Abstract
In the United States, veterinary use of mupirocin is primarily limited to the treatment of canine pyoderma caused by methicillin-resistant Staphylococcus pseudintermedius (MRSP). In this study, only 1 of 581 S. pseudintermedius isolates tested was resistant to mupirocin and carried the high-level mupirocin resistance gene, ileS2, on a plasmid.
TEXT
Staphylococcus pseudintermedius is the primary bacterial pathogen isolated from canine pyoderma and also causes postsurgical infections in dogs (1, 2). Methicillin resistance and multidrug resistance are increasing in S. pseudintermedius, thus limiting the options for therapeutic treatment of canine skin infections (2). Mupirocin is a bacteriostatic antibiotic that reversibly binds to isoleucyl-tRNA synthetase to disrupt protein synthesis and is widely used to eliminate nasal carriage of methicillin-resistant Staphylococcus aureus (MRSA) in human MRSA carriers (3). Mupirocin has been used on only a limited basis in veterinary medicine but is approved in the United States for the treatment of bacterial skin infections and superficial pyoderma in dogs (4).
In S. aureus, two levels of mupirocin resistance have been identified. Low-level mupirocin resistance occurs due to a point mutation to the chromosomal ileS gene that encodes the native isoleucyl-tRNA synthetase. The MIC for mupirocin for staphylococci carrying the low-level resistance is ≥8 μg/ml but ≤256 μg/ml (5). Conversely, high-level mupirocin resistance (MIC of ≥512 μg/ml) is usually conferred by the plasmid-borne ileS2, although a chromosomal location of ileS2 has been reported (5). Recently, ileS2 plasmid-mediated mupirocin resistance was found in a mupirocin-resistant, methicillin-susceptible S. pseudintermedius strain isolated from a dog in Croatia (6). The goal of the present study was to determine the prevalence of mupirocin resistance in S. pseudintermedius isolated from patients presented to a veterinary hospital in Texas.
In this study, 581 isolates of S. pseudintermedius were screened for phenotypic low-level mupirocin resistance. Isolates were collected from veterinary patients, predominantly dogs (n = 446), but also included isolates from cats (n = 9). Some patients were cultured at multiple sites and contributed more than one isolate, and of these, 21 patients contributed more than two isolates. The isolates included a historical collection of 403 isolates from clinical infections and contained both methicillin-resistant S. pseudintermedius (MRSP) isolates (n = 153) and methicillin-susceptible S. pseudintermedius (MSSP) isolates (n = 250). The isolates from clinical infections were collected from the following anatomic sites: skin (n = 96), external ear canal (n = 31), wounds (n = 79), postoperative infections (n = 33), urine or the urinary tract (n = 87), and other sources (n = 77). Additional isolates were collected during a study of MRSP prevalence in canine patients without clinical staphylococcal infection that presented for elective orthopedic procedures. The MRSP prevalence study yielded 178 S. pseudintermedius isolates (13 MRSP and 165 MSSP isolates) collected from the nares or perineum of 129 dogs.
All isolates were presumptively identified as S. pseudintermedius at the time of collection based on Gram stain reaction, colony color, and biochemical tests. Tests measured the ability of the isolates to produce hemolysis on Trypticase soy agar supplemented with 5% sheep blood agar (blood agar plates) (BD Diagnostic Systems, Sparks, MD), to produce coagulase, to produce catalase, and to grow on salt-mannitol agar. Isolates were also tested for resistance to polymyxin B, ability to utilize trehalose, and production of urease. At the time of initial collection, isolates were tested for antimicrobial susceptibility using commercially available systems (GPS card, Vitek; bioMérieux, Durham, NC; COMPAN1F and COMPAN2F panels, Trek Sensititre; Trek Diagnostics, Cleveland, OH) and additionally tested for methicillin resistance by oxacillin disk diffusion testing and PCR for the presence of mecA. The mecA PCR used a previously published protocol with methicillin-resistant S. aureus (ATCC 33591) and methicillin-susceptible S. aureus (ATCC 29213 or ATCC 25923) as positive and negative controls, respectively (7). Isolates were stored frozen in 10% glycerol at −80°C in 96-well deep-well plates and later inoculated aseptically using a 96-pin replicator onto Mueller-Hinton agar (BD Diagnostic Systems) and onto Mueller-Hinton agar supplemented with 8 μg/ml mupirocin (Sigma-Aldrich, St. Louis, MO) (here referred to as the mupirocin plate) to screen for low-level resistance to mupirocin. Pseudomonas aeruginosa (ATCC 27853) was used as a positive control for mupirocin resistance (8). The bacterial concentrations were not standardized prior to screening. Plates were incubated for 24 h at 37°C and then examined.
Colonies were screened by PCR for the presence of the ileS2 gene using the previously published primers mupA and mupB to amplify a 458-bp fragment of the ileS2 gene (9) and primers M1 and M2 to amplify a 237-bp fragment of the gene (Table 1) (10). A total reaction volume of 50 μl was used with the final concentrations of reagents as follows: 2.5 mM MgCl2, 0.2 mM deoxynucleoside triphosphates (dNTPs), 2.5 pmol of each primer, and 2.5 U Taq polymerase per reaction (Lucigen, Middleton, WI). Three to five colonies isolated from the mupirocin plate were used for colony PCR. Reactions were run in a 2720 thermal cycler (Applied Biosystems, Grand Island, NY) using the following settings: 95°C for 5 min; 35 cycles of 95°C for 30 s, 57°C for 30 s, and 72°C for 30 s, followed by 72°C for 7 min; and then a hold at 4°C. Negative controls included water with no-template DNA and DNA from a known mupirocin-sensitive, methicillin-sensitive S. aureus strain (ATCC 29213). No positive control was available. The products were then run on a 2% agarose gel for 2 h at 70 V, visualized with GelRed (Phenix Research, Candler, NC), and compared to a 100-bp molecular size marker (Invitrogen, Grand Island, NY).
TABLE 1.
Primers used in this study
| Target gene | Primer | Sequence (5′ to 3′) | Reference |
|---|---|---|---|
| ileS2 | mupA | TATATTATGCGATGGAAGGTTGG | 9 |
| mupB | AATAAAATCAGCTGGAAAGTGTTG | 9 | |
| M1 | GTTTATCTTCTGATGCTGAG | 10 | |
| M2 | CCCCAGTTACACCGATATAA | 10 | |
| IS257-ileS2 junctions | IS257F | GGCATGGCGAAAATCCGTAG | 11 |
| IS257R | TGGCGTATTGATGAGACGTACATC | 11 | |
| ileS2-3′ | TCGGTGTAACTGGGGAATTA | 11 | |
| ileS2-5′ | CCATGTCAACCCAGTATCC | 11 | |
| ileS | ileS-F1 | CGTGACCGTGGCGAATGGGT | 6 |
| ileS-R1 | GTATGCGGAATGATTGGCG | 6 | |
| mecA | mecA F | CTCAGGTACTGCTATCCACC | 7 |
| mecA R | CACTTGGTATATCTTCACC | 7 |
Plasmid purification was performed using the QIAprep Spin Miniprep plasmid purification kit according to the manufacturer's instructions (Qiagen, Valencia, CA). Samples were evaluated for concentration and quality using a NanoDrop spectrometer (Thermo Scientific, Waltham, MA) prior to PCR testing. PCR was used to evaluate the IS257-ileS2 spacer regions using a previously published molecular classification system (11). The primers IS257F, ileS2-5′, ileS2-3′, and IS257R (Table 1) were used in various combinations as previously described under the following conditions: 94°C for 5 min; 30 cycles of 94°C for 30 s, 60°C for 40 s, and 72°C for 60 s; then 72°C for 10 min; and then a hold at 4°C (11). PCR was performed to identify the native ileS gene using the primers ileS-F1 and ileS-R1 (Table 1) (6). Conditions for the thermal cycler were as follows: 94°C for 5 min; 30 cycles of 94°C for 30 s, 55°C for 30 s, 72°C for 60 s; then 72°C for 7 min; and a hold at 4°C.
PCR products were purified using either the QIAprep gel purification kit (Qiagen, Valencia, CA) or the Zymoclean gel DNA recovery kit (Zymo Research, Irvine, CA) according to the manufacturers' protocols. Purified PCR products were then cloned into pT7Blue plasmid vector using the Novagen pT7Blue Perfectly Blunt cloning kit (EMD Chemicals, Inc., Darmstadt, Germany) according to the manufacturer's protocol. Resultant plasmids containing the upstream IS257-ileS2 junction, the downstream ileS2-IS257 junction, and the 945-bp fragment of the native ileS gene were submitted to the Texas A&M Gene Technologies Laboratory for sequencing. Resultant sequences were compared to sequences in GenBank (JX186508, JX186509, JX186511, JX186512, JX186513, and JX186514) using MEGA5.1 software (6, 12).
Of the 581 isolates tested, only one isolate was resistant to mupirocin. The isolate, 39-045, was originally cultured from the nares of a healthy, 1-year-old, castrated, male, Bernese mountain dog presenting for an orthopedic evaluation. This isolate was pan-susceptible to all antimicrobials tested using the COMPAN2F drug panel and negative for the presence of the mecA gene via PCR analysis. The prevalence of mupirocin resistance in dogs without clinical staphylococcal infections that presented for elective orthopedic procedures was 1 in 129, or 0.8%. An additional 194 S. pseudintermedius isolates were collected from 158 dogs with clinical infections during the same period of collection (22 September 2010 to 8 February 2012), resulting in a total of 372 S. pseudintermedius isolates from 287 dogs. The prevalence of mupirocin-resistant S. pseudintermedius in dogs cultured between 22 September 2010 and 8 February 2012 was therefore 1 in 287 dogs, or 0.3%.
The mupirocin-resistant isolate was analyzed for the presence of high-level mupirocin resistance by plasmid DNA isolation followed by PCR amplification of two different regions of the plasmid-borne ileS2 gene. The presence of a 458-bp band with mupA and mupB primers and a 237-bp band with M1 and M2 primers indicates that the isolate contains the ileS2 gene (Fig. 1).
FIG 1.
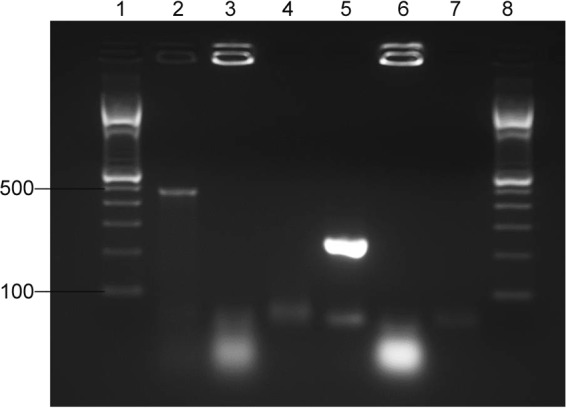
Detection of ileS2 using PCR. Lanes 1 to 3 include PCR products amplified with mupA and mupB primers (9). Lanes 4 to 6 include PCR products amplified with M1 and M2 primers (10). The molecular size marker used in lanes 1 and 8 was a 100-bp DNA ladder (Invitrogen, Grand Island, NY). Numbers at left are molecular sizes in bp. Template DNA used for PCR was plasmid DNA from isolate 39-045 (lanes 2 and 5) or genomic DNA from ATCC 29213 (lanes 3 and 6). Water was substituted for DNA in lanes 4 and 7.
To further determine the structural type of the plasmid, PCR for the IS257-ileS2 spacer regions was performed according to a previously published molecular classification system (11). The fragments are similar to the amplification for structural group S2 ileS2 plasmids found in S. aureus pattern II, with bands sized between 1,000 bp and 1,650 bp for primers ileS2-3′ and IS257F and between 2,000 and 3,054 bp for primers IS257R and ileS2-5′ (Fig. 2). This structural group is similar to the structure previously reported for the plasmid-borne ileS2 gene identified in S. pseudintermedius isolated from a dog with pyoderma in Croatia (6). The resultant PCR products were sequenced and compared to the previously published ileS2 sequences from S. pseudintermedius, JX186508 and JX186509 (6). Sequences from this study were deposited in GenBank as KJ000545, KJ000546, and KJ000547. Comparison of JX186509 with KJ000545 using MEGA5.1 software indicated 99% similarity between the two sequences. Comparison of JX186508 with KJ000546 and KJ000547 indicated 100% and 99% similarity between the sequences, respectively.
FIG 2.
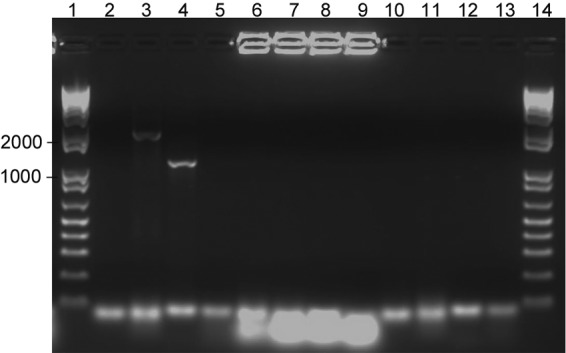
Detection of the ileS2-IS257 junctions in isolate 39-045 using ileS2-5′, IS257F, ileS2-3′, and IS257R primers (11). Primer pairs for each reaction were as follows: PCR 1, ileS2-5′ and IS257F; PCR 2, ileS2-5′ and IS257R; PCR 3, ileS2-3′ and IS257F; and PCR 4, ileS2-3′ and IS257R. Lanes 2, 6, and 10 are products from PCR 1; lanes 3, 7, and 11 are products from PCR 2; lanes 4, 8, and 12 are products from PCR 3; lanes 5, 9, and 13 are products from PCR 4; and lanes 1 and 14 are a 1-kb DNA ladder (Invitrogen, Grand Island, NY). Template DNA in lanes 2 to 5 is plasmid DNA from isolate 39-045. Template DNA in lanes 6 to 9 is genomic DNA from ATCC 29213 used as a negative control. In lanes 10 to 13, water was substituted for template DNA as a negative control. No positive controls were available. Numbers at left are sizes in bases.
To determine whether isolate 39-045 had both an ileS mutation and the ileS2 plasmid simultaneously, PCR amplification of the chromosomal ileS gene was also performed using previously published primers (6). The resultant 945-bp product was sequenced and analyzed using MEGA5.1 software, and the sequence was deposited in GenBank as KJ000544. Analysis indicated a 99% similarity between isolate 39-045 and the previously published sequences of the S. pseudintermedius chromosomal ileS gene: JX186511, JX186512, JX186513, and JX186514 (6).
In summary, this study found that the prevalence of mupirocin resistance in S. pseudintermedius isolated from dogs was 0.3% (1/287) or 0.8% (1/129) in healthy dogs without active, clinical staphylococcal infections. While no mupirocin-resistant isolates were found in our collection of isolates from dogs with clinical disease, the presence of plasmid-mediated mupirocin resistance is of concern as previous work has demonstrated that mupirocin resistance can be transmitted from one species of Staphylococcus to another in vivo (13). Increased rates of methicillin resistance and multidrug resistance in S. pseudintermedius and approval of mupirocin for use in dogs have made mupirocin an attractive alternative for topical use in canine pyoderma (2). This could result in increased mupirocin resistance in S. pseudintermedius over time. Although our study found only one mupirocin-resistant S. pseudintermedius isolate, 36.5% of U.S. households own a dog (14), and there is the potential for transmission of mupirocin resistance from canine isolates of S. pseudintermedius to human isolates of S. aureus or vice versa. This could have implications for public health. For these reasons, mupirocin resistance should be monitored and carefully considered before mupirocin is used in canine patients.
Nucleotide sequence accession numbers.
Sequences from this study were deposited in GenBank as KJ000544, KJ000545, KJ000546, and KJ000547.
ACKNOWLEDGMENTS
Funds provided by the Texas A&M University College of Veterinary Medicine and Biomedical Sciences Postdoctoral Research Grant to R. M. Gold helped support this study.
We thank Courtney Brake, Elena Gart, and Jacob Villegas for technical assistance in the lab.
Footnotes
Published ahead of print 29 January 2014
REFERENCES
- 1.Fulham KS, Lemarie SL, Hosgood G, Dick HL. 2011. In vitro susceptibility testing of meticillin-resistant and meticillin-susceptible staphylococci to mupirocin and novobiocin. Vet. Dermatol. 22:88–94. 10.1111/j.1365-3164.2010.00921.x [DOI] [PubMed] [Google Scholar]
- 2.van Duijkeren E, Catry B, Greko C, Moreno MA, Pomba MC, Pyorala S, Ruzauskas M, Sanders P, Threlfall EJ, Torren-Edo J, Torneke K. 2011. Review on methicillin-resistant Staphylococcus pseudintermedius. J. Antimicrob. Chemother. 66:2705–2714. 10.1093/jac/dkr367 [DOI] [PubMed] [Google Scholar]
- 3.Dacre JE, Emmerson AM, Jenner EA. 1983. Nasal carriage of gentamicin and methicillin resistant Staphylococcus aureus treated with topical pseudomonic acid. Lancet ii:1036. [DOI] [PubMed] [Google Scholar]
- 4.Dechra Veterinary Products. 2014. Muricin ointment 2%, product insert. Dechra Veterinary Products, Overland Park, KS [Google Scholar]
- 5.Ramsey MA, Bradley SF, Kauffman CA, Morton TM. 1996. Identification of chromosomal location of mupA gene, encoding low-level mupirocin resistance in staphylococcal isolates. Antimicrob. Agents Chemother. 40:2820–2823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Matanovic K, Perez-Roth E, Pintaric S, Seol Martinec B. 2013. Molecular characterization of high-level mupirocin resistance in Staphylococcus pseudintermedius. J. Clin. Microbiol. 51:1005–1007. 10.1128/JCM.02904-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sakoulas G, Gold HS, Venkataraman L, DeGirolami PC, Eliopoulos GM, Qian Q. 2001. Methicillin-resistant Staphylococcus aureus: comparison of susceptibility testing methods and analysis of mecA-positive susceptible strains. J. Clin. Microbiol. 39:3946–3951. 10.1128/JCM.39.11.3946-3951.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jones RN, Li Q, Kohut B, Biedenbach DJ, Bell J, Turnidge JD. 2006. Contemporary antimicrobial activity of triple antibiotic ointment: a multiphased study of recent clinical isolates in the United States and Australia. Diagn. Microbiol. Infect. Dis. 54:63–71. 10.1016/j.diagmicrobio.2005.08.009 [DOI] [PubMed] [Google Scholar]
- 9.Anthony RM, Connor AM, Power EG, French GL. 1999. Use of the polymerase chain reaction for rapid detection of high-level mupirocin resistance in staphylococci. Eur. J. Clin. Microbiol. Infect. Dis. 18:30–34. 10.1007/s100960050222 [DOI] [PubMed] [Google Scholar]
- 10.Nunes EL, dos Santos KR, Mondino PJ, Bastos MDC, Giambiagi-deMarval M. 1999. Detection of ileS-2 gene encoding mupirocin resistance in methicillin-resistant Staphylococcus aureus by multiplex PCR. Diagn. Microbiol. Infect. Dis. 34:77–81. 10.1016/S0732-8893(99)00021-8 [DOI] [PubMed] [Google Scholar]
- 11.Perez-Roth E, Armas-Gonzalez E, Alcoba-Florez J, Mendez-Alvarez S. 2011. PCR-based amplification of heterogeneous IS257-ileS2 junctions for molecular monitoring of high-level mupirocin resistance in staphylococci. J. Antimicrob. Chemother. 66:471–475. 10.1093/jac/dkq493 [DOI] [PubMed] [Google Scholar]
- 12.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28:2731–2739. 10.1093/molbev/msr121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hurdle JG, O'Neill AJ, Mody L, Chopra I, Bradley SF. 2005. In vivo transfer of high-level mupirocin resistance from Staphylococcus epidermidis to methicillin-resistant Staphylococcus aureus associated with failure of mupirocin prophylaxis. J. Antimicrob. Chemother. 56:1166–1168. 10.1093/jac/dki387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.American Veterinary Medical Association. 2012. U.S. pet ownership and demographics sourcebook (2012). American Veterinary Medical Association, Schaumburg, IL [Google Scholar]


