Abstract
Vibrio cholerae serogroup O139 was first identified in 1992 in India and Bangladesh, in association with major epidemics of cholera in both countries; cases were noted shortly thereafter in China. We characterized 211 V. cholerae O139 isolates that were isolated at multiple sites in China between 1993 and 2012 from patients (n = 92) and the environment (n = 119). Among clinical isolates, 88 (95.7%) of 92 were toxigenic, compared with 47 (39.5%) of 119 environmental isolates. Toxigenic isolates carried the El Tor CTX prophage and toxin-coregulated pilus A gene (tcpA), as well as the Vibrio seventh pandemic island I (VSP-I) and VSP-II. Among a subset of 42 toxigenic isolates screened by multilocus sequence typing (MLST), all were in the same sequence type as a clinical isolate (MO45) from the original Indian outbreak. Nontoxigenic isolates, in contrast, generally lacked VSP-I and -II, and fell within13 additional sequence types in two clonal complexes distinct from the toxigenic isolates. In further pulsed-field gel electrophoresis (PFGE) (with NotI digestion) studies, toxigenic isolates formed 60 pulsotypes clustered in one group, while the nontoxigenic isolates formed 43 pulsotypes which clustered into 3 different groups. Our data suggest that toxigenic O139 isolates from widely divergent geographic locations, while showing some diversity, have maintained a relatively tight clonal structure across a 20-year time span. Nontoxigenic isolates, in contrast, exhibited greater diversity, with multiple clonal lineages, than did their toxigenic counterparts.
INTRODUCTION
Vibrio cholerae isolates that can produce cholera toxin are the causative agent of cholera. Among more than 200 serogroups of V. cholerae so far identified, epidemic cholera has been confined almost exclusively to isolates within serogroups O1and O139 (1, 2). The current seventh pandemic of cholera originated from Indonesia in 1961; the causative agent is V. cholerae O1 of the El Tor biotype, and it is the most extensive in geographic spread and duration of any reported cholera pandemic to date. In 1992, a newly identified serogroup, designated O139 Bengal, was recognized as the cause of epidemic cholera in India, with subsequent spread across multiple Asian countries (3, 4). However, over the last few years, El Tor O1 isolates have reestablished their prominence and now share this locally with O139 isolates (5). The first outbreak in China caused by serogroup O139 isolates occurred in 1993 in Xinjiang (6), followed by outbreaks in multiple provinces across subsequent years. O139 isolates have continued to coexist with O1 isolates in China across this time period.
When they first emerged in India and Bangladesh, O139 isolates were shown to be virtually identical to O1 El Tor isolates, with the substitution of a 35-kb region of DNA encoding the O139 surface polysaccharide for a 22-kb region that included the rfb region in O1 isolates (7). O139 isolates carried all “standard” virulence factors found in O1 isolates, including the elements of CTX prophage and Vibrio seventh pandemic island I (VSP-I) and VSP-II. Interestingly, in contrast to O1 isolates, V. cholerae O139 was shown to be encapsulated (8), with the polysaccharide capsule having the same repeating subunits as the O antigen. However, little is known about possible evolutionary changes in these isolates across the 20-year time period since they were first introduced into China, nor is there a good understanding of the relationship between clinical and environmental isolates.
Several molecular subtyping techniques have been widely used to depict genetic relatedness and for molecular epidemiological studies among V. cholerae isolates, including pulsed-field gel electrophoresis (PFGE) and multilocus sequence typing (MLST) (9, 10). In this study, we characterized 211 isolates of V. cholerae O139 from 1993 to 2012, originating from clinical and environmental sources in different provinces/regions in China. PCR and sequencing tests were used to character the virulence-associated genes and their genotypes, with MLST and PFGE used to determine overall genetic relatedness and clonality.
MATERIALS AND METHODS
Bacterial isolates and serological identification.
A total of 211 V. cholerae O139 isolates (92 clinical and 119 environmental) isolated in China were included in this study (Fig. 1). Isolates were selected from each year and each province where cholera was occurring, with dates of isolation from1993 to 2012. Where multiple isolates from a single province and year were available for study, isolates were selected randomly for inclusion in the current study. There was no overlap between the O139 isolates reported in this study and isolates that have been reported in previous studies (11, 12). A listing of isolates, with source and year of isolation, is provided in Table 1. All of the isolates were screened for the oxidase reaction and were identified by a slide agglutination test using specific polyvalent antisera against V. cholerae O139 (S&A Reagents Lab, Bangkok, Thailand).
FIG 1.
Geographical locations of sampling sites and numbers of O139 isolates collected from different sources. Top, colored provinces represent areas with sampling sites (both environmental and patient), with the number of isolates shown in brackets. Bottom, numbers of different isolates (both environmental and patient) are also given by year.
TABLE 1.
Sources of V. cholerae O139 isolates used in this study
| Yr of isolation (n) | Source and province (n) |
|
|---|---|---|
| Patient | Environment | |
| 1993 (3) | Xinjiang (3) | |
| 1994 (6) | Guangdong (1), Beijing (2), Shanghai (1), Zhejiang (1), Jiangxi (1) | |
| 1995 (4) | Fujian (1), Guangdong (2) | Guangdong (1) |
| 1996 (2) | Guangdong (1), Zhejiang (1) | |
| 1997 (2) | Zhejiang (1), Liaoning (1) | |
| 1998 (4) | Anhui (1), Jiangxi (3) | |
| 1999 (4) | Jiangxi (1), Jiangsu (1) | Jiangxi (2) |
| 2000 (7) | Guangxi (1), Jiangxi (3) | Jiangxi (1), Guangxi (2) |
| 2001 (11) | Guangxi (1), Guangdong (1), Sichuan (1), Liaoning (1) | Guangdong (1), Jiangxi (6) |
| 2002 (9) | Guangdong (2), Chongqing (1), Jiangxi (1), Gansu (1) | Jiangxi (4) |
| 2003 (14) | Chongqing (3), Guangdong (1), Zhejiang (1), Jiangxi (2), Jiangsu (2) | Jiangxi (4), Chongqing (1) |
| 2004 (8) | Chongqing (2), Guangdong (1), Liaoning (1), Jiangxi (2), Sichuan (2) | |
| 2005 (29) | Beijing (1), Chongqing (1), Fujian (1), Guangdong (1), Hunan (1), Jiangsu (2), Jiangxi (1), Sichuan (2), Shandong (2), Zhejiang (1) | Jiangxi (6), Zhejiang (2), Guangxi (1), Fujian (1), Jiangsu (1), Chongqing (1).Sichuan (1), Hunan (1), Shanghai (2) |
| 2006 (53) | Anhui (1), Chongqing (3), Jiangsu (2), Guangdong (2), Sichuan (2), Henan (1) | Guangdong (14), Sichuan (7), Fujian (1), Guangxi (3), Henan (1), Jiangxi (16) |
| 2007 (22) | Anhui (1), Sichuan (2), Jiangxi (1) | Guangdong (4), Sichuan (3), Anhui (1), Jiangxi (10) |
| 2008 (12) | Jiangxi (2) | Jiangxi (9), Guizhou (1) |
| 2009 (12) | Yunnan (4), Shandong (1) | Guizhou (1), Jiangxi (6) |
| 2010 (5) | Liaoning (2) | Guangdong (3) |
| 2011 (1) | Jiangsu (1) | |
| 2012 (3) | Jiangsu (2) | Jiangsu (1) |
| Total (211) | 92 | 119 |
PCR template preparation.
Genomic DNA was extracted using a genomic DNA purification kit (Tiangen Biotech, Beijing, China) in accordance with the manufacturer's instructions. Extracted DNAs were dissolved in Tris-EDTA (TE) buffer and stored at −20°C until used as PCR templates.
PCR assays and sequencing analysis.
PCR assays were carried out using conventional PCR amplification; the target genes included the cholera toxin B subunit gene (ctxB) (13), the repeat-in-toxin gene (rtxC) (14), eight types of rstR, including rstRET (15), rstRclass (16), rstRcalc (17), rstR-4** (16), rstR-5 (16), rstR-6 (18), rstR-232 (19), and rstRVC06-18 (this study), and classical and El Tor-specific tcpA (20). PCR screening was performed for five genes (VC0175, VC0178, VC0180, VC0183, and VC0185) in the VSP-I cluster and eight genes (VC0490, VC0493, VC0498, VC0502, VC0504, VC0512, VC0514, and VC0516) in the VSP-II cluster (21). Reference isolates included V. cholerae N16961 (7th pandemic O1 El Tor) and V. cholerae MO45 (an O139 isolate from India, 1993). Table S1 in the supplemental material shows the sequences used for primer design and their origins.
To identify the different types of ctxB and rstR, a commercial company (TaKaRa, Dalian, China) was employed for the sequencing of PCR products. Comparison analyses of the ctxB sequences were conducted with BioEdit software (Ibis Biosciences, Carlsbad, CA, USA). ClustalW was used to perform multiple alignments of the nucleotide or predicted amino acid sequences for ctxB and rstR. The reference sequences of different types of ctxB and rstR were accessed from GenBank.
MLST.
Of 211 tested O139 isolates, we selected 76 isolates, including 42 toxigenic isolates and 34 nontoxigenic isolates, for MLST. The isolates were selected based on their different gene characters (types of ctxB, rstR, and VSP-I/II), different years, and different provinces/regions; all 76 isolates represented the different characters of O139 isolates in China, from both clinical and environmental sources (see Table S2 in the supplemental material). Seven housekeeping genes (adk, gyrB, metE, mdh, pntA, purM, and pyrC) were targeted for MLST analysis, based on a previous study (22); primers are listed in Table S1 in the supplemental material. The PCR products were visualized on an agarose gel, and direct sequencing was performed in both directions. Contiguous nucleotide sequences were assembled by using MEGA software, and eBURST (23) was used to identify clonal complexes (CCs), which are defined using the difference of one out of the seven genes typed. A minimum-spanning tree using the allelic difference between isolates of the seven housekeeping genes was constructed using BioNumerics (Applied Math). Reference isolates N16961 and MO45 were included in the MLST analysis.
PFGE.
We used the PulseNet 1-day standardized PFGE protocol for V. cholerae (24). Cell suspensions were placed in polystyrene tubes (Falcon; 12 by 75 mm), and their optical density was adjusted to 3.8 to 4.2 using a Densimat photometer (bioMérieux, France). V. cholerae slices were digested using 30 U per slice of NotI (New England BioLabs, Ipswich, MA, USA) for 4 h at 37°C. Electrophoresis was performed using a CHEF-DRIII system (Bio-Rad Laboratories, Hercules, CA, USA). Images were captured on a Gel Doc 2000 system (Bio-Rad Laboratories) and converted to TIFF files. The TIFF files were analyzed using the BioNumerics version 5.1 software (Applied Maths).
Nucleotide sequence accession numbers.
The nucleotide sequences of the rstR genes from representative isolates have been deposited in GenBank under accession numbers KJ023705 to KJ023707. Representative sequences of each sequence type (ST1 to -14) determined by MLST have been deposited in GenBank under accession numbers KJ020751 to KJ020848.
RESULTS
Identification of virulence-associated genes.
Among the 211 O139 isolates tested, 135 (64.0%) carried the El Tor ctxB gene. Among clinical patient isolates, 88 (95.7%) of 92 isolates were toxigenic, compared with 47 (39.5%) of 119 environmental isolates. Findings of PCR screening for other virulence genes are summarized in Table 2. tcpA+ isolates accounted for 137 (64.9%) of the 211 isolates (92.7% of clinical and 40% of environmental isolates). In addition, 204 (96.7%) of the 211 isolates carried the rtxC gene; the seven rtxC-negative isolates included one from a patient in 2003, one from a soft-shelled turtle in 2005, and five from environmental sources, isolated during 2001 to 2007.
TABLE 2.
Presence of different virulence-associated genes among O139 V. cholerae isolates used in this study
| No. of isolates | Presence of genea |
|||||
|---|---|---|---|---|---|---|
| CTX prophage |
tcpA | rtxC | VSP-I | VSP-II | ||
| rstR | ctxB | |||||
| 83 | E | E | E | + | + | + |
| 3 | − | − | − | + | − | − |
| 2 | E + C | E | E | + | + | + |
| 1 | rstR6 | E | E | + | + | + |
| 1 | E | E | E | − | + | + |
| 1 | E | E | E | + | − | + |
| 1 | − | − | E | + | + | + |
| Environmental | ||||||
| 64 | − | − | − | + | − | − |
| 42 | E | E | E | + | + | + |
| 3 | E | − | − | + | − | − |
| 3 | − | − | − | − | − | − |
| 2 | E | E | E | − | + | + |
| 1 | E + C | − | E | + | − | +b |
| 1 | E | E | E | − | − | + |
| 1 | E | E | E | + | − | − |
| 1 | E | E | − | + | + | + |
| 1 | − | − | E | + | − | − |
+, positive; −, negative; C, classical; E, El Tor.
Positive for six ORFs (VC0490, VC0493, VC0498, VC0502, VC0512, and VC0516) in VSP-II.
The majority of toxigenic isolates carried VSP-I (97.8%, 132/135) and VSP-II (99.3%, 134/135). Three isolates (one from a patient and two environmental) were negative for five of the tested open reading frames (ORFs) of VSP-I (Table 2; see Table S2 in the supplemental material), indicative of the close correlation between VSP-I/II and carriage of the ctx genes (P < 0.0001). In contrast, among all 76 nontoxigenic isolates, the positive rates for VSP-I and VSP-II were 1.3% (1/76) and 2.6% (2/76), respectively. It is notable that VSP-I/II was found in one pre-CTX (ctxB negative but rstR positive) environmental isolate (19); this isolate was negative for five ORFs in VSP-I but positive for six ORFs (VC0490, VC0493, VC0498, VC0502, VC0512, and VC0516) in VSP-II.
MLST.
Among 76 isolates selected, 14 different sequence types (STs) were obtained (Fig. 2; see Table S2 in the supplemental material). All 42 toxigenic isolates, although from 15 provinces in China, had the same ST as MO45 and N16961 and were exclusively grouped into one ST (ST1). While the 34 nontoxigenic isolates were divided into 13 STs, ST13 was predominant, having 13 isolates, from water in two provinces between 1995 and 2009; the remaining 12 STs included a total of 21 isolates, with each ST including one to six isolates. eBURST analysis demonstrated that the STs formed three clonal complexes (CCs). CC1 contained ST1 and ST2, which included all toxigenic isolates and one pre-CTX isolate (GD2002005) (see Table S2 in the supplemental material) from a patient in 2002; the two STs differed at the pyrC locus. ST13, together with the seven other STs (ST6 to ST9, ST11, ST12, and ST14), formed CC2. CC3 contained two STs (ST4 and ST5), which differed at the metE locus (see Table S2 in the supplemental material).
FIG 2.
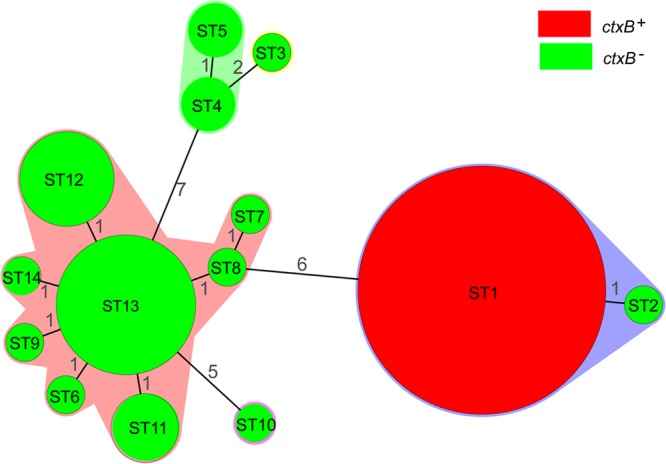
Minimum-spanning tree of 76 V. cholerae O139 isolates in China based on MLST. Each ST is represented by a circle sized in proportion to the number of isolates represented by that ST; the colors of the halos surrounding the STs denote types that belong to the same clonal complex, and the number of allelic difference between STs is indicated on the branches. The detailed MLST profiles can be seen in Table S1 in the supplemental material.
PFGE patterns of the isolates.
All 211 isolates were differentiated into 103 distinct pulsotypes (PTs) on the basis of PFGE using NotI endonuclease digestion (Fig. 3; see Table S2 in the supplemental material). The unweighted-pair group method using average linkages (UPGMA) clustered the PTs into three major groups, A, B, and C. All 135 toxigenic isolates except one clustered into group A, comprised 60 PTs, and exhibited 83% similarity, while all 76 nontoxigenic isolates except one clustered into two groups (B and C), comprised 43 PTs, and showed 72% similarity. Isolates with the same PT appeared in different years and in different provinces/regions: for example, among toxigenic isolates in group A, those in PTs CN0002, CN0005, and CN0047 (containing 25, 10, and 10 isolates, respectively) came from multiple provinces. At the same time, multiple PTs were found in the same province at the same time: for example, in group A, PTs CN0221, CN0066, and CN0219 appeared in Jiangxi in 2007, and in group B, PTs CN0234, CN0233, and CN0226 appeared in Jiangxi in 2009. PTs CN0091 and CN0079 (group B) and CN0133 (group C), from water isolates, and CN0052, CN0078, and CN0100 (group A), from patient isolates, all appeared in Guangdong in 2006. The first outbreak isolates of China, XJ1993209 and XJ93023 from Xinjiang in 1993 and BJ1994002 from Beijing in 1994, also had different PTs (Fig. 2; see Table S2 in the supplemental material).
FIG 3.
Dendrogram constructed from the PFGE profiles generated from NotI-digested genomic DNA of V. cholerae O139 isolates in China. AH, Anhui; BJ, Beijing; CQ, Chongqing; FJ, Fujian; GD, Guangdong; GS, Gansu; GX, Guangxi; GZ, Guizhou; HA, Henan; HN, Hunan; JS, Jiangsu; JX, Jiangxi; LN, Liaoning; SC, Sichuan; SD, Shandong; SH, Shanghai; XJ, Xinjiang; YN, Yunnan; ZJ, Zhejiang. The scale shows percent similarity.
DISCUSSION
Using MLST as a marker for phylogenic analysis, we found remarkably little evidence of divergence among toxigenic O139 isolates across almost 20 years, with all such isolates within a single ST, together with the reference O139 isolate from the early days of the initial O139 epidemic in India. Not unexpectedly, virtually all clinical patient isolates were toxigenic; however, we also found that approximately 40% of environmental isolates were toxigenic and were within the “toxigenic” ST. Isolates within this ST carried a full complement of El Tor virulence factors (in keeping with the El Tor O1 isolate[s] from which they have been hypothesized to have evolved), including the El Tor ctxB gene and VSP-I and -II. There was evidence of variability in ribotype, as has been previously described in studies of O139 isolates in China (11). These observations suggest that despite ongoing genetic changes (as reflected in the diversity in ribotype patterns), there has been remarkable stability in the housekeeping genes that form the basis for the MLST analysis. Given the close phylogenetic link with the original Indian isolate, it would also appear likely that the progenitor isolate was introduced from India, possibly by an infected patient, either at the time of or shortly after the original O139 epidemic.
A different pattern emerges among the nontoxigenic isolates. These isolates were from environmental sources, generally lacked other virulence genes, did not carry VSP-I and -II, and showed remarkable divergence in phylogeny: among 42 isolates for which MLST data were obtained, 13 STs, in two clonal complexes, were identified. These STs/CCs were clearly distinct from ST1/CC1, where all of the toxigenic isolates clustered. Two hypotheses might explain these results. It is possible that clinical O139 isolates lost virulence elements such as the CTX prophage when introduced into the environment; however, based on our observations, they would also have had to have lost VSP-I and VSP-II and undergone substantial changes in housekeeping genes, as reflected in the diversity of STs noted. Alternatively, nontoxigenic/nonvirulent environmental isolates, from diverse sources, may have acquired the genetic cassette responsible for biosynthesis of the O139 surface polysaccharide (capsule and O antigen) (7). In keeping with the latter hypothesis, we know that the O139 biosynthetic cassette is movable, as its introduction into a “standard” El Tor O1 isolate appears to have been responsible for the original emergence of the epidemic O139 V. cholerae (25). There is also increasing evidence that O-antigen biosynthetic cassettes are movable among V. cholerae isolates, possibly in association with a “jump-start” region adjacent to the rsb complex (26).
PFGE also clustered the toxigenic and nontoxigenic isolates into different groups, consistent with earlier reports from our group (12) and in keeping with the findings with MLST. However, PFGE further divided those groups into different PTs. Forty-six (34.1%) of toxigenic isolates harbored unique PTs, including the three isolates first identified in Xinjiang in 1993 and isolates from multiple provinces and regions in subsequent years. These results are consistent with the initial emergence of toxigenic O139 isolates in India and Bangladesh during 1992 to 1993 (4), whereas different ribotypes and PFGE patterns were reported during 1995 to 1996 in Bangladesh (27). Unlike the epidemics in Bangladesh and India, where explosive epidemics and reemergence were observed (28, 29), O139 cholera in China has primarily involved local outbreaks, often in the setting of food-borne outbreaks (30–33) associated with consumption of seafood contaminated by V. cholerae. The results obtained in this study indicate that there is a relatively high rate of toxigenicity among environmental V. cholerae O139 isolates, suggesting that there is a continuing risk for acquisition of human infections from environmental reservoirs, including seafood.
Toxigenic V. cholerae O1 from the early part of the El Tor (7th) pandemic carried ctxB of what was designated the El Tor type (ctxB-ET). However, ctxB-ET has subsequently been supplanted, at a global level, by ctxB matching that from classical (6th pandemic) strains (ctxB-CL) (34), with a suggestion that these strains have increased virulence compared with strains carrying ctxB-ET (35). For almost a decade, V. cholerae isolates carrying ctxB-ET were considered extinct, with the exception of a very recent report (36). Furthermore, variant ctxB genotypes were found in O1 El Tor isolates in Haiti (37) and the O139 isolates during 1998 to 2005 in Bangladesh (38). Recently, we found that the ctxB-CL type of V. cholerae O1 El Tor appeared in 1992 and replaced the prototype El Tor from 2002 to 2010 in China (39), with other El Tor variants that differed from known ctxB genotypes also identified (40). Interestingly, we found no evidence of a shift to the ctxB-CL type among Chinese O139 isolates included in this study. Factors responsible for the persistence of the ctxB-ET variant in these strains are uncertain; however, one might hypothesize that there was insufficient environmental (or human) exposure to ctxB-CL genes circulating in O1 strains for gene transfer to have occurred.
In summary, our data highlight the persistence of toxigenic V. cholerae O139 in clinical and environmental settings across almost 20 years in China. As reflected in their common ST, these isolates remain closely related genetically. Reservoirs responsible for persistence of these isolates across time and space remain to be identified. At the same time, there are avirulent, nontoxigenic environmental isolates, from diverse backgrounds, that appear to have acquired the O139 biosynthetic gene complex. While there remains a theoretical possibility that these nontoxigenic environmental isolates will acquire sufficient virulence determinants to become human pathogens, their importance in public health is uncertain.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the National High Technology R&D Program of China (2006AA02Z425), the NSFC of China (30872260), and the State Key Laboratory for Infectious Diseases Prevention and Control (2011SKLID201). J.G.M. was supported, in part, by U.S. Public Health Service grant RO1AI097405.
Footnotes
Published ahead of print 22 January 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JCM.03354-13.
REFERENCES
- 1.Kaper JB, Morris JG, Jr, Levine MM. 1995. Cholera. Clin. Microbiol. Rev. 8:48–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Morris JG., Jr 2011. Cholera—modern pandemic disease of ancient lineage. Emerg. Infect. Dis. 17:2099–2104. 10.3201/eid1711.111109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Albert MJ, Siddique AK, Islam MS, Faruque AS, Ansaruzzaman M, Faruque SM, Sack RB. 1993. Large outbreak of clinical cholera due to Vibrio cholerae non-O1 in Bangladesh. Lancet 341:704. 10.1016/0140-6736(93)90481-U [DOI] [PubMed] [Google Scholar]
- 4.Ramamurthy T, Garg S, Sharma R, Bhattacharya SK, Nair GB, Shimada T, Takeda T, Karasawa T, Kurazano H, Pal A, Takeda Y. 1993. Emergence of novel strain of Vibrio cholerae with epidemic potential in southern and eastern India. Lancet 341:703–704. 10.1016/0140-6736(93)90480-5 [DOI] [PubMed] [Google Scholar]
- 5.Haley BJ, Chen A, Grim CJ, Clark P, Diaz CM, Taviani E, Hasan NA, Sancomb E, Elnemr WM, Islam MA, Huq A, Colwell RR, Benediktsdottir E. 2012. Vibrio cholerae in an historically cholera-free country. Environ. Microbiol. Rep. 4:381–389. 10.1111/j.1758-2229.2012.00332.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu J, Gao S, Gao T, Qi G, Cao X, Duan G, Jiang M, Zhou B, Nu E, Ni Sha 1993. Diarrhea outbreak by Vibrio cholerae O139 in Keping Xinjiang. Dis. Surveill. 8:238–239 (In Chinese) [Google Scholar]
- 7.Comstock LE, Johnson JA, Michalski JM, Morris JG, Jr, Kaper JB. 1996. Cloning and sequence of a region encoding a surface polysaccharide of Vibrio cholerae O139 and characterization of the insertion site in the chromosome of Vibrio cholerae O1. Mol. Microbiol. 19:815–826. 10.1046/j.1365-2958.1996.407928.x [DOI] [PubMed] [Google Scholar]
- 8.Johnson JA, Salles CA, Panigrahi P, Albert MJ, Wright AC, Johnson RJ, Morris JG., Jr 1994. Vibrio cholerae O139 synonym Bengal is closely related to Vibrio cholerae El Tor but has important differences. Infect. Immun. 62:2108–2110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kam KM, Luey CK, Tsang YM, Law CP, Chu MY, Cheung TL, Chiu AW. 2003. Molecular subtyping of Vibrio cholerae O1 and O139 by pulsed-field gel electrophoresis in Hong Kong: correlation with epidemiological events from 1994 to 2002. J. Clin. Microbiol. 41:4502–4511. 10.1128/JCM.41.10.4502-4511.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee JH, Han KH, Choi SY, Lucas ME, Mondlane C, Ansaruzzaman M, Nair GB, Sack DA, von Seidlein L, Clemens JD, Song M, Chun J, Kim DW. 2006. Multilocus sequence typing (MLST) analysis of Vibrio cholerae O1 El Tor isolates from Mozambique that harbour the classical CTX prophage. J. Med. Microbiol. 55:165–170. 10.1099/jmm.0.46287-0 [DOI] [PubMed] [Google Scholar]
- 11.Qu M, Xu J, Ding Y, Wang R, Liu P, Kan B, Qi G, Liu Y, Gao S. 2003. Molecular epidemiology of Vibrio cholerae O139 in China: polymorphism of ribotypes and CTX elements. J. Clin. Microbiol. 41:2306–2310. 10.1128/JCM.41.6.2306-2310.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li BS, Tan HL, Wang DC, Deng XL, Chen JD, Zhong HJ, Ke BX, Ke CW, Kan B. 2011. Phenotypic and genotypic characterization of Vibrio cholerae O139 clinical and aquatic isolates in China. Curr. Microbiol. 62:950–955. 10.1007/s00284-010-9802-3 [DOI] [PubMed] [Google Scholar]
- 13.Kumar P, Jain M, Goel AK, Bhadauria S, Sharma SK, Kamboj DV, Singh L, Ramamurthy T, Nair GB. 2009. A large cholera outbreak due to a new cholera toxin variant of the Vibrio cholerae O1 El Tor biotype in Orissa, Eastern India. J. Med. Microbiol. 58:234–238. 10.1099/jmm.0.002089-0 [DOI] [PubMed] [Google Scholar]
- 14.Chow KH, Ng TK, Yuen KY, Yam WC. 2001. Detection of RTX toxin gene in Vibrio cholerae by PCR. J. Clin. Microbiol. 39:2594–2597. 10.1128/JCM.39.7.2594-2597.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Waldor MK, Mekalanos JJ. 1996. Lysogenic conversion by a filamentous phage encoding cholera toxin. Science 272:1910–1914. 10.1126/science.272.5270.1910 [DOI] [PubMed] [Google Scholar]
- 16.Mukhopadhyay AK, Chakraborty S, Takeda Y, Nair GB, Berg DE. 2001. Characterization of VPI pathogenicity island and CTXphi prophage in environmental strains of Vibrio cholerae. J. Bacteriol. 183:4737–4746. 10.1128/JB.183.16.4737-4746.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Davis BM, Kimsey HH, Chang W, Waldor MK. 1999. The Vibrio cholerae O139 Calcutta bacteriophage CTXphi is infectious and encodes a novel repressor. J. Bacteriol. 181:6779–6787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li M, Kotetishvili M, Chen Y, Sozhamannan S. 2003. Comparative genomic analyses of the vibrio pathogenicity island and cholera toxin prophage regions in nonepidemic serogroup strains of Vibrio cholerae. Appl. Environ. Microbiol. 69:1728–1738. 10.1128/AEM.69.3.1728-1738.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maiti D, Das B, Saha A, Nandy RK, Nair GB, Bhadra RK. 2006. Genetic organization of pre-CTX and CTX prophages in the genome of an environmental Vibrio cholerae non-O1, non-O139 strain. Microbiology 152:3633–3641. 10.1099/mic.0.2006/000117-0 [DOI] [PubMed] [Google Scholar]
- 20.Rivera IN, Chun J, Huq A, Sack RB, Colwell RR. 2001. Genotypes associated with virulence in environmental isolates of Vibrio cholerae. Appl. Environ. Microbiol. 67:2421–2429. 10.1128/AEM.67.6.2421-2429.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.O'Shea YA, Reen FJ, Quirke AM, Boyd EF. 2004. Evolutionary genetic analysis of the emergence of epidemic Vibrio cholerae isolates on the basis of comparative nucleotide sequence analysis and multilocus virulence gene profiles. J. Clin. Microbiol. 42:4657–4671. 10.1128/JCM.42.10.4657-4671.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Luo Y, Ye J, Jin D, Ding G, Zhang Z, Mei L, Octavia S, Lan R. 2013. Molecular analysis of non-O1/non-O139 Vibrio cholerae isolated from hospitalised patients in China. BMC Microbiol. 13:52. 10.1186/1471-2180-13-52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Feil EJ, Li BC, Aanensen DM, Hanage WP, Spratt BG. 2004. eBURST: inferring patterns of evolutionary descent among clusters of related bacterial genotypes from multilocus sequence typing data. J. Bacteriol. 186:1518–1530. 10.1128/JB.186.5.1518-1530.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cooper KL, Luey CK, Bird M, Terajima J, Nair GB, Kam KM, Arakawa E, Safa A, Cheung DT, Law CP, Watanabe H, Kubota K, Swaminathan B, Ribot EM. 2006. Development and validation of a PulseNet standardized pulsed-field gel electrophoresis protocol for subtyping of Vibrio cholerae. Foodborne Pathog. Dis. 3:51–58. 10.1089/fpd.2006.3.51 [DOI] [PubMed] [Google Scholar]
- 25.Garg P, Aydanian A, Smith D, J Glenn MJ, Nair GB, Stine OC. 2003. Molecular epidemiology of O139 Vibrio cholerae: mutation, lateral gene transfer, and founder flush. Emerg. Infect. Dis. 9:810–814. 10.3201/eid0907.020760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gonzalez-Fraga S, Pichel M, Binsztein N, Johnson JA, Morris JG, Jr, Stine OC. 2008. Lateral gene transfer of O1 serogroup encoding genes of Vibrio cholerae. FEMS Microbiol. Lett. 286:32–38. 10.1111/j.1574-6968.2008.01251.x [DOI] [PubMed] [Google Scholar]
- 27.Faruque SM, Ahmed KM, Siddique AK, Zaman K, Alim AR, Albert MJ. 1997. Molecular analysis of toxigenic Vibrio cholerae O139 Bengal strains isolated in Bangladesh between 1993 and 1996: evidence for emergence of a new clone of the Bengal vibrios. J. Clin. Microbiol. 35:2299–2306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mitra R, Basu A, Dutta D, Nair GB, Takeda Y. 1996. Resurgence of Vibrio cholerae O139 Bengal with altered antibiogram in Calcutta, India. Lancet 348:1181. 10.1016/S0140-6736(05)65326-3 [DOI] [PubMed] [Google Scholar]
- 29.Mukhopadhyay AK, Basu A, Garg P, Bag PK, Ghosh A, Bhattacharya SK, Takeda Y, Nair GB. 1998. Molecular epidemiology of reemergent Vibrio cholerae O139 Bengal in India. J. Clin. Microbiol. 36:2149–2152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang J, Chang Z, Zhong H, Wang D, Xu J, Kan B, Ran L, Wang Z. 2007. Investigation on status of pollution of vibrio cholerae in seafood and aquatic products in 12 provinces of China in 2005. Zhonghua Yu Fang Yi Xue Za Zhi 41:208–211 [PubMed] [Google Scholar]
- 31.Tang X, Liu L, Ma H, Zhu B, Hao C, Wu X, Fei N, Zhu X, Zhang L. 2010. Outbreak of cholera associated with consumption of soft-shelled turtles, Sichuan province, China, 2009. Zhonghua Liu Xing Bing Xue Za Zhi 31:1050–1052. 10.3760/cma.j.issn.0254-6450.2010.09.021 [DOI] [PubMed] [Google Scholar]
- 32.Li H, Wang X, Cui Z, Zhou H, Xia S, Kan B. 2008. Analysis of the source of a cholera outbreak caused by O139 Vibrio cholerae. Dis. Surveill. 23:218–220 (In Chinese) [Google Scholar]
- 33.Qiu X, Wu J, Gao Y, Shi G. 2013. Investigation on a foodborne cholera O139 outbreak in a rural banquet and the turtle markets. Pract. Prevent. Med. 20:171–174 (In Chinese.) 10.3969/j.issn.1006-3110.2013.02.014 [DOI] [Google Scholar]
- 34.Safa A, Nair GB, Kong RY. 2010. Evolution of new variants of Vibrio cholerae O1. Trends Microbiol. 18:46–54. 10.1016/j.tim.2009.10.003 [DOI] [PubMed] [Google Scholar]
- 35.Naha A, Chowdhury G, Ghosh-Banerjee J, Senoh M, Takahashi T, Ley B, Thriemer K, Deen J, Seidlein LV, Ali SM, Khatib A, Ramamurthy T, Nandy RK, Nair GB, Takeda Y, Mukhopadhyay AK. 2013. Molecular characterization of high-level-cholera-toxin-producing El Tor variant Vibrio cholerae strains in the Zanzibar Archipelago of Tanzania. J. Clin. Microbiol. 51:1040–1045. 10.1128/JCM.03162-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rashed SM, Iqbal A, Mannan SB, Islam T, Rashid MU, Johura FT, Watanabe H, Hasan NA, Huq A, Stine OC, Sack RB, Colwell RR, Alam M. 2013. Vibrio cholerae O1 El Tor and O139 Bengal strains carrying ctxB(ET), Bangladesh. Emerg. Infect. Dis. 19:1713–1715. 10.3201/eid1910.130626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kumar P, Mishra DK, Deshmukh DG, Jain M, Zade AM, Ingole KV, Yadava P. 18 September 2013. Haitian variant ctxB producing Vibrio cholerae O1 with reduced susceptibility to ciprofloxacin is persistent in Yavatmal, Maharashtra, India after causing a cholera outbreak. Clin. Microbiol. Infect. 10.1111/1469-0691.12393 [DOI] [PubMed] [Google Scholar]
- 38.Bhuiyan NA, Nusrin S, Alam M, Morita M, Watanabe H, Ramamurthy T, Cravioto A, Nair GB. 2009. Changing genotypes of cholera toxin (CT) of Vibrio cholerae O139 in Bangladesh and description of three new CT genotypes. FEMS Immunol. Med. Microbiol. 57:136–141. 10.1111/j.1574-695X.2009.00590.x [DOI] [PubMed] [Google Scholar]
- 39.Zhang P, Li F, Liang W, Li J, Kan B, Wang D. 18 December 2013. The 7th pandemic Vibrio cholerae O1 El Tor isolate in China has undergone genetic shifts. J. Clin. Microbiol. 10.1128/JCM.03121-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang P, Zhou H, Kan B, Wang D. 2013. Novel ctxB variants of Vibrio cholerae O1 isolates, China. Infect. Genet. Evol. 20C:48–53. 10.1016/j.meegid.2013.08.004 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.




