 95% CI = 1.1 to 5.07; P = 0.04), ciprofloxacin (30% versus 22%; OR = 1.36; 95% CI = 0.92 to 2.02; P = 0.09), and tobramycin (30% versus 15%; OR = 1.98; 95% CI = 1.29 to 3.04; P < 0.01). MDR and PDR were uncommon and not significantly different among PES strains relative to unique strains. Within the CACFC cohort, antimicrobial nonsusceptibility was observed to increase over the course of the study in both PES and unique isolates for each period, from 1980 to 1989, 1990 to 1999, 2000 to 2009, and 2010 to present, with rates of nonsusceptibility to ≥1 class of drugs of 5% (1/20 isolates), 12% (11/62 isolates), 41% (50/123 isolates), and 45% (60/133 isolates), respectively (P < 0.001), and rates of nonsusceptibility to ≥2 classes of drugs of 5% (1/20 isolates), 6% (4/62 isolates), 15% (18/123 isolates), and 18% (24/133 isolates), respectively (P = 0.05). In particular, this was driven by the emergence of resistance to ciprofloxacin and tobramycin (data not shown).
FIG 5.
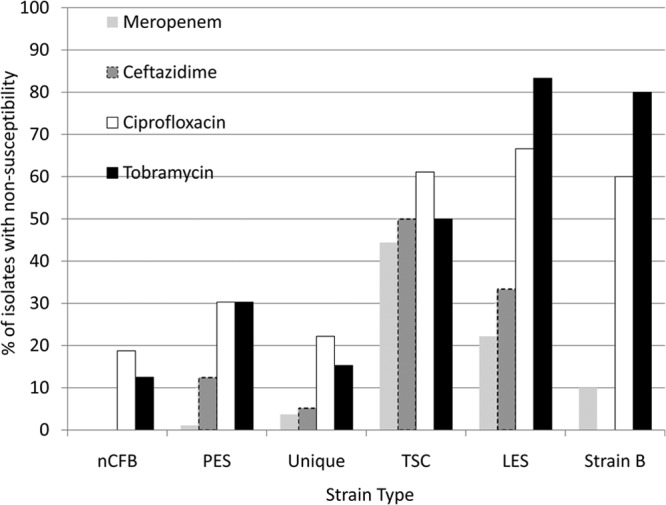
Antibiotic nonsusceptibility of the isolates within the collection to four common antipseudomonal antibacterial agents. TSC, transmissible strain collection representing global strains other than LES/strain B.
DISCUSSION
Transmissible strains of P. aeruginosa have been well documented for European CF populations, but within North America, few studies have been attempted.
Only strain A (LES), strain B, Houston-1, and now PES have been described to date in North America (9, 31, 32). The mechanisms by which these strains are capable of being transmitted from one patient to another are poorly understood, but cough aerosol is the most commonly presumed mechanism (16, 33). Furthermore, phenotypic characteristics of these transmissible strains enabling patient-to-patient spread are unclear (15).
Here we describe a novel clonal strain, PES, found in 29% of our adult CF population. PES has been present in the CACFC for ≥25 years and continues to be responsible for chronic infection with each generation graduating to our adult CF clinic. While a few strains of P. aeruginosa generally observed infrequently in the CF population have also been found in the environment (with the notable exception of clone C, a highly prevalent environmental isolate), the vast majority of prevalent CF strains have not been found in environmental reservoirs, supporting the theory of patient-to-patient spread (10, 29, 30, 34). Given the large geographical area in which patients with PES reside, the diversity of their ecological environments, the potential for these patients to have interacted through common clinics and summer camps, the fact that it has not previously been identified as an environmental source, and the inability to identify this isolate in local patients with nCFB, it is exceedingly unlikely that PES represents a common environmental source of infection. Rather, we believe that PES is a newly recognized transmissible P. aeruginosa strain relevant to patients with CF residing in the Prairie Provinces of Canada.
While only a minority of patients in this cohort were referred from other prairie-based CF clinics, PES was present in many of these individuals at the time of enrollment at the CACFC. These data suggest that PES is prevalent in other prairie-based Canadian CF clinics. Furthermore, MLST sequence type 192 was also recently identified in a strain collection from Vancouver, Canada, in strain A097, a minor clone not previously thought to represent a prevalent transmissible strain (32; D. Speert and J. Zlosnik, personal communication). In this context, PES has now been identified in at least four western Canadian provinces. Cross-Canadian prevalence studies similar to those in the United Kingdom and Australia are required to fully gauge the extent of transmissible strains within Canada (20, 35).
We observed a PES superinfection risk of 1.5% per patient-year of follow-up through 25 years in a cohort of adults with P. aeruginosa-positive sputum cultures managed with traditionally conservative infection control strategies. Because our entire clinic cohort was not assessed (i.e., we excluded those without P. aeruginosa infection), this number overestimates the true measure of incident infections. These low rates of acquisition are in accord with the work of Aaron et al. during their 3-year prospective study involving 446 CF adults in Ontario, as they observed an incidence of 7 cases per 1,000 person-years (9).
Unfortunately, little information exists regarding the risk and mechanism of acquisition of transmissible P. aeruginosa strains. This is largely owing to the fact that the majority of studies are prospective in nature and thus unable to determine when the initial transmission event occurred (13, 17, 28, 36–39). Critically, the bulk of transmission studies are concentrated in the pediatric patient age group (17, 28, 36, 38–42). Our study provides an important longitudinal perspective to studies of transmissible P. aeruginosa infection in adult populations. Despite being a retrospective analysis, our study used prospectively collected strains, which allowed a unique assessment of the natural history of infection transmission potential. This is different from other studies, where point prevalence assessments of clonality limit the ability to infer the timing of acquisition. The key finding of our study was that almost 90% of adult patients infected with PES were infected prior to enrolling in an adult CF clinic. Those who acquired PES as adults did so within 1 to 2 years of enrolling at the CACFC. This finding suggests that infection control standards might need to be adapted to recognize the heightened risk of infection in the younger years. Our decision to increase infection control standards to include standard patient segregation took place prior to this study, in keeping with international standards of care. It is not clear, however, nor do we advocate that infection control standards such as the universal patient segregation recently adopted by the majority of CF clinics could be reduced in adult clinics (43).
In this study, we observed significant movement of patients to and from our clinic during the 25-year study period, including those with chronic infections with P. aeruginosa strains such as PES and LES. We were surprised to find that other epidemic strains of P. aeruginosa occurred rarely in our patient cohort (including only one patient with LES). Nevertheless, the local epidemiology with respect to potential transmissible P. aeruginosa strains is likely to exist in a state of flux, as new, potentially transmissible strains may be introduced into a naive region. As such, point prevalence studies showing no or low levels of cross infection may be falsely reassuring, since there is a theoretical concern that a transmissible strain may be introduced into a community and spread (44–47). Therefore, a means by which CF patients with chronic P. aeruginosa infection can be assessed routinely for infection with clonal strains is required to ensure that critical metrics, such as incidence and prevalence, can be tracked over time.
Drug resistance is a common feature of transmissible P. aeruginosa strains (13, 23, 31, 37, 39). In this study, the newly identified transmissible strain PES was observed to have higher rates of nonsusceptibility than those of unique isolates. Generally, this drug resistance was observed against the aminoglycoside and fluoroquinolone classes. However, relative to other transmissible strains, PES appears to have a narrow nonsusceptibility profile. MDR and PDR were uncommon within the CACFC.
Resistance to antipseudomonal antibiotics has increased over time in CF patient isolates (1, 48). This is true for epidemic strains as well. Ashish et al. observed an increase in resistance to all common antipseudomonals in LES between 2004 and 2008 (49). In the present study, PES isolates collected through more than 3 decades showed the same trends, albeit to a lesser degree.
It will be important to review the clinical impact of PES on outcomes. While several strains have been shown to have a deleterious impact on outcome measures of CF as diverse as quality of life, treatment burden, exacerbation frequency, and progression to end-stage lung disease, others appear to have a negligible effect (9, 13, 14, 20, 50). The long duration of matched clinical and microbiologic follow-up from this study should enable longitudinal comparisons over time between those infected with PES and nonclonal strains through multiple cohort years to see if differences persist or change over time. The observed increased incidence of aminoglycoside and fluoroquinolone nonsusceptibility in PES during our study may indicate an increased treatment requirement and treatment burden in those infected with PES. PES has been demonstrated to be capable of tremendous within-patient phenotypic diversity, as measured by colony morphology, motility, protease production, auxotrophy, siderophore production, antibiotic resistance, quorum sensing, and growth patterns, similar to the within-patient heterogeneity of LES (51, 52).
The optimal P. aeruginosa strain typing method for use in the longitudinal analysis of individuals with CF has been the subject of recent research. Whereas other studies have assessed techniques such as random amplified polymorphic DNA PCR (RAPD-PCR) (25), enterobacterial repetitive intergenic consensus PCR (ERIC PCR) (24), and variable-number tandem repeat (VNTR) analysis (53) in combination with MLST and PFGE, we used a complementary approach. Whereas others have reported PFGE to have markedly discrepant profiles for up to 47% of paired patient isolates collected less than 1 year apart, we observed a high degree of conservation in our clinic cohort, as 87% (57/66 patients) of our patients had concordant profiles for up to 25 years. Of those few with discrepant PFGE types over time, 66% represented new infections, and only three patients with significantly discrepant PFGE profiles had strains with differences identified to have occurred through genomic changes. Furthermore, in these strains, mutations in at least one MLST locus were also evident, conferring different strain types despite a probable ancestral relationship (54). Given the low cost, high availability, and speed with which it can be accomplished, this study confirms that PFGE continues to have a useful role in longitudinal P. aeruginosa strain typing studies.
This study has several limitations. Our focus on morphologically distinct populations of P. aeruginosa did not enable sufficient sampling depths to identify minor strains during coinfections, particularly following superinfection events. However, once PES was identified during serial assessments of patients with displacement events, unique strains were no longer observed. Furthermore, in a small sample of patients infected with PES for which morphotypic diversity was explored in great depth, coinfections were not observed (52). The identification of a high degree of concordance in infecting strains at FE and RE does not strictly rule out the possibility of a transient infection with other strains during time points not assessed. However, the clinical impact of such a transient infection is unlikely to be significant, as exacerbations are not attributed to newly acquired strains (55). While PES isolates were not observed with different PFGE profiles, this remains a possibility that may have led to an underestimate of prevalence.
This study represents the longest observational study to date of chronic P. aeruginosa infection and the study of transmissible strains among individuals with CF. By assessing up to 25 years of data for individual patient follow-up, we conclusively demonstrated that for the vast majority of individuals, chronic infections remain stable. We have identified a new epidemic strain, PES, which is both pervasive and persistent in our own cohort and possibly throughout the Prairie Provinces of Canada. Detailed studies on the clinical impact of PES, the circumstances surrounding infection acquisition, and the microbiological factors that confer epidemic transmission of PES are under way.
ACKNOWLEDGMENTS
We thank the generosity of Shawn Aaron for providing the reference collection of described transmissible strains and for his kind advice. We thank Lauren Ruttle-Soon for her assistance in organizing the antimicrobial nonsusceptibility data.
This project was supported by grants from the Department of Medicine Alternate Relationship Plan Cost Deferral Initiative and Cystic Fibrosis Canada to M.D.P., M.G.S., and H.R.R.
The conception and design of the study were done by M.D.P., C.D.S., C.H.M., D.G.S., M.G.S., and H.R.R.; analysis and interpretation were done by M.D.P., B.A.G., C.D.S., K.A.S., J.D., S.P., and M.L.W.; and drafting of the manuscript for important intellectual content was done by M.D.P. and C.D.S.
Footnotes
Published ahead of print 22 January 2014
REFERENCES
- 1.Emerson J, McNamara S, Buccat AM, Worrell K, Burns JL. 2010. Changes in cystic fibrosis sputum microbiology in the United States between 1995 and 2008. Pediatr. Pulmonol. 45:363–370. 10.1002/ppul.21198 [DOI] [PubMed] [Google Scholar]
- 2.Lambiase A, Raia V, Del Pezzo M, Sepe A, Carnovale V, Rossano F. 2006. Microbiology of airway disease in a cohort of patients with cystic fibrosis. BMC Infect. Dis. 6:4. 10.1186/1471-2334-6-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Razvi S, Quittell L, Sewall A, Quinton H, Marshall B, Saiman L. 2009. Respiratory microbiology of patients with cystic fibrosis in the United States, 1995 to 2005. Chest 136:1554–1560. 10.1378/chest.09-0132 [DOI] [PubMed] [Google Scholar]
- 4.Henry RL, Mellis CM, Petrovic L. 1992. Mucoid Pseudomonas aeruginosa is a marker of poor survival in cystic fibrosis. Pediatr. Pulmonol. 12:158–161. 10.1002/ppul.1950120306 [DOI] [PubMed] [Google Scholar]
- 5.Kerem E, Corey M, Gold R, Levison H. 1990. Pulmonary function and clinical course in patients with cystic fibrosis after pulmonary colonization with Pseudomonas aeruginosa. J. Pediatr. 116:714–719. 10.1016/S0022-3476(05)82653-8 [DOI] [PubMed] [Google Scholar]
- 6.Kerem E, Corey M, Stein R, Gold R, Levison H. 1990. Risk factors for Pseudomonas aeruginosa colonization in cystic fibrosis patients. Pediatr. Infect. Dis. J. 9:494–498. 10.1097/00006454-199007000-00008 [DOI] [PubMed] [Google Scholar]
- 7.Kosorok MR, Zeng L, West SE, Rock MJ, Splaingard ML, Laxova A, Green CG, Collins J, Farrell PM. 2001. Acceleration of lung disease in children with cystic fibrosis after Pseudomonas aeruginosa acquisition. Pediatr. Pulmonol. 32:277–287. 10.1002/ppul.2009.abs [DOI] [PubMed] [Google Scholar]
- 8.Li Z, Kosorok MR, Farrell PM, Laxova A, West SE, Green CG, Collins J, Rock MJ, Splaingard ML. 2005. Longitudinal development of mucoid Pseudomonas aeruginosa infection and lung disease progression in children with cystic fibrosis. JAMA 293:581–588. 10.1001/jama.293.5.581 [DOI] [PubMed] [Google Scholar]
- 9.Aaron SD, Vandemheen KL, Ramotar K, Giesbrecht-Lewis T, Tullis E, Freitag A, Paterson N, Jackson M, Lougheed MD, Dowson C, Kumar V, Ferris W, Chan F, Doucette S, Fergusson D. 2010. Infection with transmissible strains of Pseudomonas aeruginosa and clinical outcomes in adults with cystic fibrosis. JAMA 304:2145–2153. 10.1001/jama.2010.1665 [DOI] [PubMed] [Google Scholar]
- 10.Kidd TJ, Ritchie SR, Ramsay KA, Grimwood K, Bell SC, Rainey PB. 2012. Pseudomonas aeruginosa exhibits frequent recombination, but only a limited association between genotype and ecological setting. PLoS One 7:e44199. 10.1371/journal.pone.0044199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schelstraete P, Van Daele S, De Boeck K, Proesmans M, Lebecque P, Leclercq-Foucart J, Malfroot A, Vaneechoutte M, De Baets F. 2008. Pseudomonas aeruginosa in the home environment of newly infected cystic fibrosis patients. Eur. Respir. J. 31:822–829. 10.1183/09031936.00088907 [DOI] [PubMed] [Google Scholar]
- 12.Kelly NM, Fitzgerald MX, Tempany E, O'Boyle C, Falkiner FR, Keane CT. 1982. Does pseudomonas cross-infection occur between cystic-fibrosis patients. Lancet ii:688–690 [DOI] [PubMed] [Google Scholar]
- 13.Bradbury R, Champion A, Reid DW. 2008. Poor clinical outcomes associated with a multi-drug resistant clonal strain of Pseudomonas aeruginosa in the Tasmanian cystic fibrosis population. Respirology 13:886–892. 10.1111/j.1440-1843.2008.01383.x [DOI] [PubMed] [Google Scholar]
- 14.Jones AM, Dodd ME, Morris J, Doherty C, Govan JR, Webb AK. 2010. Clinical outcome for cystic fibrosis patients infected with transmissible Pseudomonas aeruginosa: an 8-year prospective study. Chest 137:1405–1409. 10.1378/chest.09-2406 [DOI] [PubMed] [Google Scholar]
- 15.Fothergill JL, Walshaw MJ, Winstanley C. 2012. Transmissible strains of Pseudomonas aeruginosa in cystic fibrosis lung infections. Eur. Respir. J. 40:227–238. 10.1183/09031936.00204411 [DOI] [PubMed] [Google Scholar]
- 16.Clifton IJ, Peckham DG. 2010. Defining routes of airborne transmission of Pseudomonas aeruginosa in people with cystic fibrosis. Expert Rev. Respir. Med. 4:519–529. 10.1586/ers.10.42 [DOI] [PubMed] [Google Scholar]
- 17.Cheng K, Smyth RL, Govan JR, Doherty C, Winstanley C, Denning N, Heaf DP, van Saene H, Hart CA. 1996. Spread of beta-lactam-resistant Pseudomonas aeruginosa in a cystic fibrosis clinic. Lancet 348:639–642. 10.1016/S0140-6736(96)05169-0 [DOI] [PubMed] [Google Scholar]
- 18.Farrell PM, Rosenstein BJ, White TB, Accurso FJ, Castellani C, Cutting GR, Durie PR, Legrys VA, Massie J, Parad RB, Rock MJ, Campbell PW., 3rd 2008. Guidelines for diagnosis of cystic fibrosis in newborns through older adults: Cystic Fibrosis Foundation consensus report. J. Pediatr. 153:S4–S14. 10.1016/j.jpeds.2008.05.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee TW, Brownlee KG, Conway SP, Denton M, Littlewood JM. 2003. Evaluation of a new definition for chronic Pseudomonas aeruginosa infection in cystic fibrosis patients. J. Cyst. Fibros. 2:29–34. 10.1016/S1569-1993(02)00141-8 [DOI] [PubMed] [Google Scholar]
- 20.Scott FW, Pitt TL. 2004. Identification and characterization of transmissible Pseudomonas aeruginosa strains in cystic fibrosis patients in England and Wales. J. Med. Microbiol. 53:609–615. 10.1099/jmm.0.45620-0 [DOI] [PubMed] [Google Scholar]
- 21.Tenover FC, Arbeit RD, Goering RV, Mickelsen PA, Murray BE, Persing DH, Swaminathan B. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33:2233–2239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Curran B, Jonas D, Grundmann H, Pitt T, Dowson CG. 2004. Development of a multilocus sequence typing scheme for the opportunistic pathogen Pseudomonas aeruginosa. J. Clin. Microbiol. 42:5644–5649. 10.1128/JCM.42.12.5644-5649.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Beaudoin T, Aaron SD, Giesbrecht-Lewis T, Vandemheen K, Mah TF. 2010. Characterization of clonal strains of Pseudomonas aeruginosa isolated from cystic fibrosis patients in Ontario, Canada. Can. J. Microbiol. 56:548-557. 10.1139/W10-043 [DOI] [PubMed] [Google Scholar]
- 24.Kidd TJ, Grimwood K, Ramsay KA, Rainey PB, Bell SC. 2011. Comparison of three molecular techniques for typing Pseudomonas aeruginosa isolates in sputum samples from patients with cystic fibrosis. J. Clin. Microbiol. 49:263–268. 10.1128/JCM.01421-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Waters V, Zlosnik JE, Yau YC, Speert DP, Aaron SD, Guttman DS. 2012. Comparison of three typing methods for Pseudomonas aeruginosa isolates from patients with cystic fibrosis. Eur. J. Clin. Microbiol. Infect. Dis. 31:3341–3350. 10.1007/s10096-012-1701-z [DOI] [PubMed] [Google Scholar]
- 26.CLSI. 2010. Performance standards for antimicrobial susceptibility testing; 19th informational supplement. M100-S19, vol 29, no 31-156 Clinical and Laboratory Standards Institute, Wayne, PA [Google Scholar]
- 27.Parkins MD, Rendall JC, Elborn JS. 2012. Incidence and risk factors for pulmonary exacerbation treatment failures in patients with cystic fibrosis chronically infected with Pseudomonas aeruginosa. Chest 141:485–493. 10.1378/chest.11-0917 [DOI] [PubMed] [Google Scholar]
- 28.Armstrong DS, Nixon GM, Carzino R, Bigham A, Carlin JB, Robins-Browne RM, Grimwood K. 2002. Detection of a widespread clone of Pseudomonas aeruginosa in a pediatric cystic fibrosis clinic. Am. J. Respir. Crit. Care Med. 166:983–987. 10.1164/rccm.200204-269OC [DOI] [PubMed] [Google Scholar]
- 29.Romling U, Kader A, Sriramulu DD, Simm R, Kronvall G. 2005. Worldwide distribution of Pseudomonas aeruginosa clone C strains in the aquatic environment and cystic fibrosis patients. Environ. Microbiol. 7:1029–1038. 10.1111/j.1462-2920.2005.00780.x [DOI] [PubMed] [Google Scholar]
- 30.Romling U, Wingender J, Muller H, Tummler B. 1994. A major Pseudomonas aeruginosa clone common to patients and aquatic habitats. Appl. Environ. Microbiol. 60:1734–1738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Luna RA, Millecker LA, Webb CR, Mason SK, Whaley EM, Starke JR, Hiatt PW, Versalovic J. 2013. Molecular epidemiological surveillance of multidrug-resistant Pseudomonas aeruginosa isolates in a pediatric population of patients with cystic fibrosis and determination of risk factors for infection with the Houston-1 strain. J. Clin. Microbiol. 51:1237–1240. 10.1128/JCM.02157-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Speert DP, Campbell ME, Henry DA, Milner R, Taha F, Gravelle A, Davidson AG, Wong LT, Mahenthiralingam E. 2002. Epidemiology of Pseudomonas aeruginosa in cystic fibrosis in British Columbia, Canada. Am. J. Respir. Crit. Care Med. 166:988–993. 10.1164/rccm.2203011 [DOI] [PubMed] [Google Scholar]
- 33.Wainwright CE, France MW, O'Rourke P, Anuj S, Kidd TJ, Nissen MD, Sloots TP, Coulter C, Ristovski Z, Hargreaves M, Rose BR, Harbour C, Bell SC, Fennelly KP. 2009. Cough-generated aerosols of Pseudomonas aeruginosa and other Gram-negative bacteria from patients with cystic fibrosis. Thorax 64:926–931. 10.1136/thx.2008.112466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dinesh SD, Grundmann H, Pitt TL, Romling U. 2003. European-wide distribution of Pseudomonas aeruginosa clone C. Clin. Microbiol. Infect. 9:1228–1233. 10.1111/j.1469-0691.2003.00793.x [DOI] [PubMed] [Google Scholar]
- 35.Kidd TJ, Ramsay KA, Hu H, Marks GB, Wainwright CE, Bye PT, Elkins MR, Robinson PJ, Rose BR, Wilson JW, Grimwood K, Bell SC. 2013. Shared Pseudomonas aeruginosa genotypes are common in Australian cystic fibrosis centres. Eur. Respir. J. 41:1091–1100. 10.1183/09031936.00060512 [DOI] [PubMed] [Google Scholar]
- 36.Hoiby N, Pedersen SS. 1989. Estimated risk of cross-infection with Pseudomonas aeruginosa in Danish cystic fibrosis patients. Acta Paediatr. Scand. 78:395–404. 10.1111/j.1651-2227.1989.tb11099.x [DOI] [PubMed] [Google Scholar]
- 37.Jones AM, Govan JR, Doherty CJ, Dodd ME, Isalska BJ, Stanbridge TN, Webb AK. 2001. Spread of a multiresistant strain of Pseudomonas aeruginosa in an adult cystic fibrosis clinic. Lancet 358:557–558. 10.1016/S0140-6736(01)05714-2 [DOI] [PubMed] [Google Scholar]
- 38.Pedersen SS, Jensen T, Pressler T, Hoiby N, Rosendal K. 1986. Does centralized treatment of cystic fibrosis increase the risk of Pseudomonas aeruginosa infection? Acta Paediatr. Scand. 75:840–845. 10.1111/j.1651-2227.1986.tb10299.x [DOI] [PubMed] [Google Scholar]
- 39.Pedersen SS, Koch C, Hoiby N, Rosendal K. 1986. An epidemic spread of multiresistant Pseudomonas aeruginosa in a cystic fibrosis centre. J. Antimicrob. Chemother. 17:505–516. 10.1093/jac/17.4.505 [DOI] [PubMed] [Google Scholar]
- 40.Brimicombe RW, Dijkshoorn L, van der Reijden TJ, Kardoes I, Pitt TL, van den Broek PJ, Heijerman HG. 2008. Transmission of Pseudomonas aeruginosa in children with cystic fibrosis attending summer camps in The Netherlands. J. Cyst. Fibros. 7:30–36. 10.1016/j.jcf.2007.04.002 [DOI] [PubMed] [Google Scholar]
- 41.Pedersen SS, Jensen T, Hoiby N, Koch C, Flensborg EW. 1987. Management of Pseudomonas aeruginosa lung infection in Danish cystic fibrosis patients. Acta Paediatr. Scand. 76:955–961. 10.1111/j.1651-2227.1987.tb17271.x [DOI] [PubMed] [Google Scholar]
- 42.Petersen NT, Hoiby N, Mordhorst CH, Lind K, Flensborg EW, Bruun B. 1981. Respiratory infections in cystic fibrosis patients caused by virus, chlamydia and mycoplasma—possible synergism with Pseudomonas aeruginosa. Acta Paediatr. Scand. 70:623–628. 10.1111/j.1651-2227.1981.tb05757.x [DOI] [PubMed] [Google Scholar]
- 43.Conway S. 2008. Segregation is good for patients with cystic fibrosis. J. R. Soc. Med. 101(Suppl 1):S31–S35. 10.1258/jrsm.2008.s18007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Agarwal G, Kapil A, Kabra SK, Das BK, Dwivedi SN. 2005. Characterization of Pseudomonas aeruginosa isolated from chronically infected children with cystic fibrosis in India. BMC Microbiol. 5:43. 10.1186/1471-2180-5-43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Leao R, Carvalho-Assef APD, Ferreira AG, Folescu TW, Barth AL, Pitt TL, Marques EA. 2010. Comparison of the worldwide transmissible Pseudomonas aeruginosa with isolates from Brazilian cystic fibrosis patients. Braz. J. Microbiol. 41:1079–1081. 10.1590/S1517-83822010000400028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schmid J, Ling LJ, Leung JL, Zhang N, Kolbe J, Wesley AW, Mills GD, Brown PJ, Jones DT, Laing RT, Pattemore PK, Taylor DR, Grimwood K. 2008. Pseudomonas aeruginosa transmission is infrequent in New Zealand cystic fibrosis clinics. Eur. Respir. J. 32:1583–1590. 10.1183/09031936.00099508 [DOI] [PubMed] [Google Scholar]
- 47.Speert DP. 2002. Molecular epidemiology of Pseudomonas aeruginosa. Front. Biosci. 7:e354–e361. 10.2741/speert [DOI] [PubMed] [Google Scholar]
- 48.Merlo CA, Boyle MP, Diener-West M, Marshall BC, Goss CH, Lechtzin N. 2007. Incidence and risk factors for multiple antibiotic-resistant Pseudomonas aeruginosa in cystic fibrosis. Chest 132:562–568. 10.1378/chest.06-2888 [DOI] [PubMed] [Google Scholar]
- 49.Ashish A, Shaw M, Winstanley C, Ledson MJ, Walshaw MJ. 2012. Increasing resistance of the Liverpool epidemic strain (LES) of Pseudomonas aeruginosa (Psa) to antibiotics in cystic fibrosis (CF)—a cause for concern? J. Cyst. Fibros. 11:173–179. 10.1016/j.jcf.2011.11.004 [DOI] [PubMed] [Google Scholar]
- 50.Ashish A, Shaw M, McShane J, Ledson MJ, Walshaw MJ. 2012. Health-related quality of life in cystic fibrosis patients infected with transmissible Pseudomonas aeruginosa strains: cohort study. JRSM Short Rep. 3:12. 10.1258/shorts.2011.011119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mowat E, Paterson S, Fothergill JL, Wright EA, Ledson MJ, Walshaw MJ, Brockhurst MA, Winstanley C. 2011. Pseudomonas aeruginosa population diversity and turnover in cystic fibrosis chronic infections. Am. J. Respir. Crit. Care Med. 183:1674–1679. 10.1164/rccm.201009-1430OC [DOI] [PubMed] [Google Scholar]
- 52.Workentine ML, Sibley CD, Glezerson B, Purighalla S, Norgaard-Gron JC, Parkins MD, Rabin HR, Surette MG. 2013. Phenotypic heterogeneity of Pseudomonas aeruginosa populations in a cystic fibrosis patient. PLoS One 8:e60225. 10.1371/journal.pone.0060225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Turton JF, Turton SE, Yearwood L, Yarde S, Kaufmann ME, Pitt TL. 2010. Evaluation of a nine-locus variable-number tandem-repeat scheme for typing of Pseudomonas aeruginosa. Clin. Microbiol. Infect. 16:1111–1116. 10.1111/j.1469-0691.2009.03049.x [DOI] [PubMed] [Google Scholar]
- 54.Garcia-Castillo M, Maiz L, Morosini MI, Rodriguez-Banos M, Suarez L, Fernandez-Olmos A, Baquero F, Canton R, del Campo R. 2012. Emergence of a mutL mutation causing multilocus sequence typing–pulsed-field gel electrophoresis discrepancy among Pseudomonas aeruginosa isolates from a cystic fibrosis patient. J. Clin. Microbiol. 50:1777–1778. 10.1128/JCM.05478-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Aaron SD, Ramotar K, Ferris W, Vandemheen K, Saginur R, Tullis E, Haase D, Kottachchi D, St Denis M, Chan F. 2004. Adult cystic fibrosis exacerbations and new strains of Pseudomonas aeruginosa. Am. J. Respir. Crit. Care Med. 169:811–815. 10.1164/rccm.200309-1306OC [DOI] [PubMed] [Google Scholar]