Abstract
A molecular diagnostic technique based on real-time PCR was developed for the simultaneous detection of three of the most frequent causative agents of fungal opportunistic pneumonia in AIDS patients: Pneumocystis jirovecii, Histoplasma capsulatum, and Cryptococcus neoformans/Cryptococcus gattii. This technique was tested in cultured strains and in clinical samples from HIV-positive patients. The methodology used involved species-specific molecular beacon probes targeted to the internal transcribed spacer regions of the rDNA. An internal control was also included in each assay. The multiplex real-time PCR assay was tested in 24 clinical strains and 43 clinical samples from AIDS patients with proven fungal infection. The technique developed showed high reproducibility (r2 of >0.98) and specificity (100%). For H. capsulatum and Cryptococcus spp., the detection limits of the method were 20 and 2 fg of genomic DNA/20 μl reaction mixture, respectively, while for P. jirovecii the detection limit was 2.92 log10 copies/20 μl reaction mixture. The sensitivity in vitro was 100% for clinical strains and 90.7% for clinical samples. The assay was positive for 92.5% of the patients. For one of the patients with proven histoplasmosis, P. jirovecii was also detected in a bronchoalveolar lavage sample. No PCR inhibition was detected. This multiplex real-time PCR technique is fast, sensitive, and specific and may have clinical applications.
INTRODUCTION
HIV infection remains a threat for many people, as it is estimated that almost 34 million adults and children worldwide are infected with the virus, according to WHO/UNAIDS (1). Although mortality associated with HIV infection has decreased in recent years due to the development of highly active antiretroviral therapy (HAART), the incidence and mortality rates of non-AIDS-associated infections remain high, particularly in nondeveloped countries (1). Opportunistic pneumonias are one of the major causes of pulmonary complications among AIDS patients. In fact, almost 70% of people infected with HIV suffer at least one pulmonary complication during the course of the infection (2). Although in early HIV infection the agents that cause pneumonia are similar to those found in the general population, when the number of CD4+ T cells decreases, opportunistic pneumonias are associated with high morbidity and mortality (3–5). Tuberculosis and Pneumocystis jirovecii pneumonia (PCP; the abbreviation PCP reflects the previous nomenclature for the Pneumocystis species causing opportunistic pneumonia in humans, P. carinii; P. jirovecii is now recognized as the species that infects humans) are the most commonly reported AIDS-defining illnesses (6). Moreover, opportunistic pneumonias due to Histoplasma capsulatum and Cryptococcus spp. in immunocompromised patients involve high morbidity and mortality in areas where the virus is not endemic or in those regions where the infection is not very common. P. jirovecii has been described as the main cause of pulmonary complication in AIDS patients in developed countries; in fact, in the United States it has been estimated that 75% of HIV-positive persons suffer PCP during their lifetime (7, 8). Clinical diagnosis of PCP is based mainly on microscopic procedures performed directly on the clinical sample and usually involves immunofluorescence microscopy (IFA) or use of Grocott's methenamine silver stain (9, 10). Recently, some reports have described the utility of real-time PCR (rt-PCR) protocols to increase the sensitivity and specificity of PCP diagnosis (11–17), and some of these tests have been commercialized (18). Moreover, a recent study showed the utility of rt-PCR assays for the early diagnosis and treatment of non-HIV-infected patients diagnosed with PCP, for whom a definitive diagnosis could be performed using rt-PCR techniques that detected the low fungal burden present in the patients (19).
Histoplasmosis is a mycosis that is endemic in the United States and Africa that can produce disseminated infection associated with high morbidity and mortality in HIV patients (20–22). Due to travel and immigration, this infection has increased in recent years in areas where H. capsulatum mycosis is not endemic (23). Although culture is the reference procedure for the diagnosis of histoplasmosis, it is time-consuming (requiring 3 to 4 weeks of incubation) (24). In fact, the sensitivity and specificity of microscopy-based approaches are very low, and fungal structures are only observed when the infection is advanced. Moreover, serology has shown significant limitations in AIDS patients; it may be negative in 50% of such patients (25). Antigen detection could be useful for these patients, although it is only available in the United States (26). Several rt-PCR protocols with varied sensitivities and specificities have been described for detection of DNA from Histoplasma capsulatum in clinical samples (27–29). Recently, a comparison of different protocols for detection of H. capsulatum DNA by PCR showed that the sensitivity of the assay depended on both the selected target region and the type of amplification assay (conventional or rt-PCR) (24). Herein, the most sensitive and specific protocols were based on the amplification of the internal transcribed spacer (ITS) by rt-PCR (30), and the less sensitive protocols were based on the amplification of monocopy genes (Hc100p and SCAR220) by conventional PCR (31, 32).
In Africa, cryptococcosis, caused by Cryptococcus neoformans var. neoformans or C. neoformans var. grubii, is one of the main causes of mortality associated with HIV infection (33). However, in the northwest United States, Cryptococcus gattii is the primary causative agent of cryptococcosis and can also affect immunocompetent people; it has been classified endemic in some regions of the eastern United States, and several cases of cryptococcosis outside that area where the disease is endemic have been reported recently (34–36). This infection can produce both meningitis and pulmonary complications in these patients (37, 38). Routine clinical diagnosis of cryptococcosis is based on culture and detection of a capsular antigen, with varied sensitivity and specificity (39, 40). However, molecular methods based on PCR and new technologies for antigen detection also have been described in recent years, although validation of these methods is pending (41–44).
Fungal pneumonias are clinically and radiologically similar to each other, and definitive diagnosis must be reached to initiate the appropriate therapy (45). Moreover, multiplex real-time PCR (Mrt-PCR) protocols allow the detection of mixed infections that cannot be detected by routine protocols. Mixed infections involving H. capsulatum, C. neoformans, and P. jirovecii have been described (46–48).
The aim of this study was the development of a Mrt-PCR for the early detection of fungal opportunistic pneumonia due to H. capsulatum, C. neoformans, C. gattii, and P. jirovecii in clinical samples from HIV-positive patients.
MATERIALS AND METHODS
Control strains and plasmids.
DNA from different strains was used to standardize the Mrt-PCR assay. All the strains belonged to the Collection of the Mycology Reference Laboratory, Spanish National Center of Microbiology. To standardize the technique, we used the strains C. neoformans var. neoformans CNM-CL 2132, H. capsulatum CNM-CM 2721, and a plasmid that included the sequence target of P. jirovecii (pSG1).
To assess the specificity of the technique, the following strains of yeast and molds were used: Candida albicans (CNM-CL 5719), Candida parapsilosis (CNM-CL5683), Candida tropicalis (CNM-CL 5742), Candida glabrata (CNM-CL 5533), Candida guilliermondii (CNM-CL 7127), Candida krusei (CNM-CL 7057), Coccidioides posadasii (CNM-CM 2912), Paracoccidioides brasiliensis (CNM-CM 2908), Aspergillus fumigatus (CNM-CM AF237), Aspergillus terreus (CNM-CM 3508), Aspergillus flavus (CNM-CM 3509), Aspergillus niger (CNM-CM 3551), Fusarium verticillioides (CNM-CM 2975), Fusarium oxysporum (CNM-CM 2914), Fusarium solani (CNM-CM 3035), Scedosporium prolificans (CNM-CM1627), Scedosporium apiospermum (CNM-CM 3169), Rhizopus oryzae (CNM-CM 3020), and Rhizopus microsporum (CNM-CM 4244), Mucor circinelloides (CNM-CM 2437). In addition, we employed mouse genomic DNA (Promega, Madrid, Spain) and human genomic DNA (Promega, Madrid, Spain).
Design of primers and probes.
Primers and molecular beacon probes were designed to specifically amplify a region of the ITS of the rDNA from Cryptococcus neoformans/C. gattii, Histoplasma capsulatum var. capsulatum and H. capsulatum var. duboisii, and Pneumocystis jirovecii by Mrt-PCR. Beacon Designer 5.0 software (Premier Biosoft, Palo Alto, CA) was used for primer and probe design. An internal control was included, as described previously (49). The primers and probes designed were subjected to a BLAST search within the GenBank sequence database (http://blast.ncbi.nlm.nih.gov/Blast.cgi) and in the database of the Department of Mycology of the Spanish National Center for Microbiology (which contains more than 8,000 distinct sequences), to avoid cross-homology with other microorganisms. The primer and probe sequences are shown in Table 1.
TABLE 1.
Sequences of primers and probes designed for the multiplex real-time PCR assay
| Species and its primers and probe | Sequencea |
|---|---|
| Pneumocystis jirovecii | |
| OliPJMB1 (f) | 5′-CCCTAGTGTTTTAGCATTTTTC-3′ |
| OLIPJMB2 (r) | 5′-CTGCAATTCACACTACTTATCG-3′ |
| Probe PJ-MB1 | 5′-HEX-CGCGATACCTTTGGCGAGGCAAGCAAATCGCG-BHQ1-3′ |
| Histoplasma capsulatumb | |
| Cryptococcus neoformans | |
| OLI CRYPTO 1 2 (f) | 5′-CCTGTTGGACTTGGATTTGG-3′ |
| OLI CRYPTO 2 (r) | 5′-AGCAAGCCGAAGACTACC-3′ |
| Probe MB CRYPTO | 5′-Cyan 500-CGCGATCATTACGCCGGGCTGACAGGTAATCAGATCGCG-BHQ1-3′ |
| Jellyfish (internal control) | |
| Oli1-icjf1 (f) | 5′-GCCTGGTGCAAAAATTGCTTATC-3′ |
| Oli1-icjf2 (r) | 5′-CTAAGACAAGTGTGTTTATGGTATTG-3′ |
| Probe CIJF-MB | 5′-Cy5-CGCGATGCTGTTCTTCCGCCACTTCCAATCGCG-BHQ2-3′ |
BHQ1, black hole quencher 1.
Sequences for the primers and probe for H. capsulatum are included in patent PCT/ES2009070340.
Multiplex real-time PCR assay.
The amplification was carried out in a LightCycler 480 system (Roche Diagnostics, Mannheim, Germany). PCRs were performed in a 20-μl final volume containing 2× SensiMix DNA (Quantance; Ecogen, Madrid, Spain) used according to the manufacturer's instructions, 0.5 μM each primer for each one of the species, 0.25 μM internal control primer, 0.2 μM each species-specific probe, 0.1 μM internal control probe, 2 fg of the internal control plasmid (pICJF), and 2 μl of DNA extracted from the sample, used as the template. The PCR conditions were as follows: an initial step of 95°C for 10 min, followed by 50 cycles of 95°C for 25 s, 50°C for 30 s, and 72°C for 5 s, with a cooling cycle at 40°C for 30 s. Results were considered positive when the product amount increased until its fluorescence intensity exceeded the background, as determined by second-derivate analysis; these results were expressed in terms of the quantification cycle (Cq). Each experiment was performed in duplicate and included quantification standards as well as negative controls. Subsequently, a color compensation experiment was performed to prevent cross talk between dyes.
Standardization.
Standard curves for H. capsulatum and C. neoformans were constructed based on the results of five PCR repetitions with dilutions of genomic DNA ranging from 20 ng to 2 fg/20 μl of reaction mixture. The strains CNM-CL 2132 and CNM-CM 2721 were used to construct the standard curves. For P. jirovecii, a fragment of 175 bp containing the ITS1 target region was cloned into a pGEMT-Easy plasmid (Promega, Madrid, Spain) according to the manufacturer's instructions. A 10-fold serial dilution of the plasmid clone (pSG1), from 20 ng (2.92 × 108 log10 copies/20-μl reaction mixture) to 2 fg of pDNA/20-μl reaction mixture (2.92 log10 copies/20-μl reaction mixture), was used to construct the standard curve. Cq values for each dilution series were determined in duplicate in three different experiments.
Regression lines were constructed by plotting the logarithm of the initial template concentration versus the corresponding Cq value. If this line exhibited a linear regression coefficient value of >0.980, the standard curve was then used to determine the sensitivity, primer efficiencies, and reproducibility of the assay. In order to evaluate the specificity, 2 ng of DNA/20 μl of reaction mixture from each of the other mold and yeast species, as well as human and mouse genomic DNA, was included in the PCR assay (control strains).
DNA extraction.
DNA extraction from cultures of H. capsulatum was performed under biosafety level 3 conditions in compliance with Spanish law (Real decreto 664/1997) and following the method described by Buitrago et al. (23). DNA extraction from C. neoformans was performed as previously described by Tang et al. (50). Finally, genomic DNA of P. jirovecii was extracted from bronchoalveolar lavage (BAL) samples from 16 patients with proven P. jirovecii pneumonia. DNA extraction was performed following the recommendations from reference 15 with modifications. For that purpose, 1.5 ml of BAL fluid was centrifuged at 10,000 rpm for 10 min, the supernatant was discharged, and then the pellet was diluted in 200 μl of supernatant. After that, DNA was extracted using a Qiamp DNA minikit (Qiagen) according to manufacturer's instructions in a 50-μl elution volume. Two microliters of this DNA was used for the Mrt-PCR assays. DNA extractions from other specimens were also performed by using the Qiamp DNA minikit, without modifications. DNAs from paraffin-embedded tissues were isolated as described previously (51).
The DNA extraction efficiency was also determined. For that purpose, 5 μl of the plasmid clone or genomic DNA, containing from 10 ng to 1 fg, was added to 200 μl of serum from healthy patients. Then, DNA extraction was performed by using the Qiamp DNA minikit (Qiagen), and 2 μl was used for the Mrt-PCR protocol. The percentage of DNA recovered was determined for each of the dilutions. The experiments were run in triplicate.
Multiplex real-time assay of cultured clinical strains.
The Mrt-PCR assay was initially validated by using cultures from 13 clinical strains of Cryptococcus spp. complex (C. neoformans var. neoformans [n = 3], C. neoformans var. grubii [n = 3], C. gattii [n = 4], and hybrids [n = 3]) and 10 H. capsulatum strains belonging to the Mold Collection of the Mycology Department of the National Center for Microbiology. All the strains included in the study were previously identified by sequencing the ITS region from the rDNA (52). For P. jirovecii validation, clones from 4 different BAL samples were evaluated in triplicate. Two microliters of the extracted DNA was employed to perform the Mrt-PCR assay.
Mrt-PCR assay of clinical samples.
The utility of the Mrt-PCR was also evaluated by using clinical samples. Forty-three clinical specimens from 40 HIV-positive patients with proven infection (according to the EORTC/MSG criteria) were evaluated by Mrt-PCR. Because the EORTC/MSG criteria do not include infection caused by Pneumocystis jirovecii, proven PCP was considered when the fungus was visualized by methenamine silver stain and the patient showed clinical signs of pneumonia. Patients and clinical specimens are summarized in Table 2. Two microliters of DNA from each sample was used for the Mrt-PCR.
TABLE 2.
Mrt-PCR results for clinical samples from patients with proven opportunistic fungal pneumonia
| Patient no. | Proven infection | Sample | PCR result | Cp(s) | Amt of DNAc |
|---|---|---|---|---|---|
| 1 | Histoplasmosis | BAL | Positive | 27.54 | 7 × 10−3 |
| 2 | Histoplasmosis | Bone marrow | Negative | ||
| Blood | Positive | 29.46 | 1 × 10−3, 8 × 10−3 | ||
| 3a | Histoplasmosis | Bronchiaspirate | Positive | 29.18, 28.8b | 2.2 × 10−3, 3.37 log10 |
| 4 | Histoplasmosis | Bone marrow | Positive | 29.33 | 1.9 × 10−3 |
| Serum | Negative | ||||
| 5 | Histoplasmosis | Blood | Positive | 29.53 | 1.7 × 10−3 |
| BAL | Positive | 29 | 2.5 × 10−3 | ||
| 6 | Histoplasmosis | Biopsy | Positive | 27.2 | 8.9 × 10−3 |
| 7 | Histoplasmosis | Biopsy | Positive | 29.3 | 2 × 10−3 |
| 8 | Cryptococcosis | Blood | Positive | 40.97 | 2.5 × 10−3 |
| 9 | Cryptococcosis | Biopsy | Negative | ||
| 10 | PCP | BAL | Positive | 27.5 | 5 × 102 log10 |
| 11 | PCP | BAL | Positive | 36.75 | 1.42 log10 |
| 12 | PCP | BAL | Positive | 29.93 | 1 × 102 log10 |
| 13 | PCP | BAL | Positive | 27.7 | 4.4 × 102 log10 |
| 14 | PCP | BAL | Positive | 27.51 | 5 × 102 log10 |
| 15 | PCP | BAL | Positive | 25.61 | 1.7 × 103 log10 |
| 16 | PCP | BAL | Positive | 33.28 | 12 log10 |
| 17 | PCP | BAL | Positive | 33.05 | 14 log10 |
| 18 | PCP | BAL | Positive | 32.92 | 16 log10 |
| 19 | PCP | BAL | Positive | 33.16 | 13 log10 |
| 20 | PCP | BAL | Positive | 33.15 | 14 log10 |
| 21 | PCP | BAL | Positive | 33.83 | 9 log10 |
| 22 | PCP | BAL | Positive | 33.57 | 10 log10 |
| 23 | PCP | BAL | Positive | 33.37 | 12 log10 |
| 24 | PCP | BAL | Positive | 32.44 | 22 log10 |
| 25 | PCP | BAL | Positive | 33.22 | 13 log10 |
| 26 | Cryptococcosis | BAL | Positive | 28.12 | 8.6 × 10−3 |
| 27 | Cryptococcosis | BAL | Positive | 28.94 | 5.1 × 10−3 |
| 28 | Histoplasmosis | Biopsy | Positive | 19.35 | 2.1 × 10−3 |
| 29 | Histoplasmosis | Biopsy | Positive | 27.14 | 9.3 × 10−3 |
| 30 | Histoplasmosis | Sputum | Positive | 25.46 | 3 × 10−2 |
| 31 | Histoplasmosis | Sputum | Positive | 26.45 | 1.5 × 10−2 |
| 32 | Histoplasmosis | Sputum | Positive | 26.79 | 1.1 × 10−2 |
| 33 | Histoplasmosis | Serum | Positive | 26.97 | 1 × 10−2 |
| 34 | Histoplasmosis | Bone marrow | Negative | ||
| 35 | Cryptococcosis | BAL | Positive | 26.9 | 1.8 ×10−2 |
| 36 | PCP | BAL | Positive | 28.02 | 3.65 × 102 log10 |
| 37 | PCP | BAL | Positive | 27.69 | 4.50 × 102 log10 |
| 38 | PCP | BAL | Positive | 27.26 | 5.9 × 102 log10 |
| 39 | PCP | BAL | Positive | 25.28 | 2 × 103 log10 |
| 40 | PCP | BAL | Positive | 26.7 | 8.410 × 102 log10 |
Patient number 3 showed a mixed infection with Histoplasma capsulatum and Pneumocystis jirovecii.
The first and second Cps correspond to H. capsulatum and P. jirovecii, respectively.
Values shown are in ng/μl except where specified as “log10,” indicating log10 numbers of copies/μl.
Twenty-nine specimens from the following groups of patients were included to assess the specificity of the technique: HIV+ patients from areas where histoplasmosis is endemic, HIV+ patients from areas of endemicity and with viral or bacterial pneumonia, and HIV+ patients with invasive fungal infection (IFI) caused by other fungal species; patients with IFI and other underlying diseases were included as negative controls (Table 3).
TABLE 3.
Summary of control populations used to determine background colonization levels and the specificity of the Mrt-PCR assay
| Population | Control sample no. | Specimen | PCR result | Species causing IFI |
|---|---|---|---|---|
| HIV+ patients from areas of endemicity | 1 | Bone narrow | Negative | |
| 2 | Biopsy | Negative | ||
| 3 | Biopsy | Negative | ||
| 4 | Bone narrow | Negative | ||
| 5 | Whole Blood | Negative | ||
| 6 | Whole Blood | Negative | ||
| HIV+ patients from areas of endemicity and with viral or bacterial pneumonia | 7 | BAL | Negative | |
| 8 | BAL | Negative | ||
| 9 | BAL | Negative | ||
| 10 | Biopsy | Negative | ||
| HIV+ patients with invasive fungal infection caused by other fungal species | 11 | Biopsy | Negative | Lichtheimia corymbifera |
| 12 | Biopsy | Negative | Rhizomucor pusillus | |
| 13 | BAL | Negative | Cryptococcus laurentii | |
| 14 | BAL | Negative | Cryptococcus humicola | |
| 15 | BAL | Negative | Aspergillus flavus, Penicillium spp., and Cryptococcus albidus | |
| Non-HIV-infected patients with invasive fungal infection | 16 | Biopsy | Negative | Coccidioides immitis |
| 17 | Biopsy | Negative | Scedosporium apiospermum | |
| 18 | Biopsy | Negative | Bipolaris spp. | |
| 19 | BAL | Negative | Aspergillus fumigatus | |
| 20 | BAL | Negative | Aspergillus fumigatus | |
| 21 | BAL | Negative | Aspergillus fumigatus | |
| 22 | BAL | Negative | Aspergillus fumigatus | |
| 23 | BAL | Negative | Aspergillus fumigatus | |
| 24 | Ascitic fluid | Negative | Aspergillus fumigatus | |
| 25 | Cerebrospinal fluid | Negative | Aspergillus fumigatus | |
| 26 | BAL | Negative | Aspergillus fumigatus | |
| 27 | Biopsy | Negative | Aspergillus fumigatus | |
| 28 | Biopsy | Negative | Aspergillus fumigatus | |
| 29 | BAL | Negative | Aspergillus fumigatus |
RESULTS
Assay standardization in vitro.
The Mrt-PCR assay developed was able to reliably detect C. neoformans, H. capsulatum, and P. jirovecii DNA. The sizes of the amplicons generated were 175 bp for P. jirovecii, 106 bp for H. capsulatum, and 139 bp for C. neoformans.
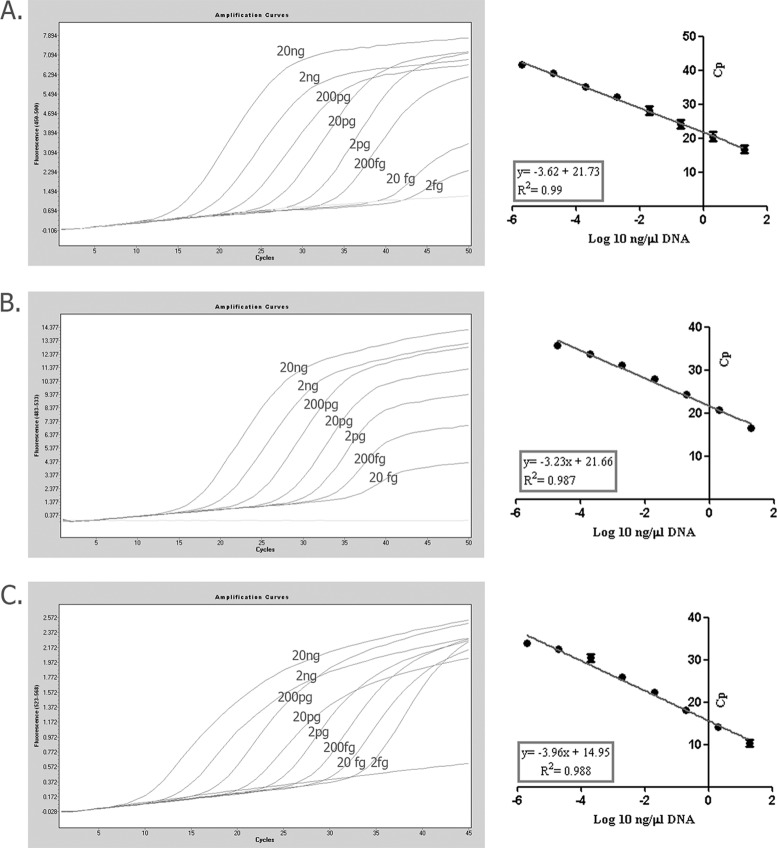
No cross-reactivity to other fungi or either human or mouse DNA was detected (control materials). In addition, no cross-reactivity between them was observed. The detection limits of the assay were 2 and 20 fg of genomic DNA per 20 μl of PCR mixture for C. neoformans and H. capsulatum, respectively, and 2.69 log10 copies/20 μl of reaction mixture for P. jirovecii (Fig. 1). Quantification was linear for all the fungal species included in the assay, and the standard curve generated showed a high coefficient of determination (R2 = 0.98 to 0.99). The average coefficient of variation was 2.18% for H. capsulatum DNA, 3.93% for C. neoformans DNA, and 3.35% for 2 replicates from two different plasmids of P. jirovecii (Table 4). Each of the species was detected in their corresponding fluorophore channels (Fig. 2). The mean crossing point (Cp) value for the internal control was 33 ± 0.6. The calculated average DNA extraction efficiencies from clinical samples were 79.8%, 78%, and 96.5% for H. capsulatum, C. neoformans, and P. jirovecii, respectively.
FIG 1.
Quantification standard curve obtained by using serial 1:10 dilutions of different concentrations of DNA from Pneumocystis jirovecii (A), Histoplasma capsulatum (B), and Cryptococcus neoformans (C). Error bars represent the percent coefficient of variation. Standard curves were constructed after a color compensation experiment.
TABLE 4.
Overview of standardization of the Mrt-PCR assay with serial dilutions of a plasmid clone containing the target for P. jirovecii or with genomic DNA of Histoplasma capsulatum or Cryptococcus neoformans
| Amt of DNA/μl |
H. capsulatum |
C. neoformans |
P. jirovecii |
||||||
|---|---|---|---|---|---|---|---|---|---|
| Cq | SD | %CV | Cq | SD | %CV | Cq | SD | %CV | |
| 10 ng | 16.53 | 0.52 | 3.15 | 16.78 | 1.17 | 6.97 | 10.26 | 0.803 | 7.834 |
| 1 ng | 20.656 | 0.50 | 2.46 | 20.494 | 1.36 | 6.64 | 14.205 | 0.716 | 5.047 |
| 100 pg | 24.304 | 0.59 | 2.43 | 24.128 | 1.21 | 5.02 | 18.055 | 0.733 | 4.06 |
| 10 pg | 27.986 | 0.66 | 2.36 | 28.172 | 1.27 | 4.53 | 22.3075 | 0.769 | 3.450 |
| 1 pg | 31.158 | 0.51 | 1.66 | 32.156 | 0.93 | 2.89 | 25.87 | 0.222 | 0.858 |
| 100 fg | 33.758 | 0.48 | 1.43 | 35.058 | 0.96 | 2.74 | 30.4925 | 0.921 | 3.022 |
| 10 fg | 35.644 | 0.29 | 0.83 | 39.1 | 0.88 | 2.25 | 32.47 | 0.621 | 1.914 |
| 1 fg | 16.53 | 0.52 | 3.15 | 41.76 | 0.16 | 0.38 | Negative | Negative | Negative |
FIG 2.
Dependence of fluorescence signal on the number of cycles in the multiplex real-time PCR assay for the patient with mixed infection. (A) Fluorescence detection in the 6-carboxyfluorescein (FAM) fluorescence channel, based on positive signal for the Histoplasma capsulatum molecular beacon probe. (B) Fluorescence detection in the Cyan 500 fluorescence channel. The positive signal is for the Cryptococcus neoformans/C. gattii molecular beacon probe. (C) Fluorescence detection in the 6-carboxy-hexachlorofluorescein (HEX) fluorescence channel. The positive signal is for the Pneumocystis jirovecii molecular beacon probe. (D) Fluorescence detection of Cy5 dye as end label to the specific pICJP molecular beacon probe.
Results with clinical cultured strains.
Results were positive for all clinical isolates tested. For the four P. jirovecii clones, the mean (± standard deviation) Cq obtained was 14.03 ± 0.71; the mean Cq was 26.27 ± 2.04 for all the clinical strains of H. capsultaum and 20.47 ± 1.34 for the C. neoformans/C. gattii isolates. No differences among Cq values for the different Cryptococcus spp. were detected.
Results with clinical samples.
A total of 43 clinical samples from 40 patients with HIV as the underlying disease were analyzed by Mrt-PCR. The source of the clinical samples was very heterogeneous (Table 2). Our Mrt-PCR assay was positive for 37/40 patients (92.5% sensitivity) and 39/43 clinical samples (90.7% sensitivity); the estimated clinical specificity was 100% for both analyses. The assay was negative in 3 cases: in 2 patients diagnosed with histoplasmosis and in 1 patient diagnosed with cryptococcosis. All patients with proven PCP showed a positive Mrt-PCR result. All the samples from patients included as controls showed negative PCR results. The average amount of DNA in each of the clinical samples is shown in Table 2. No inhibition of the PCR was detected with any of the clinical samples tested.
Although proven histoplasmosis was demonstrated by Mrt-PCR for case number 3, P. jirovecii infection was also detected (Fig. 1). Results were verified by sequencing the PCR products.
DISCUSSION
Opportunistic fungal pneumonias are one of the most frequent pulmonary complications in HIV-positive patients (4). In this study, we evaluated the utility of a single-tube Mrt-PCR assay for the diagnosis and identification of four of the most frequent causes of opportunistic fungal pneumonia in AIDS patients. Clinical symptomatology of these infections is based on the observation of abnormalities in chest X-rays, but in patients with proven or suspected HIV infection such findings are also related to Mycobacterium tuberculosis infection (45). Therefore, tuberculosis could overshadow other opportunistic infections, such as cryptococcosis and pulmonary histoplasmosis, delaying the diagnosis and correct treatment (53).
Molecular techniques have been proven to be very useful for the diagnosis of fungal infections (54). However, reports describing Mrt-PCR approaches for the diagnosis of opportunistic pneumonias are scarce (55). In fact, this is the first report to describe an Mrt-PCR assay for the simultaneous detection of P. jirovecii, H. capsulatum, and C. neoformans.
Our assay proved to be species specific (100%), with high reproducibility (r2 > 0.98) and sensitivity (100%) for each of the species. The Mrt-PCR was also validated by using clinical samples from 40 patients with proven infection diagnosed using classical techniques. Although respiratory samples (bronchoalveolar lavage fluid, pulmonary biopsy tissue, sputum, or pleural fluid) are the most appropriate samples for the diagnosis of opportunistic pneumonias, in patients with advanced immunosuppression systemic infection may develop, and other samples, such as blood, serum, or bone marrow biopsy specimens can be used for definitive diagnosis. Our assay was able to detect 92.5% of the infections in patients with proven infection. The Mrt-PCR yielded a negative result for four clinical samples (from serum, bone marrow, and biopsy) for three patients. These negative clinical samples were stored for more than 5 years at −20°C, which may have decreased the sensitivity of the assay (56).
Interestingly, for one of the patients diagnosed with histoplasmosis, P. jirovecii DNA was detected. In fact, coinfections may occur, and accurate diagnosis and prompt appropriate antifungal treatment are required. Several reports of cases of mixed infections have been published, and for all of them the definitive diagnosis was based on microbiological evidence. Bava et al. (47) reported a case in which a patient had mixed infection due to Cryptococcus and Pneumocystis. Mixed infections of H. capsulatum and C. neoformans (46), as well as of P. jirovecii and H. capsulatum, have also been reported in individuals from areas (48). Finally, coinfection with H. capsulatum and M. tuberculosis is frequent in some areas where histoplasmosis is endemic and may involve important implications for treatment (57).
To our knowledge, only one PCR-based protocol has been previously reported for the detection of tuberculosis and PCP in HIV-infected patients (55). rt-PCR approaches for the detection of C. neoformans/C. gattii, H. capsulatum, and P. jirovecii separately have been widely described based on the amplification of different targets, with varied sensitivity and specificity. Most of the rt-PCR approaches for the diagnosis of cryptococcosis are based on the amplification of monocopy genes (43, 58), which may decrease the sensitivity of the technique. Our technique allows detection of both C. neoformas spp. complex and C. gattii, but it cannot differentiate species. Although the identification at species level is important for epidemiological studies, clinical signs and treatment are similar for infections caused by either species (59–61). Moreover, a majority of the real-time protocols for the detection of PCP are based on nested PCR, which may decrease the specificity of the technique due to the possibility of false-positive results (58). Although there are several articles describing rt-PCR assays for the detection of H. capsulatum based on the amplification of multicopy regions, such as ITS, the use of molecular beacon probes can improve the sensitivity of the technique due to their small size and sequence specificity (23). The technique we have described in this article can detect several fungi in a single-tube reaction. The limit to detection of more fungal species is determined by the real-time PCR equipment.
In conclusion, this is the first report to describe an Mrt-PCR for the diagnosis of opportunistic fungal pneumonias. This approach could be very useful for differential diagnosis with tuberculosis, as clinical signs are similar, and also to detect mixed infections that could appear in patients with advanced immunosuppression living in areas of endemicity. Moreover, this assay could be useful for the clinical management of other immunocompromised patients at risk for these fungal infections (e.g., patients under corticosteroid therapy or chemotherapy or those treated with tumor necrosis factor). Although Mrt-PCR assays are more expensive than conventional approaches (the approximate cost is around 100 € per PCR determination), they provide a fast and specific diagnosis, as they allow the detection of mixed infection and avoid delays in appropriate antifungal therapy. Moreover, our Mrt-PCR could be implemented in clinical settings in developing countries and in countries that receive immigrant populations from those regions. Further studies with a larger number of samples are warranted.
ACKNOWLEDGMENTS
We warmly thank Frannk Hodkings for his careful reading and editing of the manuscript.
This work was supported by research project PI11/00412 from Spanish Fondo de Investigaciones Sanitarias of the Instituto de Salud Carlos III. S.G. and C.V. are supported by research fellowships from the Fondo de Investigaciones Biomedicas of the Spanish Ministry of Science and Innovation (FI10/00464 and FI12/00095, respectively).
We declare no conflicts of interest.
Footnotes
Published ahead of print 29 January 2014
REFERENCES
- 1.Huang L, Cattamanchi A, Davis JL, den Boon S, Kovacs J, Meshnick S, Miller RF, Walzer PD, Worodria W, Masur H, International HIV-Associated Opportunistic Pneumonias (IHOP) Study, Lung HIV Study 2011. HIV-associated Pneumocystis pneumonia. Proc. Am. Thorac Soc. 8:294–300. 10.1513/pats.201009-062WR [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gingo MR, Balasubramani GK, Kingsley L, Rinaldo CR, Jr, Alden CB, Detels R, Greenblatt RM, Hessol NA, Holman S, Huang L, Kleerup EC, Phair J, Sutton SH, Seaberg EC, Margolick JB, Wisniewski SR, Morris A. 2013. The impact of HAART on the respiratory complications of HIV infection: longitudinal trends in the MACS and WIHS cohorts. PLoS One 8(3):e58812. 10.1371/journal.pone.0058812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rosen MJ. 1992. Pulmonary complications of HIV infection. Mt. Sinai J. Med. 59:263–270 [PubMed] [Google Scholar]
- 4.Rosen MJ. 1994. Pneumonia in patients with HIV infection. Med. Clin. North Am. 78:1067–1079 [DOI] [PubMed] [Google Scholar]
- 5.Rosen MJ. 2008. Pulmonary complications of HIV infection. Respirology 13:181–190. 10.1111/j.1440-1843.2007.01167.x [DOI] [PubMed] [Google Scholar]
- 6.Schneider E, Whitmore S, Glynn KM, Dominguez K, Mitsch A, McKenna MT, Centers for Disease Control and Prevention 2008. Revised surveillance case definitions for HIV infection among adults, adolescents, and children aged <18 months and for HIV infection and AIDS among children aged 18 months to <13 years—United States, 2008. MMWR Recommend. Rep. 57(RR-10):1–12 [PubMed] [Google Scholar]
- 7.Hay JW, Osmond DH, Jacobson MA. 1988. Projecting the medical costs of AIDS and ARC in the United States. J. Acquir. Immune Defic. Syndr. 1:466–485 [PubMed] [Google Scholar]
- 8.McKinnell JA, Cannella AP, Kunz DF, Hook EW, III, Moser SA, Miller LG, Baddley JW, Pappas PG. 2012. Pneumocystis pneumonia in hospitalized patients: a detailed examination of symptoms, management, and outcomes in human immunodeficiency virus (HIV)-infected and HIV-uninfected persons. Transpl. Infect. Dis. 14:510–518. 10.1111/j.1339-3062.2012.00739.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Calderon EJ, Gutierrez-Rivero S, Durand-Joly I, Dei-Cas E. 2010. Pneumocystis infection in humans: diagnosis and treatment. Expert Rev. Anti Infect. Ther. 8:683–701. 10.1586/eri.10.42 [DOI] [PubMed] [Google Scholar]
- 10.Carmona EM, Limper AH. 2011. Update on the diagnosis and treatment of Pneumocystis pneumonia. Ther. Adv. Respir. Dis. 5:41–59. 10.1177/1753465810380102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Summah H, Zhu YG, Falagas ME, Vouloumanou EK, Qu JM. 2013. Use of real-time polymerase chain reaction for the diagnosis of Pneumocystis pneumonia in immunocompromised patients: a meta-analysis. Chin. Med. J. 126:1965–1973. 10.3760/cma.j.issn.0366-6999.20122506 [DOI] [PubMed] [Google Scholar]
- 12.Tasaka S, Tokuda H. 2013. Recent advances in the diagnosis of Pneumocystis jirovecii pneumonia in HIV-infected adults. Expert Opin. Med. Diagn. 7:85–97. 10.1517/17530059.2012.722080 [DOI] [PubMed] [Google Scholar]
- 13.Fillaux J, Berry A. 2013. Real-time PCR assay for the diagnosis of Pneumocystis jirovecii pneumonia. Methods Mol. Biol. 943:159–170. 10.1007/978-1-60327-4_11 [DOI] [PubMed] [Google Scholar]
- 14.Fillaux J, Malvy S, Alvarez M, Fabre R, Cassaing S, Marchou B, Linas MD, Berry A. 2008. Accuracy of a routine real-time PCR assay for the diagnosis of Pneumocystis jirovecii pneumonia. J. Microbiol. Methods 75:258–261. 10.1016/j.mimet.2008.06.009 [DOI] [PubMed] [Google Scholar]
- 15.Botterel F, Cabaret O, Foulet F, Cordonnier C, Costa JM, Bretagne S. 2012. Clinical significance of quantifying Pneumocystis jirovecii DNA by using real-time PCR in bronchoalveolar lavage fluid from immunocompromised patients. J. Clin. Microbiol. 50:227–231. 10.1128/JCM.06036-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.van Oosterhout JJ, Laufer MK, Perez MA, Graham SM, Chimbiya N, Thesing PC, Alvarez-Martinez MJ, Wilson PE, Chagomerana M, Zijlstra EE, Taylor TE, Plowe CV, Meshnick SR. 2007. Pneumocystis pneumonia in HIV-positive adults, Malawi. Emerg. Infect. Dis. 13:325–328 http://wwwnc.cdc.gov/eid/article/13/2/06-0462/htm [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dalpke AH, Hofko M, Zimmermann S. 2013. Development and evaluation of a real-time PCR assay for detection of Pneumocystis jirovecii on the fully automated BD MAX platform. J. Clin. Microbiol. 51:2337–2343. 10.1128/JCM.00616-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Orsi CF, Gennari W, Venturelli C, La Regina A, Pecorari M, Righi E, Machetti M, Blasi E. 2012. Performance of 2 commercial real-time polymerase chain reaction assays for the detection of Aspergillus and Pneumocystis DNA in bronchoalveolar lavage fluid samples from critical care patients. Diagn. Microbiol. Infect. Dis. 73:138–143. 10.1016/j.diagmicrobio.2012.03.001 [DOI] [PubMed] [Google Scholar]
- 19.Asai N, Motojima S, Ohkuni Y, Matsunuma R, Nakashima K, Iwasaki T, Nakashita T, Otsuka Y, Kaneko N. 2012. Early diagnosis and treatment are crucial for the survival of Pneumocystis pneumonia patients without human immunodeficiency virus infection. J. Infect. Chemother. 18:898–905. 10.1007/s10156-012-0441-4 [DOI] [PubMed] [Google Scholar]
- 20.Hernandez SL, Lopez De Blanc SA, Sambuelli RH, Roland H, Cornelli C, Lattanzi V, Carnelli MA. 2004. Oral histoplasmosis associated with HIV infection: a comparative study. J. Oral Pathol. Med. 33:445–450. 10.1111/j.1600-0714.2004.00200.x-i1 [DOI] [PubMed] [Google Scholar]
- 21.Huber F, Nacher M, Aznar C, Pierre-Demar M, El Guedj M, Vaz T, Vantilcke V, Mahamat A, Magnien C, Chauvet E, Carme B, Couppie P. 2008. AIDS-related Histoplasma capsulatum var. capsulatum infection: 25 years experience of French Guiana. AIDS 22:1047–1053. 10.1097/QAD.0b013e3282ffde67 [DOI] [PubMed] [Google Scholar]
- 22.Couppie P, Aznar C, Carme B, Nacher M. 2006. American histoplasmosis in developing countries with a special focus on patients with HIV: diagnosis, treatment, and prognosis. Curr. Opin. Infect. Dis. 19:443–449. 10.1097/01.qco.0000244049.15888.b9 [DOI] [PubMed] [Google Scholar]
- 23.Buitrago MJ, Canteros CE, Frias De Leon G, Gonzalez A, Marques-Evangelista De Oliveira M, Munoz CO, Ramirez JA, Toranzo AI, Zancope-Oliveira R, Cuenca-Estrella M. 2013. Comparison of PCR protocols for detecting Histoplasma capsulatum DNA through a multicenter study. Rev. Iberoam. Micol. 30:256–260. 10.1016/j.riam.2013.03.004 [DOI] [PubMed] [Google Scholar]
- 24.Buitrago MJ, Cuenca-Estrella M. 2012. Current epidemiology and laboratory diagnosis of endemic mycoses in Spain. Enferm Infecc Microbiol. Clin. 30:407–413 (In Spanish.) 10.1016/j.eimc.2011.09.014 [DOI] [PubMed] [Google Scholar]
- 25.Deepe GS. 2000. Histoplasma capsulatum, p 2718–2733 In Mandell GL, Bennett JE, Dolin R. (ed), Principles and practice of infectious diseases, 7th ed. Churchill Livingston, Philadelphia, PA [Google Scholar]
- 26.Wheat LJ, Freifeld AG, Kleiman MB, Baddley JW, McKinsey DS, Loyd JE, Kauffman CA, Infectious Diseases Society of America 2007. Clinical practice guidelines for the management of patients with histoplasmosis: 2007 update by the Infectious Diseases Society of America. Clin. Infect. Dis. 45:807–825. 10.1086/521259 [DOI] [PubMed] [Google Scholar]
- 27.Bialek R, Feucht A, Aepinus C, Just-Nubling G, Robertson VJ, Knobloch J, Hohle R. 2002. Evaluation of two nested PCR assays for detection of Histoplasma capsulatum DNA in human tissue. J. Clin. Microbiol. 40:1644–1647. 10.1128/JCM.40.5.1644-1647.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bialek R, Ernst F, Dietz K, Najvar LK, Knobloch J, Graybill JR, Schaumburg-Lever G. 2002. Comparison of staining methods and a nested PCR assay to detect Histoplasma capsulatum in tissue sections. Am. J. Clin. Pathol. 117:597–603 [DOI] [PubMed] [Google Scholar]
- 29.Martagon-Villamil J, Shrestha N, Sholtis M, Isada CM, Hall GS, Bryne T, Lodge BA, Reller LB, Procop GW. 2003. Identification of Histoplasma capsulatum from culture extracts by real-time PCR. J. Clin. Microbiol. 41:1295–1298. 10.1128/JCM.41.3.1295-1298.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Buitrago MJ, Berenguer J, Mellado E, Rodriguez-Tudela JL, Cuenca-Estrella M. 2006. Detection of imported histoplasmosis in serum of HIV-infected patients using a real-time PCR-based assay. Eur. J. Clin. Microbiol. Infect. Dis. 25:665–668. 10.1007/s10096-006-0207-y [DOI] [PubMed] [Google Scholar]
- 31.Porta A, Colonna-Romano S, Callebaut I, Franco A, Marzullo L, Kobayashi GS, Maresca B. 1999. An homologue of the human 100-kDa protein (p100) is differentially expressed by Histoplasma capsulatum during infection of murine macrophages. Biochem. Biophys. Res. Commun. 254:605–613 [DOI] [PubMed] [Google Scholar]
- 32.Frias De Leon MG, Arenas Lopez G, Taylor ML, Acosta Altamirano G, Reyes-Montes Mdel R. 2012. Development of specific sequence-characterized amplified region markers for detecting Histoplasma capsulatum in clinical and environmental samples. J. Clin. Microbiol. 50:673–679. 10.1128/JCM.05271-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Park BJ, Wannemuehler KA, Marston BJ, Govender N, Pappas PG, Chiller TM. 2009. Estimation of the current global burden of cryptococcal meningitis among persons living with HIV/AIDS. AIDS 23:525–530. 10.1097/QAD.0b013e328322ffac [DOI] [PubMed] [Google Scholar]
- 34.Byrnes EJ, Heitman J. 2009. Cryptococcus gattii outbreak expands into the northwestern United States with fatal consequences. F1000 Biol. Rep. 1:pii. 10.3410/B1-62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fyfe M, MacDougall L, Romney M, Starr M, Pearce M, Mak S, Mithani S, Kibsey P. 2008. Cryptococcus gattii infections on Vancouver Island, British Columbia, Canada: emergence of a tropical fungus in a temperate environment. Can. Commun. Dis. Rep. 34:1–12 [PubMed] [Google Scholar]
- 36.Marr KA. 2012. Cryptococcus gattii as an important fungal pathogen of western North America. Expert Rev. Anti Infect. Ther. 10:637–643. 10.1586/eri.12.48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Govender NP, Patel J, van Wyk M, Chiller TM, Lockhart SR, Group for Enteric, Respiratory and Meningeal Disease Surveillance in South Africa (GERMS-SA) 2011. Trends in antifungal drug susceptibility of Cryptococcus neoformans isolates obtained through population-based surveillance in South Africa in 2002–2003 and 2007–2008. Antimicrob. Agents Chemother. 55:2606–2611. 10.1128/AAC.00048-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lortholary O, Sitbon K, Dromer F, French Cryptococcosis Study G 2005. Evidence for human immunodeficiency virus and Cryptococcus neoformans interactions in the pro-inflammatory and anti-inflammatory responses in blood during AIDS-associated cryptococcosis. Clin. Microbiol. Infect. 11:296–300. 10.1111/j.1469-0691.2005.01074.x [DOI] [PubMed] [Google Scholar]
- 39.Liechty CA, Solberg P, Were W, Ekwaru JP, Ransom RL, Weidle PJ, Downing R, Coutinho A, Mermin J. 2007. Asymptomatic serum cryptococcal antigenemia and early mortality during antiretroviral therapy in rural Uganda. Trop. Med. Int. Health 12:929–935. 10.1111/j.1365-3156.2007.01874.x [DOI] [PubMed] [Google Scholar]
- 40.Jarvis JN, Percival A, Bauman S, Pelfrey J, Meintjes G, Williams GN, Longley N, Harrison TS, Kozel TR. 2011. Evaluation of a novel point-of-care cryptococcal antigen test on serum, plasma, and urine from patients with HIV-associated cryptococcal meningitis. Clin. Infect. Dis. 53:1019–1023. 10.1093/cid/cir613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Carvalho VG, Terceti MS, Dias AL, Paula CR, Lyon JP, de Siqueira AM, Franco MC. 2007. Serotype and mating type characterization of Cryptococcus neoformans by multiplex PCR. Rev. Inst. Med. Trop. Sao Paulo 49:207–210. 10.1590/S0036-46652007000400002 [DOI] [PubMed] [Google Scholar]
- 42.Diaz MR, Fell JW. 2005. Use of a suspension array for rapid identification of the varieties and genotypes of the Cryptococcus neoformans species complex. J. Clin. Microbiol. 43:3662–3672. 10.1128/JCM.43.8.3662-3672.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Veron V, Simon S, Blanchet D, Aznar C. 2009. Real-time polymerase chain reaction detection of Cryptococcus neoformans and Cryptococcus gattii in human samples. Diagn. Microbiol. Infect. Dis. 65:69–72. 10.1016/j.diagmicrobio.2009.05.005 [DOI] [PubMed] [Google Scholar]
- 44.Hansen J, Slechta ES, Gates-Hollingsworth MA, Neary B, Barker AP, Bauman S, Kozel TR, Hanson KE. 2013. Large-scale evaluation of the immuno-mycologics lateral flow and enzyme-linked immunoassays for detection of cryptococcal antigen in serum and cerebrospinal fluid. Clin. Vaccine Immunol. 20:52–55. 10.1128/CVI.00536-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Reynolds JH, Banerjee AK. 2012. Imaging pneumonia in immunocompetent and immunocompromised individuals. Curr. Opin. Pulm. Med. 18:194–201. 10.1097/MCP.0b013e328351f953 [DOI] [PubMed] [Google Scholar]
- 46.Aronis ML, dos Santos RP, Goldani LZ. 2011. Disseminated Histoplasma capsulatum and Cryptococcus neoformans co-infection in patients with AIDS. Mycopathologia 172:233–236. 10.1007/s11046-011-9422-x [DOI] [PubMed] [Google Scholar]
- 47.Javier B, Susana L, Santiago G, Alcides T. 2012. Pulmonary coinfection by Pneumocystis jirovecii and Cryptococcus neoformans. Asian Pac. J. Trop. Biomed. 2:80–82. 10.1016/S2221-1691(11)60195-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tschudy J, Michail S. 2010. Disseminated histoplasmosis and pneumocystis pneumonia in a child with Crohn disease receiving infliximab. J. Pediatr. Gastroenterol. Nutr. 51:221–222. 10.1097/MPG.0b013e3181c2c10d [DOI] [PubMed] [Google Scholar]
- 49.Bernal-Martinez L, Buitrago MJ, Castelli MV, Rodriguez-Tudela JL, Cuenca-Estrella M. 2013. Development of a single tube multiplex real-time PCR to detect the most clinically relevant Mucormycetes species. Clin. Microbiol. Infect. 19:E1–E7. 10.1111/j.1469-0691.2012.03976.x [DOI] [PubMed] [Google Scholar]
- 50.Tang CM, Cohen J, Krausz T, Van Noorden S, Holden DW. 1993. The alkaline protease of Aspergillus fumigatus is not a virulence determinant in two murine models of invasive pulmonary aspergillosis. Infect. Immun. 61:1650–1656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Buitrago MJ, Aguado JM, Ballen A, Bernal-Martinez L, Prieto M, Garcia-Reyne A, Garcia-Rodriguez J, Rodriguez-Tudela JL, Cuenca-Estrella M. 2013. Efficacy of DNA amplification in tissue biopsy samples to improve the detection of invasive fungal disease. Clin. Microbiol. Infect. 19:E271–277. 10.1111/1469-0691.12110 [DOI] [PubMed] [Google Scholar]
- 52.White TJ, Bruns S, Lee S, Taylor J. 2011. Amplification and direct sequencing of fungal ribosomal genes for phylogenetics, p 315–324 In Innis MA, Gelfand DH, Sninsky JJ, White TJ. (ed), PCR protocols: a guide to methods and applications. Academic Press, San Diego, CA [Google Scholar]
- 53.Kyeyune R, den Boon S, Cattamanchi A, Davis JL, Worodria W, Yoo SD, Huang L. 2010. Causes of early mortality in HIV-infected TB suspects in an East African referral hospital. J. Acquir. Immune Defic. Syndr. 55:446–450. 10.1097/qai.0b013e3181eb611a [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cuenca-Estrella M, Bassetti M, Lass-Florl C, Racil Z, Richardson M, Rogers TR. 2011. Detection and investigation of invasive mould disease. J. Antimicrob. Chemother. 66(Suppl. 1):i15–i24. 10.1093/jac/dpkq438 [DOI] [PubMed] [Google Scholar]
- 55.Boondireke S, Mungthin M, Tan-ariya P, Boonyongsunchai P, Naaglor T, Wattanathum A, Treewatchareekorn S, Leelayoova S. 2010. Evaluation of sensitivity of multiplex PCR for detection of Mycobacterium tuberculosis and Pneumocystis jirovecii in clinical samples. J. Clin. Microbiol. 48:3165–3168. 10.1128/JCM.00323-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lau A, Halliday C, Chen SC, Playford EG, Stanley K, Sorrell TC. 2010. Comparison of whole blood, serum, and plasma for early detection of candidemia by multiplex-tandem PCR. J. Clin. Microbiol. 48:811–816. 10.1128/JCM.01650-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Agudelo CA, Restrepo CA, Molina DA, Tobon AM, Kauffman CA, Murillo C, Restrepo A. 2012. Tuberculosis and histoplasmosis co-infection in AIDS patients. Am. J. Trop. Med. Hyg. 87:1094–1098. 10.4269/ajtmh.2012.12-0292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Qishui O, Ling J, Ni L, Bin Y, Wen L. 2012. Comparison of real-time florescence quantitative PCR measurements of VAD1 mRNA with three conventional methods in diagnosis and follow-up treatment of Cryptococcus neoformans infection. Mycoses 55:326–332. 10.1111/j.1439-0507.2011.02100.x [DOI] [PubMed] [Google Scholar]
- 59.Antinori S. 2013. New insights into HIV/AIDS-associated cryptococcosis. ISRN AIDS 2013:471363. 10.1155/2013/471363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bratton EW, El Husseini N, Chastain CA, Lee MS, Poole C, Sturmer T, Weber DJ, Juliano JJ, Perfect JR. 2013. Approaches to antifungal therapies and their effectiveness among patients with cryptococcosis. Antimicrob. Agents Chemother. 57:2485–2495. 10.1128/AAC.01800-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Perfect JR, Dismukes WE, Dromer F, Goldman DL, Graybill JR, Hamill RJ, Harrison TS, Larsen RA, Lortholary O, Nguyen MH, Pappas PG, Powderly WG, Singh N, Sobel JD, Sorrell TC. 2010. Clinical practice guidelines for the management of cryptococcal disease: 2010 update by the Infectious Diseases Society of America. Clin. Infect. Dis. 50:291–322. 10.1086/649858 [DOI] [PMC free article] [PubMed] [Google Scholar]