Abstract
Hepatitis E virus (HEV) is a major cause of acute viral hepatitis in people in many developing countries and is also endemic in many industrialized countries. Mammalian HEV (mHEV) isolates can be divided into at least four recognized major genotypes. Several nucleic acid amplification techniques have been developed for mHEV detection, with great differences in sensitivity. The aim of this study was to compare the performances of two singleplex real-time reverse transcriptase (RT) PCR assays for broad detection of all four mHEV genotypes (assays A and B) and two duplex real-time RT-PCR assays for detection and differentiation of mHEV genotypes 3 and 4 (assays C and D). RNAs extracted from 28 fecal samples from pigs experimentally inoculated with HEV genotype 3 and 186 fecal samples from commercial pigs with unknown HEV exposure were tested by all four assays. In experimental samples, HEV RNA was detected in 96.4% (assay A), 39.2% (assay B), 14.2% (assay C), and 0% (assay D) of the samples. In field samples with unknown HEV exposure, HEV RNA was detected in 67.2% (assay A), 36.4% (assay B), 1.1% (assay C), and 0.5% (assay D) of the samples. The assays showed overall poor agreement (κ = 0.19 to 0.03), with differences in detection rates between assays (P < 0.01). Assays A and B, which broadly detect HEV genotypes 1 to 4, had significantly higher detection rates for HEV RNA than the duplex assays C and D, which were both designed to detect and differentiate between HEV genotypes 3 and 4.
INTRODUCTION
Hepatitis E virus (HEV) is the causative agent of hepatitis E in humans (1). HEV infection in pregnant women may cause particularly severe illness, with a mortality rate of 10 to 20%, and recently, there have been numerous reports of persistent and chronic HEV infection in immunocompromised patients, such as organ transplant recipients (2). Currently, HEV is classified in the genus Hepevirus in the family Hepeviridae (3). The virus is a nonenveloped, positive-sense, single-stranded RNA virus that carries three open reading frames (ORFs). ORF1 encodes nonstructural proteins, ORF2 encodes the viral capsid, and ORF3, which overlaps with ORF2, encodes a multifunctional small protein (3).
HEV has been identified in several animal species, including domestic pigs, chickens, deer, wild boars, mongooses, rabbits, rats, ferrets, bats, and fish (4), and based on the host tropism, the strains genetically identified thus far can be clustered into mammalian HEV (mHEV), avian HEV (aHEV), and piscine HEV (pHEV) strains. Within mHEV, there are at least four recognized genotypes capable of infecting humans. Genotypes 1 and 2 are associated with epidemics and restricted to humans in developing countries, whereas genotypes 3 and 4 can infect a wide variety of species, including humans and pigs, and are associated with sporadic and cluster cases of human hepatitis E in both developing and industrialized countries (2). While mHEV genotype 3 has worldwide distribution (5), genotype 4 was reported in Asia (5) and more recently in Europe (6, 7). In humans, infections with genotypes 1 and 2 are mainly transmitted via consumption of water contaminated with feces, while infections with genotypes 3 and 4 appear to occur primarily by food-borne zoonotic transmission through the consumption of raw or undercooked meat from pigs, wild boars, or deer (4).
Due to its implication in public health and pork safety, several nucleic acid amplification techniques and immunoassays have been developed for mHEV detection; however, a reliable diagnostic procedure for mHEV is still needed (1, 8). Serological studies comparing immunoassays widely used for mHEV diagnosis found 2.9- to 6.5-fold variation in anti-HEV antibody detection rates (9–11), and only two of six commonly available IgM anti-HEV detection assays had sensitivities and specificities above 95% (11). Due to this overall low sensitivity, a combination of antibody detection and nucleic acid detection has been suggested for optimizing mHEV diagnosis (12, 13).
Considering the heterogeneity of mHEV strains circulating in humans and other animal species, several conventional reverse transcriptase (RT) PCR and real-time RT-PCR assays have been developed for the detection of HEV RNA in various types of samples, including sera, feces, and environmental samples (14–19). Comparisons of RT-PCR assays have shown a 10- to 1,000-fold variation in sensitivity when samples were tested in parallel in the same laboratory (20, 21). In a blinded study to investigate the performance of conventional and real-time RT-PCR assays in 20 laboratories that performed HEV RNA detection on a regular basis, variations in sensitivity on the order of 100- to 1,000-fold were found using a standard panel of HEV genotype 3 and 4 strains (12). Currently, a real-time RT-PCR designed in 2006 (17) is the most widely used assay for detection of HEV infection in humans (12, 22), primarily based on the reported high sensitivity (limit of detection, 4 genome equivalents [GE] of the HEV genome) and its ability to detect all four recognized mHEV genotypes that are capable of infecting humans (23).
Although real-time RT-PCR assays targeting conserved regions can provide accurate detection of the HEV genomes and yield results more rapidly than conventional RT-PCR, commonly, a second molecular method, such as sequencing or subtyping, is required to further characterize strains. Recently, a real-time duplex RT-PCR assay for detection and identification of HEV genotypes 3 and 4 in amounts as small as 50 GE copies per reaction has been reported (24). This assay, targeting the ORF2-ORF3 overlap region, was designed to allow sensitive and rapid detection of the zoonotic HEV genotypes to potentially facilitate epidemiological investigations and to better understand outbreak situations. The aim of this study was to compare the performances of two singleplex real-time RT-PCR assays for broad detection of all 4 recognized mHEV genotypes (assays A and B) and two duplex real-time RT-PCR assays for detection and differentiation of mHEV genotypes 3 and 4 (assays C and D). One singleplex and one duplex real-time RT-PCR assay had been previously described (17, 24), while the other assays were developed in-house.
MATERIALS AND METHODS
Experimental samples.
The experimental protocol was approved by the Virginia Polytechnic and State University Institutional Animal Care and Use Committee and by the Virginia Polytechnic and State University Institutional Biosafety Committee. Twenty-eight serial fecal samples were collected daily from two pigs experimentally inoculated with human HEV genotype 3 strain US-2 (GenBank accession number AF060669) or swine HEV genotype 3 strain Meng (GenBank accession number AF082843) from 2 to 14 days postinoculation (p.i.). The fecal samples were suspended in saline (10% [wt/vol]), and the fecal suspensions were stored at −80°C until use.
Field samples.
A total of 186 pig fecal samples were chosen arbitrarily from routine diagnostic cases submitted during May 2013 to the Iowa State University Veterinary Diagnostic Laboratory (ISU-VDL). These samples originated on 86 farms located in 12 U.S. states: Iowa, Illinois, Indiana, Minnesota, Missouri, North Carolina, North Dakota, Nebraska, Ohio, South Dakota, Texas, and Wisconsin. The fecal samples were obtained from different age groups of pigs: suckling (1 to 2 weeks of age), nursery (3 to 7 weeks of age), and grow-finish (8 to 25 weeks of age) pigs.
Sample processing and RNA extraction.
Fecal samples of ∼1 g were resuspended in phosphate-buffered saline (PBS), vigorously vortexed, and centrifuged at 1,500 × g for 10 min to obtain a final 10% (wt/vol) fecal suspension in PBS. Viral RNA extraction was carried out on 50 μl of the fecal suspension using a MagMax 96 Viral Isolation kit (Ambion, Foster City, CA, USA) according to the manufacturer's instructions on an automated extraction platform (KingFisher Flex; Thermo Fisher Scientific). Negative controls, using DNA- and RNA-free sterile water as a sample, and positive controls, using fecal suspensions from pigs experimentally infected with either mHEV genotype 3 or 4, were added to each extraction plate. The extracted RNA was stored at −80°C until use.
Primers and probes.
All the primers and probes used in this study are listed in Table 1. The primers and probes from assays B and D developed in this study were designed manually based on a multiple-sequence alignment of mHEV genotypes 1 to 4 in GenBank. The sequences were aligned using CLUSTAL W within DNASTAR (Lasergene 8). Specifically, a pair of primers (HEV5606F/HEV5427DR) and a probe (HEVGenP) located in the conserved ORF2-ORF3 overlap region and broadly reactive with mHEV genotypes 1 to 4 were selected (assay B). Additionally, probes specific for the detection of mHEV genotype 3 or 4 (HEVg3 and HEVg4; assay D) were selected in the same region. Oligonucleotide primers/probes were analyzed for the absence of possible hairpins and dimers by Primer Express software (version 3.0; Applied Biosystems).
TABLE 1.
Primers and probes used in this study
| Assay | Amplified region | Primer and probe | Sequence (5′–3′) | Annealing temp (°C) | Locationa | Reference |
|---|---|---|---|---|---|---|
| A | ORF2/3 | JVHEVF | GGTGGTTTCTGGGGTGAC | 60 | 5311–5328 | 17 |
| JVHEVR | AGGGGTTGGTTGGATGAA | 5363–5380 | ||||
| JVHEVP | FAM-TGATTCTCAGCCCTTCGC-BHQ | 5334–5351 | ||||
| B | ORF2/3 | HEV5306F | GTTGATTCTCAGCCCTTCGC | 60 | 5332–5325 | This study |
| HEV5427DR | TGGGMYTGRTCDCGCCAAG | 5427–5445 | ||||
| HEVGenP | FAM-CCCCTATATTCATCCAACCAACCCCTT-BHQ | 5329–5355 | ||||
| C | ORF2/3 | HEV-uni-F | TATTCATCCAACCAACCCCTT | 60 | 5335–5355 | 24 |
| HEV-uni-R | GTCDCGCCAAGYGGAGC | 5453–5471 | ||||
| HEV-3-CY5 | QUASAR 670-GCCGATGTCGTTTCACAA-BHQ | 5386–5403 | ||||
| HEV-4-FAM | FAM-CGCATCTGACATWCCARCCGC-BHQ | 5371–5391 | ||||
| D | ORF2/3 | HEV5306F | GTTGATTCTCAGCCCTTCGC | 60 | 5332–5351 | This study |
| HEV5427DR | TGGGMYTGRTCDCGCCAAG | 5453–5471 | ||||
| HEVg3 | QUASAR 670-TYGTWYCACAAYCCGGGGCTGG-BHQ | 5393–5414 | ||||
| HEVg4 | FAM-CATCYGACATWYCARCCGCMGCCG-BHQ | 5373–5396 | ||||
| Conventional nested RT-PCR | ORF2 | 3156N | AATTATGCYCAGTAYCGRGTTG | 55 | 5737–5732 | 28 |
| 3157N | CCCTTRTCYTGCTGMGCATTCTC | 6445–6467 | ||||
| 3158N | GTWATGCTYTGCATWCATGGCT | 6022–6043 | ||||
| 3159N | AGCCGACGAAATCAATTCTGTC | 6348–6369 |
Construction of plasmid DNA standards for the real-time RT-PCRs.
Plasmid DNA standards were constructed by amplifying a genomic region at nucleotide positions 5311 to 5471 of a genotype 3 human HEV strain (US-2) (25) and a region at nucleotide positions 5285 to 5445 of a genotype 4 human HEV strain (TW6196E) (26) using primers JVHEVF and HEV5427DR (described in Table 1). Conventional RT-PCRs were carried out in a total volume of 20 μl using the Qiagen OneStep RT-PCR kit (Qiagen, Valencia, CA, USA) according to the manufacturer's recommendations. Purified PCR products were cloned into the pGEM-T Vector (Promega, Madison, WI, USA) and transfected into Escherichia coli TOP10 bacteria (Invitrogen, Foster City, CA, USA) following the instructions in the cloning kit manual. Sequencing was performed on recombinant plasmids in both directions using the AB 3730xl DNA Analyzer (Applied Biosystems, Foster City, CA, USA) at the Iowa State University DNA Facility (Ames, IA, USA). The recombinant plasmid stocks were quantified using the NanoDrop ND-1000 spectrophotometer according to the manufacturer's instructions (NanoDrop Technologies Inc., Wilmington, DE, USA) and converted into genome copy numbers. The total numbers of genome copies in the plasmid stock were calculated as follows: copy number = [(concentration of linearized plasmid)/(molar mass)] × (6.023 × 1023). The plasmid DNA was used to generate standard curves using 101 to 108 GE copies of plasmid. The GE titers of HEV were determined based on the standard curve.
Real-time RT-PCR assays.
The real-time RT-PCRs were carried out in 96-well plates using the TaqMan One-Step RT-PCR Master Mix Reagent (Applied Biosystems, Foster City, CA, USA) in a 25-μl volume comprising 5 μl of extracted RNA and 20 μl of master mix according to the manufacturers' recommendation. All four assays (Table 1) were performed on the same day, and the same nucleic acid extracts were utilized. Singleplex assay B, capable of detecting mHEV genotypes 1 to 4, and duplex assay D, capable of detecting and differentiating mHEV genotypes 3 and 4, both utilized the same forward and reverse primers. The concentrations of the primers and probe or probes (duplex assays) were 400 and 200 nM for assay A, 800 and 200 nM for assay B, 400 and 200 nM for assay C, and 800 and 400 nM for assay D. One-step RT-PCR amplification was performed on an ABI 7500 real-time PCR instrument (Applied Biosystems, Foster City, CA, USA) under the following conditions: 15 min at 50°C for the RT reaction, 10 min at 95°C followed by 45 cycles at 95°C for 15 s for denaturation, and 60°C for 45 s for annealing and extension. A sample was considered negative if the cycle threshold (CT) was ≥41 amplification cycles. Quality control of the real-time RT-PCR process included negative (nuclease-free water) and positive (HEV RNA and HEV plasmid DNA) controls added to each PCR plate.
Efficiency, limit of detection, and intra-assay and interassay precision of the RT-PCR assays.
Verifications of assay sensitivity, specificity, and precision were performed as proposed previously (27). Standard curves of mHEV genotypes 3 and 4 ranging from 101 to 108 copies of HEV plasmid DNA were used to determine the efficiency, limit of detection, and intra-assay and interassay precision of the real-time PCR assays. For assays A and B, which do not differentiate HEV genotypes, standard curves for genotypes 3 and 4 were tested separately. For assays C and D, which differentiate between HEV genotypes 3 and 4, the standard curves were tested as duplex assays or separately in singleplex assays. The amplification efficiency (E) for each assay was calculated according to the following formula to determine the performance of quantitative PCR (qPCR): E = [10−1/S] − 1, where S indicates the slope of the regression line. The limit of detection of each assay and the intra-assay variation were assessed with the standard curves tested in triplicate. The limit of detection was specified as the smallest amount of DNA standard that could be detected with 100% probability. The interassay variation was determined by three independent runs of the standard curves in triplicate.
Conventional nested RT-PCR.
Twenty field samples tested by all four real-time PCR assays that presented discrepant results, defined as a sample that exhibited a positive result in one assay but a negative result in another assay, were arbitrarily chosen for sequencing confirmation using a nested RT-PCR assay based on a partial HEV ORF2 fragment. Conventional nested RT-PCRs were performed using previously described primers (28) (Table 1). Briefly, for the first PCR, 6 μM (each) primers 3156N and 3157N and a Qiagen OneStep RT-PCR kit (Qiagen, Valencia, CA, USA) were used. The thermal cycler conditions for the first reaction were as follows: 50°C for 30 min; 95°C for 15 min; 35 cycles of 94°C for 30 s, 55°C for 30 s, and 72°C for 1 min; and a final elongation step at 72°C for 10 min. The second reaction was performed with 0.2 μM (each) primers 3158N and 3159N and ReadyMix Taq PCR Mix (Sigma-Aldrich, St. Louis, MO, USA). The thermal cycler conditions for the second PCR were as follows: 95°C for 5 min; 35 cycles at 94°C for 30 s, 55°C for 30 s, and 72°C for 30 s; and a final elongation step at 72°C for 7 min. The 348-bp second-round PCR products were visualized after electrophoresis on a 1% agarose gel.
Sequencing and phylogenetic analysis.
Sequencing of PCR products from HEV RNA-positive samples was performed directly on both strands at the Iowa State University DNA Facility, Ames, IA, USA. The sequences were aligned with published data using BLAST at the National Center for Biotechnology Information (NCBI) (http://www.ncbi.nlm.nih.gov/). The sequences were compiled using Lasergene software and the Clustal W alignment algorithm (DNAStar, Madison, WI, USA). The nucleotide distance of the sequences was evaluated by neighbor joining (NJ) using Lasergene MegAlign. Confidence in the NJ tree was estimated by bootstrap replicates.
Statistical analysis.
Inter- and intra-assay variances were computed using the CT values, standard deviations, and coefficients of variation of the standard curves. The variance was analyzed by a one-way repeated-measures analysis of variance, followed by Bonferroni's test for pairwise comparison. Cochran's Q test for matched data, followed by McNemar's test for pairwise comparisons, was used to determine whether the proportions of RT-PCR-positive samples were significantly different between assays. Differences between groups were considered significant if the P value was <0.05. A kappa index was performed to determine the agreement of positive and negative results between assays. The strength of agreement was scored as follows, as previously described (37): ≤0, poor; 0.01 to 0.2, slight; 0.21 to 0.4, fair; 0.41 to 0.60, moderate; 0.61 to 0.80, substantial; and 0.81 to 1, almost perfect. Statistical analyses were performed using SAS version 9.2 (SAS Institute, Inc., Cary, NC, USA).
Nucleotide sequence accession numbers.
The sequences reported in this paper have been deposited in the GenBank database under accession numbers KF719308 to KF719310.
RESULTS
For assays C and D, which differentiate between HEV genotypes 3 and 4, there was no difference between the standard curves tested as duplex or singleplex assays regarding PCR efficiency, limits of detection, and intra- or interassay precision; therefore, only the results of the duplex assays are presented. A purine-pyrimidine mismatch was identified in the probes used to detect HEV genotype 4 for both assays C (base 19) and D (base 17) compared to the strain used as a control (5389 C/A in the virus/probes).
Evaluation of real-time RT-PCR assays.
Standard curves were established for each real-time PCR assay using the HEV genotype 3 and 4 DNA controls serially diluted from 1 × 108 to 1 × 101 copies and amplified in triplicate. The efficiency, regression coefficient, slope, and intercept for each assay are shown in Table 2. Assays A and B had similar performance regardless of the HEV genotype used, while assays C and D showed a decrease in efficiency of at least 11% when HEV genotype 4 was used (Table 2).
TABLE 2.
Efficiency, regression coefficient, slope, and intercept for real-time PCR assays A, B, C, and D
| Variable | Valuea |
|||||||
|---|---|---|---|---|---|---|---|---|
| A |
B |
C |
D |
|||||
| HEV-3 | HEV-4 | HEV-3 | HEV-4 | HEV-3 | HEV-4 | HEV-3 | HEV-4 | |
| Efficiency (%) | 95.9 | 98.7 | 88.2 | 90.7 | 95.6 | 82.1 | 93.3 | 82.3 |
| Regression coefficient | 0.992 | 0.99 | 0.991 | 0.992 | 0.990 | 0.993 | 0.990 | 0.991 |
| Slope | −3.422 | −3.352 | −3.641 | −3.565 | −3.433 | −3.843 | −3.493 | −3.833 |
| Intercept | 42.7 | 42.1 | 43.0 | 42.4 | 47.82 | 50.88 | 43.6 | 48.7 |
The values were obtained by quantification of serially diluted plasmid DNA containing a HEV genotype 3 (HEV-3) or 4 (HEV-4) ORF2 and ORF3 overlapping region from 1 × 108 to 1 × 101 copies of genome equivalents per reaction.
Limit of detection and inter- and intra-assay precision of the four real-time PCR assays.
For each assay, interassay precision was assessed by calculating the standard deviation and coefficient of variation of the CT obtained for each standard dilution tested in three independent runs, and the coefficient of variation was found to be <7% for all assays (data not shown). Intra-assay precision was assessed by calculating the standard deviation and coefficient of variation of the CT obtained for each standard dilution tested in triplicate, and the coefficient of variation was found to be <4% (Table 3). The limit of detection of each assay was specified as the lowest recognized concentration of genotype 3 or 4 mHEV DNA control serially diluted from 1 × 105 to 1 × 101 in triplicate (Table 3). However, the GE copy numbers do not reflect the number of RNA molecules, since the efficiency of the RT reaction was not directly determined.
TABLE 3.
Limits of detection of four real-time PCR assays and intra-assay precision results of 10-fold dilutions of HEV genotype 3 or 4 DNA plasmid controls tested in triplicate
| DNA copy no.a | Assay A |
Assay B |
Assay C |
Assay D |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| No. of detected samples/total | Mean CT | SD | CV%b | Detected samples | Mean CT | SD | CV%b | No. of detected samples/total | Mean CT | SD | CV%b | No. of detected samples/total | Mean CT | SD | CV%b | |
| HEV-3 | ||||||||||||||||
| 100,000 | 3/3 | 24.55 | 0.47 | 1.86 | 3/3 | 25.30 | 0.94 | 3.56 | 3/3 | 30.28 | 0.85 | 2.73 | 3/3 | 26.12 | 0.41 | 1.53 |
| 10,000 | 3/3 | 28.58 | 0.37 | 1.24 | 3/3 | 28.09 | 0.64 | 2.05 | 3/3 | 33.48 | 0.68 | 1.91 | 3/3 | 29.98 | 0.48 | 1.54 |
| 1,000 | 3/3 | 31.37 | 0.34 | 1.09 | 3/3 | 32.77 | 0.16 | 0.48 | 3/3 | 37.34 | 0.78 | 2.09 | 3/3 | 32.80 | 0.94 | 2.88 |
| 100 | 3/3 | 34.91 | 0.23 | 0.65 | 3/3 | 36.35 | 1.32 | 3.53 | 3/3 | 40.78 | 1.17 | 2.90 | 3/3 | 36.74 | 0.59 | 1.64 |
| 10 | 3/3 | 37.16 | 0.73 | 1.95 | 2/3 | 39.31 | 1.81 | 4.60 | 0/3 | 1/3 | 39.92 | |||||
| HEV-4 | ||||||||||||||||
| 100,000 | 3/3 | 23.78 | 0.66 | 2.77 | 3/3 | 25.86 | 0.18 | 0.69 | 3/3 | 31.85 | 0.17 | 0.53 | 3/3 | 30.77 | 0.90 | 2.82 |
| 10,000 | 3/3 | 28.44 | 0.39 | 1.36 | 3/3 | 29.25 | 0.72 | 2.31 | 3/3 | 35.86 | 0.12 | 0.32 | 3/3 | 34.03 | 0.33 | 0.93 |
| 1,000 | 3/3 | 31.59 | 0.68 | 2.16 | 3/3 | 32.81 | 0.09 | 0.24 | 1/3 | 39.54 | 1/3 | 38.21 | ||||
| 100 | 3/3 | 34.53 | 0.56 | 1.64 | 3/3 | 36.24 | 0.58 | 1.53 | 0/3 | 0/3 | ||||||
| 10 | 3/3 | 37.36 | 0.13 | 0.35 | 2/3 | 40.22 | 0.51 | 1.26 | 0/3 | 0/3 | ||||||
Number of plasmid DNA copies per reaction.
CV%, coefficient of variation.
Variation of detection limits was in the order of 10- to 1,000-fold among assays. Assay A was able to detect 101 GE copies of the plasmid HEV DNA per reaction (4 × 103 copies ml−1), assay B detected all 102-GE-copy dilutions (4 × 104 copies ml−1) and occasionally the 101-GE-copy dilution, and assays C and D were able to detect all HEV DNA standards down to the 104-GE-copy dilutions (4 × 106 copies ml−1) (Table 3). An impact of the HEV genotype was observed for assays C and D, which could detect all 102-GE-copy dilutions of genotype 3 but only 1 of 3 103-GE-copy dilutions of genotype 4. Assay A and B detection was genotype independent.
Detection of HEV RNA in experimental samples with known mHEV exposure by the four real-time RT-PCR assays.
Detection of mHEV RNA in experimental samples evaluated in parallel using the same RNA extracts is shown in Fig. 1. The GE titers of mHEV were determined from the HEV genotype 3 standard curve included in each run and for each assay. Considering RNA detection over time, assay A presented the highest rate of cumulative positive detection (96.4%; 27/28) (P < 0.05), followed by assay B (39.2%; 11/28), which presented a positive detection rate higher than those of assays C (14.2%; 4/28) and D (0/28) (P < 0.05). The detected viral-RNA loads ranged from 3.62 to 7.16 log10 HEV GE copies ml−1 in 10% fecal samples for assay A; from 3.22 to 5.26 log10 HEV GE copies ml−1 in 10% fecal samples for assay B, and from 4.68 to 4.81 log10 HEV GE copies ml−1 for assay C (Fig. 1). In order to further investigate the reason for the low detection rates found with assay C and the lack of detection of any positive sample with assay D, the primer and probe sequences from each assay were compared to the genome sequence of each of the HEV strains used. Mismatches were not identified for any primer or probe (data not shown), indicating that the detection rates achieved were due to intrinsic differences in the limits of detection for each assay.
FIG 1.
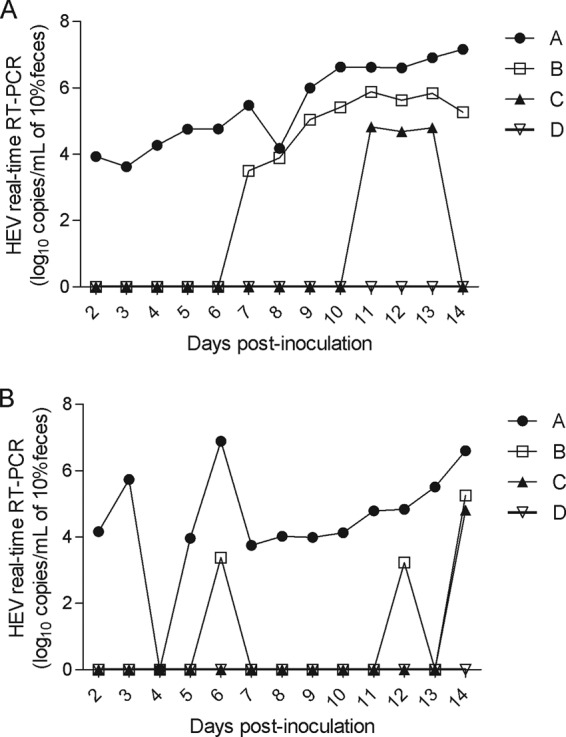
Comparison of four real-time RT-PCR assays (A, B, C, and D) in detecting and quantifying HEV RNA on fecal samples after experimental inoculation of pigs with HEV genotype 3 strain US-2 (A) or Meng (B).
Detection of HEV RNA in field samples with unknown mHEV exposure by each of the four real-time RT-PCR assays.
The rates of HEV RNA detection with the four assays on field samples are summarized in Table 4. The overall detection rates of HEV RNA-positive samples regardless of age were 67.2% (125/186) for assay A, 36.4% (68/136) for assay B, 1.1% (2/136) for assay C, and 0.5% (1/136) for assay D. The assays showed overall poor agreement (κ = 0.19 to 0.03), with differences in detection rates between assays (P < 0.01). Assay A presented the highest HEV RNA detection rate (P < 0.01). All positive samples with assays C and D were also positive with assays A and B. Regarding positive samples with assay B, 80.8% (55/68) were also positive by assay A, indicating that assay B identified an additional 7.0% (13/186) positive samples that were not identified by assay A. However, assay A identified an additional 37.6% (70/186) positive samples that were not identified by assay B (κ = 0.19; P < 0.01).
TABLE 4.
Detection rates for HEV RNA in fecal samples collected from pigs of unknown HEV status by real-time RT-PCR assays A, B, C, and D
| Age group | No. tested | No. (%) positivea |
|||||
|---|---|---|---|---|---|---|---|
| A | B | C |
D |
||||
| HEV-3 | HEV-4 | HEV-3 | HEV-4 | ||||
| Suckling | 46 | 25 (54.3) A | 17 (36.9) A | 0 B | 0 B | 0 B | 0 B |
| Nursery | 86 | 62 (72.0) A | 24 (27.9) B | 0 C | 0 C | 0 C | 0 C |
| Grow-finish | 54 | 38 (70.3) A | 27 (49.1) B | 2 (3.6) C | 0 C | 1 (1.8) C | 0 C |
| Total | 186 | 125 (67.2) A | 68 (36.4) B | 2 (1.1) C | 0 C | 1 (0.5) C | 0 C |
Different uppercase letters within each column indicate significant differences (P < 0.05) within assays.
Further evaluation of 20 field samples with discrepant results among real-time RT-PCR assays.
Twenty samples with CT values lower than 36 on the real-time RT-PCR assay were arbitrarily selected for amplification with a conventional nested RT-PCR assay, followed by sequencing to further verify the results (Table 5). Twelve of 20 samples positive in at least one real-time RT-PCR assay were also positive in the nested RT-PCR, and 3/12 samples that were successfully sequenced were determined to be mHEV genotype 3 (GenBank accession no. KF719308, KF719309, and KF719310) (Table 5).
TABLE 5.
Detection of HEV RNA by conventional nested RT-PCR and real-time RT-PCR assays in 20 pig field samples
| Sample | Detectiona |
||||
|---|---|---|---|---|---|
| Real-time RT-PCR assay |
Conventional nested RT-PCR | ||||
| A | B | C | D | ||
| 19482 | − | 35.69 | − | − | + (w) |
| 19702-C | − | 36.18 | − | − | + (w) |
| 19714 | − | 36.00 | − | − | + (w) |
| 19762-B | 33.02 | 34.46 | − | − | +b |
| 19775 | 36.97 | − | − | − | − |
| 19913 | 33.04 | 35.51 | 33.62 | − | +b |
| 19903-A | − | 35.63 | − | − | − |
| 19912-B | 36.06 | − | − | − | + (w) |
| 19955 | 37.41 | 36.06 | − | − | − |
| 20354-A | 35.99 | 38.99 | − | − | − |
| 20361-A | 36.40 | − | − | − | + |
| 20383-B | 36.39 | 33.32 | − | − | − |
| 20513 | 36.13 | − | − | − | − |
| 20517 | 36.88 | − | − | − | + |
| 20613 | − | 36.78 | − | − | + (w) |
| 20467-E | 36.59 | 38.05 | − | − | + |
| 20468-D | 36.31 | 37.84 | − | − | +b |
| 20777-B | 36.52 | 35.86 | − | − | − |
| 20792-B | 37.23 | 36.46 | − | − | + (w) |
| 20855 | 35.93 | − | − | − | − |
−, HEV RNA-negative sample; +, HEV RNA-positive sample; w, weak reaction.
HEV genotype 3 by sequencing.
DISCUSSION
In this study, two singleplex real-time RT-PCR assays for detection of all four known mammalian HEV genotypes without differentiation and two duplex real-time RT-PCR assays for detection and differentiation of HEV genotypes 3 and 4 were evaluated. All the assays were compared on the same real-time RT-PCR instrument on the same day, using the same RT-PCR enzymes, standard curves, and nucleic acid extracts. Under these conditions, singleplex assays A and B, designed to broadly detect HEV genotypes 1 to 4, showed significantly better performance (P < 0.01) than duplex assays C and D, which both allow detection and differentiation of HEV genotypes 3 and 4. On field samples, the singleplex real-time RT-PCR assays detected at least 34-fold more positive samples than the duplex real-time RT-PCR assays.
The design of a broadly reactive assay for detection of mHEV genotypes is a complex and challenging task due to the heterogeneity among the various HEV strains (5, 29). The sensitivities of real-time assays can vary widely depending on the target region and HEV genotype (20, 21). Previous comparison of conventional and real-time RT-PCR assays to detect HEV RNA have shown that targeting a more conserved region, such as ORF2/3, appears more reliable than the use of degenerate primers and probes targeting a less conserved region, such as ORF2 (20, 21, 30). In this study, all the real-time RT-PCR assays used target the overlap region of ORF2 and ORF3; however, with the exception of assay A, the assays used degenerate primers and probes. Degeneracies may reduce the sensitivity and specificity of an assay due to factors such as a lower effective concentration of each primer or difficulties in estimating the suitable annealing temperatures and primer lengths (31), which could partially explain the poor results achieved with assays B, C, and D compared with assay A in the present study. Analysis of the full-length genomes of various human and animal mHEV strains revealed that the HEV genomes vary even in the conserved regions (5, 29), and this genetic variability complicates reliable detection of different mHEV genotypes and subtypes. In fact, recent studies found polymorphisms in the region of the probe-binding site of the most widely used real-time RT-PCR assay for HEV detection (assay A) (17), and a modification of the probe and subsequent increase of the melting temperature restored detection of the polymorphic strains (22, 23). Employment of more than one set of primers targeting different regions of the HEV genome could increase the likelihood of HEV detection (30). The same strategy has been used for detection of other highly variable RNA viruses, such as influenza A virus (32) and porcine reproductive and respiratory syndrome virus (PRRSV) (33, 34).
The best PCR performance on experimental samples was observed for assay A (27/28), followed by assays B (11/28) and C (4/28). Assay D could not detect a single positive sample. In PCR comparisons similar to those presented here, assay A was determined to be the most suitable, reproducible, and reliable assay for the detection of HEV RNA (12, 20, 21). It is well recognized that fecal samples, such as those used in this study, could contain metabolic compounds that possibly interfere with RT-PCR. The addition of an internal control to monitor the presence of such inhibitors would ensure the reliability of negative results. Although such a control was not included in this study, all assays used the same nucleic acid extracts, and issues with the viral-RNA extraction recovery can therefore be excluded. No mismatch could be found for any primer or probe compared to the mHEV strains used to infect the pigs, indicating that the detection rates achieved were due to intrinsic differences in the limits of detection for each assay. Moreover, it is worth noting that the singleplex assay B and the duplex assay D developed in the present study used the same primer pair, and the difference in the positive detection rates between them (11/28 versus 0/28; P < 0.01) are likely due to the differences in the nucleotide compositions of the targeted regions of the probes.
HEV genome variability may also influence the quantification of its RNA. Comparison of the analytical sensitivities of the assays based on the detection of the plasmid DNA standards showed that the sensitivities of assays A and B were independent of the HEV genotype (3 or 4), and assay A was 10-fold more sensitive than assays B, C, and D based on genotype 3 standard curve detection. However, the sensitivities of assays C and D for HEV genotype 4 were 100-fold lower than the sensitivities yielded by using HEV genotype 3 within the same assay, which could be partially explained by a single mismatch in the probe region. Although a study has reported that probes with up to two mismatches showed little variation in PCR efficiency and nucleic acid quantification compared to probes that were fully complementary (35), another study has shown that a single mismatch in the probe-binding region resulted in a quantification error of up to 33% (36). Due to the use of plasmid DNA for quantification, reverse transcription reaction, as one of the crucial steps of RT-PCR, was dismissed, and the limit of detection presented here cannot be fully compared to assays in which there is use of RNA standards.
In summary, real-time RT-PCR assays A and B, which broadly detect HEV genotypes 1 to 4, showed better results for RNA detection than the duplex assays C and D, which were both designed to detect and differentiate between HEV genotypes 3 and 4. Assay A presented the overall best performance among the tested assays.
Footnotes
Published ahead of print 15 January 2014
REFERENCES
- 1.Purcell RH, Emerson SU. 2008. Hepatitis E: an emerging awareness of an old disease. J. Hepatol. 48:494–503. 10.1016/j.jhep.2007.12.008 [DOI] [PubMed] [Google Scholar]
- 2.Kamar N, Bendall R, Legrand-Abravanel F, Xia NS, Ijaz S, Izopet J, Dalton HR. 2012. Hepatitis E. Lancet 379:2477–2488. 10.1016/S0140-6736(11)61849-7 [DOI] [PubMed] [Google Scholar]
- 3.Meng XJ, Anderson D, Arankalle VA, Emerson SU, Harrison TJ, Jameel S, Okamoto H. 2012. Hepeviridae, p 1021–1028 In King AMQ, Adams MJ, Carstens EB, Lefkowitz EJ. (ed), Virus taxonomy: 9th Report of the International Committee on Taxonomy of Viruses. Academic Press, London, United Kingdom [Google Scholar]
- 4.Meng XJ. 2013. Zoonotic and foodborne transmission of hepatitis E virus. Semin. Liver Dis. 33:41–49. 10.1055/s-0033-1338113 [DOI] [PubMed] [Google Scholar]
- 5.Lu L, Li C, Hagedorn CH. 2006. Phylogenetic analysis of global hepatitis E virus sequences: genetic diversity, subtypes and zoonosis. Rev. Med. Virol. 16:5–36. 10.1002/rmv.482 [DOI] [PubMed] [Google Scholar]
- 6.Colson P, Romanet P, Moal V, Borentain P, Purgus R, Benezech A, Motte A, Gerolami R. 2012. Autochthonous infections with hepatitis E virus genotype 4, France. Emerg. Infect. Dis. 18:1361–1364. 10.3201/eid1808.111827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hakze-van der Honing RW, van Collie E, Antonis AF, van der Poel WH. 2011. First isolation of hepatitis E virus genotype 4 in Europe through swine surveillance in the Netherlands and Belgium. PLoS One 6:e22673. 10.1371/journal.pone.0022673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kumar S, Subhadra S, Singh B, Panda BK. 2013. Hepatitis E virus: the current scenario. Int. J. Infect. Dis. 17:e228–e233. 10.1016/j.ijid.2012.11.026 [DOI] [PubMed] [Google Scholar]
- 9.Wenzel JJ, Preiss J, Schemmerer M, Huber B, Jilg W. 2013. Test performance characteristics of Anti-HEV IgG assays strongly influence hepatitis E seroprevalence estimates. J. Infect. Dis. 207:497–500. 10.1093/infdis/jis688 [DOI] [PubMed] [Google Scholar]
- 10.Rossi-Tamisier M, Moal V, Gerolami R, Colson P. 2013. Discrepancy between anti-hepatitis E virus immunoglobulin G prevalence assessed by two assays in kidney and liver transplant recipients. J. Clin. Virol. 56:62–64. 10.1016/j.jcv.2012.09.010 [DOI] [PubMed] [Google Scholar]
- 11.Drobeniuc J, Meng J, Reuter G, Greene-Montfort T, Khudyakova N, Dimitrova Z, Kamili S, Teo CG. 2010. Serologic assays specific to immunoglobulin M antibodies against hepatitis E virus: pangenotypic evaluation of performances. Clin. Infect. Dis. 51:e24–e27. 10.1086/654801 [DOI] [PubMed] [Google Scholar]
- 12.Baylis SA, Hanschmann KM, Blumel J, Nubling CM. 2011. Standardization of hepatitis E virus (HEV) nucleic acid amplification technique-based assays: an initial study to evaluate a panel of HEV strains and investigate laboratory performance. J. Clin. Microbiol. 49:1234–1239. 10.1128/JCM.02578-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huang S, Zhang X, Jiang H, Yan Q, Ai X, Wang Y, Cai J, Jiang L, Wu T, Wang Z, Guan L, Shih JW, Ng MH, Zhu F, Zhang J, Xia N. 2010. Profile of acute infectious markers in sporadic hepatitis E. PLoS One 5:e13560. 10.1371/journal.pone.0013560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Legrand-Abravanel F, Mansuy JM, Dubois M, Kamar N, Peron JM, Rostaing L, Izopet J. 2009. Hepatitis E virus genotype 3 diversity, France. Emerg. Infect. Dis. 15:110–114. 10.3201/eid1501.080296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Enouf V, Dos RG, Guthmann JP, Guerin PJ, Caron M, Marechal V, Nicand E. 2006. Validation of single real-time TaqMan PCR assay for the detection and quantitation of four major genotypes of hepatitis E virus in clinical specimens. J. Med. Virol. 78:1076–1082. 10.1002/jmv.20665 [DOI] [PubMed] [Google Scholar]
- 16.Ahn JM, Rayamajhi N, Gyun KS, Sang YH. 2006. Comparison of real-time reverse transcriptase-polymerase chain reaction and nested or commercial reverse transcriptase-polymerase chain reaction for the detection of hepatitis E virus particle in human serum. Diagn. Microbiol. Infect. Dis. 56:269–274. 10.1016/j.diagmicrobio.2006.04.010 [DOI] [PubMed] [Google Scholar]
- 17.Jothikumar N, Cromeans TL, Robertson BH, Meng XJ, Hill VR. 2006. A broadly reactive one-step real-time RT-PCR assay for rapid and sensitive detection of hepatitis E virus. J. Virol. Methods 131:65–71. 10.1016/j.jviromet.2005.07.004 [DOI] [PubMed] [Google Scholar]
- 18.Meng J, Dai X, Chang JC, Lopareva E, Pillot J, Fields HA, Khudyakov YE. 2001. Identification and characterization of the neutralization epitope(s) of the hepatitis E virus. Virology 288:203–211. 10.1006/viro.2001.1093 [DOI] [PubMed] [Google Scholar]
- 19.Gyarmati P, Mohammed N, Norder H, Blomberg J, Belak S, Widen F. 2007. Universal detection of hepatitis E virus by two real-time PCR assays: TaqMan and Primer-Probe Energy Transfer. J. Virol. Methods 146:226–235. 10.1016/j.jviromet.2007.07.014 [DOI] [PubMed] [Google Scholar]
- 20.Ward P, Poitras E, Leblanc D, Letellier A, Brassard J, Plante D, Houde A. 2009. Comparative analysis of different TaqMan real-time RT-PCR assays for the detection of swine Hepatitis E virus and integration of Feline calicivirus as internal control. J. Appl. Microbiol. 106:1360–1369. 10.1111/j.1365-2672.2008.04104.x [DOI] [PubMed] [Google Scholar]
- 21.Mokhtari C, Marchadier E, Haim-Boukobza S, Jeblaoui A, Tesse S, Savary J, Roque-Afonso AM. 2013. Comparison of real-time RT-PCR assays for hepatitis E virus RNA detection. J. Clin. Virol. 58:36–40. 10.1016/j.jcv.2013.06.038 [DOI] [PubMed] [Google Scholar]
- 22.Baylis SA, Blumel J, Mizusawa S, Matsubayashi K, Sakata H, Okada Y, Nubling CM, Hanschmann KM. 2013. World Health Organization International Standard to harmonize assays for detection of hepatitis E virus RNA. Emerg. Infect. Dis. 19:729–735. 10.3201/eid1905.121845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Garson JA, Ferns RB, Grant PR, Ijaz S, Nastouli E, Szypulska R, Tedder RS. 2012. Minor groove binder modification of widely used TaqMan probe for hepatitis E virus reduces risk of false negative real-time PCR results. J. Virol. Methods 186:157–160. 10.1016/j.jviromet.2012.07.027 [DOI] [PubMed] [Google Scholar]
- 24.Zhang X, Li A, Shuai J, Dai Y, Zhu Z, Wu S, He Y. 2013. Validation of an internally controlled multiplex real time RT-PCR for detection and typing of HEV genotype 3 and 4. J. Virol. Methods 193:432–438. 10.1016/j.jviromet.2013.07.007 [DOI] [PubMed] [Google Scholar]
- 25.Schlauder GG, Dawson GJ, Erker JC, Kwo PY, Knigge MF, Smalley DL, Rosenblatt JE, Desai SM, Mushahwar IK. 1998. The sequence and phylogenetic analysis of a novel hepatitis E virus isolated from a patient with acute hepatitis reported in the United States. J. Gen. Virol. 79:447–456 [DOI] [PubMed] [Google Scholar]
- 26.Feagins AR, Opriessnig T, Huang YW, Halbur PG, Meng XJ. 2008. Cross-species infection of specific-pathogen-free pigs by a genotype 4 strain of human hepatitis E virus. J. Med. Virol. 80:1379–1386. 10.1002/jmv.21223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rabenau HF, Kessler HH, Kortenbusch M, Steinhorst A, Raggam RB, Berger A. 2007. Verification and validation of diagnostic laboratory tests in clinical virology. J. Clin. Virol. 40:93–98. 10.1016/j.jcv.2007.07.009 [DOI] [PubMed] [Google Scholar]
- 28.Huang FF, Haqshenas G, Guenette DK, Halbur PG, Schommer SK, Pierson FW, Toth TE, Meng XJ. 2002. Detection by reverse transcription-PCR and genetic characterization of field isolates of swine hepatitis E virus from pigs in different geographic regions of the United States. J. Clin. Microbiol. 40:1326–1332. 10.1128/JCM.40.4.1326-1332.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Smith DB, Purdy MA, Simmonds P. 2013. Genetic variability and the classification of hepatitis E virus. J. Virol. 87:4161–4169. 10.1128/JVI.02762-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vasickova P, Kralik P, Slana I, Pavlik I. 2012. Optimisation of a triplex real time RT-PCR for detection of hepatitis E virus RNA and validation on biological samples. J. Virol. Methods 180:38–42. 10.1016/j.jviromet.2011.12.007 [DOI] [PubMed] [Google Scholar]
- 31.Rose TM, Henikoff JG, Henikoff S. 2003. CODEHOP (COnsensus-DEgenerate Hybrid Oligonucleotide Primer) PCR primer design. Nucleic Acids Res. 31:3763–3766. 10.1093/nar/gkg524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hoffmann B, Harder T, Lange E, Kalthoff D, Reimann I, Grund C, Oehme R, Vahlenkamp TW, Beer M. 2010. New real-time reverse transcriptase polymerase chain reactions facilitate detection and differentiation of novel A/H1N1 influenza virus in porcine and human samples. Berl. Munch. Tierarztl. Wochenschr. 123:286–292 [DOI] [PubMed] [Google Scholar]
- 33.Wernike K, Hoffmann B, Dauber M, Lange E, Schirrmeier H, Beer M. 2012. Detection and typing of highly pathogenic porcine reproductive and respiratory syndrome virus by multiplex real-time RT-PCR. PLoS One 7:e38251. 10.1371/journal.pone.0038251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kleiboeker SB, Schommer SK, Lee SM, Watkins S, Chittick W, Polson D. 2005. Simultaneous detection of North American and European porcine reproductive and respiratory syndrome virus using real-time quantitative reverse transcriptase-PCR. J. Vet. Diagn. Invest. 17:165–170. 10.1177/104063870501700211 [DOI] [PubMed] [Google Scholar]
- 35.Yao Y, Nellaker C, Karlsson H. 2006. Evaluation of minor groove binding probe and Taqman probe PCR assays: influence of mismatches and template complexity on quantification. Mol. Cell Probes 20:311–316. 10.1016/j.mcp.2006.03.003 [DOI] [PubMed] [Google Scholar]
- 36.Süß B, Flekna G, Wagner M, Hein I. 2009. Studying the effect of single mismatches in primer and probe binding regions on amplification curves and quantification in real-time PCR. J. Microbiol. Methods 76:316–319. 10.1016/j.mimet.2008.12.003 [DOI] [PubMed] [Google Scholar]
- 37.Landis JR, Koch GG. 1977. An application of hierarchical kappa-type statistics in the assessment of majority agreement among multiple observers. Biometrics 33:363–374 [PubMed] [Google Scholar]


