Abstract
This report describes a short, on-plate formic acid (FA) extraction method for the identification of clinical yeast isolates using matrix-assisted laser desorption ionization–time-of-flight mass spectrometry (MALDI-TOF MS). A total of 41.1% (78/190) and 63.7% (121/190) of yeasts were identified using species log score thresholds of >2.0 and >1.9, respectively. Overall, 97.4% (185/190) of yeasts were identified in combination with conventional FA extraction.
TEXT
Matrix-assisted laser desorption ionization–time-of-flight mass spectrometry (MALDI-TOF MS) has been shown to be a highly successful method for the identification of yeast isolates in the microbiology setting (1–5). Due to their robust fungal cell wall, comprised of glucans and chitins, yeasts require formic acid protein extraction prior to MALDI-TOF MS analysis (2). This extraction process extends the turnaround time to results by 10 to 15 min. A more time-efficient approach is to apply formic acid directly on yeast analytes smeared onto the MALDI-TOF MS target plate prior to the addition of matrix. This study aimed to evaluate on-plate formic acid (FA) extraction as a rapid approach for the identification of clinical yeast isolates using MALDI-TOF MS.
A total of 190 clinical isolates were selected from a library of isolates stored over an 8-year period by the United Kingdom Clinical Mycology Network (UKCMN) regional laboratory at the Royal Free London NHS Foundation Trust. Isolates were passaged twice on CHROMagar Candida to ensure purity. Cultures were processed using on-plate and conventional FA extraction in parallel. Conventional FA extraction was performed per the manufacturer's instruction. For on-plate FA extraction, small amounts of yeast colony were inoculated in duplicate onto the target plate in thin smears using a wooden applicator. A 1-μl volume of 100% FA was added to each spot and allowed to dry, and then 1 μl of α-cyano-4-hydroxy-cinnamic-acid matrix was immediately added to each spot. MALDI-TOF MS analysis was performed on a Microflex LT platform (Bruker Daltonics) with Flex Control software (version 3.0) using default settings. Spectra were analyzed using Biotyper 3.0 software. Species log score thresholds of >2.0 and >1.9 were applied to analyses.
As a gold standard, internal transcribed spacer (ITS) rRNA sequence-based identification was performed on all isolates. DNA was extracted from yeast suspensions at a McFarland standard of 4 and lysed mechanically by bead beating in combination with a Promega Wizard genomic DNA kit per the manufacturer's instruction (omitting the lyticase treatment). Amplification of the ITS1-5.8S-ITS2 region was performed using previously described ITS primers ITS1 (5′ TCC GTA GGT GAA CCT GCG G 3′) and ITS4 (5′ TCC TCC GCT TAT TGA TAT GC 3′) (6). PCRs consisted of 2 U of HotStar Taq polymerase (Qiagen), 1.5 mM MgCl2, a 0.3 mM concentration of each deoxynucleoside triphosphate, a 0.5 mM concentration of each primer, and 5 μl of template DNA. Following enzyme activation at 94°C for 15 min, reactions were subjected to 40 thermal cycles of 94°C (60 s), 50°C (60 s), and 72°C (180 s) on a GeneAmp PCR system 9700 thermocycler. DNA amplification was detected by gel electrophoresis, and sequence-based analysis was performed as detailed by Jenkins et al. (7). The sequences were identified by matching against sequences within GenBank.
In total, 190 isolates comprised of 19 yeast species were included in this study, as detailed in Table S1 in the supplemental material. Commonly encountered species Candida albicans, C. parapsilosis, C. krusei, C. glabrata, C. tropicalis, and C. guilliermondii accounted for 87.9% (167/190) of the isolates, while 12.1% (23/190) consisted of less commonly isolated species. The success rate of each processing method was calculated from the first attempt at identification along with the overall success rate after repeat analysis (presented in Table 1). On-plate FA extraction successfully identified 41.1% (78/190) of yeast isolates to the species level with a log score > 2.0 and 33.7% (64/190) to the genus level only (log score > 1.7), and 25.3% (48/190) remained unidentified. By reducing the log score species threshold to >1.9, the success rate for species identification increased to 64% (121/190), with 11% (21/190) remaining as genus identifications only. Conventional FA extraction successfully identified 83.7% (159/190) of yeast isolates at the species level with a log score > 2.0 and 13.7% (26/190) to the genus level only (log score > 1.7), and 2.6% (5/190) were unidentified on the first attempt. By lowering the species log score threshold to >1.9, the success rate for conventional FA extraction increased to 90% (171/190), with 7.4% (14/190) remaining identifications only to the genus level. Repeat analysis using conventional FA extraction enabled the remaining 7.4% (14/190) identified only to the genus level to be identified at the species level. Comparing MALDI-TOF MS results with ITS rRNA sequencing, 100% (185/185) agreement was observed for isolates identified by MALDI-TOF MS; 2.6% (5/190) of the isolates returned a result of “no identification” with a log score < 1.7 despite the fact that high-quality MALDI-TOF MS spectra had been obtained. ITS rRNA sequencing returned identifications of the following rare yeast species: Cyberlindnera fabianii (1/5), Debaryomyces nepalensis (1/5), and Meyerozyma caribbica (3/5).
TABLE 1.
MALDI-TOF MS identification success rates using on-plate and conventional formic acid extraction on first attempt and overall after repeat analysis
| Extraction method | Species log score | % (no. of isolates identified to indicated level/total no. of isolates) |
||
|---|---|---|---|---|
| Species | Genus | No IDa | ||
| Conventional | >2.0 | 83.7 (159/190) | 13.7 (26/190) | 2.6 (5/190) |
| >1.9 | 90 (171/190) | 7.4 (14/190) | 2.6 (5/190) | |
| On plate | >2.0 | 41.1 (78/190) | 33.7 (64/190) | 25.3 (48/190) |
| >1.9 | 63.7 (121/190) | 11 (21/190) | 25.3 (48/190) | |
| Overall success rate | 97.4 (185/190) | 0 | 2.6 (5/190) | |
ID, identification.
Following the evaluation study, MALDI-TOF MS was implemented for yeast identification in January 2012. Identifications collected over a 12-month period from 1 January 2012 to 31 December 2012 for 297 clinical yeast isolates demonstrated that MALDI-TOF MS successfully identified 99.7% (296/297) of the isolates from culture. One isolate could not be identified by MALDI-TOF MS despite the fact that high-quality spectra had been obtained. That isolate was identified as Cyberlindnera fabianii by ITS rRNA sequence analysis by the PHE Mycology reference laboratory, Bristol, United Kingdom.
This study demonstrated that on-plate FA extraction combined with a reduced species log score threshold of >1.9 can decrease turnaround time to results for a significant proportion of yeasts in the routine diagnostic setting, as seen for 63.7% (121/190) of isolates in this study. There was no association between yeast species and failed identification using on-plate FA extraction. The 36.3% (69/190) of isolates requiring conventional FA extraction included 11 of the 19 species. Yeast misidentification was not observed as a result of the reducing the log score threshold for species identification. Similar observations have been reported in three recent studies using 70% FA on-plate extraction and a species log score threshold of ≥1.7 (8–10). In two of the studies, the mean log score thresholds for on-plate FA extraction (1.78 and 1.941) were significantly lower than those obtained using conventional FA extraction (2.0 and 2.223), but all log score values associated with misidentification by MALDI-TOF MS were below 1.7 (8, 10). When utilizing on-plate FA extraction, it is necessary to reduce the species log score threshold to achieve suitable identification success rates (11, 12).
The Bruker Biotyper 3.0 database consists of reference spectra generated using conventional FA extraction. On-plate FA extraction generates spectra with fewer peaks than conventional FA extraction (11). This contributes to the lower success rate using on-plate FA extraction, as spectra generated from the two extraction methods differ. The overall effect is a reduced log score, as fewer peaks are matched between the analyte and reference spectra. In this study, identification of 36.3% (69/190) of the isolates to the species level with on-plate FA extraction failed despite reduction of the species log score threshold to >1.9. In a recent study using the Vitek MS system, a success rate of 95.8% was achieved for yeast identification using the manufacturer's recommended 25% FA on-plate extraction method (12, 13). The Vitek MS database is built from reference spectra obtained using the same 25% FA on-plate extraction procedure. Improved consistency between MS spectra obtained from clinical isolates and those contained within the Vitek MS reference database could explain this improved success (11, 13) and is an important consideration in evaluating new reference databases.
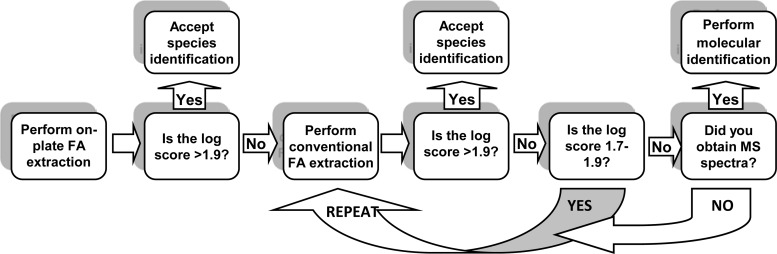
Based on data from this study, our center routinely tests yeast smears prepared from primary cultures using on-plate FA extraction, and conventional FA extraction is performed on isolates that fail to be identified. The overall workflow for yeast identification in our laboratory is indicated in Fig. 1. Molecular identification is rarely required and was used only for 0.3% (1/297) of isolates encountered in our routine practice over a 12-month period, but it is the most appropriate option when MALDI-TOF MS fails to identify yeast. The fact that six isolates were not identified by MALDI-TOF MS in this study demonstrates that not all species or strains are represented in the Biotyper 3.0 database. This finding has been replicated in a recent study identifying uncommonly encountered yeast isolates which reported an overall success rate of 84.5%, concluding that a result of “no identification” indicated that the yeast species was not contained within the MALDI-TOF database (14). For rare yeast species not identified by MALDI-TOF MS, molecular methods should be utilized to obtain the correct identification. In this study, 2.6% (5/190) of the isolates included in the validation were not identified by MALDI-TOF MS. Molecular identification confirmed that the original identifications obtained using biochemical profiling were incorrect (unpublished data), a recognized concern when using biochemical identification methods to identify rare yeast species (11, 12, 15). One example, a Cyberlindnera fabianii isolate, was originally identified as C. pelliculosa using biochemical profiling. During the 12-month prospective evaluation of MALDI-TOF MS, a second Cyberlindnera fabianii isolate was identified by ITS rRNA sequencing after failing to be identified by the use of MALDI-TOF MS. This highlights that the isolation of rare yeast species was not detected in past years due to misidentification by biochemical methods. Using MALDI-TOF MS, it is now possible to detect the isolation of rare species, as they will not be misidentified.
FIG 1.
Diagrammatic representation of the work flow for yeast identification in our laboratory setting.
In summary, this report supports previous findings that MALDI-TOF MS using Bruker Biotyper 3.0 software is a highly accurate and successful method for the identification of clinical yeast isolates. Maintaining a streamlined diagnostic service is essential in modern-day microbiology, and any intervention that can reduce sample processing time has obvious benefits. On-plate FA extraction contributes to this and in our setting has optimized workflow.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the National Institute for Health Research (NIHR) as part of the Chief Scientific Officers NIHR Ph.D. funded Fellowship scheme.
We wish to acknowledge Mark Fraser at the Mycology Reference Laboratory, Public Health England, Bristol, United Kingdom, for taxonomy guidance.
C.C.K. has received honoraria from Astellas, Gilead, MSD, and Pfizer.
Footnotes
Published ahead of print 29 January 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JCM.03489-13.
REFERENCES
- 1.Bader O, Weig M, Taverne-Ghadwal L, Lugert R, Gross U, Kuhns M. 2011. Improved clinical laboratory identification of human pathogenic yeasts by matrix-assisted laser desorption ionization time-of-flight mass spectrometry. Clin. Microbiol. Infect. 17:1359–1365. 10.1111/j.1469-0691.2010.03398.x [DOI] [PubMed] [Google Scholar]
- 2.Marklein G, Josten M, Klanke U, Muller E, Horre R, Maier T, Wenzel T, Kostrzewa M, Bierbaum G, Hoerauf A, Sahl HG. 2009. Matrix-assisted laser desorption ionization–time of flight mass spectrometry for fast and reliable identification of clinical yeast isolates. J. Clin. Microbiol. 47:2912–2917. 10.1128/JCM.00389-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rosenvinge FS, Dzajic E, Knudsen E, Malig S, Andersen LB, Lovig A, Arendrup MC, Jensen TG, Gahrn-Hansen B, Kemp M. 2013. Performance of matrix-assisted laser desorption-time of flight mass spectrometry for identification of clinical yeast isolates. Mycoses 56:229–235. 10.1111/myc.12000 [DOI] [PubMed] [Google Scholar]
- 4.Sendid B, Ducoroy P, Francois N, Lucchi G, Spinali S, Vagner O, Damiens S, Bonnin A, Poulain D, Dalle F. 2013. Evaluation of MALDI-TOF mass spectrometry for the identification of medically-important yeasts in the clinical laboratories of Dijon and Lille hospitals. Med. Mycol. 51:25–32. 10.3109/13693786.2012.693631 [DOI] [PubMed] [Google Scholar]
- 5.Mancini N, De Carolis E, Infurnari L, Vella A, Clementi N, Vaccaro L, Ruggeri A, Posteraro B, Burioni R, Clementi M, Sanguinetti M. 2013. Comparative evaluation of the Bruker Biotyper and Vitek MS matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) mass spectrometry systems for identification of yeasts of medical importance. J. Clin. Microbiol. 51:2453–2457. 10.1128/JCM.00841-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.White TJ, Bruns T, Lee S, Taylor J. 1990. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics, p 315–322 In Innis MA, Gelfand DH, Sninsky JJ, White TJ. (ed), PCR protocols—a guide to methods and applications. Academic Press, New York, NY [Google Scholar]
- 7.Jenkins C, Ling CL, Ciesielczuk HL, Lockwood J, Hopkins S, McHugh TD, Gillespie SH, Kibbler CC. 2012. Detection and identification of bacteria in clinical samples by 16S rRNA gene sequencing: comparison of two different approaches in clinical practice. J. Med. Microbiol. 61(Pt 4):483–488. 10.1099/jmm.0.030387-0 [DOI] [PubMed] [Google Scholar]
- 8.Cassagne C, Cella AL, Suchon P, Normand AC, Ranque S, Piarroux R. 2013. Evaluation of four pretreatment procedures for MALDI-TOF MS yeast identification in the routine clinical laboratory. Med. Mycol. 51:371–377. 10.3109/13693786.2012.720720 [DOI] [PubMed] [Google Scholar]
- 9.Theel ES, Schmitt BH, Hall L, Cunningham SA, Walchak RC, Patel R, Wengenack NL. 2012. Formic acid-based direct, on-plate testing of yeast and Corynebacterium species by Bruker Biotyper matrix-assisted laser desorption ionization–time of flight mass spectrometry. J. Clin. Microbiol. 50:3093–3095. 10.1128/JCM.01045-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Van Herendael BH, Bruynseels P, Bensaid M, Boekhout T, De Baere T, Surmont I, Mertens AH. 2012. Validation of a modified algorithm for the identification of yeast isolates using matrix-assisted laser desorption/ionisation time-of-flight mass spectrometry (MALDI-TOF MS). Eur. J. Clin. Microbiol. Infect. Dis. 31:841–848. 10.1007/s10096-011-1383-y [DOI] [PubMed] [Google Scholar]
- 11.Bader O. 2013. MALDI-TOF-MS-based species identification and typing approaches in medical mycology. Proteomics 13:788–799. 10.1002/pmic.201200468 [DOI] [PubMed] [Google Scholar]
- 12.Posteraro B, De Carolis E, Vella A, Sanguinetti M. 2013. MALDI-TOF mass spectrometry in the clinical mycology laboratory: identification of fungi and beyond. Expert. Rev. Proteomics 10:151–164. 10.1586/epr.13.8 [DOI] [PubMed] [Google Scholar]
- 13.Iriart X, Lavergne RA, Fillaux J, Valentin A, Magnaval JF, Berry A, Cassaing S. 2012. Routine identification of medical fungi by the new Vitek MS matrix-assisted laser desorption ionization–time of flight system with a new time-effective strategy. J. Clin. Microbiol. 50:2107–2110. 10.1128/JCM.06713-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dhiman N, Hall L, Wohlfiel SL, Buckwalter SP, Wengenack NL. 2011. Performance and cost analysis of matrix-assisted laser desorption ionization-time of flight mass spectrometry for routine identification of yeast. J. Clin. Microbiol. 49:1614–1616. 10.1128/JCM.02381-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vervaeke S, Vandamme K, Boone E, De Laere E, Swinne D, Surmont I. 2008. A case of Candida lambica fungemia misidentified as Candida krusei in an intravenous drug abuser. Med. Mycol. 46:853–856. 10.1080/13693780802342552 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.